Abstract
A 26‐color staining panel was developed to profile human antigen‐specific T cells in an intracellular cytokine staining (ICS) assay using peptide pools to various antigens of interest. In addition to multiple functional markers, the panel includes differentiation/activation markers and markers to assess γδ, mucosal‐associated invariant T, and NK T cells as well as conventional NK cells. Panel optimization was performed using previously cryopreserved PBMC from healthy adults, and then, expression of key functional markers in the panel was cross‐validated against a validated ICS assay used in the HIV Vaccine Trials Network (HVTN). The panel is currently being used to evaluate the responses to tuberculosis and malaria vaccine candidates in volunteers from different geographic areas. © 2019 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: cytometry, human PBMC, T cells, MAIT cells, γδ T cells, NK cells, memory, intracellular cytokines
Short abstract

Despite extensive research and clinical testing, highly effective vaccines against complex pathogens such as HIV, tuberculosis (TB), and malaria have not yet been developed. An impediment to this development is the lack of known immune correlates of protection to the pathogens. Multicolor flow cytometry, and in particular, new technologies allowing up to 28‐color flow cytometry, could enable identification of novel immune correlates of risk or protection.
Within our HIV Vaccine Trials Network (HVTN) Laboratory Center, intracellular cytokine staining (ICS) is used to quantify and profile T‐cell responses to vaccine candidates. The laboratories adhere to good clinical laboratory practices (GCLP), and thus, our ICS assays are validated for the primary functional markers. A prerequisite of applying this new panel in vaccine studies was to retain the same sensitivity for expression of the functional biomarkers IFN‐γ, IL‐2, TNF‐α, and CD40L as in prior ICS assays widely used within the HVTN 1, 2.
Because this staining panel was designed to elucidate potential immune correlates in a TB vaccine efficacy clinical trial, markers proposed for the panel were prioritized based on the types of immune responses that could be important for control of TB (in order of priority): 1) lineage and viability: CD3, CD4, CD8, CD14, viability; 2) functional markers: IFN‐γ, IL‐2, TNF‐α, CD40L, IL‐17, granzyme A, Th2 cytokines (IL‐4/IL‐13 combined for one detector); 3) memory markers: CCR7, CD45RA; 4) additional lineage markers: γδ TCR, CD56, CD16; 5) MAIT cells: CD161, CD26, Vα7.2; 6) activation: HLA‐DR; 7) chemokine receptor for helper T cell classification: CXCR3, CCR6; 8) additional functional markers: IL‐22, perforin; and 9) differentiation markers: KLRG‐1, PD‐1.
Although the BD FACSymphony instrument currently includes capability for 28 markers, designing panels that use all available detectors can be challenging. Because we were initially concerned that there is a limited choice for conjugated reagents for the G710 detector, and the fluorochrome commonly used for this detector (PE‐Cy5.5) is spectrally similar to fluorochromes detected in the B710 detector (PerCP‐Cy5.5 and BB700), our aim was to develop a panel for 27 markers (or colors). Many panel versions were tested, and one detector (B610 for detection of BB630) was shown to cause many spreading issues into other detectors, as assessed by fluorescence minus one (FMO) testing. The B610 detector was therefore not used, although additional optimization would likely have enabled use of this detector. Thus, the final panel included 26 markers (Tables 1 and 2). Among the list of markers above, PD‐1 was considered lower priority and was dropped. Figure 1 shows an example of the staining profile for PBMC stimulated with staphylococcal enterotoxin B. Although this panel was initially developed for peptide pool antigens, other types of antigens such as recombinant proteins or whole pathogens have also be tested using this panel. Further developmental strategies and details for the panel may be found in the online material.
Table 1.
Summary table for application of OMIP‐056
| Purpose | Characterization of antigen‐specific CD4+, CD8+, γδ, MAIT, and NK T cells and NK cells |
|---|---|
| Species | Human |
| Cell types | Cryopreserved PBMC |
| Cross‐references | OMIP‐014, OMIP‐025 |
MAIT, mucosal‐associated invariant T cells; NK, natural killer; PBMC, peripheral blood mononuclear cells.
Table 2.
