Abstract
Dysfunction of tight junctions is a critical step during the initial stage of tumor progression. Trophoblast cell surface antigen 2 (Trop-2) belongs to the family of tumor-associated calcium signal transducer (TACSTD) and is required for the stability of claudin-7 and claudin-1, which are often dysregulated or lost in carcinogenesis. Here, we investigated the effects of Trop-2 phosphorylation on cell motility. Analyses using HCT116 cells expressing WT Trop-2 (HCT116/WT) or Trop-2 alanine-substituted at Ser-303 (HCT116/S303A) or Ser-322 (HCT116/S322A) revealed that Trop-2 is phosphorylated at Ser-322. Furthermore, coimmunoprecipitation and Transwell assays indicated that Trop-2 S322A interacted with claudin-7 the strongest, and a phosphomimetic variant, Trop-2 S322E, the weakest and that HCT116/S322E cells have the highest motility and HCT116/S322A cells the lowest. All cell lines had similar levels of claudin-7 mRNA, but levels of claudin-7 protein were markedly decreased in the HCT116/S322E cells, suggesting posttranscriptional control of claudin-7. Moreover, claudin-7 was clearly localized to cell–cell borders in HCT116/S322A cells but was diffusely distributed on the membrane and partially localized in the cytoplasm of HCT116/S322E and HCT116/WT cells. These observations suggested that Trop-2 phosphorylation plays a role in the decrease or mislocalization of claudin-7. Using protein kinase C (PKC) inhibitors and PKC-specific siRNAs, we found that PKCα and PKCδ are responsible for Trop-2 phosphorylation. Of note, chemical PKC inhibition and PKCα- and PKCδ-specific siRNAs reduced motility. In summary, our findings provide evidence that Trop-2 is phosphorylated at Ser-322 by PKCα/δ and that this phosphorylation enhances cell motility and decreases claudin-7 localization to cellular borders.
Keywords: protein kinase C (PKC), protein phosphorylation, cancer, cell motility, cell adhesion, tight junction, cell signaling, claudin-7, transmembrane glycoprotein, trophoblast cell surface antigen 2 (Trop-2)
Introduction
At the initial stage of epithelial tumors, disorder of the epithelial sheet architecture with its polarity may be a key step to tumor progression. Trophoblast cell surface antigen 2 (Trop-2)3 and epithelial cell adhesion molecule (EpCAM) belong to the tumor-associated calcium signal transducer (TACSTD) gene family (1). They have been reported to directly interact with claudin-1 and claudin-7 (2–4), suggesting the involvement of these molecules in the function of tight junctions. From the spatial-temporal view on tumor progression, there is a possibility that these molecules may be correlated with the disorder of tight junctions, resulting in the loss of cell polarity.
Trop-2 is a 36-kDa transmembrane glycoprotein that is highly expressed in a variety of epithelial cancer cells (1, 5–7) and originally found on normal and malignant trophoblast cells (8, 9). Many studies showed that increased Trop-2 expression in tumor tissues is correlated with biological aggressiveness and poor survival of patients with various cancer types (1, 5, 10, 11). Thus, Trop-2 has been identified as an oncogene leading to invasiveness and tumorigenesis (12). Overexpression of EpCAM is also frequently observed in many types of carcinomas (13–15). On the other hand, it has been reported that loss of Trop-2 promotes carcinogenesis and features of epithelial-to-mesenchymal transition in squamous cell carcinomas (16) and that a mutation of the Trop-2 gene impairs the function of tight junctions through decreased expression and altered subcellular localization of tight junction proteins, claudin-1 and claudin-7, in gelatinous drop-like corneal dystrophy (GDLD) corneas (2, 17, 18). In EpCAM knockout mice, the barrier function of the intestinal epithelium was impaired (3). These individual studies are not comprehensive. Thus, the function of Trop-2 remains obscure.
Claudins constitute a family of 27 distinct transmembrane proteins that are composed of four transmembrane domains and two extracellular loops, which are involved in the homophilic and heterophilic interactions with adjacent claudins (19). The abnormal regulation of claudins, including both increased and decreased expression levels of specific claudins, has been reported in various human epithelial cancers (20–25). Claudin-7 is normally expressed in bronchial epithelial cells of human lungs but is either down-regulated or disrupted in lung cancer (23). Furthermore, claudin-7 is reduced in breast cancer cells and head and neck squamous cells (24, 25). If the stability and proper subcellular localization of claudin-7 are maintained through interaction with Trop-2 and/or EpCAM, as described above, there may be a certain relationship between the Trop-2/EpCAM and claudin-7 expression levels. However, in these studies, it was not determined whether the expression level of Trop-2 and/or EpCAM is related to the level of claudin-7. If Trop-2 overexpressed in various epithelial cancers is normally functional, it is supposed to maintain the function of tight junctions. However, this is not the case. It seems to be difficult to explain this contradiction only by the change in the level of Trop-2 expression. Thus, we investigated whether posttranslational modification of Trop-2 has any relation with its biological function. We focused on the modification of Trop-2, such as phosphorylation and the subsequent causal change of cell behavior.
In the present study, we demonstrated that Trop-2 is phosphorylated at Ser-322 by protein kinase Cα (PKCα) and PKCδ. Phosphorylation of Trop-2 decreased the interaction with claudin-7, leading to posttranscriptional down-modulation and/or mislocalization of claudin-7, and reciprocally increased the cell motility. Consistently, treatment of WT Trop-2–expressing HCT116 cells (HCT116/WT) with PKC inhibitors, BIM-I and Gö6983, and with PKCα and PKCδ siRNAs induced a less motile phenotype.
Results
Phosphorylation of Trop-2 and identification of phosphorylation site
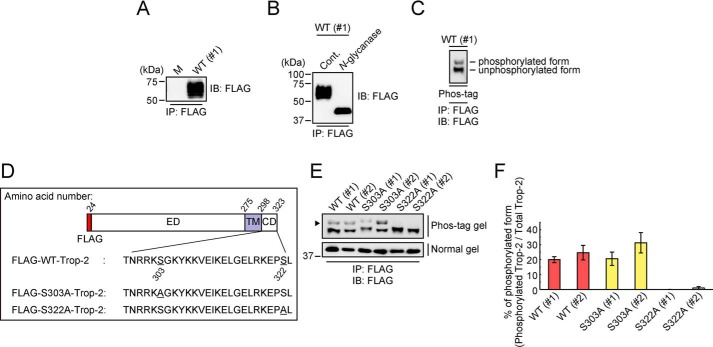
To investigate the tumor-promoting activity of Trop-2, FLAG-tagged WT Trop-2 was introduced into a human colorectal cancer cell line, HCT116 cells (HCT116/WT). HCT116 cells expressed a negligible level of endogenous Trop-2 (data not shown). Expression of Trop-2 was confirmed by flow cytometry (Fig. S1A) and SDS-PAGE, followed by immunoblotting. A broad band with a molecular mass of 50–75 kDa was detected, probably due to heterogeneity of glycosylation (Fig. 1A). In fact, Trop-2 treated with N-glycanase gave a clear band with a molecular mass of ∼40 kDa (Fig. 1B).
Figure 1.
Phosphorylation of Trop-2. A, FLAG-tagged Trop-2 was immunoprecipitated (IP) from lysates of HCT116/M (M) and HCT116/WT (WT: #1) cells and subjected to SDS-PAGE, followed by immunoblotting (IB). B, FLAG-tagged Trop-2 immunoprecipitated from a lysate of HCT116/WT (#1) cells with anti-FLAG magnetic beads was treated with or without N-glycanase and then subjected to SDS-PAGE, followed by immunoblotting. C, after N-glycanase treatment, FLAG-tagged Trop-2 obtained from a lysate of HCT116/WT (#1) cells as described above was subjected to SDS-PAGE using a Phos-tag–containing gel (Phos-tag SDS-PAGE), followed by immunoblotting. The top and bottom bands represent phosphorylated and unphosphorylated FLAG-tagged Trop-2, respectively. D, schematic model of WT and mutated FLAG-tagged Trop-2. ED, ectodomain; TM, transmembrane domain; CD, cytoplasmic domain. E, after N-glycanase treatment, FLAG-tagged Trop-2 immunoprecipitated from lysates of HCT116/WT (#1 and #2), HCT116/S303A (S303A: #1 and #2), and HCT116/S322A (S322A: #1 and #2) cells as described above was subjected to Phos-tag (top) and ordinary (bottom) SDS-PAGE, followed by immunoblotting. The arrowhead indicates phosphorylated FLAG-tagged Trop-2. F, intensities of the bands in the Phos-tag gel panel in E were measured, and then the ratio of phosphorylated to total FLAG-tagged Trop-2 was calculated (means ± S.E. (error bars), n = 4).
