Abstract
Bifidobacterium infantis, a Gram-positive bacterium, is one of the commonly used probiotics. We previously showed that B. infantis modified host defense systems and extended the lifespan of the nematode Caenorhabditis elegans. In the present study, we showed that the lifespan extension caused by B. infantis was enhanced in animals having a mutation in the tol-1 gene that encodes the sole C. elegans homolog of Toll-like receptors (TLRs). Meanwhile, lifespan increased by other probiotic bacteria, such as Bacillus subtilis or Clostridium butyricum, was not affected in the tol-1 mutant animals. A microarray analysis revealed that the expression of innate immune response-related genes was significantly increased in the tol-1 mutant. Worms with the tol-1 mutation exhibited reduced leaving behavior from the B. infantis lawn, while canonical downstream factors trf-1/TRAF and ikb-1/IκB appeared to not be involved. In conclusion, C. elegans tol-1/TLR regulates B. infantis-induced longevity and also regulates behavior against B. infantis.
Keywords: Caenorhabditis elegans, Toll-like receptor, longevity, behavior, Bifidobacterium infantis
INTRODUCTION
Probiotic bacteria are defined as living microorganisms that exert beneficial effects on human health when ingested in adequate amounts [1]. Metchnikoff, who first proposed the concept of probiotic bacteria in 1907, hypothesized that lactic acid bacteria (LAB) were important for promoting human health and longevity. Bifidobacteria, which is one of the LAB in a broad sense, has been commonly used as a probiotic. Among them, Bifidobacterium longum subsp. infantis (B. infantis), an infant microbe, has been reported to exert a variety of probiotic properties, such as anti-microbial activities [2], immunomodulatory effects [3, 4], and symptom improvement in irritable bowel syndrome (IBS) [5].
Caenorhabditis elegans has been used extensively as an experimental animal model, especially for basic studies on senescence. However, the creature has also been used to study the influence of food and nutrition on senescence and host defense since we reported the beneficial effects of LAB on the worm [6]. The appeal of this organism is due to its ease of cultivation, abundance of genetic tools, and short and reproducible lifespan [7]. We have previously shown that feeding of B. infantis extends lifespan via the p38 MAPK pathway in C. elegans [8]. In the course of the mutant analyses, we noticed that the mutation in the tol-1 gene, encoding the C. elegans sole homolog of Toll-like receptors (TLRs), appeared to enhance the lifespan extension caused by B. infantis.
TLRs are pattern-recognition receptors that respond to diverse pathogen-associated molecular patterns (PAMPs) and initiate innate immune signals. The first Toll was discovered in Drosophila melanogaster [9,10,11] and was later found in diverse species from the nematode C. elegans to mammals, including humans [12, 13]. Canonical TLR signaling negatively regulates the transcriptional inhibitor IκB/Cactus and, in turn, NF-κB/Relish, a master regulator of the inflammatory response, and drives expression of pathogen-defense molecules, such as antimicrobial peptides in insects and interferons and cytokines in mammals [14]. In contrast to mammals and flies, C. elegans lacks NF-κB-like transcription factors [15]. Nevertheless, Tenor et al. demonstrated that TOL-1 has roles in defense responses to pathogenic Gram-negative and Gram-positive bacteria; tol-1 mutants showed increased susceptibility to certain Gram-negative bacteria, including Salmonella enterica, but increased resistance to Gram-positive bacteria such as Enterococcus faecalis [16]. Pujol et al. reported that TOL-1 is required for the avoidance of Serratia marcescens [17]. Subsequently, Brandt and Ringstad showed that TOL-1 in a pair of BAG chemosensory neurons is required for avoidance of S. marcescens [18]. The role of TOL-1 in response to probiotic bacteria, however, remains poorly understood. In the present study, we described the role of TOL-1 in longevity and behavior elicited by a probiotic, B. infantis.
