Abstract
Dynamic changes in protein structure can be monitored using a fluorescent probe and a dark quencher. This approach is contingent upon the ability to precisely introduce a fluorophore/quencher pair into two specific sites of a protein of interest. Despite recent advances, there is continued demand for new and convenient approaches to site-selectively label proteins with such optical probes. We have recently developed a chemoselectively rapid azo-coupling reaction (CRACR) for site-specific protein labeling, which relies on rapid coupling between a genetically encoded 5-hydroxytryptophan residue and various aromatic diazonium ions. Here we report that the product of this conjugation reaction, a highly chromophoric biaryl-azo group, is a potent fluorescence quencher. The absorption properties of this azo product can be tuned by systematically altering the structure of the aryldiazonium species. We identified a particular ‘quenchergenic’ aryldiazonium that, upon conjugation, efficiently quenches the fluorescence of the green fluorescent protein (GFP), a widely used genetically encoded fluorescent probe that can be terminally attached to target proteins. This fluorophore/quencher pair was used to evaluate the protein-labeling kinetics of CRACR, as well as to monitor the proteolysis of a fusion protein.
Keywords: bioconjugation, unnatural amino acid, genetic code expansion, site-specific protein modification, fluorescence quencher, FRET, proteolysis
Graphical Abstract
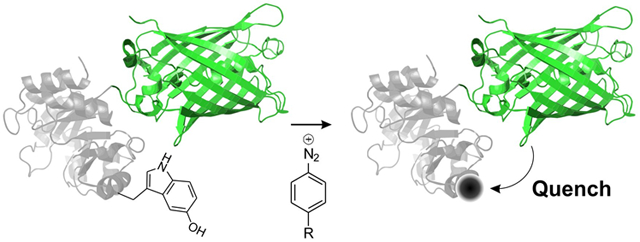
Proteins often depend on dynamic structural changes to accomplish their biological roles, and the ability to monitor such processes is essential to fully understand protein function.1 Additionally, the ability to detect the dynamic changes of engineered proteins and peptides in response to specific cues serves as the basis for the development of designer biological sensors.2 One popular strategy for detecting such dynamic changes takes advantage of a fluorescent probe and a non-fluorescent dye (a ‘dark quencher’), where the fluorescence of the former is quenched by the latter in a distance-dependent manner.3 Such fluorophore/quencher pairs have been frequently used in numerous applications such as for detecting protease activity,4 development of sensors,5 and in real-time PCR.6
To use a fluorophore/quencher pair for detecting dynamic changes in protein structure, it is essential to selectively attach them at strategic locations. Several approaches have been developed to attach such optical probes onto proteins with site-selectivity, which include labeling an engineered cysteine residue (if no other accessible cysteine present),7 unique protein and peptide tags,8 terminal modification,9 etc. Genetically encoded non-canonical amino acids (ncAAs), which can be site-specifically incorporated into proteins in response to nonsense codons, provide a versatile alternative strategy for labeling proteins with optical probes at predefined locations.10 A number of ncAAs with uniquely reactive bioconjugation handles have been genetically encoded, which can be chemoselectively labeled post-translationally.10-11 Recent advances in this technology has enabled the incorporation of up to two distinct ncAAs into the same protein, followed by their labeling with different optical probes.12
Typically, biophysical studies rely on labeling proteins with pre-synthesized optical probe(s) using one of the methods described above. However, it is attractive to design an alternative strategy, where a chemoselective reaction is used to construct an optically active probe directly on the protein from optically inactive reagents. Indeed, it has been possible to develop ‘fluorogenic’ labeling reactions, where the conjugation of a non-fluorescent or weakly fluorescent reagent onto a protein results in an entity that is highly fluorescent.13 For example, the inverse-electron demand Diels-Alder cycloaddition between tetrazine and alkenes has been used for this purpose; while the tetrazine reagent is non-fluorescent and a dark quencher, the resulting diazines can be fluorescent.13c Despite their attractive nature, only a limited collection of ‘probe-genic’ labeling strategies are currently available. Here we report a chemoselective protein labeling strategy at genetically incorporated 5-hydroxytryptophan residues, that results in the formation of efficient fluorescence quenchers. Quenchers with different absorption properties can be generated simply by using structurally diverse aryldiazonium reagents. We developed a particular ‘quenchergenic’ aryldiazonium reagent that efficiently quenches the fluorescence of the green fluorescent protein (GFP) and demonstrated the use of this fluorescence/quencher pair for monitoring a proteolysis reaction in vitro.
