Abstract
BACKGROUND
Chronic lung allograft dysfunction (CLAD) is the main limitation to long-term survival after lung transplantation. Because effective therapies are lacking, early identification and mitigation of risk factors is a pragmatic approach to improve outcomes. Acute cellular rejection (ACR) is the most pervasive risk factor for CLAD, but diagnosis requires transbronchial biopsy, which carries risks. We hypothesized that gene expression in the bronchoalveolar lavage (BAL) cell pellet (CP) could replace biopsy and inform on mechanisms of CLAD.
METHODS
We performed RNA sequencing on BAL CPs from 219 lung transplant recipients with A-grade ACR (n = 61), lymphocytic bronchiolitis (n = 58), infection (n = 41), or no rejection/infection (n = 59). Differential gene expression was based on absolute fold difference >2.0 and Benjamini-adjusted p-value ≤0.05. We used the Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resource for pathway analyses. For classifier modeling, samples were randomly split into training (n = 154) and testing sets (n = 65). A logistic regression model using recursive feature elimination and 5-fold cross-validation was trained to optimize area under the curve (AUC).
RESULTS
Differential gene expression identified 72 genes. Enriched pathways included T-cell receptor signaling, natural killer cell–mediated cytotoxicity, and cytokine–cytokine receptor interaction. A 4-gene model (AUC = 0.72) and classification threshold defined in the training set exhibited fair performance in the testing set; accuracy was 76%, specificity 82%, and sensitivity 60%. In addition, classification as ACR was associated with worse CLAD-free survival (hazard ratio = 2.42; 95% confidence interval = 1.29–4.53).
CONCLUSIONS
BAL CP gene expression during ACR is enriched for immune response pathways and shows promise as a diagnostic tool for ACR, especially ACR that is a precursor of CLAD.
KEYWORDS: Lung transplant, Acute cellular rejection, bronchoalveolar lavage, gene expression, transcriptome, genomic classifier
See Related Editorial, page 856
Alt-text: Unlabelled box
Lung transplant remains a viable treatment option for select patients with advanced lung diseases, but long-term outcomes remain disappointing. The main limitation to better long-term survival is chronic lung allograft dysfunction (CLAD), which affects more than half of the lung recipients by 5 years post-transplant.1 Currently, there are no proven effective treatments. Therefore, preventive strategies are key, including risk factor identification and mitigation. The principal risk factor for CLAD is acute rejection or, more precisely, A-grade acute cellular rejection (ACR), which is diagnosed by transbronchial biopsy (TBBX) exhibiting perivascular mononuclear cell infiltrates that can extend into the interstitium.2 Although definitive data are lacking, it is widely accepted that treatment of A-grade ACR with augmented immune suppression is an important strategy for reducing the risk of CLAD. However, unnecessary treatment increases the risk of opportunistic infections and malignancy, which makes an accurate diagnosis important. Airway inflammation, also known as lymphocytic bronchiolitis (LB), is variably also termed B-grade ACR.2 However, the frequent coexistence with airway infection, as well as reported refractoriness to corticosteroid treatment,3 are sources of controversy for inclusion of LB as ACR.
Although considered the current gold standard, the utility of TBBX to diagnose A-grade ACR is limited by several factors. First, TBBX has been associated with a 4% incidence of pneumothorax and 3% incidence of major bleeding.4 In lung transplant recipients specifically, the incidence of a major complication of bronchoscopy was 2.3%, with TBBX being the major risk factor.5 Besides patient safety, TBBX is associated with a relatively high rate of sampling error. In a recent multicenter study, approximately 8% of TBBXs yielded an inadequate sample unable to be graded for A-grade ACR, and another 26% were assessed as sub-optimal (fewer than 5 pieces of well-expanded alveolated lung).6 Even when alveolated tissue is obtained, affected areas may be missed by the relatively small volume of tissue sampled by TBBX. Finally, there is well-described variability in interobserver interpretation of TBBX for A-grade ACR, with κ values ranging from 0.183 to 0.479.6, 7, 8 Furthermore, the interobserver agreement for B-grade ACR, or LB, was even worse, with κ values ranging from −0.042 to 0.465.6, 7, 8 This limited ability to safely and reliably diagnose ACR may affect clinicians’ ability to mitigate risk and prevent CLAD.
Bronchoalveolar lavage (BAL) is routinely performed concurrently with TBBX to rule out infection. As compared with TBBX, BAL is safer and samples a relatively large area of the lung. We hypothesized that gene expression in the BAL cell pellet (CP) could replace TBBX for the diagnosis of ACR and improve risk stratification for progression to CLAD. Participants in this study provided written informed consent for enrollment in the University of California, Los Angeles (UCLA) lung transplant outcomes registry and biorepository approved by the UCLA institutional review board. This study was sponsored by a Clinical Trials in Organ Transplant ancillary studies grant.
Methods
We have enrolled lung transplant recipients into the UCLA lung transplant outcomes registry and biorepository study since 2001. Lung recipients at UCLA undergo surveillance bronchoscopy at 1, 3, 6, and 12 months post-transplant and when clinically indicated. We collected and banked any leftover BAL fluid from all surveillance and “for cause” bronchoscopies.
The BAL procedure was performed according to a standardized protocol using three 60-ml aliquots of isotonic saline instilled into a sub-segmental bronchus of either the right middle lobe or left lingula. Retrieved BAL fluid was pooled and then split into a 15-ml clinical specimen and a research specimen with the remaining volume. The research samples were immediately placed on ice for transport to the lab and were processed within 6 hours of collection. BAL fluid was filtered through sterile gauze, and cells were separated from fluid by centrifugation. Cells were washed twice with phosphate-buffered saline and lysed in TRIzol (Invitrogen, Carlsbad, CA).
TBBX specimens were graded for ACR by experienced thoracic pathologists according to the standard International Society for Heart and Lung Transplantation criteria.2, 9, 10 Briefly, peri-vascular (A-grade) infiltrates were scored 0–4. However, before 2008, our pathologists only graded peri-airway infiltrates (LB or B-grade) as absent (B0) or present (B1). In 2008, our center adopted the revised nomenclature for B-grade rejection (B0, B1R, and B2R). Because the specimens used in this study span this change in nomenclature, we could only consider LB as present or absent. Biopsy results were based upon chart review of clinical read only. Slides were not re-reviewed. A-grade ACR episodes were also categorized as either spirometrically significant (SSAR) or non-SSAR based on a ≥10% decline in forced expiratory volume in one second (FEV1) from the baseline, defined as the highest of the 2 preceding FEV1 measurements, as described by Davis et al.11 A-grade ACR episodes without a paired FEV1 measurement between 0 and 14 days before the biopsy could not be classified as SSAR or non-SSAR. CLAD was defined as a sustained drop in FEV1 by at least 20% from the average of the 2 best post-transplant measurements, consistent with published criteria.12 For each potential CLAD case, we reviewed available clinical data to exclude alternative causes of FEV1 decline other than CLAD.