Reagents used for OMIP‐056
| Detector | Fluorochrome | Specificity | Clone | Purpose |
|---|---|---|---|---|
| B515 | FITC | Perforin | B‐D48 | Function |
| B660 | BB660 | CD14 | MϕP9 | Monocytes |
| B710 | BB700 | IL2 | MQ1‐17H12 | Function |
| G575 | PE | IL22 | 22URTI | Function |
| G610 | PE‐Dazzle 594 | KLRG1 (MAFA) | SA231A2 | Differentiation |
| G660 | PE‐Cy5 | CXCR3 | 1C6/CXCR3 | T helper class |
| G710 | PE‐Cy5.5 | CD56 | CMSSB | NK, NKT |
| G780 | PE‐Cy7 | CD154 | 24–31 | Function |
| R660 | APC | IL4 | MP4‐25D2 | Function |
| IL13 | JES10‐5A2 | |||
| R710 | Alx700 | Granzyme A | CB9 | Function |
| R780 | APC‐Cy7 | TCRvα7.2 | 3C10 | MAIT |
| U395 | BUV395 | CD3 | UCHT1 | T cell lineage |
| U450 | UViD | Viability | NA | Viability |
| U500 | BUV496 | CD45RA | HI100 | Differentiation |
| U570 | BUV563 | CD8 | RPA‐T8 | T cell lineage |
| U660 | BUV661 | HLA‐DR | G46‐6 | Activation |
| U730 | BUV737 | IL17a | N49‐653 | Function |
| U780 | BUV805 | CD4 | RPA‐T4 | T cell lineage |
| V450 | V450 | IFNy | B27 | Function |
| V510 | BV510 | TCR γδ | 11F2 | γδ T cell lineage |
| V570 | BV570 | CD16 | 3G8 | NK, NKT |
| V610 | BV605 | CCR6 (CD196) | 11‐A9 | T helper class |
| V655 | BV650 | CD161 | DX12 | MAIT |
| V710 | BV711 | CD26 | M‐A261 | MAIT |
| V750 | BV750 | TNFα | MAb11 | Function |
| V780 | BV785 | CCR7 (CD197) | G043H7 | Differentiation |
APC, allophycocyanin; Alx, Alexa; BB, brilliant blue, BUV, brilliant ultraviolet; BV, brilliant violet; Cy, cyanine; FITC, fluorescein isothiocyanate; PE, R‐phycoerythrin; UViD, LIVE/DEAD fixable ultraviolet dead cell stain.
Figure 1.
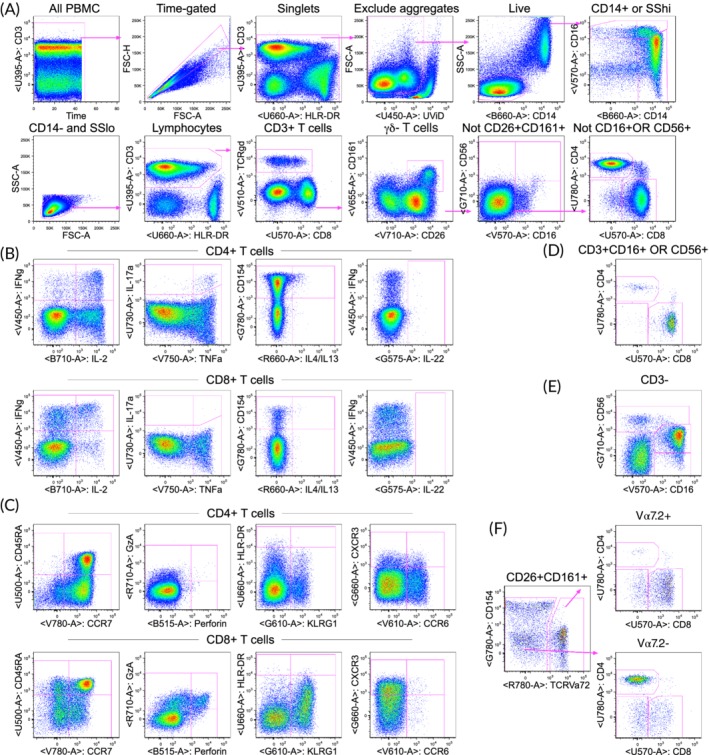
Example of the staining and gating strategy for PBMC stimulated with staphylococcal enterotoxin B (SEB). PBMC from a healthy adult were stimulated for 6 h with SEB. (A) Gating hierarchy to identify lineages. Initial gating on time (seconds) to exclude any events early in collection if there are pressure fluctuations, singlet gating on forward scatter height vs. area, exclusion of aggregates (only one example shown, but several sequential gates on various parameters are used), and live cell gating. Monocytes are gated as either CD14+ or high for side scatter and the upper right graph shows three monocyte subsets based on CD14 vs. CD16. Non‐monocytes are gated as CD14‐SSlo and then scatter gated on lymphocytes. The gating scheme avoids any overlapping subsets as shown in supplemental figure 4. Thus, conventional CD4+ and CD8+ T cells are gated as CD3+, then γδ‐, not CD26 + CD161+ (containing MAIT cells), and not CD16+ OR CD56+ (containing NKT cells). (B) Functional markers for CD4+ and CD8+ T cells. A gate is applied for each cytokine, and Boolean gates are created to identify cells expressing different combinations of markers. The single function gates are sometimes chosen vs. a parameter that displays some FMO spreading to allow for angled gates. Most gates are copied, applied to