Recently, we reported the development of a unique E. coli derived tryptophanyl-tRNA synthetase (EcTrpRS)/tRNA pair that can be evolved in an engineered E. coli strain ATMW1 to charge a variety of tryptophan analogs, including 5-hydroxytryptophan (5HTP).14 ATMW1 was generated from a precursor strain by functionally substituting its endogenous EcTrpRS/tRNA pair with a yeast-derived tryptophanyl pair.14 The ability to liberate the endogenous tryptophanyl-pair from its housekeeping duties in E. coli enabled its use in the resulting ATMW1 strain as an orthogonal nonsense suppressor. The engineered EcTrpRS/tRNA pairs are also orthogonal in eukaryotic cells, allowing their use for ncAA incorporation into virtually any protein.14 The ability to genetically encode electron-rich aromatic amino acids such as 5HTP presented the opportunity to develop new chemoselective protein modification strategies that take advantage of their unique reactivity. Indeed, we have recently developed an oxidative coupling reaction that enables selective attachment of various aromatic amines to the 5HTP residue.15 Additionally, we have shown that various aryldiazonium ions react rapidly with the 5HTP residue, leading to the development of a chemoselective rapid azo-coupling reaction (CRACR) for site-specific protein labeling.16
5HTP does not have significant absorption in the visible range of the spectrum, but upon reaction with aryldiazonium reagents, it generates intensely chromophoric azo-compounds. Given aromatic azo-dyes are the most frequently used scaffolds for the development of commonly used dark quenchers (e.g., Dabcyl, Black Hole Quencher®, etc.),3 we envisioned that the coupling product of CRACR may also be used as a fluorescence quencher. Structurally diverse aryldiazoniums can be readily generated from the corresponding amines, which can be used to optimize the optical properties of the azo-coupling product with 5HTP. To test this hypothesis, we synthesized the azo-coupling product of a model 5-hydroxyindole with three different aryldiazoniums: 4-carboxyphenyldiazonium (4CDZ), 4-nitrophenyldiazonium (4NDZ) and 4-methoxyphenyldiazonium (4MDZ) (Figure 1A). As expected, all three compounds were highly chromophoric, with absorption maxima between 460 – 480 nm, but the more electron deficient aryldiazoniums 4CDZ and 4NDZ produced azo dyes with more red-shifted spectra (Figure 1B). The molar extinction coefficient of these dyes at 480 nm were found to be between 6,100 and 12,700 M−1cm−1 (Figure S1). The 4CDZ-derived azo product had the strongest absorptivity, and we focused on further evaluating its potential use as a fluorescence quencher.
Figure 1.
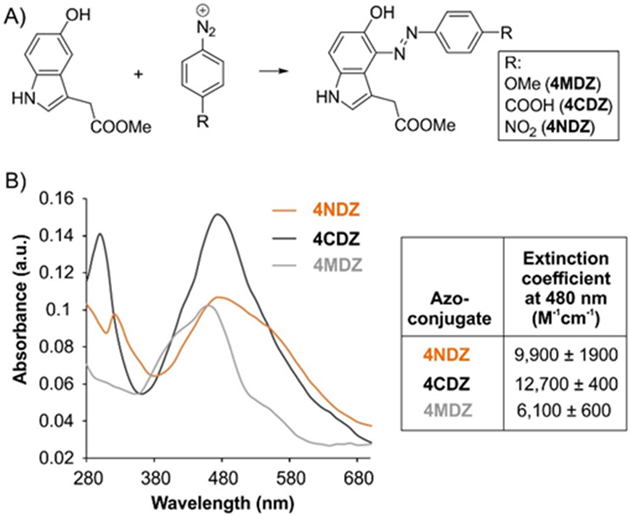
Azo dyes derived from 5-hydroxyindole are highly chromophoric. A) Structures of the azo compounds synthesized, and B) their normalized absorption spectrum, as well as molar extinction coefficient at 480 nm.