There are multiple possible phosphorylation sites (Ser-303, Tyr-306, and Ser-322) in the cytoplasmic domain of Trop-2. To distinguish phosphorylated Trop-2 from the unphosphorylated form, N-glycanase–treated Trop-2 was subjected to SDS-PAGE using Phos-tag–containing acrylamide gels (Phos-tag SDS-PAGE), followed by immunoblotting. Two bands were clearly detected, indicating the phosphorylation of Trop-2 in HCT116/WT cells (Fig. 1C). In addition, no phosphorylated Trop-2 was detected with anti-phosphorylated tyrosine antibodies (data not shown). To determine the phosphorylation site, mutated Trop-2 with Ala instead of Ser-303 or Ser-322 was generated using a site-directed mutagenesis kit (Fig. 1D) and expressed in HCT116 cells (HCT116/S303A and HCT116/S322A). A similar expression level of mutated Trop-2 was confirmed by flow cytometry (Fig. S1, B and C). Mutated Trop-2 was prepared from the lysates of each transfected cell type and subjected to Phos-tag SDS-PAGE as described above. Mutated Trop-2 prepared from HCT116/S303A cells gave two bands like those of WT Trop-2, whereas only a single band corresponding to unphosphorylated Trop-2 was detected for HCT116/S322A cells, indicating that the Ser-322 residue was phosphorylated (Fig. 1, E and F).
Different interaction between Trop-2 and claudin-7 depending on Trop-2 phosphorylation
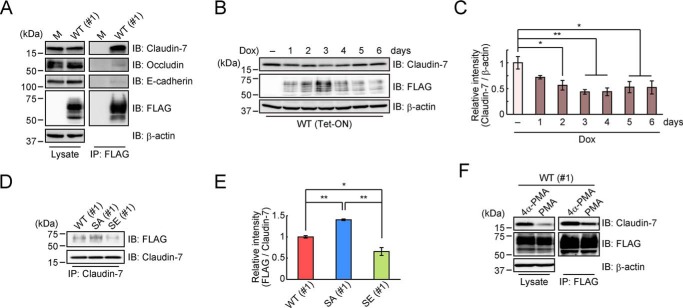
It has been reported that Trop-2 directly binds to claudin-7 and claudin-1 and that the interaction is required for claudin-7 and claudin-1 to be stabilized or to prevent their degradation (2). To confirm this interaction between Trop-2 and claudin-7 in epithelial cancer cells, Trop-2 was immunoprecipitated from lysates of HCT116/WT cells and then subjected to SDS-PAGE, followed by immunoblotting. It was claudin-7, but not occludin or E-cadherin, was coimmunoprecipitated with Trop-2, indicating a specific interaction of Trop-2 with claudin-7 (Fig. 2A). Claudin-1 was not detected due to its low expression in HCT116 cells (data not shown). To determine whether there is a causal relationship between Trop-2 and claudin-7, we established HCT116 cells, in which expression of Trop-2 can be controlled through the Tet-On system. Expression of WT Trop-2 started at 1 day after treatment with doxycycline and continued for a week. Unexpectedly, the level of claudin-7 decreased reciprocally in response to the increased expression of Trop-2, and thereafter expression of claudin-7 tended to recover gradually with a decrease of Trop-2 (Fig. 2, B and C). This result is incompatible with the report that Trop-2 is responsible for the maintenance of tight junction proteins as described above. Related to this discrepancy, we focused on the phosphorylation of Trop-2. In addition to HCT116/S322A cells in which cell phosphorylation of Trop-2 is blocked, we also prepared HCT116 cells expressing phosphomimetic (Glu instead of Ser-322) Trop-2 (HCT116/S322E). A similar expression level of mutated Trop-2 was confirmed by flow cytometry (Fig. S1D). For further experiments, clone #1 (#1) was used unless otherwise stated.
Figure 2.
Interaction of Trop-2 with claudin-7. A, lysates (left) of HCT116/M (M) and HCT116/WT (WT: #1) cells and the immunoprecipitates (IP) (right) obtained from the lysates with anti-FLAG magnetic beads were subjected to SDS-PAGE, followed by immunoblotting (IB), and detection of claudin-7, occludin, E-cadherin, and FLAG-tagged Trop-2. β-Actin served as a loading control. B and C, HCT116 cells were stably transfected with both a Tet-ON regulator plasmid and a response plasmid–inserted FLAG-tagged Trop-2 gene. Lysates of the cells treated with or without 10 μg/ml doxycycline (Dox) for 1–6 days were subjected to SDS-PAGE, followed by immunoblotting. β-Actin served as a loading control. The intensity of the claudin-7 and β-actin bands in B was measured, and the ratio of claudin-7 to β-actin in each Dox-treated cell is indicated, with that in Dox-nontreated cells being taken as 1 (means ± S.E. (error bars), n = 3; *, p < 0.05; **, p < 0.01). D and E, immunoprecipitates from the lysates of HCT116/WT (#1), HCT116/S322A (SA: #1), and HCT116/S322E (SE: #1) cells with anti-claudin-7 antibodies were subjected to SDS-PAGE, followed by immunoblotting. Histograms show the intensity of the FLAG-tagged Trop-2 bands, each density being normalized as to that of claudin-7, and the value for HCT116/WT (#1) cells was taken as 1 (means ± S.E., n = 3; *, p < 0.05; **, p < 0.01). F, after treatment of HCT116/WT (#1) cells with PMA or 4α-PMA for 2 h, their lysates (left) and claudin-7 coimmunoprecipitated with anti-FLAG magnetic beads (right) were subjected to SDS-PAGE, followed by immunoblotting, and detection with anti-claudin-7 and anti-FLAG antibodies. β-Actin served as a loading control.
Next, coimmunoprecipitation assays were performed to investigate the interaction of WT and mutated Trop-2 with claudin-7. Claudin-7 was immunoprecipitated from a lysate of each type of Trop-2–expressing cells. As described below, the protein level of claudin-7 was different among these cells. Thus, the amount of each cell lysate used for immunoprecipitation was adjusted so that a similar amount of claudin-7 was immunoprecipitated. It is notable that a lower amount of Trop-2 was coimmunoprecipitated with claudin-7 from the lysates of HCT116/WT and HCT116/S322E cells compared with HCT116/S322A cells, suggesting that phosphorylation of Trop-2 leads to a decrease in the interaction with claudin-7 (Fig. 2, D and E). Although it is uncertain whether the interaction was affected by the negative charge of amino acid 322 directly or indirectly, there is a possibility that phosphorylation of the Ser-322 residue leads to regulation of the stability of claudin-7 as a downstream event. In this context, because Trop-2 phosphorylation was elevated by treatment of HCT116/WT cells with phorbol myristate acetate (PMA), a PKC and protein kinase D (PKD) activator (data not shown), coimmunoprecipitation assays were carried out using PMA-treated HCT116/WT cells. Not only coimmunoprecipitated claudin-7 with Trop-2, but also total cellular claudin-7 was decreased by PMA treatment, probably due to the down-modulation of its stability (Fig. 2F).