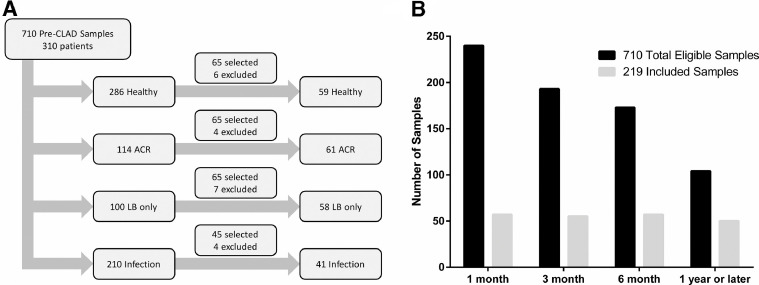
We searched our biorepository for BAL CP samples, collected before a diagnosis of CLAD or from patients who never developed CLAD, who were paired with a TBBX graded for both A- and B-grade rejection scores. Out of 1,548 pre-CLAD samples in our repository, 660 did not have a paired TBBX and were excluded. An additional 178 samples were excluded because the paired biopsy was not gradable for A-grade (AX) (n = 12) or not gradable for B-grade (BX) (n = 166) rejection. We identified a total of 710 eligible BAL samples from 310 patients. Samples were then categorized into the following groups: (1) healthy (n = 286) without A-grade ACR, LB, or infection; (2) A-grade ACR (n = 114) defined as A-grade of A1 or greater, with or without concurrent LB, and without infection; (3) LB (n = 100) defined as the presence of small airway inflammation without concurrent A-grade ACR or infection; and (4) infection (n = 210) defined as a positive culture for a potential pathogen. Blinded to CLAD and mortality outcomes, we then selected 65 samples each from the healthy, A-grade ACR, and LB categories and 45 additional samples from the infection group for RNA isolation. In addition, samples were selected to achieve relative balance across the following post-transplant time-frame windows: 1 month (16–60 days post-transplant), 3 months (61–135 days post-transplant), 6 months (136–270 days post-transplant), and 1 year or later (>271 days post-transplant). No more than 1 sample per patient was included.
Total RNA was isolated using TRIzol/chloroform extraction, resuspended in RNase-free water, and purified using the miRNeasy Mini kit (Qiagen Inc, Valencia, CA). RNA samples were then bio-analyzed with the Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA), and samples were excluded if RNA quantity was too low (<100 ng) or if the RNA integrity number indicated severe degradation (<3.0). This excluded 21 samples (6 healthy, 4 infection, 7 LB, and 4 A-grade ACR) for insufficient quantity (n = 16) or for degraded RNA (n = 5).
RNA sequencing (RNASeq) libraries were prepared with Clontech SMARTer Stranded Total RNASeq (Pico) Kit (Takara Bio, Kusatsu, Japan). The key steps include first-strand synthesis, template switching, adapter ligation, cleavage of ribosomal complementary DNA, and polymerase chain reaction amplification. The library qualities were evaluated using the Agilent 2100 BioAnalyzer and then sequenced using Illumina Hiseq3000 (SR 1 × 50 run) (Illumina, San Diego, CA). After demultiplexing with Illumina Bcl2fastq2 v 2.17 and initial data quality check with Illumina SAV, the raw reads were mapped to the latest UCSC transcript set using Bowtie2 version 2.1.0, and the gene expression level was estimated using RSEM v1.2.15.
Statistical methods
Bioconductor package LIMMA (linear models for microarray data)13, 14 (Bioconductor, Buffalo, NY) was used for differential gene expression analysis for normalized log2-transformed counts of RNASeq data. To avoid overinterpretation, we only included genes with at least 1 read per million mapped reads in at least 30 samples. LIMMA was used in conjunction with voom, which weighs the mean-variance relationship of the log-counts needed for accurate generalized linear modeling. A candidate list of differentially expressed genes were identified based on an absolute fold change >2.0 and Benjamini–Hochberg adjusted p-value of LIMMA's moderated t-test (p < 0.05). For functional annotation and pathway enrichment analysis, the candidate probes were analyzed in the Database for Annotation, Visualization and Integrated Discovery (DAVID),15 and processes and pathways were selected based on Benjamini–Hochberg adjusted p-values < 0.05.16 Principal component analysis (PCA)17 was used to visualize the separation of the 2 groups. Further unsupervised hierarchical clustering of differentially expressed probes was done by applying the Ward's minimum variance criterion linkage method18 with Euclidean distance and presented in a heatmap.
For the construction of a classification model, we first split the 219 subjects randomly into a training set and a testing set with a ratio of 70:30. We summarized subject characteristics as means with standard deviation, medians with interquartile ranges, or proportions. Characteristics were compared between training and testing sets using Student's t-test, Mann–Whitney, Fisher's exact, or chi-square testing as appropriate. A logistic regression model was fit using recursive feature elimination with 5-fold cross-validation to reduce the number of predictors and thus achieve a parsimonious model.19 In the model fitting process, a receiver operator characteristic area under the curve (AUC) was the metric used for selecting the optimal model. Threshold selection was based on model performance metrics in the training set. We then validated the threshold in the testing set. In addition, Cox proportional hazard models and Kaplan–Meier curves were fit for CLAD-free survival and compared between groups diagnosed with A-grade ACR on biopsy vs all other clinical conditions, and between predicted ACR vs no predicted ACR using the genomic classifier model.
Most statistical analyses were conducted using the Bioconductor suite of packages,20 and Package “caret”21 in the R statistical software environment version 3.3.1.22 Kaplan–Meier analyses for CLAD-free survival were performed using GraphPad Prism version 6.05 for Windows (GraphPad Software, La Jolla, CA, www.graphpad.com).
The data discussed in this publication are available in the Mendeley Data open research data repository.