MATERIALS AND METHODS
Bacterial strain and culture conditions
Escherichia coli OP50 was grown on tryptone soya broth (TSB) and tryptone soya agar (TSA) (Nissui Pharmaceutical, Tokyo, Japan) at 37°C. Similarly, Bacillus subtilis NBRC3134 and Staphylococcus aureus NBRC13276 were cultured using TSB and TSA. B. infantis ATCC15697 was cultured using GAM broth (Nissui) and TOS propionate agar (Yakult Pharmaceutical Industry, Tokyo, Japan). Clostridium butyricum MIYAIRI 588 (CBM 588), which was kindly provided by Miyarisan Pharmaceutical, was cultured using GAM broth and GAM agar (Nissui Pharmaceutical, Japan). Cultivated bacteria were scraped and weighed. Aliquots (100 mg wet weight) of bacteria were suspended in 0.5 ml of M9 buffer (5 mM potassium phosphate, 1 mM CaCl2, and 1 mM MgSO4) and used in the lifespan assays; aliquots at 66.7 mg/ml were used in the behavioral assays.
C. elegans strains and culture conditions
The wild-type C. elegans strain Bristol N2 and the following derivative mutant strains were obtained from the Caenorhabditis Genetics Center: IG10 tol-1(nr2033) I, NS2937 trf-1(nr2014) III, and NS3026 ikb-1(nr2027) I. C. elegans strains were maintained using standard techniques [19]. For gene expression analyses and lifespan assays, synchronized animals were prepared as follows: eggs were prepared from adult C. elegans by exposure to a sodium hypochlorite/sodium hydroxide solution. The egg suspension was incubated in M9 buffer for one day at 25°C to allow hatching and synchronization, and the resulting suspension of synchronized L1-stage worms was centrifuged at 156 × g for 1 min. After removing the supernatant by aspiration, the remaining larvae were transferred onto mNGM plates (1.7% (w/v) agar, 50 mM NaCl, 1 mM CaCl2, 5 µg/ml cholesterol, 25 mM KH2PO4, 1 mM MgSO4) covered with 10 mg OP50. Transferred worms were cultured at 25°C for two days until they reached the young adult stage (referred to as three-day-old animals). For behavioral assays, synchronized L1-stage worms were transferred onto NGM plates where OP50 was seeded at least two weeks before assays.
Determination of C. elegans lifespan
Lifespan assays were performed as follows. Synchronized three-day-old animals (35 animals per plate) were placed on 5-cm mNGM plates covered with 10 mg of bacteria, and the plates were incubated at 25°C. Supplementation with astaxanthin was performed as previously described [20]. Animals were transferred daily to fresh plates for the first four days and thereafter transferred every second day. The numbers of live and dead animals were scored every day. An animal was considered dead when it failed to respond to a gentle touch with a worm picker. Animals that crawled off the plate or died from internal hatching were considered lost and not included in the analysis. Worm survival was calculated by the Kaplan-Meier method, and survival differences were tested for significance using the log-rank test with Microsoft Excel supplemented with the add-in software Statcel 4 (OMS, Tokorozawa, Japan).
Mean lifespan (MLS) was estimated using the formula [21]
,
where dj is the number of worms that died in the age interval (xj to xj+1) and N is the total number of worms. The standard error (SE) of the estimated mean lifespan was calculated using the following equation:
.
Maximum lifespan was calculated as the mean lifespan of the longest-living 15% of the worms in each group.
Microarray analysis
Three-day-old hermaphrodites of the wild-type or the tol-1(nr2033) mutant worms were cultured for one day on mNGM plates covered with OP50. Microarray expression profiling was performed with the wild-type and tol-1 mutant worms. Approximately 100 worms in each group were collected by a worm picker and soaked in RNAlater solution (Qiagen). Total RNA was isolated using an RNeasy Lipid Tissue kit (Qiagen).