The 4CDZ-derived azo compound absorbs strongly between 430 – 530 nm, indicating that fluorophores emitting in this range could be suitable partners for this quencher (Figure 1B). The green fluorescent protein (GFP) is a widely used genetically encoded fluorescent protein which emits in this range.17 It can be readily introduced at either termini of a target protein, and such fusions are typically well tolerated. To evaluate if the 5HTP-4CDZ azo-conjugate can quench the fluorescence of GFP, we first site-specifically incorporated the 5HTP residue into the surface-exposed 151 position of superfolder GFP (sfGFP). A plasmid harboring the sfGFP-151-UGA gene was co-transformed into ATMW1 strain with a second plasmid encoding an engineered EcTrpRS/tRNAUCA pair, which was previously engineered to incorporate 5HTP (Figure 2A). The protein expression was performed in rich media in the presence of 1 mM 5HTP and the resulting sfGFP-151–5HTP was purified using a C-terminal polyhistidine tag by immobilized metal-ion affinity chromatography (IMAC). ESI-MS analysis was used to verify the incorporation of 5HTP into the desired position, and to demonstrate that sfGFP-151–5HTP could be readily modified by 4CDZ under ambient conditions (Figure 2B). To investigate if the 5HTP-4CDZ azo conjugate indeed quenches the sfGFP fluorescence, we monitored the fluorescence of sfGFP-151–5HTP before and after the conjugation reaction. A dramatic attenuation of sfGFP fluorescence was observed both by fluorescence spectroscopy, as well as visual inspection under trans-illumination, indicating efficient quenching by the azo-conjugate (Figure 2C). We have previously shown that the azo-linkages generated through CRACR can be reduced by dithionite. Reduction of the azo group would destroy the quencher chromophore, and should restore sfGFP fluorescence. Indeed, treatment of the sfGFP-151–5HTP-4CDZ conjugate with dithionite resulted in restoration of its fluorescence, confirming our hypothesis (Figure 2C).
Figure 2.
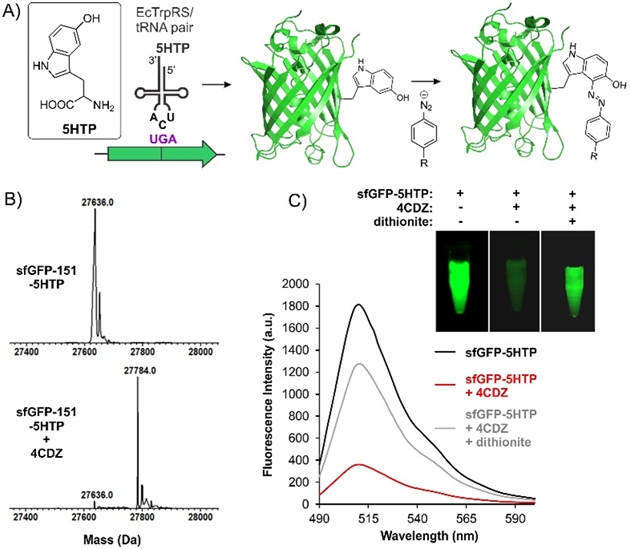
5HTP-4CDZ azo dye can quench sfGFP fluorescence. A) The scheme for expressing sfGFP-151-5HTP and its subsequent labeling with 4CDZ. B) ESI-MS analysis of sfGFP-151-5HTP before and after treatment with 4CDZ confirm selective labeling. C) Normalized fluorescence spectra of sfGFP-151-5HTP before and after 4CDZ labeling show robust quenching of fluorescence. Reduction of the azo-linkage by dithionite results in a significant recovery of the sfGFP fluorescence. Images of these samples under trans-illumination are also shown.