Results
Patient characteristics
Of the 310 lung transplant recipients with eligible BAL samples, the final study cohort included 219 unique subjects with 1 BAL sample each (Figure 1 ). The clinical characteristics of the 91 eligible patients from whom we did not include a sample were similar to the final study cohort (Table S1 in the Supplementary Material available online at www.jhltonline.org). By design, selected samples were enriched for later time points post-transplant were more likely to have histopathology positive for LB and A-grade ACR and were less likely to have infection diagnosed or be healthy (Supplementary Table S2 online). In addition, the characteristics of samples excluded for RNA quantity or quality were similar to the final study cohort,with the exception of the measures of RNA quantity and quality (Supplementary Table S2 online). Among the samples included in the final study cohort, the characteristics of those with histopathologic A-grade ACR were similar to those without A-grade ACR on biopsy (Table 1 ). Training and testing sets were also similar (Supplementary Table S3 online).
Figure 1.
Eligible and Included Samples (a) Sample selection flow diagram. (b) Distribution of included samples over time, relative to total eligible samples in biorepository. ACR, acute cellular rejection; CLAD, chronic lung allograft dysfunction; LB, lymphocytic bronchiolitis.
Table 1.
Characteristics of Subjects With A-Grade ACR and Other Histopathology
| Total cohort |
Training set |
Test set |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | A-grade ACR | Other | p-value | A-grade ACR | Other | p-value | A-grade ACR | Other | p-value |
| Sex, n (%) | 0.54 | 0.86 | 0.55 | ||||||
| Male | 40 (66) | 96 (61) | 30 (65) | 74 (62) | 10 (67) | 22 (56) | |||
| Female | 21 (34) | 62 (39) | 16 (35) | 45 (38) | 4 (33) | 17 (44) | |||
| Age, mean (SD) | 58.7 (11.0) | 60.1 (11.3) | 0.41 | 59.3 (11.1) | 59.7 (11.4) | 0.85 | 56.9 (10.9) | 61.4 (10.8) | 0.17 |
| Pre-transplant disease, n (%) | 0.80 | 0.93 | 0.28 | ||||||
| Restrictive | 38 (62) | 92 (58) | 27 (59) | 74 (62) | 11 (73) | 18 (46) | |||
| Obstructive | 15 (25) | 43 (27) | 12 (26) | 30 (25) | 3 (20) | 13 (33) | |||
| CF/bronchiectasis | 2 (3) | 10 (6) | 2 (4) | 5 (4) | 0 (0) | 5 (13) | |||
| Other | 6 (10) | 13 (8) | 5 (11) | 10 (8) | 1 (7) | 3 (8) | |||
| Transplant type, n (%) | 0.76 | 0.73 | 1.00 | ||||||
| Bilateral | 34 (56) | 84 (53) | 25 (54) | 61 (51) | 9 (60) | 23 (59) | |||
| Single | 27 (44) | 74 (47) | 21 (46) | 58 (49) | 6 (40) | 16 (41) | |||
| CMV serostatus, n (%) | 0.41 | 0.55 | 0.78 | ||||||
| R+/D+ | 32 (52) | 70 (44) | 24 (52) | 50 (42) | 8 (53) | 20 (51) | |||
| R+/D- | 14 (23) | 32 (20) | 10 (22) | 24 (20) | 4 (27) | 8 (21) | |||
| R-/D+ | 8 (13) | 36 (23) | 7 (15) | 29 (24) | 1 (7) | 7 (18) | |||
| R-/D- | 7 (12) | 20 (13) | 5 (11) | 16 (13) | 2 (13) | 4 (10) | |||
| Days to biopsy, mean (SD) | 170 (207) | 230 (253) | 0.10 | 157 (212) | 235 (264) | 0.07 | 213 (194) | 216 (219) | 0.96 |
| Indication for biopsy, n (%) | 0.85 | 1.00 | 1.00 | ||||||
| Surveillance | 49 (80) | 124 (78) | 38 (83) | 97 (82) | 11 (73) | 27 (69) | |||
| For cause | 12 (20) | 34 (22) | 8 (17) | 22 (18) | 4 (27) | 12 (31) | |||
| Induction, n (%) | 1.00 | 0.61 | 0.37 | ||||||
| ATG | 32 (52) | 82 (52) | 23 (50) | 65 (55) | 9 (60) | 17 (44) | |||
| Basiliximab | 29 (48) | 76 (48) | 23 (50) | 54 (45) | 6 (40) | 22 (56) | |||
| Tacrolimus trough, mean (SD) | 10.6 (5.4) | 9.9 (3.9) | 0.32 | 10.7 (5.6) | 10.1 (4.1) | 0.46 | 10.5 (4.9) | 9.5 (3.3) | 0.45 |
| FEV1 % of baseline, median (IQR) | 100 (93–100) | 99 (90–100) | 0.06 | 100 (92–100) | 99 (90–100) | 0.13 | 100 (95–100) | 99 (88–100) | 0.23 |
| TBBX histopathology, n (%) | . | . | . | ||||||
| A1 | 38 (62) | . | 29 (63) | . | 9 (60) | . | |||
| A2 | 17 (28) | . | 12 (26) | . | 5 (33) | . | |||
| A3 | 6 (10) | . | 5 (11) | . | 1 (7) | . | |||
| Lymphocytic bronchiolitis | 27 (44) | 70 (44) | 22 (48) | 52 (44) | 5 (33) | 18 (46) | |||
| Infection, n (%) | . | 41 (26) | . | . | 34 (29) | . | . | 7 (18) | |
| Respiratory virusa | . | 10 (6) | . | 8 (7) | . | 2 (5) | |||
| Bacterialb | . | 19 (12) | . | 18 (15) | . | 1 (3) | |||
| Fungalc | . | 12 (8) | . | 8 (7) | . | 4 (10) | |||
ACR, acute cellular rejection; ATG, anti-thymocyte globulin; CF, cystic fibrosis; CMV, cytomegalovirus; FEV1, forced expiratory volume in one second; IQR, intraquartile range; SD, standard deviation; TBBX, transbronchial biopsy
3 coronavirus, 3 parainfluenza, 2 rhinovirus, 1 respiratory syncytial virus, 1 influenza A.
