Abstract
The cyclic dinucleotide (CDN)-stimulator of interferon genes (STING) pathway plays an important role in the detection of viral and bacterial pathogens in animals. Previous studies have shown that the metazoan second messenger cyclic [G(2′,5′)pA(3′,5′)p] (2′,3′-cGAMP) generated by cyclic GMP-AMP synthase cGAS binds STING with high affinity compared with bacterial CDNs such as c-di-GMP, c-di-AMP, and 3′,3′-cGAMP. Despite recent progress indicating that the CDN-binding domain (CBD) of dimeric STING binds asymmetric 2′,3′-cGAMP preferentially over symmetric 3′,3′-CDNs, it remains an open question whether STING molecules, such as human STING, adopt a symmetric dimeric conformation to efficiently engage its asymmetric ligand. Here, structural studies of the CBD from porcine STING (STINGCBD) in complex with CDNs at 1.76–2.6 Å resolution revealed that porcine STINGCBD, unlike its human and mouse counterparts, can adopt an asymmetric ligand-binding pocket to accommodate the CDNs. We observed that the extensive interactions and shape complementarity between asymmetric 2′,3′-cGAMP and the ligand-binding pocket make it the most preferred ligand for porcine STING and that geometry constraints limit the binding between symmetric 3′,3′-CDN and porcine STING. The ligand-discrimination mechanism of porcine STING observed here expands our understanding of how the CDN–STING pathway is activated and of its role in antiviral defense.
Keywords: signaling, pathogen-associated molecular pattern (PAMP), pattern recognition receptor (PRR), cyclic diadenosine monophosphate (c-di-AMP), interferon
Introduction
The innate immunity system forms the first line of defense against bacterial and viral infections through pattern recognition receptors (PRRs)5 that detect pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) (1–4). DNAs in the cytosol originating from a bacterial or viral invasion are PAMPs that are recognized by the cytosolic double-strand DNA (dsDNA)-sensing pathways (5). A major cytosolic DNA sensor is cyclic GMP-AMP (cGAS), which synthesizes cyclic [G(2′,5′)pA(3′,5′)p] (2′,3′-cGAMP) upon activation by binding of dsDNA (6). Numerous studies have established that cGAMP functions as the second messenger to bind and activate the endoplasmic reticulum (ER)-located stimulator of interferon genes (STING) (also known as MITA (7, 8), MPYS (9), ERIS (10), and TMEM173 (11)). Ligand-activated STING is quickly translocated from ER to the ER–Golgi intermediate compartment to form punctate structures (12–14). STING uses its C-terminal tail to recruit and activate the TBK1 kinase, which phosphorylates the transcription factor IRF3 (15, 16). Phosphorylated IRF3 becomes an active dimer and then enters the nucleus to induce the expression of type I IFNs, which trigger the host antiviral action.
STING also functions as the direct PRR for other CDNs (17), such as c-di-GMP, c-di-AMP, and 3′,3′-cGAMP, which were first found in bacteria, having important roles in biofilm formation, motility, and virulence (18, 19). The cGAS product 2′,3′-cGAMP is unique in that it not only has two different purine moieties (guanine and adenine) but also contains mixed phosphodiester linkages in which the hydroxyl group of ribose in guanosine was connected at 2′- and 5′-position. In contrast, ribose moieties in all the bacterial CDNs are connected at 3′- and 5′-positions. 2′,3′-cGAMP as the endogenous ligand has been shown previously to have higher binding affinity for STING than the bacterial second messengers (20, 21).