Although we have previously kinetically characterized the azo-coupling reaction between free 5HTP and 4CDZ,16 it is presumable that the 5-hydroxyindole residue on a protein may react somewhat differently. The quenchergenic aspect of CRACR that we describe here should enable facile measurement of the kinetics of this labeling reaction on sfGFP, by monitoring the concomitant decrease in its fluorescence. We evaluated the rate of labeling of sfGFP-151–5HTP using 4CDZ under pseudo-first order conditions (Figure 3), revealing a second-order rate constant of 38 ± 2 M−1s−1, which is roughly five-fold lower than the rate-constant observed for free 5HTP (190 M−1s−1) that we previously reported.16 This is not surprising, given that the microenvironment of the 5HTP residue on a protein may sterically hinder the approach of the aryldiazonium reagent. A better understanding of the labeling kinetics of this chemoselective reaction on a protein would be helpful for designing future applications.
Figure 3.
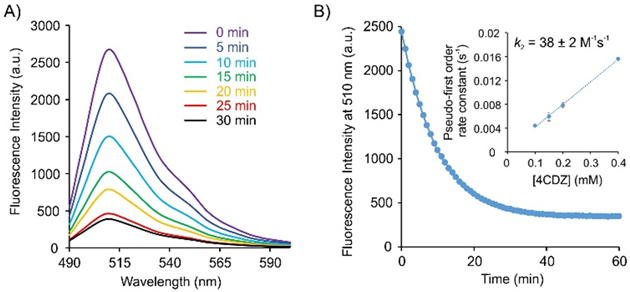
Evaluating the labeling kinetics of sfGFP-151-5HTP by CRACR using the associated fluorescence quenching. A) Fluorescence spectra of a reaction mixture consisting of 5 μM sfGFP-151-5HTP and 50 μM 4CDZ at room temperature in 100 mM phosphate buffer over time. B) Fluorescence intensity of the same reaction monitored over time yields the pseudo-first order rate constant. (Inset) Plotting the resulting pseudo-first order rate constant at various 4CDZ concentrations against the corresponding [4CDZ] provides the second order rate constant (k2) of the labeling reaction.
Proteolysis reactions are associated with numerous important biochemical pathways, and much effort has been devoted to develop assays for monitoring these reactions.1b, 4, 18 We envisioned that the fluorophore/quencher pair we describe here can be useful for monitoring proteolysis reactions. If GFP and 5HTP are incorporated on the two sides of the scissile peptide bond, and the 5HTP residue is functionalized using our quenchergenic 4CDZ reagent, the proteolysis reaction can be monitored by the gain of GFP fluorescence as the fluorophore and the quencher are cleaved apart (Figure 4B). To test this notion, we constructed an enhanced GFP (EGFP) construct N-terminally fused with a SUMO domain for expression in mammalian cells.19 The N-terminal SUMO can be proteolytically removed using the highly selective SUMO protease.19b We also mutated the surface exposed Glu52 residue to a UGA codon to direct the incorporation of 5HTP at this site, which can be subsequently converted to a dark quencher through its conjugation with 4CDZ. A single plasmid was constructed encoding the 5HTP-specific EcTrpRS/tRNAUCA pair, as well as the SUMO-EGFP construct (wild-type or mutant), for expression in mammalian cells (Figure 4A). HEK293T cells were transfected with this plasmid in the presence of 1 mM 5HTP in the media. Expression of full-length proteins could be readily monitored using the fluorescence of EGFP by fluorescence microscopy (Figure S2). The wild-type and 52–5HTP mutant of SUMO-EGFP were purified from HEK293T cells by IMAC using a C-terminal polyhistidine tag and ESI-MS analysis was used to confirm expected masses for both (Figure S3 and S4). Treatment with 4CDZ led to the expected modification of only SUMO-52–5HTP-EGFP (Figure 4C), but not the wild-type SUMO-EGFP (Figure S5), as revealed by ESI-MS analysis. Fluorescence spectroscopy was used to demonstrate that the 4CDZ modification of SUMO-52–5HTP-EGFP was indeed associated with significant attenuation of EGFP fluorescence (Figure 4D). The degree of quenching was weaker than what was observed with sfGFP-151–5HTP (Figure 2C), which is expected, since the distance between the quencher and the GFP chromophore is significantly longer in the SUMO-EGFP construct. Next, we treated the 4CDZ-modified SUMO-52–5HTP-EGFP construct with SUMO-protease. As expected, proteolytic removal of SUMO-52–5HTP-4CDZ led to a time-dependent recovery of the EGFP fluorescence, which reached completion at around 90 minutes (Figure 4E). SDS-PAGE analysis of the proteolysis reaction at this time confirmed complete cleavage of the 4CDZ-modified SUMO-52–5HTP-EGFP construct (Figure 4F). These observations demonstrate that the terminal GFP and the site-specifically incorporated 5HTP-4CDZ quencher can be used effectively to monitor proteolysis reactions.