13 Pseudomonas aeruginosa, 3 Escherichia coli, 2 Haemophilus influenzae, 1 Klebsiella pneumoniae
8 Aspergillus fumigatus, 3 Aspergillus niger, 1 Aspergillus nidulans
Differential gene expression analyses
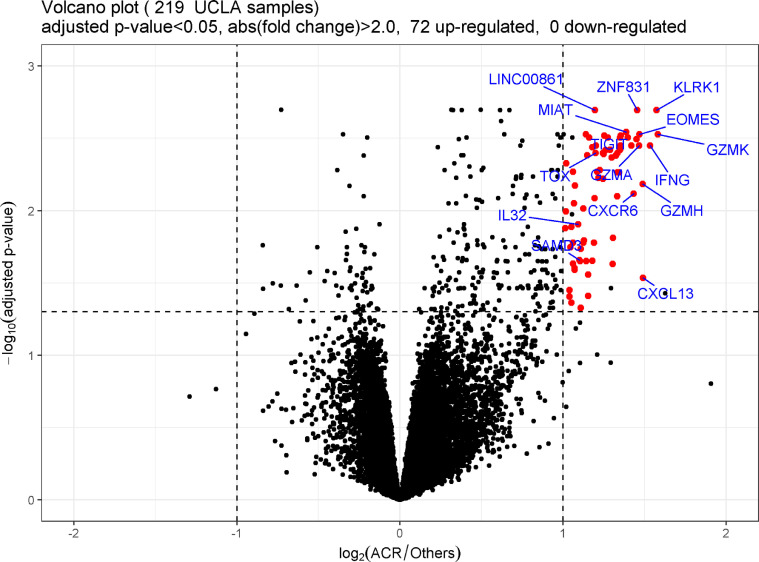
After removing the transcripts with low read count across samples, 16,081 out of 25,343 transcripts remained. Differential gene expression analysis identified 72 genes, all of which were upregulated with A-grade ACR (Figure 2 , Table 2 ).
Figure 2.
Volcano plot demonstrating gene expression in bronchoalveolar lavage (BAL) cell pellet (CP) from lung transplant recipients with A-grade acute cellular rejection (ACR) compared with other non–A-grade ACR clinical conditions. Each dot on the graph corresponds to a gene. The fold difference in expression between A-grade ACR and others is graphed on the x axis (logarithm to the base 2-fold changes). The p-values for each gene are graphed on the y axis (negative logarithm to the base 10 of the t-test Benjamini–Hochberg adjusted p-values). The vertical dashed lines correspond to absolute fold difference of -2.0 and 2.0. The horizontal dashed line corresponds to a Benjamini-adjusted p-value of 0.05. Differentially expressed genes (n = 72) are labeled red (selected interesting genes are labeled with official gene symbol as well). Additional black dots in the significance range (n = 5) represent chimera transcripts removed from the gene list. No genes were expressed at significantly lower levels during A-grade ACR. UCLA, University of California, Los Angeles.
Table 2.
Differentially Expressed Genes During ACR
| Gene symbol | FC | p-value | Adjusted p-value | Gene symbol | FC | p-value | Adjusted p-value |
|---|---|---|---|---|---|---|---|
| GZMK | 2.99 | 0.0000 | 0.0030 | TRAT1 | 2.30 | 0.0000 | 0.0035 |
| KLRK1 | 2.98 | 0.0000 | 0.0020 | TOX | 2.30 | 0.0000 | 0.0040 |
| IFNG | 2.90 | 0.0000 | 0.0035 | LINC00861 | 2.29 | 0.0000 | 0.0020 |
| CXCL13 | 2.81 | 0.0004 | 0.0292 | CD3E | 2.29 | 0.0000 | 0.0082 |
| GZMH | 2.81 | 0.0000 | 0.0065 | ITM2A | 2.28 | 0.0001 | 0.0166 |
| EOMES | 2.77 | 0.0000 | 0.0030 | TBX21 | 2.27 | 0.0002 | 0.0222 |
| GZMA | 2.76 | 0.0000 | 0.0035 | ATP1A3 | 2.27 | 0.0000 | 0.0036 |
| ZNF831 | 2.74 | 0.0000 | 0.0020 | ITK | 2.24 | 0.0000 | 0.0031 |
| TIGIT | 2.73 | 0.0000 | 0.0032 | ZNF683 | 2.23 | 0.0004 | 0.0277 |
| CXCR6 | 2.70 | 0.0000 | 0.0076 | MS4A1 | 2.22 | 0.0007 | 0.0388 |
| SIRPG | 2.68 | 0.0000 | 0.0035 | ETS1 | 2.22 | 0.0000 | 0.0042 |
| CD8A | 2.64 | 0.0000 | 0.0031 | PRF1 | 2.21 | 0.0002 | 0.0224 |
| MIAT | 2.62 | 0.0000 | 0.0029 | RASGRP1 | 2.20 | 0.0000 | 0.0030 |
| FCRL3 | 2.56 | 0.0000 | 0.0030 | GZMM | 2.19 | 0.0001 | 0.0160 |
| P2RY10 | 2.55 | 0.0000 | 0.0035 | IL2RB | 2.18 | 0.0001 | 0.0166 |
| CD8B | 2.55 | 0.0000 | 0.0038 | UBASH3A | 2.18 | 0.0001 | 0.0097 |
| KLRC4 | 2.55 | 0.0000 | 0.0031 | B3GAT1 | 2.16 | 0.0002 | 0.0183 |
| FASLG | 2.54 | 0.0000 | 0.0054 | PDCD1 | 2.16 | 0.0009 | 0.0470 |
| KLRC2 | 2.52 | 0.0000 | 0.0040 | NKG7 | 2.15 | 0.0002 | 0.0224 |
| SH2D2A | 2.52 | 0.0000 | 0.0054 | SAMD3 | 2.15 | 0.0002 | 0.0220 |
| CTLA4 | 2.52 | 0.0000 | 0.0080 | IL32 | 2.13 | 0.0001 | 0.0124 |
| CXCR3 | 2.51 | 0.0000 | 0.0042 | THEMIS | 2.10 | 0.0000 | 0.0067 |
| LAG3 | 2.50 | 0.0000 | 0.0042 | NELL2 | 2.10 | 0.0003 | 0.0257 |
| VCAM1 | 2.48 | 0.0001 | 0.0154 | TSPAN5 | 2.10 | 0.0003 | 0.0250 |
| GZMB | 2.47 | 0.0003 | 0.0234 | CPNE7 | 2.09 | 0.0001 | 0.0089 |
| SH2D1A | 2.46 | 0.0000 | 0.0043 | APBA2 | 2.09 | 0.0003 | 0.0233 |
| CD27 | 2.44 | 0.0000 | 0.0038 | SLA2 | 2.09 | 0.0000 | 0.0054 |
| GPR171 | 2.42 | 0.0000 | 0.0031 | TMEM204 | 2.09 | 0.0001 | 0.0166 |
| ZAP70 | 2.40 | 0.0000 | 0.0038 | TTC24 | 2.07 | 0.0001 | 0.0130 |
| LEF1 | 2.39 | 0.0000 | 0.0030 | JAKMIP1 | 2.07 | 0.0008 | 0.0433 |
| GPR174 | 2.38 | 0.0000 | 0.0041 | GRAPH2 | 2.07 | 0.0002 | 0.0179 |
| CD96 | 2.37 | 0.0000 | 0.0040 | AFAP1L2 | 2.06 | 0.0006 | 0.0355 |
| LCK | 2.37 | 0.0000 | 0.0060 | TOX2 | 2.05 | 0.0007 | 0.0393 |
| CD247 | 2.34 | 0.0000 | 0.0053 | PYHIN1 | 2.03 | 0.0000 | 0.0047 |
| BCL11B | 2.33 | 0.0000 | 0.0059 | GPR18 | 2.03 | 0.0001 | 0.0101 |
| ABCD2 | 2.31 | 0.0000 | 0.0054 | KIAA0125 | 2.02 | 0.0001 | 0.0132 |
ACR, acute cellular rejection; FC, fold change.