The STING protein consists of four transmembrane helices in the N terminus that is followed by the cytosolic CDN-binding domain (CBD). A flexible tail at the C terminus of STING is important for recruiting and activating TBK1 and IRF3. In the past few years, a myriad of apo-STING and STING–CDNs complex structures from different species has been determined, revealing a dimeric architecture that forms the CDN-binding pocket between the two protomers (22). In the apo-form, the angle between two protomers is relatively large, and the ligand-binding pocket is open. Ligand binding induces conformational changes characterized by a reduced angle between the two protomers and the closure of the “lid” β-strand that encloses the ligand in the deep pocket formed between the two protomers. Recently, the full-length STING was solved by the cryo-EM method (23). It reveals that ligand binding not only induces these conformational changes mentioned above but results in the cytosolic domain half-turn rotation relative to the transmembrane region as well, which adds another layer of regulation of STING signaling.
Until now, all the structures of STING CBD adopt a symmetric dimeric conformation, which is intriguing considering that the asymmetric ligand 2′,3′-cGAMP shows the highest binding affinity. The mechanistic basis for ligand discrimination therefore remains elusive. Here, we determined the CBD of porcine STING complex with four CDNs: 2′,3′-cGAMP; 3′,3′-cGAMP; c-di-GMP; and c-di-AMP. Surprisingly, we found that porcine STING adopts an asymmetric conformation to bind the CDNs. Our analyses of this asymmetric conformation led to a mechanism for ligand recognition and discrimination of STING.
Results
High-affinity binding of porcine STING for 2′,3′-cGAMP
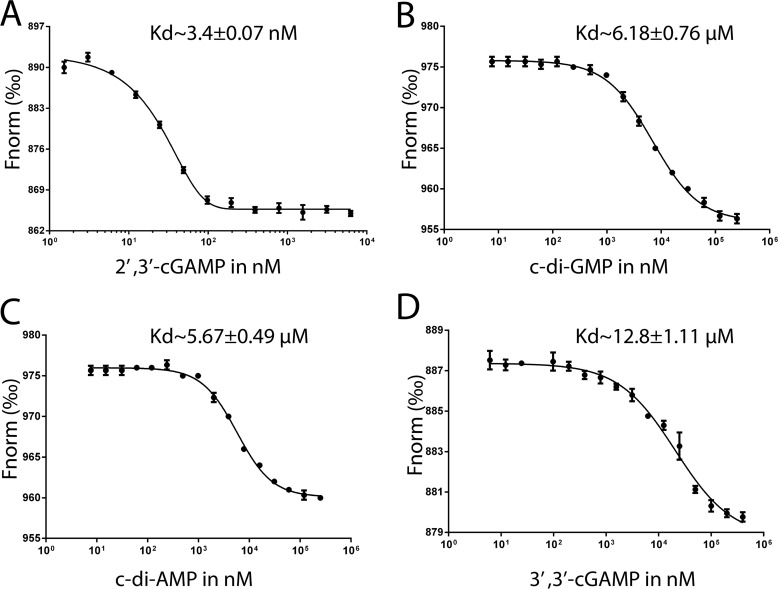
The binding affinities of natural CDNs and various isomers for STING from other species have been extensively studied in the past few years (17, 20, 21, 24–32). These studies show that, in general, the order of binding affinity for natural CDNs is 2′,3′-cGAMP > 3′,3′-cGAMP ≈ c-di-GMP > c-di-AMP. Here, we used the microscale thermophoresis (MST) assay to measure the binding affinity between porcine STING and CDNs. The results show that the binding affinity for 2′,3′-cGAMP with the dissociation constant (Kd) of 3.4 nm (Fig. 1A) is comparable with those reported previously for STING from human (4.59 nm) and sea anemone (<1 nm) (21, 31) but is substantially higher than those in mouse and rat STING (submicromolar level, 0.18 and 0.12 μm) (20, 30). The binding affinities of porcine STING for c-di-GMP and c-di-AMP are similar (∼6 μm) (Fig. 1, B and C), which are lower than their human counterpart (21). Furthermore, the binding affinity of porcine STING for 3′,3′-cGAMP ligand is the lowest with a Kd of 12.8 μm in our MST assay (Fig. 1D). These results show that, like STING from other species, porcine STING binds the endogenous ligand 2′,3′-cGAMP with the highest binding affinity.