Different expression and distribution of claudin-7 in WT and mutated Trop-2–expressing HCT116 cells
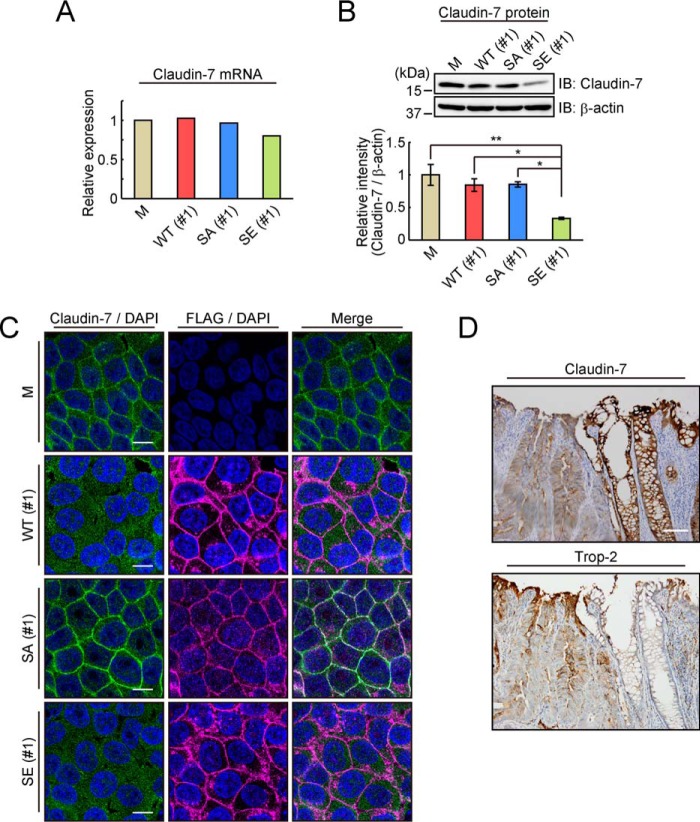
Furthermore, we examined the expression of claudin-7 in WT and mutated Trop-2–expressing HCT116 cells. Although the mRNA level of claudin-7 was substantially unchanged among these cells, the protein level of claudin-7 in HCT116/S322E cells was markedly decreased compared with that in other cells (Fig. 3, A and B). Given that the abundance of claudin-7 mRNA was maintained in HCT116/S322E cells, the reduction in the amount of claudin-7 protein appears to occur at a posttranscriptional step. Next, the distribution of claudin-7 and Trop-2 in WT and mutated Trop-2–expressing HCT116 cells were also investigated immunochemically. Generally, in the normal epithelium, claudins are clearly localized at the cell–cell borders. Forced expression of WT Trop-2 changed the distribution of claudin-7 compared with that in mock-transfected cells (HCT116/M). Although claudin-7 was concentrated at the cell membrane in HCT116/M cells, diffuse membrane staining of claudin-7 was observed in HCT116/WT cells. Its diffuse distribution was observed more prominently in HCT116/S322E cells. It is also noted that cell surface claudin-7 decreased, and a part of claudin-7 was localized in the cytoplasm. In contrast, strong expression of claudin-7 was concentrated at the cell membrane in HCT116/S322A cells like in HCT116/M cells (Fig. 3C). To further analyze the distribution of claudin-7 and Trop-2 in four types of cells as shown in Fig. 3C, fluorescent signals were scanned along the cell–cell junctions. As shown in Fig. S2, A and B, a part of the descending blue line (DAPI) shows the region of the cell–cell junctions and cytoplasm. Trop-2 is considered to be localized exclusively in the cell–cell junctions. In HCT116/S322A cells, the distribution of claudin-7 exhibited a sharp peak coinciding with that of Trop-2, indicating that both proteins are confined in the cell–cell junctions. In contrast, in HCT116/S322E and HCT116/WT cells, claudin-7 was distributed more widely with multiple peaks in the cytoplasm, and no peak coincided with that of Trop-2. In addition, claudin-7 in HCT116/S322A cells was more confined in the membrane than that in HCT116/M cells, probably due to the interaction of claudin-7 with Trop-2 in HCT116/S322A cells. These observations suggest that Trop-2 may play a causative role in localization of claudin-7.
Figure 3.
Expression and distribution of Trop-2 and claudin-7 in various types of Trop-2–expressing cells and in cancer tissues. A, levels of claudin-7 mRNA in HCT116/M (M), HCT116/WT (WT: #1), HCT116/S322A (SA: #1), and HCT116/S322E (SE: #1) cells were determined by DNA microarray analysis, and its level in HCT116/M cells was taken as 1. B, expression of claudin-7 protein in the four cell types described above was analyzed by SDS-PAGE, followed by immunoblotting, and that in HCT116/M cells was normalized as to β-actin, and the value for HCT116/M cells was taken as 1. β-Actin served as a loading control (means ± S.E. (error bars), n = 4; *, p < 0.05; **, p < 0.01). C, distribution of claudin-7 (green) and FLAG-tagged Trop-2 (magenta) in HCT116/M, HCT116/WT (#1), HCT116/S322A (#1), and HCT116/S322E (#1) cells was observed immunochemically. Nuclei were stained with DAPI (blue). Scale bars, 10 μm. D, distribution of claudin-7 and Trop-2 was observed immunohistochemically in colorectal cancer tissues. Nuclei were stained with hematoxylin. Scale bars, 100 μm.
It is interesting to investigate whether decreased expression of claudin-7 in cancer tissues is related to the expression of Trop-2. Colorectal tissue sections including both normal mucosa and dysplasia were immunostained. Intense claudin-7 staining was observed in histologically normal appearing tissues, whereas in cancerous tissues, the staining was reduced. Inversely, Trop-2 was highly expressed in cancerous tissues, and adjacent normal appearing tissues were only faintly stained (Fig. 3D).
Phosphorylation of Trop-2 by PKCα and PKCδ
As described above, phosphorylation of Trop-2 was related to the posttranslational regulation and intracellular distribution of claudin-7. In relation to the fact that Trop-2 phosphorylation was elevated by treatment with PMA, it has been reported that dysfunction of epithelial junctions is affected by PKC and PKD (26–28). Thus, we investigated which PKCs and/or PKDs phosphorylate Trop-2. Before this experiment, we prepared anti-phosphorylated Trop-2 antibodies by using the phosphorylated peptide of the Trop-2 cytoplasmic domain as an immunogen. The specificity of the antibodies was examined by immunoblotting and plate assays. Using Phos-tag SDS-PAGE, WT Trop-2 prepared from HCT116/WT cells was separated into phosphorylated and unphosphorylated Trop-2 (Fig. S3A, left). The anti-phosphorylated Trop-2 antibodies bound to the phosphorylated Trop-2 but not to unphosphorylated Trop-2 (Fig. S3A, right). Furthermore, phosphorylated and unphosphorylated Trop-2 cytoplasmic peptides were adsorbed to the plate and incubated with the anti-phosphorylated Trop-2 antibodies. The anti-unphosphorylated Trop-2 antibodies bound to both the unphosphorylated and phosphorylated Trop-2 peptide (Fig. S3B, left), whereas the anti-phosphorylated Trop-2 antibodies bound only to the phosphorylated Trop-2 peptide (Fig. S3B, right). Thus, it was clear that the anti-phosphorylated Trop-2 antibodies specifically bound to the phosphorylated Trop-2.
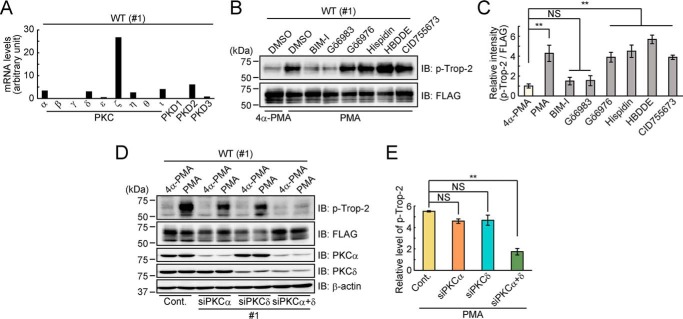
We first investigated which PKC and PKD isoforms are expressed in HCT116/WT cells. PKC isoenzymes are classified into three subclasses, “classical PKCs” (α, βI, βII, and γ), “novel PKCs” (δ, ϵ, η, and θ), and “atypical PKCs” (ζ and ι/λ) (29), and PKD family consists of three members (PKD1, -2, and -3) (30). DNA microarray analysis revealed the presence of multiple isoforms, including one classical (PKCα), three novel (PKCδ, -ϵ, and -η), and two atypical (PKCζ and -ι) isoenzymes, and PKD2 and -3 (Fig. 4A). Furthermore, PKC inhibitors with different subclass specificities and a PKD inhibitor were used to determine which PKCs and/or PKDs are responsible for Trop-2 phosphorylation. Elevated phosphorylation of Trop-2 by treatment with PMA was effectively inhibited by BIM-I and Gö6983, which are inhibitors of both classical and novel PKCs, whereas other PKC inhibitors, Gö6976, Hispidin, and HBDDE, and a PKD inhibitor, CID755673, had no effect on Trop-2 phosphorylation (Fig. 4, B and C). In addition, other kinase inhibitors, LY294002 (PI3K inhibitor), SB203580 (p38 MAPK inhibitor), PD98059 (MEK1/2 inhibitor), and SP600125 (JNK1/2/3 inhibitor), did not inhibit Trop-2 phosphorylation at all (Fig. S4).
Figure 4.