To get a visual impression of gene expression by patient group, we performed PCA using the 72 differentially expressed genes. The PCA demonstrated considerable overlap between A-grade ACR and other sample categories (Supplementary Figure S1a online), which improved modestly by focusing on A2 or greater ACR and healthy groups (Supplementary Figure S1b online). Likewise, in unsupervised hierarchical cluster analyses, A-grade ACR samples tended to cluster together, but there was still considerable misclassification (Supplementary Figure S1c online).
Functional annotation and pathway enrichment analyses
To test for and validate the biological relevance of the 72 differentially expressed genes, we compared functional annotation and pathway mapping using DAVID Bioinformatics Resources. Comparison of the biological process category of gene ontology classification indicated that the predominant processes associated with A-grade ACR included immune response, adaptive immune response, T-cell receptor (TCR) signaling pathway, and inflammatory response (Table 3 ). Similarly, Kyoto Encyclopedia of Genes and Genomes pathways significantly enriched in this gene list included TCR signaling pathway, natural killer (NK) cell–mediated cytotoxicity, cytokine–cytokine receptor interactions, and allograft rejection, among others (Table 4 ).
Table 3.
Gene Ontology: Top Biological Processes Enriched With ACR
| Term | Fold enrichment | Genes | p-value | FDR |
|---|---|---|---|---|
| Immune response | 9.8 | CXCL12, CD27, FASLG, CTLA4, GZMB, GZMH | 0.00028 | 0.0037 |
| Adaptive immune response | 24.1 | SH2D1A, CTLA4, IFNG, KLRK1 | 0.00055 | 0.0071 |
| T-cell receptor signaling pathway | 44.3 | CD27, CD3E, IFNG | 0.00190 | 0.0250 |
| Inflammatory response | 8.7 | CXCL13, CXCR3, CXCR6, CD27, AFAP1L2 | 0.00220 | 0.0290 |
ACR, acute cellular rejection; FDR, false discovery rate.
Table 4.
KEGG Pathways Enriched With ACR
| Term | Fold enrichment | Genes | p-value | FDR |
|---|---|---|---|---|
| T-cell receptor signaling pathway | 21.5 | CD247, CD3e, CD8a, CD8b, GRAP2, ITK, LCK, CTLA4, IFNG, PDCD1, ZAP70 | 2.70E-11 | 2.90E-10 |
| Natural killer cell–mediated cytotoxicity | 17.2 | CD247, FASLG, LCK, SH2D1A, GZMB, IFNG, KLRK1, PRF1, ZAP70 | 2.40E-08 | 2.60E-07 |
| Primary immunodeficiency | 27.9 | CD3E, CD8A, CD8B, LCK, ZAP70 | 2.40E-05 | 2.60E-04 |
| Cell adhesion molecules | 8.7 | CD8a, CD8b, TIGIT, CTLA4, PDCD1, VCAM1 | 4.90E-04 | 0.0052 |
| Cytokine–cytokine receptor interaction | 6.5 | CXCL13, CXCR3, CXCR6, FASLG, IFNG, IL2RB | 4.90E-04 | 0.0052 |
| Graft-versus-host disease | 24.3 | FASLG, GZMB, IFNG, PRF1 | 5.10E-04 | 0.0055 |
| Allograft rejection | 22.4 | FASLG, GZMB, IFNG, PRF1 | 6.60E-04 | 0.0070 |
ACR, acute cellular rejection; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Acute cellular rejection classifier development and performance
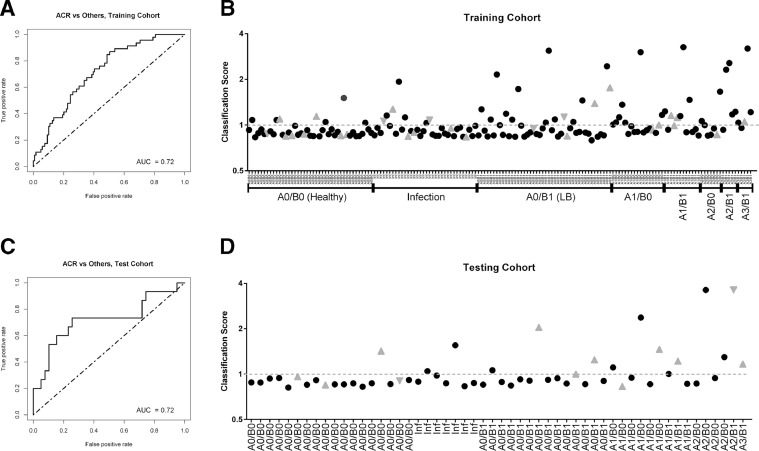
We sought to develop a stable and economic classification model for acute rejection based on our candidate list of differentially expressed target genes (Table 2). Subjects were randomly split 70:30 into a training set and a testing set. After elimination of the redundant/correlated genes by recursive feature elimination, we developed a final logistic regression model in the training set that maximized AUC and required only 4 genes: thymocyte selection–associated high mobility group box protein (TOX), sterile alpha motif domain–containing 3 (SAMD3), interleukin 32 (IL32), and killer cell lectin-like receptor K1 (KLRK1). This 4-gene model achieved an AUC of 0.72 in the training set (Figure 3 a). We then defined a classification threshold that favored specificity (76%) over sensitivity (54%), which yielded 70% classification accuracy (Figure 3b). In the independent test set of 54 samples, the model exhibited similar performance with an AUC of 0.72 (Figure 3c). The performance of the classification threshold was also similar to the training set, with specificity of 82%, sensitivity of 60%, and accuracy of 76% (Figure 3d).
Figure 3.