Figure 1.
MST assays for quantification of binding between porcine STINGCBD protein and four CDNs. The normalized change in the fluorescence of the labeled porcine STINGCBD protein was plotted against the CDN concentration. The experiments were performed in triplicate. Mean values ± S.D. (n = 3) are shown. A, 2′,3′-cGAMP. B, c-di-GMP. C, c-di-AMP. D, 3′,3′-cGAMP.
Strong activation of porcine STING by 2′,3′-cGAMP
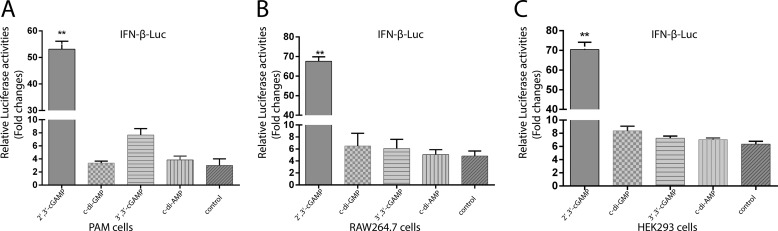
We then examined whether 2′,3′-cGAMP functions as a potent inducer for IFN-β in porcine immune cells. The IFN-β promoter along with the luciferase reporter gene were transfected into porcine alveolar macrophage (PAM) cells, which have endogenous porcine STING expression. Cells were treated with equal concentrations of CDNs. Fig. 2A shows that the 2′,3′-cGAMP stimulated the highest level of IFN-β promoter activity, which is consistent with the results obtained by using mouse and human STING (Fig. 2, B and C). These results correlate well with the in vitro ligand-binding assays, thereby suggesting that 2′,3′-cGAMP is the preferred ligand for porcine STING, as well as human and mouse STING.
Figure 2.
Cell-based luciferase assay of STING in response to CDNs. Cells were transfected with 0.5 μg of IFN-β promoter along with 0.25 μg of pRL-TK. After 18 h of transfection, PAM cells and RAW264.7 cells were incubated with 60 μm CDNs, whereas HEK293 cells were transfected with 60 μm CDNs by using lipo3000. Luciferase reporter assay was performed after an additional 24-h incubation. Values stand for the mean average of triplicate experiments. Error bars indicate S.D. A, luciferase reporter assay for PAM cells expressing endogenous porcine STING stimulated by CDNs. B, luciferase reporter assay for RAW264.7 cells expressing endogenous mouse STING stimulated by CDNs. C, luciferase reporter assay for HEK293 cells expressing endogenous human STING stimulated by CDNs.
Overall structure of porcine STINGCBD–2′,3′-cGAMP complex
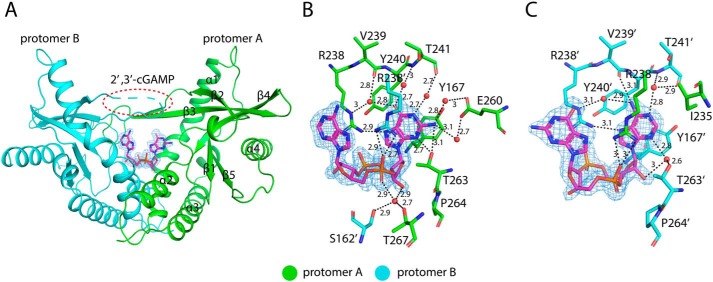
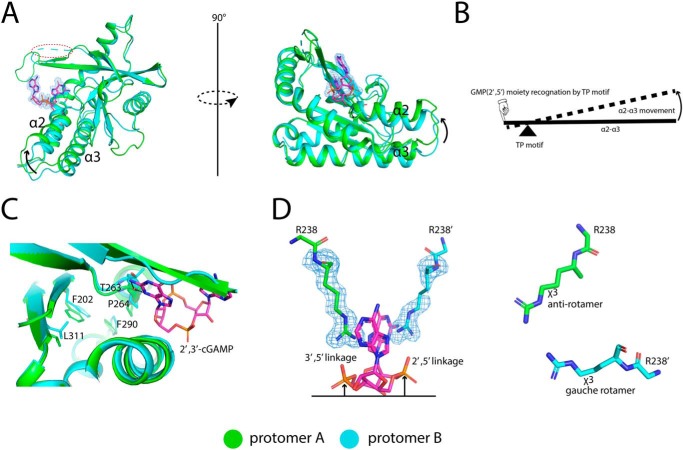
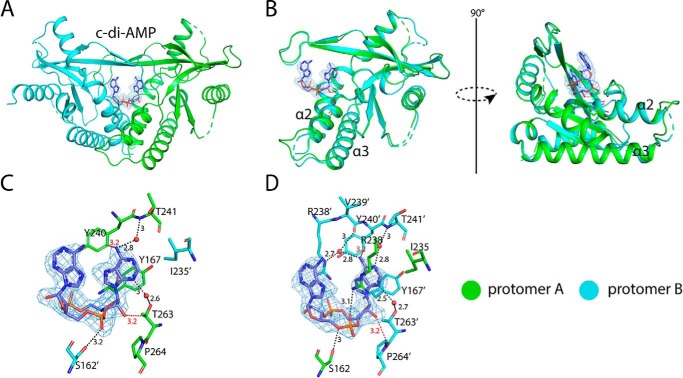