Phosphorylation of Trop-2 by PKCα and PKCδ. A, expression of PKC and PKD isoforms in HCT116/WT (WT: #1) cells was examined by DNA microarray analysis. B and C, HCT116/WT (#1) cells were treated with or without a PKC or PKD inhibitor for 1 h and subsequently with PMA or 4α-PMA for 2 h. Each cell lysate was subjected to SDS-PAGE, followed by immunoblotting. The intensities of the bands in B were determined. Histograms show the inhibitory effect of each inhibitor, the levels of phosphorylated Trop-2 being represented as the ratio of phosphorylated to total FLAG-tagged Trop-2 (p-Trop-2/FLAG), and the ratio obtained for 4α-PMA–treated cells was taken as 1 (means ± S.E. (error bars), n = 3; **, p < 0.01; NS, not significant). D and E, HCT116/WT (#1) cells were transiently transfected with PKCα siRNA (#1), PKCδ siRNA (#1), both PKCα and PKCδ siRNAs (#1), or a control siRNA and then treated with PMA or 4α-PMA for 2 h. A lysate of each cell type was subjected to SDS-PAGE, followed by immunoblotting. β-Actin served as a loading control. The intensities of the bands in D were determined, and the relative value (p-Trop-2/FLAG) was calculated as described in B and C. Histograms show the p-Trop-2/FLAG ratio in PMA-treated cells and that in 4α-PMA–treated cells (mean ± S.E., n = 3; **, p < 0.01; NS, not significant).
Based on the results of DNA microarray analysis and the effects of pharmacological PKC and PKD inhibitors on Trop-2 phosphorylation, knockdown of relevant PKCs and PKDs that might phosphorylate Trop-2 in HCT116/WT cells was performed by using PKC or PKD isoform–specific siRNAs. It was revealed that knockdown of PKCα or PKCδ was most effective in reducing the phosphorylation of Trop-2, but considerable activity of Trop-2 phosphorylation remained even after the knockdown of individual PKCs, suggesting that Trop-2 may be phosphorylated by multiple PKCs and that the activity may be retained due to compensation by other PKCs. Furthermore, we tried to examine the inhibitory effect on Trop-2 phosphorylation with various combinations of PKC knockdown. It was demonstrated that knockdown of both PKCα and PKCδ abolished the phosphorylation of Trop-2 almost completely (Fig. 4, D and E). In addition, Trop-2 phosphorylation was similarly down-modulated by using different types of PKCα and PKCδ siRNAs (Fig. S5).
Different motility of WT and mutated Trop-2–expressing cells
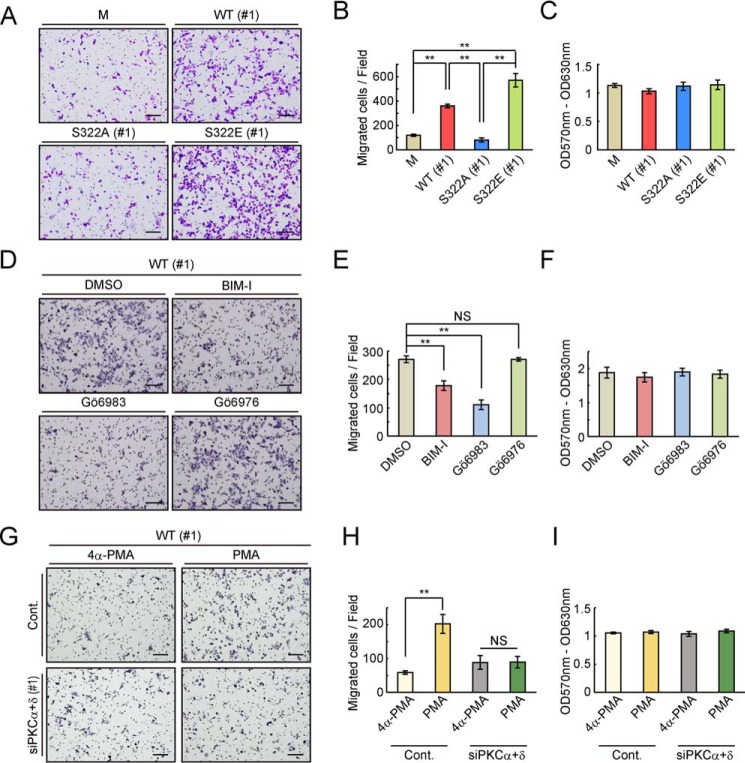
As described above, the cell–cell interaction may be regulated by downstream events after phosphorylation of the Ser-322 residue of Trop-2. Next, we performed migration assays using Transwells. HCT116/S322A cells were significantly less motile than the other Trop-2–expressing cells, which to a greater or lesser extent possess negative charges of amino acid 322 (Fig. 5, A and B). In addition, no difference in the rate of cell proliferation was observed among these cells, indicating that different numbers of migrated cells have no relation to the growth rate of these cells (Fig. 5C). The same WT and mutated Trop-2–expressing vectors were also transfected into a human pancreatic cancer cell line, PANC-1 cells, and stable transfectants (PANC-1/M, PANC-1/WT, PANC-1/S322A, and PANC-1/S322E) were prepared. Similar expression levels of Trop-2 were confirmed by flow cytometry (Fig. S6, A–C), and migration assays were performed (Fig. S6, D and E). Consistently, PANC-1/S322E cells showed the highest motility, and PANC-1/S322A cells exhibited less than half motility compared with that of PANC-1/S322E cells. Similar cell proliferation was observed among these cells (Fig. S6F). These results suggest that phosphorylation of Trop-2 is involved in the regulation of cell motility.
Figure 5.
Effects of Trop-2 phosphorylation on motility of WT and mutated Trop-2–expressing HCT116 cells. A and B, migration of HCT116/M (M, n = 7), HCT116/WT (WT: #1, n = 4), HCT116/S322A (SA: #1, n = 4), and HCT116/S322E (SE: #1, n = 3) cells was evaluated by means of Transwell assays. A representative photograph of each migrated cell type is shown in A. Scale bars, 100 μm. The number of migrated cells per field was determined and is shown as a histogram in B (means ± S.E. (error bars); **, p < 0.01). C, proliferation of HCT116/M (n = 7), HCT116/WT (#1: n = 4), HCT116/S322A (#1: n = 4), and HCT116/S322E (#1: n = 3) cells was analyzed using MTT assays (means ± S.E.). D and E, migration of HCT116/WT (#1) cells treated with or without PKC inhibitors was measured as described in A and B. A representative photograph of each migrated cell is shown in D. Scale bars, 100 μm. The number of migrated cells per field was determined as described in A and B and is shown as a histogram in E (means ± S.E., n = 3; **, p < 0.01; NS, not significant). F, Proliferation of cells was examined using MTT assays as described in C (means ± S.E., n = 3). G and H, migration of HCT116/WT (#1) cells treated with both PKCα and PKCδ siRNAs (#1) or a control siRNA and then with PMA or 4α-PMA was analyzed by using Transwells. A representative photograph of each migrated cell type is shown in G. Scale bars, 100 μm. The number of migrated cells per field was determined as described in A and B and is shown as a histogram in H (means ± S.E., n = 3; **, p < 0.01; NS, not significant). I, proliferation of cells was analyzed using MTT assays as described in C (means ± S.E., n = 3).
We next investigated the effects of down-modulation of PKCα and PKCδ on the migration of HCT116/WT cells in vitro by treatment with PKC inhibitors and both PKCα and PKCδ siRNAs. As expected, BIM-I and Gö6983 but not Gö6976 inhibited the motility of HCT116/WT cells strongly, approximately in correspondence to the inhibitory level of Trop-2 phosphorylation (Fig. 5, D and E). On the other hand, HCT116/S322E cells exhibited similar motility regardless of treatment with or without PKC inhibitors, BIM-I and Gö6983 (Fig. S7, A and B). These results suggest that PKCα- and PKCδ-mediated Trop-2 phosphorylation leads to enhancement of cell motility. Furthermore, after treatment with PMA or its analog, 4α-PMA, motility of control and both PKCα and PKCδ knockdown HCT116/WT cells was examined using Transwells. The effects of PMA on the migration of control cells were abolished by knockdown of both PKCα and PKCδ (Fig. 5, G and H). Cell growth did not change with these treatments (Fig. 5, F and I and Fig. S7C).