Acute cellular rejection (ACR) classification of bronchoalveolar lavage (BAL) cell pellet (CP) gene expression by classifier model. (a) Receiver operator characteristic (ROC) curve of classifier performance on 165 subject training set. (b) Classification scores (the predicted probability of ACR divided by 0.261) are plotted on the y axis for each subject. In the training set, the dashed line (score 1.0) denotes the classification threshold between ACR and no ACR classification. Transbronchial biopsy (TBBX) histopathology or infection diagnoses are provided for each subject on the x axis. Black closed circles represent chronic lung allograft dysfunction (CLAD)-free survivors. Gray triangles pointing up represent subjects developing CLAD within 1 year of sample. Gray triangles pointing down represent subjects who died within 1 year of sample. (c) ROC curve of classifier performance on the 54 subject testing set. (d) Classification scores in the 54 subject testing set. AUC, area under the curve.
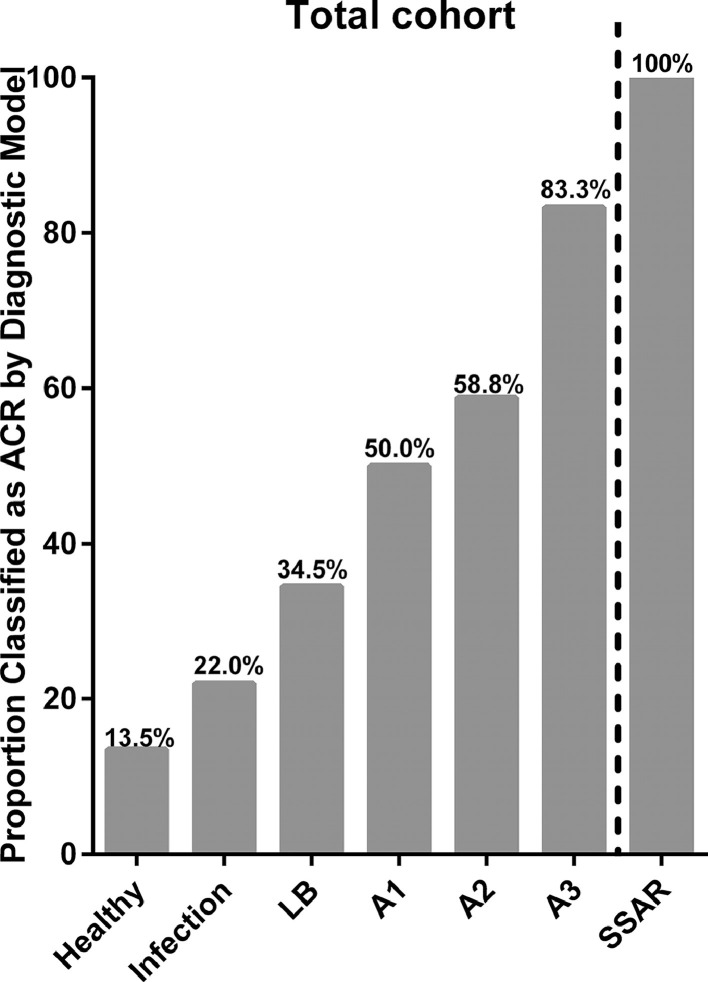
In the total cohort (training and testing sets combined), the proportion of TBBX A-grade ACR classified as ACR by our genomic analysis model increased with increasing A-grade (Figure 4 ). Similarly, 100% (6/6) of SSAR cases were classified as ACR in our model, compared with 56.2% (18/32) of A-grade ACR cases that were non-SSAR (p = 0.067).
Figure 4.
The proportion of subjects in each clinical category classified as acute cellular rejection (ACR) by the genomic classifier model. LB, lymphocytic bronchiolitis; SSAR, spirometrically significant acute rejection.
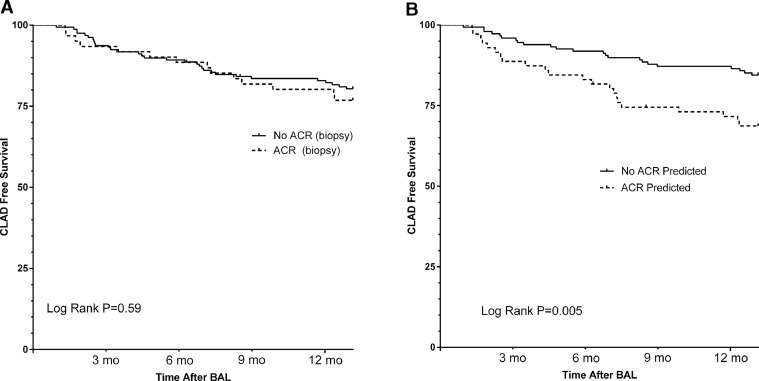
Furthermore, classification as ACR by genomic analysis was also associated with worse CLAD-free survival over the 1-year follow-up after biopsy (hazard ratio [HR] = 2.42 for CLAD or death, 95% confidence interval [CI] = 1.29–4.53) (Figure 5 ). When follow-up was extended to 5 years post-biopsy, CLAD-free survival remained worse in subjects classified as ACR by genomic analysis (Supplementary Figure S2a online). By comparison, A-grade ACR diagnosed by TBBX was neither associated with a significant difference in 1-year CLAD-free survival (HR 1.17 for CLAD or death, 95% CI = 0.59–2.30) (Figure 5) nor when follow-up was extended to 5 years post-biopsy (Supplementary Figure 2b online). In addition, the effect of genomic classification as ACR on CLAD-free survival was greatest in the subset of patients where TBBX was negative for A-grade ACR (HR = 2.88 for CLAD or death, 95% CI = 1.32–6.14) (Supplementary Figure S2c online).
Figure 5.
Kaplan–Meier estimates of chronic lung allograft dysfunction (CLAD)-free survival in the 1 year following biopsy. (a) Comparison of CLAD-free survival following the bronchoscopy procedure for patients with or without acute cellular rejection (ACR) histopathology. (b) Comparison of CLAD-free survival following the bronchoscopy procedure for patients classified as ACR or no ACR by the genomic classifier model. BAL, bronchoalveolar lavage.
Discussion
Given the importance of ACR as a modifiable risk factor for CLAD, accurate diagnosis remains a priority in lung transplantation. However, the current gold standard requires TBBX, which has drawbacks including the small volume of tissue sampled, leading to high rates of sub-optimal tissues and sampling error. Furthermore, even when biopsy samples are considered adequate, interpretation is fraught with high interobserver disagreement.6, 7, 8 Finally, TBBX is associated with a small risk of major complications that may lead to morbidity and even mortality in rare cases.5 We hypothesized that gene expression in the BAL CP could be used to diagnose ACR without the need for TBBX. Furthermore, BAL CP gene expression might improve upon TBBX because BAL samples a larger volume of lung, and a genomic classifier would remove the subjectivity of pathology interpretation. The current study demonstrates the proof of concept that gene expression informs about ACR pathogenesis, while also suggesting a potential utility to diagnose ACR and risk stratify for the development of CLAD better than TBBX.