DNA synthesis and microarray hybridization were performed by Kurabo Industries, Ltd. RNA quality (RNA integrity number (RIN) >7) was confirmed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A total of ~1 µg of RNA was used as the template for fluorescent labeling of cRNA. Labeled cRNAs were hybridized to the Affymetrix C. elegans Genome Array (containing 22,500 transcripts). Microarray data analyses were performed with MAS5.0 (Microarray Suite statistical algorithm, Affymetrix). Differential expression was analyzed by the Comparison Analysis of MAS5.0 using the Wilcoxon’s Signed Rank test. Each probe set on the experiment array (tol-1) was compared to its counterpart on the baseline array (wild type), and a ‘Change p-value’ was calculated. Probe sets that showed differential expression were assigned with ‘Change calls’ (increase, p<0.002; marginal increase, 0.002≤p<0.002667; marginal decrease, 0.997333<p≤0.998; or decrease, p>0.998). The ‘Signal Log Ratio’ was computed using a one-step Tukey’s Biweight method by taking a mean of the log ratios of probe pair intensities across the two arrays (wild-type vs. tol-1). The final data extraction was performed using DNA Microarray Viewer ver. 2 (Kurabo Industries, Ltd., Osaka, Japan).
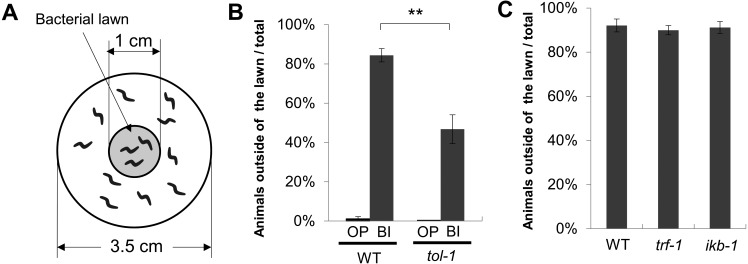
Lawn-leaving assay
Assay plates were prepared as follows. Fifty microliters of bacteria suspension was placed at the center of 3.5-cm mNGM plates (1.7% (w/v) agar, 50 mM NaCl, 1 mM CaCl2, 5 µg/ml cholesterol, 25 mM KH2PO4, 1 mM MgSO4), and after the plates were allowed to dry for 20 min, a bacteria lawn approximately 1 cm in diameter appeared. Animals at the young adult stage were collected in microfuge tubes and washed 3 times with M9 buffer (5 mM potassium phosphate, 1 mM CaCl2, 1 mM MgSO4, and 0.5 g/l gelatin). Approximately 20 animals were placed in the center of the bacteria lawn. At 10 min after placing the animals, the number of animals that were outside of the lawn and the number of those that remained on the lawn were counted. The percentage of animals that were outside of the bacterial lawn was calculated as (number of animals that were outside of the lawn) / (total number of animals) × 100%.
RESULTS
The tol-1 mutation enhanced the prolongevity caused by B. infantis
In our previous work, the mean lifespan of C. elegans increased by approximately 40% when the tol-1(nr2033) mutant worms were fed B. infantis; the same regimen increased the lifespan of the wild-type worms by approximately 20% [8]. From these observations, we hypothesized that the tol-1 mutation may enhance lifespan extension caused by B. infantis. To test this hypothesis, we determined the significant differences between the lifespans of the wild-type and tol-1 mutant animals. The lifespan of the tol-1 mutant was significantly longer than that of the wild-type animal after feeding with B. infantis (Fig. 1, Table 1). In contrast, the lifespan of the tol-1 mutant was similar to that of the wild-type animal after feeding with the standard food, E. coli OP50 (Fig. 1, Table 1). These results suggest that the mutation in the tol-1 gene enhanced the longevity caused by B. infantis. Notably, enhancement of B. infantis-elicited longevity in the tol-1 mutant animals was observed after 20 days (Fig. 1). Previously, Zhao et al. identified two classes of death: early deaths with a swollen pharynx, which is mainly caused by bacterial infection and damage to the pharynx (“early damage”), and later deaths with an atrophied pharynx (“later intrinsic causes”) [22]. Taken together with our results, tol-1 might suppress the effect of B. infantis on the reduction of later deaths due to intrinsic causes.