Figure 4.
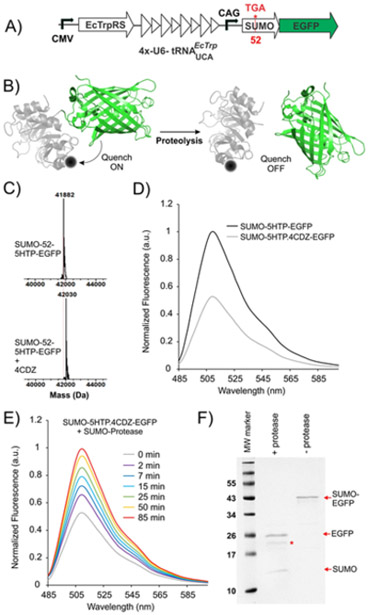
Monitoring a proteolysis reaction using 5HTP-4CDZ/GFP quencher/fluorophore pair. A) Map of the plasmid for mammalian expression of SUMO-52-5HTP-EGFP. B) The strategy for monitoring the proteolysis reaction. C) ESI-MS analysis and D) fluorescence spectra of purified SUMO-52-5HTP-EGFP before and after modification with 4CDZ. E) Normalized fluorescence spectra of the proteolysis reaction of 4CDZ-labeled SUMO-52-5HTP-EGFP (by SUMO-star protease) over time show complete recovery of fluorescence after 90 minutes. F) SDS-PAGE analysis confirms complete proteolysis at this time. Intact SUMO-EGFP, as well as cleaved SUMO and EGFP bands are shown. The band marked with * corresponds to SUMO-star protease.
In summary, here we describe a CRACR-based site-selective protein labeling strategy that results in the construction of a potent quencher functionality on proteins. We show that this quenchergenic labeling strategy can be applied to proteins expressed in both bacteria and mammalian cells. We also developed a specific quenchergenic label that can be combined with GFP to monitor dynamic changes in protein structure. A notable strength of this approach is the ability to generate a large variety of different azo-quenchers with distinct optical properties simply by using different aryldiazonium ions, which can be readily prepared from the corresponding aromatic amines. The quenchers generated by this approach should also be compatible with various small fluorescent probes, which can be simultaneously attached onto proteins or peptides using a mutually compatible strategy.
Supplementary Material
Acknowledgements
This work was supported by NIH grants R01GM126220 and R01GM124319 to A.C.