Discussion
During oncogenic transformation, tumor cells typically lose tight junctions, leading to derangement of the tissue architecture and loss of cell polarity. In fact, expression of some claudins has been shown to be either deregulated or lost in cancers (20, 22–25, 31). It is also interesting that the level of claudin-7 is reduced as an early event in colorectal carcinogenesis (31). Conversely, many reports demonstrated that Trop-2 is overexpressed in a variety of human cancers. In the colon dysplastic mucosa, Trop-2 expression was demonstrated at the earliest stages of cell transformation, and its expression level further increased after neoplastic transformation, thus suggesting the role of Trop-2 as a driver (6). Trop-2 is widely expressed in normal tissues (32), suggesting that it may play a certain biological role in epithelial tissues. One possibility is that Trop-2 is responsible for the maintenance of tight junction proteins, which is demonstrated by analysis of corneal epithelial cell disorder, GDLD (2, 18). Thus, we investigated whether posttranslational modification of Trop-2 has any relation with its biological function. As shown in Fig. 1, analyses of WT and mutated Trop-2 using Phos-tag SDS-PAGE revealed that Trop-2 was phosphorylated and that the phosphorylated site was the Ser-322 residue of Trop-2. In a previous study (33), phosphorylation of Trop-2 was analyzed by incubation of purified Trop-2 with PKC in vitro, and the phosphorylated site was revealed to be the Ser-303 residue. This discrepancy may be due to the conformational difference between purified Trop-2 and membrane-anchored Trop-2 in the cell. Other reports have also predicted that Ser-322 but not the Ser-303 residue of Trop-2 has the potential to be phosphorylated (34, 35).
It is also notable that blocking of Ser-322 phosphorylation by changing the Ser-322 residue to Ala (HCT116/S322A) decreased the migratory ability as compared with that of HCT116/WT cells. Reciprocally, HCT116 cells expressing phosphomimetic Trop-2 (HCT116/S322E) showed a higher migratory ability (Fig. 5, A and B). These results strongly suggest that phosphorylation of Trop-2 is critical for acquisition of motility potential.
The cell–cell interaction seems to be regulated by downstream events triggered by Trop-2 phosphorylation. As reported previously (2), claudin-7 was specifically coimmunoprecipitated with Trop-2 (Fig. 2A). However, the level of Trop-2 coimmunoprecipitated with claudin-7 decreased with the phosphorylation of Trop-2 (Fig. 2, D and E), suggesting that phosphorylation of Trop-2 may have a causal effect on the interaction with claudin-7, leading to the loss of claudin-7 stability. Actually, the claudin-7 protein in HCT116/S322E cells was markedly reduced (Fig. 3B), and elevated phosphorylation of Trop-2 in HCT116/WT cells by treatment with PMA reduced the level of claudin-7 (Fig. 2F). In addition, although the levels of claudin-7 protein were similar in HCT116/S322A and HCT116/WT cells, its distribution was quite different in these cells. Claudin-7 in HCT116/S322A cells seemed to be confined to the cell membrane (Fig. 3, B and C). In contrast, claudin-7 in HCT116/WT cells showed a dispersed distribution on the cell membrane, and a considerable part of claudin-7 was located in the cytoplasm. This mislocalization of claudin-7 was prominent in HCT116/S322E cells (Fig. 3C). We speculate that mislocalized claudin-7 may be nonfunctional, and thereby HCT116/WT and HCT116/S322E cells exhibit similar phenotypes. The loss of membrane-localized claudins and their relocalization to the cytoplasm have been reported for breast cancer (24) like GDLD caused by a Trop-2 mutation (2, 18) and intestinal epithelial cells of EpCAM knockout mice (3). EpCAM is the closest homologue to Trop-2, and analyses of EpCAM knockout mice showed that EpCAM contributes to the formation of functional tight junctions in the intestinal epithelium by recruiting claudin-7 to apical cell–cell junctions.
We also observed low and high intensity of immunochemically stained claudin-7 in colorectal cancer tissues and adjacent normal tissues, respectively (Fig. 3D). Inversely, Trop-2 appears to be distributed densely in cancer tissues and only faintly in adjacent normal tissues. Usami et al. (22) demonstrated that claudin-7 expression was prominently reduced at the invasive front, which is associated with the depth of invasion, stage, lymphatic vessel invasion, and lymph node metastasis. The level of claudin-7 protein in HCT116/S322E cells was markedly decreased compared with in other cells, but its mRNA level was substantially similar to that in the other cells (Fig. 3, A and B), indicating that the reduction in the amount of claudin-7 protein appears to occur at a posttranslational step. This posttranslational regulation of claudin-7 was also demonstrated in the intestines of EpCAM mutant mice (3). In this mouse model, claudin-7 protein decreased to an undetectable level in the intestines, even though the abundance of claudin-7 mRNA was maintained, suggesting similar posttranslational reduction of claudin-7 protein. Furthermore, modified mice lacking claudin-7 exhibit intestinal defects similar to those in EpCAM knockout mice (36).
We speculate that interaction of claudin-7 with Trop-2 may lead to the recruitment of claudin-7 to tight junctions, such that through a decrease of their interaction, claudin-7 becomes mislocalized and susceptible to degradation. However, there remain the questions of how and when Trop-2 loses or decreases the interaction with claudin-7. It has been reported that the (G/A)XXX(G/A) motif in the transmembrane domain supports protein–protein associations (37, 38). Interestingly, the transmembrane domains of both Trop-2 and claudin-7 contain the AXXXG motif. Thus, it is plausible that Trop-2 associates with claudin-7 through this motif in the transmembrane region. Actually, Nakatsukasa et al. (2) reported that Trop-2 interacts with claudin-7 but not claudin-4, which does not have the AXXXG motif in its transmembrane domain. Furthermore, it has been reported that Trop-2 is cleaved by tumor necrosis factor-α–converting enzyme, followed by further cleavage by γ-secretase within the transmembrane domain, resulting in shedding of the extracellular domain and accumulation of the intracellular domain in the nucleus (39). One possible scenario is that Trop-2 may be cleaved through downstream events after Trop-2 phosphorylation, and cleavage of Trop-2 may result in loss of the interaction with claudin-7. At present, there are no data on whether there is a causal relationship between the phosphorylation and cleavage of Trop-2.
From the view of tumor progression, it is important to determine the PKCs responsible for the phosphorylation of Trop-2. It is suggested that changes in the activation balance of different PKC isozymes are linked to cancer development and that the PKC balance plays a central role in the function of the adhesive zipper, which works toward stable cell–cell junctions and adhesion (40). Among PKC inhibitors, Gö6976, an inhibitor of PKCα and PKCβI, has been extensively studied and revealed to be a potential anticancer drug due to its effects on migration and invasion (40). Because many studies have been performed with a PKC activator, PMA, or a PKC inhibitor, Gö6976, elucidation of specific PKC isoforms and their substrates has been limited. Through a combination of experiments involving PKC inhibitors and PKC siRNAs, we determined that PKCα and PKCδ are responsible for the phosphorylation of Trop-2 and found that an effective inhibitor was Gö6983 but not Gö6976. It should be noted that the maintenance of cell–cell junctions through the inhibition of Trop-2 phosphorylation by Gö6983 and knockdown of both PKCα and PKCδ can induce a less motile phenotype of HCT116/WT cells (Fig. 5, D, E, G, and H). Although the Trop-2 ligand remains unknown, Trop-2 seems to be phosphorylated by external signals directly or indirectly. In this context, growth factors such as EGF elevated the phosphorylation of Trop-2, probably through the activation of PKCα and PKCδ (data not shown).
The whole cascade from external stimulation to enhancement of motility has not been elucidated. However, we demonstrated a novel mechanism underlying tumor progression through molecular cross-talk between Trop-2–PKC–claudin-7. Binding of an external component to Trop-2 or other receptors enhances the phosphorylation of Trop-2 by PKCα and PKCδ. Phosphorylation of Trop-2 results in the loss or a decrease of the interaction with claudin-7, leading to dysfunction of claudin-7 through its mislocalization.
Experimental procedures
Construction of expression plasmids
Preparation of total RNA from a human ovarian cancer cell line, OVCAR3 cells and subsequent cDNA synthesis were performed as described previously (41). The synthesized cDNA was used for amplification of the Trop-2 gene using the following primers: 5′-CCT GCA GAC CAT CCC AGA C-3′ and 5′-CTG GGA CGA TAC CGA AAT CC-3′. The PCR product was cloned into the pCR2.1®-TOPO® TA vector (Invitrogen) according to the manufacturer's instructions. After sequencing, the vector was treated with EcoO65I, and then the EcoO65I site was converted to a blunt end using a Blunting Kination Ligation Kit (Takara Bio Inc.). Thereafter, the blunt-ended vector was treated with BamHI, followed by subcloning into the pFLAG-CMVTM-3 expression vector (Sigma-Aldrich) with blunt-ended KpnI and cohesive-ended BamHI sites (WT-Trop-2). Site-directed mutagenesis of the Trop-2 gene was performed using a QuikChange II XL site-directed mutagenesis kit (Stratagene). Three types of mutations with Ala instead of Ser-303 (i) or Ser-322 (ii) and with Glu instead of Ser-322 (iii) were introduced by using the following primers, respectively: (i) 5′-CAC CAA CCG GAG AAA GGC GGG GAA GTA CAA GAA G-3′ and 5′-CTT CTT GTA CTT CCC CGC CTT TCT CCG GTT GGT G-3′, (ii) 5′-GTT GAG AAA GGA ACC GGC CTT GTA GGT ACC CGG C-3′ and 5′-GCC GGG TAC CTA CAA GGC CGG TTC CTT TCT CAA C-3′, and (iii) 5′-GTT GAG AAA GGA ACC GGA GTT GTA GGT ACC CGG CG-3′ and 5′-CGC CGG GTA CCT ACA ACT CCG GTT CCT TTC TCA AC-3′.