all lineages, and then cloned so that any changes to the gate on one lineage changes that gate for all lineages. However, the IL‐22 gate was uniquely lower for CD4 T cells (compared to other lineages) since the CD8 reagent caused some spreading into IL‐22 and thus requiring a higher gate for all other lineages that express CD8. (C) Additional functional and non‐functional markers for CD4+ and CD8+ T cells. Perforin and granzyme A are constitutive but can be examined as co‐expression with another functional marker. (D) NK T cells gated as CD16+ OR CD56+ on CD3+ γδ‐ T cells. Expression of CD4 vs. CD8 is shown, but all the other markers are also gated on the NK T cells. (E) NK cell subsets defined by CD16 vs. CD56 on CD3‐ lymphocytes. (F) MAIT cells identified as CD3+ γδ‐ CD26 + CD161+ and then Vα7.2+. The Va7.2+ cells are predominantly CD8+; however, the Vα7.2‐ cells are predominantly CD4+ and are likely not MAIT cells. For all gates, none are placed lower than that defined by FMO controls. Some gates are placed higher to improve the specificity, for example, for the functional markers based on the background as observed in the unstimulated controls (Online Fig. 6). The labels above each graph indicate the cells included in that graph.
similarity to published omips
This panel is unique in the combination of functional and phenotypic markers, but it can be considered an expansion of two of our prior ICS assays, OMIP‐014 1, and OMIP‐025 2.
funding
Grant sponsors: National Institute of Allergy and Infectious Diseases funding for the HIV Vaccine Trials Network Laboratory Center UM1 AI068618 (to MJM); Bill and Melinda Gates Foundation investments OPP1066048, OPP1088952, and OPP1099507 (to MJM); University of Washington/Fred Hutch Center for AIDS Research P30 AI027757.
Supporting information
Appendix S1: Supporting information
acknowledgments
The authors wish to thank all the individuals enrolled in the Cape Town and Seattle Assay Control cohorts, from which PBMC were used for optimization and testing of the panel. The authors are grateful to CHIL research technologists, Saleha Omarjee and Stanley Loots, for their help in the laboratory. We also thank our colleagues at the South Africa TB Vaccine Initiative (SATVI), Tom Scriba, Elisa Nemes, and Virginie Rozot, for discussions regarding appropriate markers and gating strategy. We acknowledge Stephen Voght for his help with editing.
Literature Cited
- 1. De Rosa SC, Carter DK, McElrath MJ. OMIP‐014: Validated multifunctional characterization of antigen‐specific human T cells by intracellular cytokine staining. Cytometry A 2012;81:1019–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moncunill G, Dobano C, McElrath MJ, De Rosa SC. OMIP‐025: Evaluation of human T‐ and NK‐cell responses including memory and follicular helper phenotype by intracellular cytokine staining. Cytometry A 2015;87:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information