The structure of porcine STINGCBD–2′,3′-cGAMP complex was determined at 1.8 Å resolution. The asymmetric unit contains two STINGCBD molecules (protomer A and B) that form the dimer as seen in previous structures of STINGCBD from other species (Fig. 3A). All residues except residues 228–236 located at the lid region of protomer B are modeled. Porcine STINGCBD adopts the same α + β-fold as STING from other species (22). The individual protomers make extensive interactions with each other through the α1, α2, α3, and central β-sheet region, which forms a deep U-shaped cleft. The 2′,3′-cGAMP ligand is located at the cleft using its purines pointing upward and the ribose ring sticking in the downward position (relevant to the membrane). The base, phosphate, and ribose groups all contribute to binding to STING (Fig. 3, B and C). The purine rings of 2′,3′-cGAMP stack against Tyr-167 and Arg-238 from two promoters. The phosphate moieties of 2′,3′-cGAMP are recognized by Arg-238 through charge–charge interactions and hydrogen bonds. Arg-238 also forms hydrogen bonds at the Hoogsteen edge of the nucleoside moieties. Beside the stacking interactions, the adenosine and guanosine moieties have different interaction modes with STING. For both, the interactions involve their Watson-Crick, Hoogsteen, and sugar edges (Fig. S1). All the N atoms (except the position-9 amino) and O atom in the purines interact directly or indirectly (water-mediated) with the STING protein (also see details in Table S1). These extensive interactions are consistent with the high binding affinity between ligand and porcine STINGCBD as demonstrated in our in vitro binding experiment.
Figure 3.
Structure of porcine STINGCBD in complex with 2′,3′-cGAMP. Protomers A and B are colored green and cyan, respectively. The black dashed line represents the hydrogen bond formed between ligand and protein. The values indicate the length of hydrogen bond with the unit of Å. The waters are shown as red sphere. The ligand is shown as a stick model colored in magenta. The simulated annealing omit Fo − Fc electron-density map for 2′,3′-cGAMP (blue mesh) is contoured at 3 σ. A, overall structure of porcine STINGCBD–2′,3′-cGAMP complex (side view). The red dashed circle shows the disordered lid region in protomer B. B, detailed interactions between GMP(2′,5′) moiety of 2′,3′-cGAMP and G site of STING protein. C, detailed interactions between AMP(3′,5′) moiety of 2′,3′-cGAMP and A site of STING protein.