Cell culture and transfection of plasmids
A human colorectal cancer cell line, HCT116 cells, and a human pancreatic cancer cell line, PANC-1 cells, were obtained from the American Type Culture Collection (Manassas, VA). An expression plasmid or an empty vector was transfected into the cells using FuGENE® HD Transfection Reagent (Promega) according to the manufacturer's instructions, and the following stable transfectants were prepared by selection with 600 μg/ml G418: mock cells (HCT116/M and PANC-1/M), FLAG-tagged WT Trop-2–expressing cells (HCT116/WT and PANC-1/WT), FLAG-tagged mutated Trop-2 with Ala instead of Ser-303 or Ser-322–expressing cells (HCT116/S303A or HCT116/S322A and PANC-1/S322A), and FLAG-tagged mutated Trop-2 with Glu instead of Ser-322–expressing cells (HCT116/S322E and PANC-1/S322E). These transfectants were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 4 mm l-glutamine, and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin).
Preparation of cell extracts
Subconfluent cells were lysed with solubilizing buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100) containing protease and phosphatase inhibitor cocktails (Nacalai Tesque). After solubilization, the lysates were centrifuged at 13,000 rpm at 4 °C for 10 min, and the supernatants were used as cell extracts.
Treatment with PKC, PKD, and Ser/Thr kinase inhibitors and PMA or 4α-PMA
Cells were treated with PKC, PKD, and Ser/Thr kinase inhibitors or DMSO for 1 h. The following PKC, PKD, and Ser/Thr kinase inhibitors were used: BIM-I (pan-PKC (α, βI, βII, γ, δ, and ϵ) inhibitor, 1 μm; Cell Signaling Technology), Gö6983 (pan-PKC (α, βI, βII, γ, δ, and ζ) inhibitor, 1 μm; Sigma-Aldrich), Gö6976 (PKCα and -βI inhibitor, 1 μm; Cayman Chemical), Hispidin (PKCβI and -βII inhibitor, 10 μm; Merck Millipore), HBDDE (PKCα and -γ inhibitor, 100 μm; Enzo Life Science), CID755673 (PKD inhibitor, 10 μm; Merck Millipore), LY294002 (PI3K inhibitor, 50 μm; Cell Signaling Technology), SB203580 (p38 MAPK inhibitor, 20 μm; Cell Signaling Technology), PD98059 (MEK1/2 inhibitor, 50 μm; Cell Signaling Technology), and SP600125 (JNK1/2/3 inhibitor, 25 μm; Tokyo Chemical Industry). Cells were treated with 100 ng/ml PMA or 4α-PMA for the indicated times.
Immunoprecipitation
FLAG-tagged Trop-2 and claudin-7 were immunoprecipitated from cell extracts as described above with anti-FLAG® M2 magnetic beads (Sigma-Aldrich) and with mouse anti-claudin-7 antibodies (Invitrogen) and Protein G–Sepharose 4 Fast Flow (GE Healthcare), respectively.
Immunoblotting
Cell extracts or immunoprecipitates prepared as described above were subjected to SDS-PAGE, followed by immunoblotting. After successive incubation with primary antibodies and horseradish peroxidase (HRP)-conjugated secondary antibodies, immunoreactive bands were detected by means of chemiluminescence, and the bands were quantified with ImageJ software (National Institutes of Health). The following antibodies were used as primary antibodies: HRP-conjugated mouse anti-FLAG® M2 (Sigma-Aldrich), mouse anti-claudin-7, rabbit anti-E-cadherin (Cell Signaling Technology), mouse anti-occludin (Invitrogen), mouse anti-β-actin (Sigma-Aldrich), rabbit anti-PKCα (Abcam), and rabbit anti-PKCδ (Abcam) antibodies. Rabbit anti-phosphorylated Trop-2 antibodies were prepared in our laboratory. The details are given below.
Preparation of anti-phosphorylated and unphosphorylated Trop-2 antibodies and evaluation of their specificity
KLH-conjugated peptides (Trop-2 cytoplasmic domain) with phosphorylated or unphosphorylated Ser-322 were purchased from Hokkaido System Science. These peptides were emulsified with Freund's complete (first time) and incomplete (from second time) adjuvant (Difco Laboratories) and injected subcutaneously five times into a 12-week-old female New Zealand White rabbit. After the fifth immunization, blood was taken, and an IgG fraction was prepared from the serum by protein A–Sepharose column chromatography. The IgG fraction containing antibodies against phosphorylated or unphosphorylated Trop-2 was applied to a KLH-unphosphorylated Trop-2 peptide or KLH-conjugated CNBr-activated Sepharose 4 Fast Flow column (GE Healthcare), respectively. Each passed-through fraction was used as anti-phosphorylated or unphosphorylated Trop-2 antibodies. Rabbits were handled in accordance with the guidelines of the animal committee of Kyoto Sangyo University.
Plate assays were performed to confirm the specificity of each antibody. Phosphorylated and unphosphorylated Trop-2 cytoplasmic peptides were immobilized on 96-well MaxiSorp plates (Nunc) overnight at 4 °C. After successive incubation with serially diluted anti-phosphorylated or unphosphorylated Trop-2 antibodies (0–1 μg/ml), the immune complex was determined by adding a 1-Step Ultra TMB-ELISA Substrate (Thermo Fisher Scientific Inc.) and subsequent measurement of absorbance at 450 nm. In plate assays, each experiment was performed in quadruplicate.
Flow cytometry
Cells were incubated with FITC-conjugated mouse anti-FLAG® M2 antibodies (Sigma-Aldrich) at 4 °C for 2 h. After washing with 1% BSA/PBS, expression of FLAG-tagged Trop-2 was detected using a FACSCalibur (BD Biosciences).
DNA microarray analysis
DNA microarray analysis was performed basically as described previously (42). In brief, total RNA was isolated from cells using ISOGEN (Nippon Gene), and then cyanine-3–labeled cRNA was prepared from the total RNA using a Low Input Quick Amp Labeling Kit (Agilent Technologies). After purification and fragmentation, the labeled cRNAs were hybridized to a SurePrint G3 Human Gene Expression 8×60K v3 Microarray (Agilent Technologies). After washing, the array slides were scanned with an Agilent SureScan microarray scanner (G2600D).
Migration and MTT assays
Migration and MTT assays were performed basically as described previously (41). Briefly, cells (4 × 104 cells) were suspended in serum-free medium containing 0.2% BSA and then seeded into the upper chamber of a Transwell (24-well culture plate, pore size 8.0 μm; Corning Inc.) precoated with fibronectin. In some cases, several PKC inhibitors or DMSO, and 100 ng/ml PMA or 4α-PMA was added to the cell suspension. After filling the lower wells with serum-containing medium, the cells were incubated for 20 h. The chambers were fixed with methanol and stained with hematoxylin, and then nonmigrated cells were scraped off with a cotton swab. The migration was quantified by counting the migrated cells in five randomly selected fields at a magnification of ×100.
MTT assays were performed to examine the effect of cell proliferation on cell migration assays. Cells (4 × 104 cells) were cultured in the same medium using migration assays in 96-well plates for 18 h. After adding the MTT reagent (Nacalai Tesque) and subsequent incubation for 2 h, the absorbance of wells was measured at 570 nm with a reference at 630 nm. In MTT assays, each experiment was performed with cells plated at least in duplicate.
Phos-tag SDS-PAGE analysis
FLAG-tagged Trop-2 immunoprecipitated as described above was treated with glycopeptidase F (Takara Bio Inc.) according to the manufacturer's instructions, followed by exclusion of salts and concentration using a Nanosep® centrifugal device with a 10-kDa cutoff (Pall Life Sciences). The samples were subjected to Zn2+-Phos-tag SDS-PAGE (FUJIFILM Wako Pure Chemical Corp.) according to the manufacturer's instructions, followed by immunoblotting, and detection of phosphorylated and unphosphorylated FLAG-tagged Trop-2 with HRP-conjugated mouse anti-FLAG® M2 antibodies.