Our primary finding is that A-grade ACR on TBBX is associated with a characteristic gene expression profile in BAL cells. The list of differentially expressed genes is enriched for biological processes and pathways integral to allograft rejection. For instance, the Kyoto Encyclopedia of Genes and Genomes TCR signaling pathway was enriched 21.5-fold during A-grade ACR, including genes for CD8a, CD8b, lymphocyte-specific protein tyrosine kinase (LCK), and CD3e. CD8 acts to stabilize binding of the TCR to the peptide-major histocompatibility complex, while also localizing LCK to the TCR/CD3 complex to facilitate early signaling events during T-cell activation.23 We have also previously shown that BAL CP gene expression of CD8 and LCK is associated with the development of CLAD.24 In addition, the cytokine–cytokine receptor interaction pathway was enriched 6.5-fold and included CXCR3 and interferon gamma. This finding is corroborated by previous work showing that protein concentrations of the interferon gamma–inducible CXCR3-binding chemokines in the BAL fluid are associated with A-grade ACR and other lung injury patterns, as well as with CLAD.25, 26, 27 Finally, enrichment for pathways related to cell adhesion, NK cell–mediated cytotoxicity, and allograft rejection add additional support of the biological relevance of BAL CP gene expression during A-grade ACR.
This expression profile may be efficiently simplified to an economical 4-gene classifier model that exhibits fair performance as a diagnostic biomarker for histopathologic A-grade ACR. We chose a threshold for the model that prioritized specificity over sensitivity. Our rationale was aimed at better identifying clinically significant ACR at high risk of progression to CLAD. Recognizing the well-described shortcomings of TBBX, it is not surprising that our model reclassifies cases of histopathologic A-grade ACR as no ACR and other cases of no histopathologic A-grade ACR as positive for ACR. The performance of this genomic classifier improved with increasing histopathologic A-grade, for which there is reported greater observer agreement and therefore confidence in the ACR diagnosis.6, 7, 8 In addition, our classifier model indicated ACR for all cases of SSAR, which is a strong risk factor for CLAD.11 Given the importance of ACR as a potentially modifiable CLAD risk factor, our most intriguing finding is that classification indicating ACR also discriminated risk of CLAD-free survival, especially in the year following biopsy, whereas a histopathologic diagnosis of A-grade ACR did not. Finally, the highest incidence of CLAD or death in the year after biopsy was seen in the 37 cases where histopathology was negative for A-grade ACR but where the genomic classifier predicted ACR. None of these cases were treated for rejection at the time. We can hypothesize that corticosteroid treatment could have reduced the risk of CLAD in this group, but this hypothesis requires prospective evaluation in a clinical trial.
Our study corroborates prior smaller observational studies of gene expression during ACR.28, 29 Although each study used different methods and the specific genes associated with ACR differ somewhat, the functional relevance of the differentially expressed genes were remarkably similar across studies. Specifically, increased gene expression related to T-cell activation, and cytotoxicity was common to each of these studies.28, 29 These similar conclusions are interesting, especially because our study relied on a different platform to measure gene expression: this study used RNASeq, whereas the prior studies used microarray. In studies of T-cell activation, there has been a high correlation of gene expression profiles between RNASeq and microarray platforms using the same set of samples.30 However, RNASeq was superior in detecting low abundance transcripts and demonstrated a broader dynamic range than microarray, therefore allowing the detection of more differentially expressed genes. Furthermore, performing RNASeq allows for the avoidance of technical issues found using microarray probes (e.g., cross-hybridization, non-specific hybridization, and limited detection range of individual probes). In addition, RNASeq does not rely on pre-designed detection probes; thus, there are no issues associated with probe redundancy and annotation, which simplifies the interpretation of the data.
Our final genomic classifier model included 4 genes: TOX, SAMD3, IL32, and KLRK1. TOX has been shown to be upregulated by calcineurin-mediated TCR signaling during CD4 T-cell lineage development, including CD1d-dependent NK and T regulatory CD4 T-cell sub-lineages, and has been shown to also effect CD8 T-cell development.31 The biological relevance of SAMD3 with allograft rejection is not known at this time, although the Human Protein Atlas (http://www.proteinatlas.org)32 predicts that SAMD3 is an intracellular protein that is broadly expressed in lymphoid tissues, especially spleen and lymph nodes. IL-32 is expressed by IL-2–activated T cells and NK cells and plays a role in acute graft-versus-host disease after hematopoietic cell transplantation.33 KLRK1 is a receptor expressed by NK cells and cytotoxic T lymphocytes; it mediates activation in NK cells and costimulation in T cells.34 Collectively, this 4-gene model is consistent with the paradigm of ACR and CLAD pathobiology. Specifically, ACR is characterized by activated immune cells including CD4 T cells, CD8 T cells, NK cells, and NK T cells, which drive the cytotoxicity responsible for allograft injury that eventually leads to CLAD. Future prospective studies should test whether this genomic classifier leads to earlier or more appropriate treatment of ACR, an intervention that might reduce CLAD.
There are limitations inherent to the design of this study. We examined a cross-sectional selection of samples from lung transplant recipients at a single center. It would be valuable to include longitudinal sampling to characterize the evolution of gene expression before and after ACR and through the development of CLAD. Our study cannot determine the effect of treatment on gene expression, and therefore, we do not know whether knowledge of gene expression can impact outcomes. Although our study included independent training and testing cohorts, all patients were from a single center; future studies would benefit from the inclusion of an external validation cohort. We are reassured by the fact that the biological processes and pathways enriched in our gene list are consistent with expectations. We also acknowledge that 9% of samples selected for inclusion were inadequate for RNASeq either because of degraded RNA or low RNA concentrations. This is similar to the 8% rate of inadequate TBBX reported in lung transplant recipients.6 However, the proportion of inadequate BAL samples for RNASeq could probably be reduced to nearly 0% with the use of RNA stabilization solutions and RNASeq library construction kits designed for low input. Finally, although a BAL CP genomic classifier test could reduce the complications associated with TBBX, it does not eliminate the need for invasive bronchoscopy. It is unclear whether the findings in the BAL CP would also be observed in peripheral blood.