Asymmetric ligand-binding pocket in porcine STING
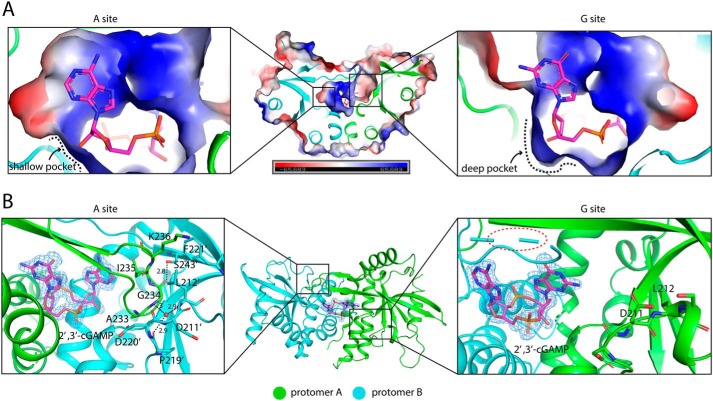
A strikingly distinctive feature of the porcine STINGCBD–2′,3′-cGAMP complex is that the two protomers adopt different conformations, resulting in an asymmetric ligand-binding pocket. Consequently, 2′,3′-cGAMP, which is also asymmetric, is held in one well-defined conformation in the ligand-binding pocket (Fig. 4). This unique binding mode is in sharp contrast with that seen in previous structures involving STING proteins from other species (22), where 2′,3′-cGAMP binds the symmetric STING dimer in two alternative modes with the adenosine and guanosine moieties switching positions.
Figure 4.
Asymmetric ligand-binding pocket in porcine STINGCBD. Protomer A and B are colored green and cyan, respectively. 2′,3′-cGAMP is shown as a magenta stick. A, cross-section of 2′,3′-cGAMP–binding pocket in porcine STINGCBD. Left panel shows the complete A site, and right panel shows the incomplete G site. Electrostatic potential is shown. B, left panel shows the interactions between hairpin tip of protomer A and protomer B. The water molecule is shown as a red sphere. The black dashed lines represent the hydrogen bond, and the values show the length of hydrogen bond with the unit of Å. The simulated annealing omit Fo − Fc electron-density map for 2′,3′-cGAMP (blue mesh) is contoured at 3 σ. Middle panel shows the top view of porcine STING-2′,3′–cGAMP complex. Right panel shows the interaction between disordered hairpin of protomer B and protomer A.
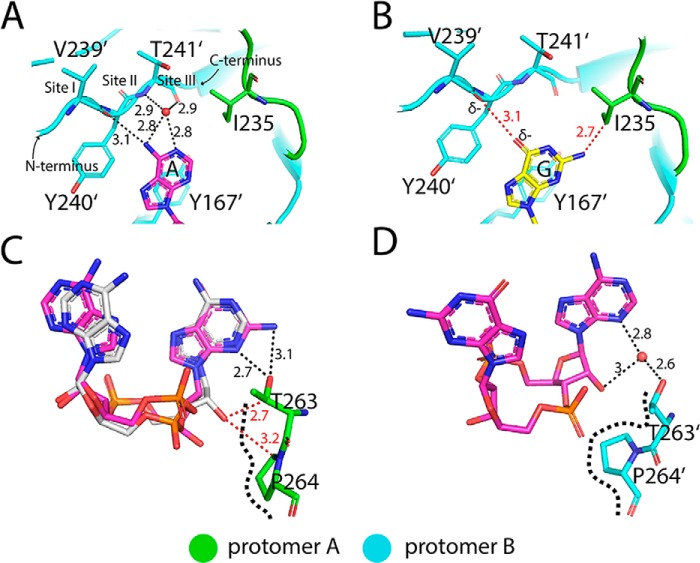
The recognition sites in porcine STINGCBD for the adenosine with 3′,5′-phosphodiester bond-linked phosphate (or AMP(3′,5′)) and guanosine with 2′,5′-phosphodiester bond-linked phosphate (or GMP(2′,5′)) moieties show very different conformations. The AMP(3′,5′) moiety recognition pocket (designated as A site) is complete with the ordered lid region and a shallow 3′,5′-phosphodiester linkage sugar edge recognition pocket (Fig. 4A, left panel). In contrast, the GMP(2′,5′) moiety recognition pocket (designated as G site) is partially exposed with the β-hairpin tip totally invisible in the density and a deep 2′,5′-phosphodiester linkage sugar edge recognition pocket (Fig. 4A, right panel).