Treatment with siRNAs
Cells were transfected with Silencer® Select siRNA of PKCα (5 nm) and/or PKCδ (10 nm) or Silencer® Select Negative Control #1 siRNA (Ambion) by reverse transfection using LipofectamineTM RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions and incubated for 3 days. The following two types of siRNAs (#1, #2) for each PKC were used: (i) PKCα siRNAs, 5′-GGCUGUACUUCGUCAUGGAtt-3′ and 5′-UCCAUGACGAAGUACAGCCga-3′(#1) and 5′-CAACGUACCCAUUCCGGAAtt-3′ and 5′-UUCCGGAAUGGGUACGUUGta-3′ (#2); (ii) PKCδ siRNAs, 5′-GGGACACUAUAUUCCAGAAtt-3′ and 5′-UUCUGGAAUAUAGUGUCCCgg-3′ (#1) and 5′-AGAAGGAUGUGGUCCUGAUtt-3′ and 5′-AUCAGGACCACAUCCUUCUtg-3′ (#2).
Immunocytochemistry
The distribution of Trop-2 and claudin-7 was determined as described previously (42). Briefly, cells were fixed with 4% paraformaldehyde in PBS, followed by treatment with 5% BSA/PBS containing 0.1% Triton X-100. After incubation with primary antibodies (rabbit anti-DDDDK (FLAG) tag (Medical and Biological Laboratories) and mouse anti-claudin-7 antibodies), the cells were stained with fluorescence-conjugated secondary antibodies and DAPI. The fluorescent signals were detected under a confocal laser-scanning fluorescence microscope (Leica) at a magnification of ×630 and quantified with LAS AF Lite software (Leica).
Immunohistochemistry
Sections of paraffin-embedded tissues were deparaffinized and rehydrated using serial Hemo-De (as a substitute for xylene; Medical Chemical Corp.) and ethanol. After washing with Wash Buffer (Dako), antigen retrieval was carried out with Target Retrieval Solution (pH 6.0) (Dako), under high-pressure conditions. The sections were washed and then blocked with REAL Peroxidase-Blocking Solution (Dako) at room temperature for 5 min. The distribution of Trop-2 was visualized as follows. Sections were successively incubated with goat anti-Trop-2 antibodies (R&D Systems) and HRP-conjugated secondary antibodies at room temperature. To detect claudin-7, the sections were successively incubated with mouse anti-claudin-7 antibodies and REAL Envision Detection Reagent Peroxidase Rabbit/Mouse (Dako). All sections were stained with diaminobenzidine and subsequently with hematoxylin. Thereafter, dehydration and clearing were performed by serial treatment with ethanol and Hemo-De. Specimens of malignant and adjacent nonmalignant tissues were obtained from cancer patients in accordance with a protocol approved by Osaka University.
Tet-On expression systems
For construction of a Tet-On response plasmid, the DNA fragment of the FLAG-containing WT Trop-2 gene was amplified from FLAG-tagged WT-Trop-2 plasmids as described above using the following primers: 5′-CCC TCG TAA AGT CGA CAT GTC TGC ACT TCT GAT CCT AGC TC-3′ and 5′-CAG TTA CAT TGG ATC CCT ACA AGC TCG GTT CCT TTC TCA AC-3′. After purification, the fragment was subcloned into the SalI/BamHI-digested pTRE3G vector (Clontech) according to the manufacturer's instructions. The Tet-On system was established as follows: first, cells were transfected with a Tet-On regulator plasmid, the pCMV-Tet3G vector (Clontech), followed by selection with 600 μg/ml G418. Next, the Tet-On response plasmid described above was transfected into the regulator plasmid–introduced cells and subsequently selected with 1 μg/ml puromycin. The double-stable cells were treated with 10 μg/ml doxycycline to induce FLAG-tagged Trop-2 expression.
Statistical analysis
All statistical analyses were conducted with SPSS Statistics 25 (IBM). Differences between three or more groups were assessed by analysis of variance, followed by Tukey's or Dunnett's test. In all analyses, differences were considered statistically significant at p < 0.05.
Author contributions
Y. M., K. A., and H. N. conceptualization; Y. M. and K. A. formal analysis; Y. M., K. A., and H. N. supervision; Y. M. funding acquisition; Y. M., K. A., K. O., S. I., and T. Y. investigation; Y. M., K. A., and H. N. writing-original draft; Y. M. and H. N. project administration; Y. M. and H. N. writing-review and editing; E. M. preparation of the human tissue specimens and pathological analysis.
Supplementary Material
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 17K14998 (to Y. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7.
- Trop-2
- trophoblast cell surface antigen 2
- BIM-I
- bisindolylmaleimide I
- DAPI
- 4′,6-diamidino-2-phenylindole
- EGF
- epidermal growth factor
- EpCAM
- epithelial cell adhesion molecule
- JNK
- c-Jun N-terminal kinase
- KLH
- keyhole limpet hemocyanin
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI3K
- phosphatidylinositol 3-kinase
- PKC
- protein kinase C
- PKD
- protein kinase D
- PMA
- phorbol myristate acetate
- PKD
- protein kinase D
- HRP
- horseradish peroxidase.
References
- 1. Cubas R., Li M., Chen C., and Yao Q. (2009) Trop2: a possible therapeutic target for late stage epithelial carcinomas. Biochim. Biophys. Acta 1796, 309–314 10.1016/j.bbcan.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 2. Nakatsukasa M., Kawasaki S., Yamasaki K., Fukuoka H., Matsuda A., Tsujikawa M., Tanioka H., Nagata-Takaoka M., Hamuro J., and Kinoshita S. (2010) Tumor-associated calcium signal transducer 2 is required for the proper subcellular localization of claudin 1 and 7: implications in the pathogenesis of gelatinous drop-like corneal dystrophy. Am. J. Pathol. 177, 1344–1355 10.2353/ajpath.2010.100149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lei Z., Maeda T., Tamura A., Nakamura T., Yamazaki Y., Shiratori H., Yashiro K., Tsukita S., and Hamada H. (2012) EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev. Biol. 371, 136–145 10.1016/j.ydbio.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 4. Wu C. J., Mannan P., Lu M., and Udey M. C. (2013) Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J. Biol. Chem. 288, 12253–12268 10.1074/jbc.M113.457499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bignotti E., Todeschini P., Calza S., Falchetti M., Ravanini M., Tassi R. A., Ravaggi A., Bandiera E., Romani C., Zanotti L., Tognon G., Odicino F. E., Facchetti F., Pecorelli S., and Santin A. D. (2010) Trop-2 overexpression as an independent marker for poor overall survival in ovarian carcinoma patients. Eur. J. Cancer 46, 944–953 10.1016/j.ejca.2009.12.019 [DOI] [PubMed] [Google Scholar]
- 6. Trerotola M., Cantanelli P., Guerra E., Tripaldi R., Aloisi A. L., Bonasera V., Lattanzio R., de Lange R., Weidle U. H., Piantelli M., and Alberti S. (2013) Upregulation of Trop-2 quantitatively stimulates human cancer growth. Oncogene 32, 222–233 10.1038/onc.2012.36 [DOI] [PubMed] [Google Scholar]
- 7. Xie J., Mølck C., Paquet-Fifield S., Butler L., Australian Prostate Cancer Bioresource, Sloan E., Ventura S., and Hollande F. (2016) High expression of TROP2 characterizes different cell subpopulations in androgen-sensitive and androgen-independent prostate cancer cells. Oncotarget 7, 44492–44504 10.18632/oncotarget.9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fornaro M., Dell'Arciprete R., Stella M., Bucci C., Nutini M., Capri M. G., and Alberti S. (1995) Cloning of the gene encoding Trop-2, a cell-surface glycoprotein expressed by human carcinomas. Int. J. Cancer 62, 610–618 10.1002/ijc.2910620520 [DOI] [PubMed] [Google Scholar]
- 9. Ripani E., Sacchetti A., Corda D., and Alberti S. (1998) Human Trop-2 is a tumor-associated calcium signal transducer. Int. J. Cancer 76, 671–676 [DOI] [PubMed] [Google Scholar]
- 10. Trerotola M., Jernigan D. L., Liu Q., Siddiqui J., Fatatis A., and Languino L. R. (2013) Trop-2 promotes prostate cancer metastasis by modulating β(1) integrin functions. Cancer Res. 73, 3155–3167 10.1158/0008-5472.CAN-12-3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeng P., Chen M. B., Zhou L. N., Tang M., Liu C. Y., and Lu P. H. (2016) Impact of TROP2 expression on prognosis in solid tumors: a systematic review and meta-analysis. Sci. Rep. 6, 33658 10.1038/srep33658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J., Day R., Dong Y., Weintraub S. J., and Michel L. (2008) Identification of Trop-2 as an oncogene and an attractive therapeutic target in colon cancers. Mol. Cancer Ther. 7, 280–285 10.1158/1535-7163.MCT-07-2003 [DOI] [PubMed] [Google Scholar]