In summary, we showed that BAL CP gene expression during histopathologic A-grade ACR is enriched for immune responses including TCR signaling, cytokine signaling, cell adhesion, and cytotoxicity. Gene expression to diagnose ACR could be a less invasive alternative to TBBX. In fact, we found that differential gene expression can be simplified to a 4-gene signature with fair performance as a surrogate for TBBX, but with potentially greater clinical implications. This study demonstrates proof of concept that BAL CP gene expression informs about the pathogenesis of ACR and risk of CLAD. A multicenter study is required to establish whether BAL CP gene expression could reduce or eliminate the need for TBBX to diagnose ACR.
Disclosure statement
None of the authors has a financial relationship with a commercial entity that has an interest in the subject of the presented manuscript or other conflicts of interest to disclose. This work was supported by grants from the National Institute of Allergy and Infectious Diseases (U01AI063594-12 and U01AI113315-1).
Supplementary data
Supplementary data associated with this article can be found in the online version at www.jhltonline.org/.
Footnotes
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.healun.2019.05.001.
Appendix. Supplementary materials
References
- 1.Weigt SS, DerHovanessian A, Wallace WD, Lynch JP, 3rd, Belperio JA. Bronchiolitis obliterans syndrome: the Achilles’ heel of lung transplantation. Semin Respir Crit Care Med. 2013;34:336–351. doi: 10.1055/s-0033-1348467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart S, Fishbein MC, Snell GI. Revision of the 1996 working formulation for the standardization of nomenclature in the diagnosis of lung rejection. J Heart Lung Transplant. 2007;26:1229–1242. doi: 10.1016/j.healun.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Ross DJ, Marchevsky A, Kramer M, Kass RM. ”Refractoriness” of airflow obstruction associated with isolated lymphocytic bronchiolitis/bronchitis in pulmonary allografts. J Heart Lung Transplant. 1997;16:832–838. [PubMed] [Google Scholar]
- 4.Pue CA, Pacht ER. Complications of fiberoptic bronchoscopy at a university hospital. Chest. 1995;107:430–432. doi: 10.1378/chest.107.2.430. [DOI] [PubMed] [Google Scholar]
- 5.Rademacher J, Suhling H, Greer M. Safety and efficacy of outpatient bronchoscopy in lung transplant recipients—a single centre analysis of 3,197 procedures. Transplant Res. 2014;3:11. doi: 10.1186/2047-1440-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace WD, Li N, Andersen CB. Banff study of pathologic changes in lung allograft biopsy specimens with donor-specific antibodies. J Heart Lung Transplant. 2016;35:40–48. doi: 10.1016/j.healun.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Arcasoy SM, Berry G, Marboe CC. Pathologic interpretation of transbronchial biopsy for acute rejection of lung allograft is highly variable. Am J Transplant. 2011;11:320–328. doi: 10.1111/j.1600-6143.2010.03382.x. [DOI] [PubMed] [Google Scholar]
- 8.Bhorade SM, Husain AN, Liao C. Interobserver variability in grading transbronchial lung biopsy specimens after lung transplantation. Chest. 2013;143:1717–1724. doi: 10.1378/chest.12-2107. [DOI] [PubMed] [Google Scholar]
- 9.Berry GJ, Brunt EM, Chamberlain D. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Lung Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant. 1990;9:593–601. [PubMed] [Google Scholar]
- 10.Yousem SA, Berry GJ, Cagle PT. Revision of the 1990 working formulation for the classification of pulmonary allograft rejection: Lung Rejection Study Group. J Heart Lung Transplant. 1996;15:1–15. [PubMed] [Google Scholar]
- 11.Davis WA, Finlen Copeland CA, Todd JL, Snyder LD, Martissa JA, Palmer SM. Spirometrically significant acute rejection increases the risk for BOS and death after lung transplantation. Am J Transplant. 2012;12:745–752. doi: 10.1111/j.1600-6143.2011.03849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estenne M, Maurer JR, Boehler A. Bronchiolitis obliterans syndrome 2001: an update of the diagnostic criteria. J Heart Lung Transplant. 2002;21:297–310. doi: 10.1016/s1053-2498(02)00398-4. [DOI] [PubMed] [Google Scholar]
- 13.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 17.Jolliffe IT. Springer-Verlag; New York: 2002. Principal component analysis. [Google Scholar]
- 18.Ward JH., Jr Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 19.Kuhn M. Building predictive models in R using the caret package. J Stat Soft. 2008;28:26. [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M, Wing J, Weston S, et al. Caret: classification and regression training: R package version6.0-73; 2016.
- 22.Team RC . R Foundation for Statistical Computing; Vienna, Austria: 2017. R: a language and environment for statistical computing. [Google Scholar]
- 23.Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE. The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc Natl Acad Sci USA. 1989;86:3277–3281. doi: 10.1073/pnas.86.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weigt SS, Wang X, Palchevskiy V. Gene expression profiling of bronchoalveolar lavage cells preceding a clinical diagnosis of chronic lung allograft dysfunction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shino MY, Weigt SS, Li N. The prognostic importance of bronchoalveolar lavage fluid CXCL9 during minimal acute rejection on the risk of chronic lung allograft dysfunction. Am J Transplant. 2018;18:136–144. doi: 10.1111/ajt.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shino MY, Weigt SS, Li N. CXCR3 ligands are associated with the continuum of diffuse alveolar damage to chronic lung allograft dysfunction. Am J Respir Crit Care Med. 2013;188:1117–1125. doi: 10.1164/rccm.201305-0861OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shino MY, Weigt SS, Li N. The prognostic importance of CXCR3 chemokine during organizing pneumonia on the risk of chronic lung allograft dysfunction after lung transplantation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gimino VJ, Lande JD, Berryman TR, King RA, Hertz MI. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am J Respir Crit Care Med. 2003;168:1237–1242. doi: 10.1164/rccm.200305-644OC. [DOI] [PubMed] [Google Scholar]
- 29.Patil J, Lande JD, Li N, Berryman TR, King RA, Hertz MI. Bronchoalveolar lavage cell gene expression in acute lung rejection: Development of a diagnostic classifier. Transplantation. 2008;85:224–231. doi: 10.1097/TP.0b013e318160268a. [DOI] [PubMed] [Google Scholar]
- 30.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlén M, Björling E, Agaton C. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Marcondes AM, Li X, Tabellini L. Inhibition of IL-32 activation by alpha-1 antitrypsin suppresses alloreactivity and increases survival in an allogeneic murine marrow transplantation model. Blood. 2011;118:5031–5039. doi: 10.1182/blood-2011-07-365247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3:1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.