Fig. 1.
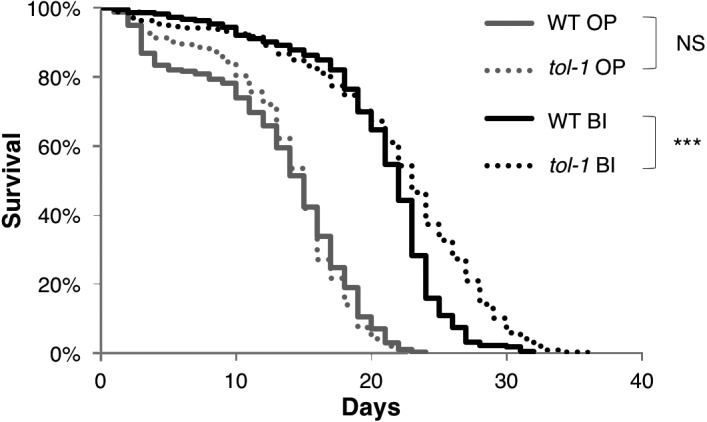
C. elegans longevity caused by feeding with B. infantis was enhanced in the tol-1 mutants.
Survival curves of the wild-type (WT) and tol-1 mutant animals fed E. coli (OP) or B. infantis (BI). The lifespan of the tol-1 mutant fed BI (black dash line) was increased compared with that of the wild-type fed BI (black solid line), while in the feeding of a standard food, E. coli OP50, the lifespan of the tol-1 mutant (gray dash line) was similar to that of the wild-type animal (gray solid line). ***p<0.001, log-rank test. NS: not significant, log-rank test. Detailed lifespan data and statistics are provided in Table 1.
Table 1. Summary of lifespan experiments.
| Strain/Food | Mean lifespan (days) | Maximum lifespan (days) | Number of animals (n) | p-value |
|---|---|---|---|---|
| ± SE (days) | ± SE (days) | |||
| N2/E. coli OP50 (OP) | 16.79 ± 0.36 | 23.96 ± 0.21 | 257 | |
| tol-1/E. coli OP50 (OP) | 17.35 ± 0.31 | 23.44 ± 0.23 | 243 | 0.57924 |
| N2/B. infantis (BI) | 24.11 ± 0.38 | 30.41 ± 0.36 | 212 | |
| tol-1/B. infantis (BI) | 25.24 ± 0.47 | 34.03 ± 0.30 | 238 | 0.00002 |
| N2/S. aureus | 7.04 ± 0.14 | 8.75 ± 0.37 | 52 | |
| tol-1/S. aureus | 7.69 ± 0.14 | 9.63 ± 0.23 | 54 | 0.04922 |
| N2/B. subtilis | 20.40 ± 0.67 | 27.70 ± 0.47 | 68 | |
| tol-1/B. subtilis | 20.08 ± 0.92 | 29.00 ± 0.43 | 67 | 0.23265 |
| N2/C. butyricum | 23.00 ± 0.45 | 28.44 ± 0.30 | 112 | |
| tol-1/C. butyricum | 22.67 ± 0.50 | 28.97 ± 0.49 | 124 | 0.75936 |
| N2/astaxanthin | 19.55 ± 0.64 | 25.61 ± 0.63 | 59 | |
| tol-1/astaxanthin | 20.63 ± 0.62 | 26.72 ± 0.40 | 60 | 0.24357 |
SE: standard error.