References
- [1] a).Henzler-Wildman K, Kern D, Nature 2007, 450, 964–972; [DOI] [PubMed] [Google Scholar]; b) King RW, Deshaies RJ, Peters JM, Kirschner MW, Science 1996, 274, 1652–1659; [DOI] [PubMed] [Google Scholar]; c) Smock RG, Gierasch LM, Science 2009, 324, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2] a).VanEngelenburg SB, Palmer AE, Current opinion in chemical biology 2008, 12, 60–65; [DOI] [PubMed] [Google Scholar]; b) Zhang J, Campbell RE, Ting AY, Tsien RY, Nature Reviews Molecular Cell Biology 2002, 3, 906; [DOI] [PubMed] [Google Scholar]; c) Ibraheem A, Campbell RE, Current opinion in chemical biology 2010, 14, 30–36. [DOI] [PubMed] [Google Scholar]
- [3] a).Crisalli P, Kool ET, Bioconjugate chemistry 2011, 22, 2345–2354; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chevalier A, Hardouin J, Renard P-Y, Romieu A, Organic letters 2013, 15, 6082–6085; [DOI] [PubMed] [Google Scholar]; c) Le Reste L, Hohlbein J, Gryte K, Kapanidis AN, Biophysical journal 2012, 102, 2658–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4] a).Bullok KE, Maxwell D, Kesarwala AH, Gammon S, Prior JL, Snow M, Stanley S, Piwnica-Worms D, Biochemistry 2007, 46, 4055–4065; [DOI] [PubMed] [Google Scholar]; b) Zheng G, Chen J, Stefflova K, Jarvi M, Li H, Wilson BC, Proceedings of the National Academy of Sciences 2007, 104, 8989–8994; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Blum G, Mullins SR, Keren K, Fonovič M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M, Nature chemical biology 2005, 1, 203; [DOI] [PubMed] [Google Scholar]; d) Blum G, Von Degenfeld G, Merchant MJ, Blau HM, Bogyo M, Nature chemical biology 2007, 3, 668; [DOI] [PubMed] [Google Scholar]; e) Taliani M, Bianchi E, Narjes F, Fossatelli M, Urbani A, Steinkühler C, De Francesco R, Pessi A, Analytical biochemistry 1996, 240, 60–67; [DOI] [PubMed] [Google Scholar]; f) Garcia-Echeverria C, Rich DH, FEBS letters 1992, 297, 100–102; [DOI] [PubMed] [Google Scholar]; g) Beekman B, van El B, Drijfhout JW, Ronday HK, TeKoppele JM, FEBS letters 1997, 418, 305–309. [DOI] [PubMed] [Google Scholar]
- [5] a).Mueller C, Grossmann TN, Angewandte Chemie 2018; [Google Scholar]; b) Tyagi S, Kramer FR, Nature biotechnology 1996, 14, 303; [DOI] [PubMed] [Google Scholar]; c) Tan W, Wang K, Drake TJ, Current opinion in chemical biology 2004, 8, 547–553; [DOI] [PubMed] [Google Scholar]; d) Zheng J, Yang R, Shi M, Wu C, Fang X, Li Y, Li J, Tan W, Chemical Society Reviews 2015, 44, 3036–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6] a).Huang S, Salituro J, Tang N, Luk K-C, Hackett J Jr, Swanson P, Cloherty G, Mak W-B, Robinson J, Abravaya K, Nucleic acids research 2007, 35, e101; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bustin S, Journal of molecular endocrinology 2002, 29, 23–39. [DOI] [PubMed] [Google Scholar]
- [7].Kim Y, Ho SO, Gassman NR, Korlann Y, Landorf EV, Collart FR, Weiss S, Bioconjugate chemistry 2008, 19, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8] a).Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, ACS chemical biology 2008, 3, 373–382; [DOI] [PubMed] [Google Scholar]; b) Gautier A, Juillerat A, Heinis C, Corrêa IR Jr, Kindermann M, Beaufils F, Johnsson K, Chemistry & biology 2008, 15, 128–136; [DOI] [PubMed] [Google Scholar]; c) Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K, Nature biotechnology 2003, 21, 86; [DOI] [PubMed] [Google Scholar]; d) Chen I, Howarth M, Lin W, Ting AY, Nature methods 2005, 2, 99; [DOI] [PubMed] [Google Scholar]; e) Fernández-Suárez M, Baruah H, Martínez-Hernández L, Xie KT, Baskin JM, Bertozzi CR, Ting AY, Nature biotechnology 2007, 25, 1483; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Martin BR, Giepmans BN, Adams SR, Tsien RY, Nature biotechnology 2005, 23, 1308. [DOI] [PubMed] [Google Scholar]