- 13. Gastl G., Spizzo G., Obrist P., Dünser M., and Mikuz G. (2000) Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet 356, 1981–1982 10.1016/S0140-6736(00)03312-2 [DOI] [PubMed] [Google Scholar]
- 14. Went P. T., Lugli A., Meier S., Bundi M., Mirlacher M., Sauter G., and Dirnhofer S. (2004) Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 35, 122–128 10.1016/j.humpath.2003.08.026 [DOI] [PubMed] [Google Scholar]
- 15. Seligson D. B., Pantuck A. J., Liu X., Huang Y., Horvath S., Bui M. H., Han K. R., Correa A. J., Eeva M., Tze S., Belldegrun A. S., and Figlin R. A. (2004) Epithelial cell adhesion molecule (KSA) expression: pathobiology and its role as an independent predictor of survival in renal cell carcinoma. Clin. Cancer Res. 10, 2659–2669 10.1158/1078-0432.CCR-1132-03 [DOI] [PubMed] [Google Scholar]
- 16. Wang J., Zhang K., Grabowska D., Li A., Dong Y., Day R., Humphrey P., Lewis J., Kladney R. D., Arbeit J. M., Weber J. D., Chung C. H., and Michel L. S. (2011) Loss of Trop2 promotes carcinogenesis and features of epithelial to mesenchymal transition in squamous cell carcinoma. Mol. Cancer Res. 9, 1686–1695 10.1158/1541-7786.MCR-11-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsujikawa M., Kurahashi H., Tanaka T., Nishida K., Shimomura Y., Tano Y., and Nakamura Y. (1999) Identification of the gene responsible for gelatinous drop-like corneal dystrophy. Nat. Genet. 21, 420–423 10.1038/7759 [DOI] [PubMed] [Google Scholar]
- 18. Takaoka M., Nakamura T., Ban Y., and Kinoshita S. (2007) Phenotypic investigation of cell junction-related proteins in gelatinous drop-like corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 48, 1095–1101 10.1167/iovs.06-0740 [DOI] [PubMed] [Google Scholar]
- 19. Günzel D., and Yu A. S. (2013) Claudins and the modulation of tight junction permeability. Physiol. Rev. 93, 525–569 10.1152/physrev.00019.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michl P., Barth C., Buchholz M., Lerch M. M., Rolke M., Holzmann K. H., Menke A., Fensterer H., Giehl K., Löhr M., Leder G., Iwamura T., Adler G., and Gress T. M. (2003) Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 63, 6265–6271 [PubMed] [Google Scholar]
- 21. Rangel L. B., Agarwal R., D'Souza T., Pizer E. S., Alò P. L., Lancaster W. D., Gregoire L., Schwartz D. R., Cho K. R., and Morin P. J. (2003) Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res. 9, 2567–2575 [PubMed] [Google Scholar]
- 22. Usami Y., Chiba H., Nakayama F., Ueda J., Matsuda Y., Sawada N., Komori T., Ito A., and Yokozaki H. (2006) Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum. Pathol. 37, 569–577 10.1016/j.humpath.2005.12.018 [DOI] [PubMed] [Google Scholar]
- 23. Lu Z., Ding L., Hong H., Hoggard J., Lu Q., and Chen Y. H. (2011) Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp. Cell Res. 317, 1935–1946 10.1016/j.yexcr.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kominsky S. L., Argani P., Korz D., Evron E., Raman V., Garrett E., Rein A., Sauter G., Kallioniemi O. P., and Sukumar S. (2003) Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 22, 2021–2033 10.1038/sj.onc.1206199 [DOI] [PubMed] [Google Scholar]
- 25. Al Moustafa A. E., Alaoui-Jamali M. A., Batist G., Hernandez-Perez M., Serruya C., Alpert L., Black M. J., Sladek R., and Foulkes W. D. (2002) Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene 21, 2634–2640 10.1038/sj.onc.1205351 [DOI] [PubMed] [Google Scholar]
- 26. Tinsley J. H., Teasdale N. R., and Yuan S. Y. (2004) Involvement of PKCδ and PKD in pulmonary microvascular endothelial cell hyperpermeability. Am. J. Physiol. Cell Physiol. 286, C105–C111 10.1152/ajpcell.00340.2003 [DOI] [PubMed] [Google Scholar]
- 27. Kyuno D., Kojima T., Yamaguchi H., Ito T., Kimura Y., Imamura M., Takasawa A., Murata M., Tanaka S., Hirata K., and Sawada N. (2013) Protein kinase Cα inhibitor protects against downregulation of claudin-1 during epithelial-mesenchymal transition of pancreatic cancer. Carcinogenesis 34, 1232–1243 10.1093/carcin/bgt057 [DOI] [PubMed] [Google Scholar]
- 28. Gan H., Wang G., Hao Q., Wang Q. J., and Tang H. (2013) Protein kinase D promotes airway epithelial barrier dysfunction and permeability through down-regulation of claudin-1. J. Biol. Chem. 288, 37343–37354 10.1074/jbc.M113.511527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steinberg S. F. (2012) Cardiac actions of protein kinase C isoforms. Physiology (Bethesda) 27, 130–139 10.1152/physiol.00009.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu Y., and Rubin C. S. (2011) Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 12, 785–796 10.1038/embor.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bornholdt J., Friis S., Godiksen S., Poulsen S. S., Santoni-Rugiu E., Bisgaard H. C., Lothe I. M., Ikdahl T., Tveit K. M., Johnson E., Kure E. H., and Vogel L. K. (2011) The level of claudin-7 is reduced as an early event in colorectal carcinogenesis. BMC Cancer 11, 65 10.1186/1471-2407-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lipinski M., Parks D. R., Rouse R. V., and Herzenberg L. A. (1981) Human trophoblast cell-surface antigens defined by monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 78, 5147–5150 10.1073/pnas.78.8.5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Basu A., Goldenberg D. M., and Stein R. (1995) The epithelial/carcinoma antigen EGP-1, recognized by monoclonal antibody RS7–3G11, is phosphorylated on serine 303. Int. J. Cancer 62, 472–479 10.1002/ijc.2910620419 [DOI] [PubMed] [Google Scholar]
- 34. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- 35. Sharma K., D'Souza R. C., Tyanova S., Schaab C., Winiewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 10.1016/j.celrep.2014.07.036 [DOI] [PubMed] [Google Scholar]
- 36. Ding L., Lu Z., Foreman O., Tatum R., Lu Q., Renegar R., Cao J., and Chen Y. H. (2012) Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 142, 305–315 10.1053/j.gastro.2011.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleiger G., Grothe R., Mallick P., and Eisenberg D. (2002) GXXXG and AXXXA: common α-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41, 5990–5997 10.1021/bi0200763 [DOI] [PubMed] [Google Scholar]
- 38. Schneider D., and Engelman D. M. (2004) Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 343, 799–804 10.1016/j.jmb.2004.08.083 [DOI] [PubMed] [Google Scholar]
- 39. Stoyanova T., Goldstein A. S., Cai H., Drake J. M., Huang J., and Witte O. N. (2012) Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via β-catenin signaling. Genes Dev. 26, 2271–2285 10.1101/gad.196451.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koivunen J., Aaltonen V., Koskela S., Lehenkari P., Laato M., and Peltonen J. (2004) Protein kinase C α/β inhibitor Go6976 promotes formation of cell junctions and inhibits invasion of urinary bladder carcinoma cells. Cancer Res. 64, 5693–5701 10.1158/0008-5472.CAN-03-3511 [DOI] [PubMed] [Google Scholar]
- 41. Akita K., Tanaka M., Tanida S., Mori Y., Toda M., and Nakada H. (2013) CA125/MUC16 interacts with Src family kinases, and over-expression of its C-terminal fragment in human epithelial cancer cells reduces cell-cell adhesion. Eur. J. Cell Biol. 92, 257–263 10.1016/j.ejcb.2013.10.005 [DOI] [PubMed] [Google Scholar]
- 42. Mori Y., Akita K., Tanida S., Ishida A., Toda M., Inoue M., Yashiro M., Sawada T., Hirakawa K., and Nakada H. (2014) MUC1 protein induces urokinase-type plasminogen activator (uPA) by forming a complex with NF-κB p65 transcription factor and binding to the uPA promoter, leading to enhanced invasiveness of cancer cells. J. Biol. Chem. 289, 35193–35204 10.1074/jbc.M114.586461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.