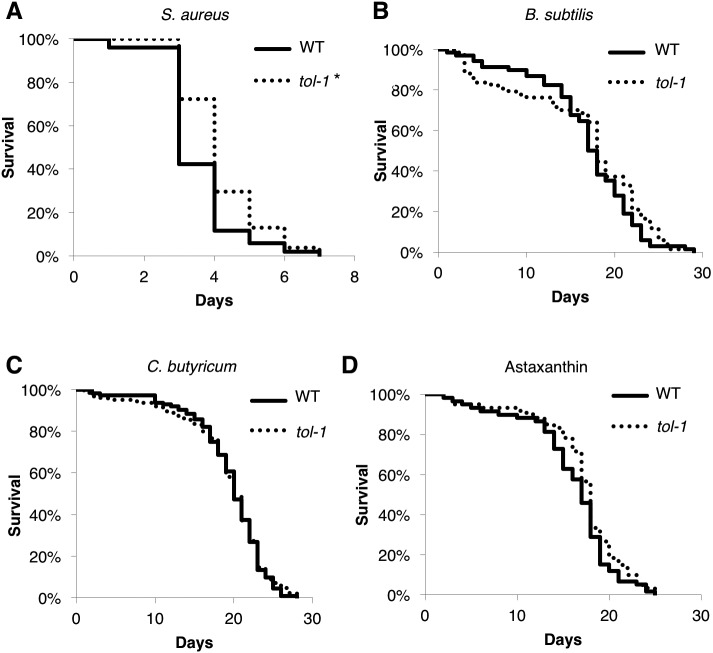
The tol-1 mutation affected lifespan modulated by certain Gram-positive bacteria
Tenor et al. showed that tol-1 mutants were more resistant to Gram-positive bacteria such as E. faecalis compared with wild-type worms [16]. We confirmed that tol-1 mutants exhibited increased resistance to Staphylococcus aureus, which is a Gram-positive bacterium known to shorten the lifespan of the worms [23] (Fig. 2A, Table 1). The lifespan of the animals fed B. subtilis was reported to be significantly longer than that of the animals fed E. coli OP50 [24], and more recently, we showed that feeding with C. butyricum extended the lifespan of C. elegans [25]. Therefore, we examined whether the tol-1 mutation enhanced the lifespan extension caused by the Gram-positive probiotic bacteria B. subtilis and C. butyricum. Neither the lifespan prolongation caused by B. subtilis nor that caused by C. butyricum was enhanced in the tol-1 mutant animals (Fig. 2B, C, Table 1). Finally, we tested whether the lifespan increase due to antioxidative compounds was extended further in the tol-1 mutants. We previously showed that oral supplementation with astaxanthin prolonged the lifespan of the worms [20]. Again, the prolonged lifespan caused by astaxanthin was not changed by the tol-1 mutation. Taken together, the tol-1 mutation appears to affect lifespan modulated by several, but not all, Gram-positive bacteria.
Fig. 2.
Neither the lifespan prolonged by Bacillus subtilis nor that prolonged by Clostridium butyricum was extended in the tol-1 mutant animals.
Survival curves of animals fed several Gram-positive bacteria or astaxanthin. There was a significant difference between the wild-type (WT) and tol-1 mutant animals when they were fed with S. aureus (A) but not when they were fed with B. subtilis (B), C. butyricum (C), or astaxanthin (D). *p<0.05, log-rank test. NS: not significant, log-rank test. Detailed lifespan data and statistics are provided in Table 1.