In the A site, Leu-212 from protomer B flips out and interacts with Phe-221 (protomer B) and Lys-236 (protomer A) through hydrophobic interactions. Leu-212 also buttresses the hairpin tip (Gly-234, Ile-235, and Lys-236) from protomer A, further stabilizing and rigidifying the β-hairpin structure, resulting in the closure of the A site (Fig. 4B, left panel). Several distinctive features in this pocket of porcine STING appear to underlie its preference for adenosine. The loop region (Val-239, Tyr-240, and Thr-241) and Tyr-167 form an adenine-binding motif, which is found in many adenylate-containing proteins to recognize the adenine ring (Fig. 5A, and Fig. S2) (33). Meanwhile, hydrophobic interactions between the aromatic C2 atom of adenine and the aliphatic side chain of Ile-235 further enhance the specific binding of adenine to the pocket. This type of specific adenine recognition mode can also be seen in the double-strand RNA–binding motif such as ADAR2, which uses the side chain of Met to specifically recognize the C2 atom of adenine from RNA (34, 35). However, guanine has an extra exocyclic–amino group at position-2 and carbonyl oxygen at position-6, which are expected to generate van der Waals repulsion (bad contact or bump) with the side chain of Ile-235 and the carbonyl group of Val-239, respectively, disfavoring its binding in this pocket (Fig. 5B).
Figure 5.
Recognition modes of AMP(3′,5′) and GMP(2′,5′) moieties by porcine STINGCBD. The black dashed lines represent the hydrogen bond, and the values show the length of hydrogen bond with the unit of Å. The red dashed line represents the repulsive van der Waals contact (bad contact or bump), and the values show the distance between nonhydrogen-bonding atoms. The black dashed curve represents the boundary of TP motif. A, adenine-binding pocket in A site. B, guanine was modeled into adenine-binding pocket, and the steric clashes will limit its binding. C, sugar edge of guanosine with 2′,5′-phosphodiester bond linkage is recognized by TP motif in G site, whereas the sugar edge of adenosine with 3′,5′ phosphodiester bond linkage will generate steric clashes. c-di-AMP and 2′,3′-cGAMP are shown as sticks colored in silver and magenta, respectively. D, recognition of sugar edge of adenosine with 3′,5′-phosphodiester bond linkage by TP motif in A site. The water is shown as a red sphere.