- [9] a).Rosen CB, Francis MB, Nature chemical biology 2017, 13, 697–705; [DOI] [PubMed] [Google Scholar]; b) Bloom S, Liu C, Kölmel DK, Qiao JX, Zhang Y, Poss MA, Ewing WR, MacMillan DW, Nature chemistry 2018, 10, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10] a).Chin JW, Nature 2017, 550, 53; [DOI] [PubMed] [Google Scholar]; b) Dumas A, Lercher L, Spicer CD, Davis BG, Chemical Science 2015, 6, 50–69; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mukai T, Lajoie MJ, Englert M, Söll D, Annual review of microbiology 2017, 71, 557–577; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Young DD, Schultz PG, ACS chemical biology 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Italia JS, Zheng Y, Kelemen RE, Erickson SB, Addy PS, Chatterjee A, Biochemical Society Transactions 2017, 45, 555–562. [DOI] [PubMed] [Google Scholar]
- [11] a).Lang K, Chin JW, Chemical reviews 2014, 114, 4764–4806; [DOI] [PubMed] [Google Scholar]; b Lang K, Chin JW, ACS Chem Biol 2014, 9, 16–20. [DOI] [PubMed] [Google Scholar]
- [12] a).Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW, Nature 2010, 464, 441–444; [DOI] [PubMed] [Google Scholar]; b) Sachdeva A, Wang K, Elliott T, Chin JW, J Am Chem Soc 2014, 136, 7785–7788; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR, Angewandte Chemie (International ed. in English) 2010, 49, 3211–3214; [DOI] [PubMed] [Google Scholar]; d) Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, Mehl RA, Chin JW, Nature chemistry 2014, 6, 393–403; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Xiao H, Chatterjee A, Choi SH, Bajjuri KM, Sinha SC, Schultz PG, Angewandte Chemie (International ed. in English) 2013, 52, 14080–14083; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Zheng Y, Addy PS, Mukherjee R, Chatterjee A, Chem Sci 2017, 8, 7211–7217; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Zheng Y, Gilgenast MJ, Hauc S, Chatterjee A, ACS chemical biology 2018, 13, 1137–1141; [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Zheng Y, Mukherjee R, Chin MA, Igo P, Gilgenast MJ, Chatterjee A, Biochemistry 2018, 57, 441–445; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Willis JCW, Chin JW, Nature chemistry 2018, 10, 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13] a).Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R, Angewandte Chemie (International ed. in English) 2010, 49, 2869–2872; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vázquez A, Dzijak R, Dračínský M, Rampmaier R, Siegl SJ, Vrabel M, Angewandte Chemie 2017, 129, 1354–1357; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shang X, Song X, Faller C, Lai R, Li H, Cerny R, Niu W, Guo J, Chemical science 2017, 8, 1141–1145; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu Y, Miao K, Dunham NP, Liu H, Fares M, Boal AK, Li X, Zhang X, Biochemistry 2017, 56, 1585–1595; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Cambray S, Bandyopadhyay A, Gao J, Chemical Communications 2017, 53, 12532–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Italia JS, Addy PS, Wrobel CJ, Crawford LA, Lajoie MJ, Zheng Y, Chatterjee A, Nature chemical biology 2017, 13, 446–450. [DOI] [PubMed] [Google Scholar]
- [15].Addy PS, Italia JS, Chatterjee A, ChemBioChem 2018, 19, 1375–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Addy PS, Erickson SB, Italia JS, Chatterjee A, Journal of the American Chemical Society 2017, 139, 11670–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsien RY, Annual review of biochemistry 1998, 67, 509–544. [DOI] [PubMed] [Google Scholar]
- [18] a).Ciechanover A, Nature reviews. Molecular cell biology 2005, 6, 79–87; [DOI] [PubMed] [Google Scholar]; b) Li J, Yuan J, Oncogene 2008, 27, 6194–6206. [DOI] [PubMed] [Google Scholar]
- [19] a).Mitchell AL, Addy PS, Chin MA, Chatterjee A, ChemBioChem 2017, 18, 511–514; [DOI] [PubMed] [Google Scholar]; b) Peroutka Iii RJ, Orcutt SJ, Strickler JE, Butt TR, Methods in molecular biology 2011, 705, 15–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