Expression of innate immune response-related genes was significantly upregulated in the tol-1 mutant
To investigate the TOL-1-dependent molecular mechanism(s) in C. elegans, we performed a microarray analysis. Comparing the transcriptional profiles of the wild-type and tol-1 mutant worms revealed genes that were upregulated and downregulated in the tol-1 mutant. Genes with | Signal Log Ratio | ≥ 1 were subjected to the enriched gene ontology (GO) terms using DAVID (https://david.ncifcrf.gov/summary.jsp). Notably, the top-ranked GO term in the upregulated genes was ‘innate immune response’ (p=1.4 × 10−4) (Table 2). We also found that the lys-3, ilys-2, ilys-3, and ilys-4 genes, which encode lysozyme, were induced in the tol-1 mutants (Table 3). ilys-2 and ilys-3 have been reported to be specifically induced by Gram-positive bacteria [26], although lys-3 is upregulated by both Gram-positive and Gram-negative bacteria and by fungi [26]. Because lysozyme catalyzes the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in peptidoglycan, which is the major component of the Gram-positive bacterial cell wall, the increased expression of lysozyme genes might be one of the mechanisms that govern enhanced resistance to Gram-positive staphylococci in the tol-1 mutants. Moreover, upregulation of the lysozymes in the tol-1 mutants might also promote digestion of the Gram-positive B. infantis cell wall. We previously reported that the cell-wall fraction of B. infantis could extend the worm’s lifespan, suggesting that crucial component(s) for longevity are included in the cell wall [8]. Taken together, effective digestion of the B. infantis cell wall might result in the enhancement of longevity caused by B. infantis in the tol-1 mutants.
Table 2. Gene Ontology (GO) enrichment for genes upregulated in the tol-1 mutants.
| GO terms | p-values |
|---|---|
| Innate immune response | 1.4 × 10–4 |
| Lipid storage | 2.2 × 10–4 |
| Endoplasmic reticulum unfolded protein response | 2.7 × 10–4 |
| Protein ubiquitination | 1.8 × 10–3 |
| Positive regulation of smooth muscle contraction | 4.0 × 10–3 |
Table 3. Lysozyme genes upregulated in the tol-1 mutants.
| Gene symbol (Ensembl name) | Log ratio | p-values |
|---|---|---|
| lys-3 (Y22F5A.6) | 2.3 | 2.0 × 10–5 |
| ilys-2 (C45G7.2) | 1 | 5.5 × 10–4 |
| ilys-3 (C45G7.3) | 0.7 | 9.7 × 10–4 |
| ilys-4 (C55F2.2) | 0.6 | 3.9 × 10–4 |
tol-1 mutants showed reduced leaving behavior from the B. infantis lawn
TOL-1 has been reported to be required for the avoidance of the pathogenic bacterium S. marcescens; in contrast to the wild-type animals, the tol-1 mutants showed reduced leaving behavior from the S. marcescens lawn [17]. We previously found that the wild-type worms exhibited leaving behavior from the B. infantis lawn due to low preference [27] despite its beneficial effects, such as lifespan extension. The lawn-leaving assay using B. infantis revealed that the tol-1 mutants displayed significantly reduced leaving behavior from the B. infantis lawn when compared with the wild-type animals (Fig. 3A, B). Brandt and Ringstad showed that the canonical TLR signaling factors trf-1/TRAF and ikb-1/IκB were also required for avoidance of S. marcescens [18]. The trf-1 and ikb-1 mutants, however, showed normal leaving behavior from the B. infantis lawn (Fig. 3C). Thus, the mechanisms underlying the leaving behavior from the B. infantis lawn are at least partially distinguishable from those underlying the leaving behavior from the S. marcescens lawn.
Fig. 3.
TOL-1, but not the canonical TLR pathway, was involved in leaving behavior from the B. infantis lawn.
(A) The numbers of worms that left the bacteria lawn and remained on the bacteria lawn were counted, and the percentage of animals that were outside of the lawn was calculated as follows: (number of animals outside of the lawn) / (total number of animals) × 100%. (B) The percentages of animals that left the bacterial lawn. The percentage of animals that left the lawn of B. infantis was significantly lower in the tol-1 mutants than in the wild-type worms. OP: E. coli OP50; BI: B. infantis. (C) trf-1 or ikb-1 mutants exhibited normal leaving behavior from the B. infantis lawn. Error bars indicate the SEM. More than three independent replicates were tested for each assay. **p<0.005, t-test.
DISCUSSION
Our results suggest that the mutation in the tol-1 gene enhanced the increased lifespan caused by B. infantis but not by other probiotic bacteria, such as B. subtilis or C. butyricum. In addition, Tenor et al. and the present study showed that tol-1 mutants exhibited increased resistance to Gram-positive bacteria, including E. faecalis and S. aureus, but decreased resistance to Gram-negative bacteria [16]. Together, the tol-1 mutation appears to affect lifespan modulated by several, but not all, Gram-positive bacteria. Although Tenor et al. discussed that TOL-1 might be more important in innate immunity to Gram-negative bacteria than in innate immunity to Gram-positive bacteria and that C. elegans might have evolved a TOL-1-dependent mechanism to enhance immunity to certain pathogens while inhibiting immunity to others [16], the molecular mechanisms that account for this have not been elucidated. Our microarray results showed that the expression of innate immune response-related genes was induced in the tol-1 mutant and that lysozyme genes were also upregulated. The increased expression of these genes might be one of the mechanisms that enhance resistance to Gram-positive bacteria in the tol-1 mutants. However, the possible mechanism likely does not explain the difference in the influence of the tol-1 mutation among probiotic bacteria because B. infantis, B. subtilis, and C. butyricum are all Gram-positive bacteria. Although the enhanced expression of the lysozyme genes in the tol-1 mutant could be effective in feeding on B. infantis, the genes regulated by TOL-1 might not play pivotal roles in B. subtilis and C. butyricum. Kim et al. reported that there were some similarities and differences in the expression patterns of defense-related genes that were induced by Gram-positive bacteria; e.g., clec-60, which encodes a C-type lectin and is known to be upregulated by S. aureus infection, was induced by Lactobacillus acidophilus NFCM, but not by B. subtilis ATCC 6633 [28]. Identification of genes affected by B. infantis but not by B. subtilis or C. butyricum may be fruitful in exploring the mechanism(s) by which TOL-1 specifically modulates the lifespan of worms in the presence of B. infantis.
We demonstrated that tol-1 mutants showed reduced leaving behavior from the B. infantis lawn. Recent findings suggest that mammalian TLRs play essential roles not only in immune responses but also in nervous system development, neurodegeneration, and diseases [29]. As shown in mammals, the C. elegans TLR is also expressed in neurons and plays roles in pathogen-related avoidance behavior [17, 18]. Considering that the tol-1 gene has been reported to be expressed in the four URY neurons, six mechanoreceptor cells (ALML/R, AVM, PLML/R, and PVM), and six interneurons (ALNL/R, AVDL/R, and two neurons in the retrovesicular ganglion that remain to be identified) [3, 17], TOL-1 might act in the neurons to regulate B. infantis-related longevity and/or behavior. In the present study, it remains to be determined whether the reduced lawn-leaving behavior was responsible for the enhancement of the prolonged lifespan caused by B. infantis. However, we previously reported that daf-16/FOXO isoform b plays a positive role in leaving behavior from the B. infantis lawn [27], and determining whether B. infantis-induced lifespan extension is enhanced by the daf-16 mutation, may give a hint.
In conclusion, the C. elegans TLR, TOL-1, regulates B. infantis-induced longevity and also regulates leaving behavior from the B. infantis lawn. TLRs are highly conserved among animal species, including humans, and B. infantis is distributed in the human gut and is also widely used as a probiotic. It would be worth pursuing the role of TLRs in innate immunity and the nervous system as well as B. infantis-induced beneficial effects, including prolongevity.
Acknowledgments
We thank the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA; supported by the National Institutes of Health-National Center for Research Resources) for providing C. elegans strains and Miyarisan Pharmaceutical for providing C. butyricum MIYAIRI 588.
References
- 1.Sanders ME. 2008. Probiotics: definition, sources, selection, and uses. Clin Infect Dis 46Suppl 2: S58–S61, discussion S144–S151. [DOI] [PubMed] [Google Scholar]
- 2.Gibson GR, Wang X. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol 77: 412–420. [DOI] [PubMed] [Google Scholar]
- 3.O’Hara AM, O’Regan P, Fanning A, O’Mahony C, Macsharry J, Lyons A, Bienenstock J, O’Mahony L, Shanahan F. 2006. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology 118: 202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheil B, MacSharry J, O’Callaghan L, O’Riordan A, Waters A, Morgan J, Collins JK, O’Mahony L, Shanahan F. 2006. Role of interleukin (IL-10) in probiotic-mediated immune modulation: an assessment in wild-type and IL-10 knock-out mice. Clin Exp Immunol 144: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O’Mahony L, Kiely B, Shanahan F, Quigley EM. 2006. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol 101: 1581–1590. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y. 2007. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against salmonella enterica serovar enteritidis. Appl Environ Microbiol 73: 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finch CE, Ruvkun G. 2001. The genetics of aging. Annu Rev Genomics Hum Genet 2: 435–462. [DOI] [PubMed] [Google Scholar]
- 8.Komura T, Ikeda T, Yasui C, Saeki S, Nishikawa Y. 2013. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 14: 73–87. [DOI] [PubMed] [Google Scholar]
- 9.Stein D, Roth S, Vogelsang E, Nüsslein-Volhard C. 1991. The polarity of the dorsoventral axis in the Drosophila embryo is defined by an extracellular signal. Cell 65: 725–735. [DOI] [PubMed] [Google Scholar]
- 10.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. 1996. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86: 973–983. [DOI] [PubMed] [Google Scholar]
- 11.Belvin MP, Anderson KV. 1996. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol 12: 393–416. [DOI] [PubMed] [Google Scholar]
- 12.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388: 394–397. [DOI] [PubMed] [Google Scholar]
- 13.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. 1998. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA 95: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libermann TA, Baltimore D. 1990. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol 10: 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore TD. 1999. The Rel/NF-kappaB signal transduction pathway: introduction. Oncogene 18: 6842–6844. [DOI] [PubMed] [Google Scholar]
- 16.Tenor JL, Aballay A. 2008. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep 9: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol 11: 809–821. [DOI] [PubMed] [Google Scholar]
- 18.Brandt JP, Ringstad N. 2015. Toll-like receptor signaling promotes development and function of sensory neurons required for a C. elegans pathogen-avoidance behavior. Curr Biol 25: 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashima N, Fujikura Y, Komura T, Fujiwara S, Sakamoto M, Terao K, Nishikawa Y. 2012. Development of a method for oral administration of hydrophobic substances to Caenorhabditis elegans: pro-longevity effects of oral supplementation with lipid-soluble antioxidants. Biogerontology 13: 337–344. [DOI] [PubMed] [Google Scholar]
- 21.Wu D, Rea SL, Yashin AI, Johnson TE. 2006. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp Gerontol 41: 261–270. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Gilliat AF, Ziehm M, Turmaine M, Wang H, Ezcurra M, Yang C, Phillips G, McBay D, Zhang WB, Partridge L, Pincus Z, Gems D. 2017. Two forms of death in ageing Caenorhabditis elegans. Nat Commun 8: 15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sifri CD, Begun J, Ausubel FM, Calderwood SB. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun 71: 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300: 1921. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Hamazaki Y, Sun S, Nishikawa Y, Kage-Nakadai E. 2018. Clostridium butyricum MIYAIRI 588 increases the lifespan and multiple-stress resistance of Caenorhabditis elegans. Nutrients 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dierking K, Yang W, Schulenburg H. 2016. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Philos Trans R Soc Lond B Biol Sci 371: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S, Ohta A, Kuhara A, Nishikawa Y, Kage-Nakadai E. 2019. daf-16/FOXO isoform b in AIY neurons is involved in low preference for Bifidobacterium infantis in Caenorhabditis elegans. Neurosci Res S0168-0102(18)30597-2. doi: 10.1016/j.neures.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 28.Kim Y, Mylonakis E. 2012. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect Immun 80: 2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okun E, Griffioen KJ, Mattson MP. 2011. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci 34: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]