The G site is partially open, as the β-hairpin tip from protomer B is disordered. The loop accommodating the β-hairpin tip adopts a different conformation with the Leu-212 buried in the hydrophobic core of protomer A (Fig. 4B, right panel). As a result, the guanine moiety of the ligand is partially exposed to solvent. As discussed above, the closed lid as seen for the adenine recognition pocket is not favorable for the binding of the guanine duo to the selectivity of Ile-235. The configuration of sugar edges of adenosine and guanosine in 2′,3′-cGAMP are quite different especially in their connection by phosphate moieties. Adenosine moiety is linked by the usual 3′,5′-phosphodiester bond with the convex-free 2′-hydroxyl group, whereas the guanosine moiety is connected by the unusual 2′,5′-phosphodiester bond with the concave-free 3′-hydroxyl group (Fig. S1B). To accommodate these two different sugar edges of the nucleoside, helix 2 from each STING protomer adopts different conformations. The hydroxyl group of Thr-263 in helix 2 from protomer A in the G site specifically forms two hydrogen bonds with the guanine moiety, whereas only one hydrogen bond can be formed if the guanine moiety was replaced by the adenine moiety because of the absence of an extra exocyclic-amino group at the C6-position. The recognition of guanine moiety by Thr-263 will generate a steric clash if the usual 3′,5′-phosphodiester linkage with convex-free 2′-hydroxyl group presents. However, the unusual 2′,5′-phosphodiester linkage with the concave- free 3′-hydroxyl group could solve this problem properly by shifting away from the side chains of Thr-263 and Pro-264 (Fig. 5C). Therefore, Thr-263 and Pro-264 from protomer A in the G site form a specific recognition motif (designated as TP motif) to engage the unique sugar edge of guanosine with 2′,5′-phosphodiester linkage. However, Thr-263 in helix 2 from protomer B in the A site could not specifically recognize the sugar edge of adenosine, which lacks the exocyclic-amino group at position-C2 of the purine ring. The indirect interaction (water-mediated hydrogen bond) between Thr-263 and the N3 atom of adenine will generate enough space resolving the possible steric clashes between the convex-free 2′-hydroxyl group and Thr-263 and Pro-264 (Fig. 5D).
Superimposing protomer B to protomer A shows helix 2 and helix 3 in protomer A tilts upward, and the hydrophobic core undergoes rearrangement in protomer A (Fig. 6, A–C). The conformational changes of helix 2 and helix 3 in protomer A during ligand recognition can be considered as the movement of a lever. Helix 2 and helix 3 function as the beam, whereas Pro-264 is the turning point. The recognition of GMP(2′,5′) moiety by the TP motif will input the force at one end of the lever and result in the movement of the beam. In addition, the phosphate moiety in the 2′,5′-linkage part sticks higher than its counterpart in the 3′,5′-linkage, and therefore, the side chain of Arg-238 is bumped up by the phosphate moiety, and its χ3 dihedral angle is forced to adopt an unfavored gauche rotamer other than the anti-rotamer in protomer A, further contributing to the asymmetric conformation of the lid region (Fig. 6D). As described above, porcine STING using complete A site specifically recognizes AMP(3′,5′) moiety, while using incomplete G site specifically recognizes GMP(2′,5′) moiety.
Figure 6.
Structural comparison between two protomers in porcine STINGCBD–2′,3′-cGAMP complex. 2′,3′-cGAMP is shown with stick colored in magenta. The simulated annealing omit Fo − Fc electron-density map for 2′,3′-cGAMP and R238 (blue mesh) is contoured at 3 σ. A. superimposition of protomer A and protomer B. The arrow indicates the tilt of α2–α3 in protomer A relative to it in protomer B. The red dashed oval shows the disordered lid region in protomer B. B, working model of the conformational changes of α2–α3 induced by GMP(2′,5′) moiety recognition. C, hydrophobic core difference between protomer A and protomer B. D, Arg-238 adopts different rotamer in protomer A and protomer B.
Structures of porcine STINGCBD complex with other CDNs reveal the binding preference
As shown above, porcine STINGCBD adopts asymmetric conformation to bind 2′,3′-cGAMP. This prompted us to explore how porcine STING binds other more symmetric 3′,3′-CDNs. We crystallized porcine STINGCBD complex with bacterial second messengers, including c-di-GMP, c-di-AMP, and 3′,3′-cGAMP. All the crystals belong to the P212121 space group with two molecules of STING protein per asymmetric unit. To our surprise, all the three 3′,3′-CDNs can induce porcine STINGCBD to adopt the similar asymmetric conformation as the 2′,3′-cGAMP does (Figs. 7–9). Besides, the c-di-AMP can induce porcine STINGCBD to form a much more symmetric conformation with all the lid region becoming totally visible (Fig. 10). However, the tilt of helix 2 and helix 3 in protomer A (comparing with that in protomer B) of the 3′,3′-CDN–porcine STINGCBD complex is not as dramatic as that seen in 2′3′-cGAMP–porcine STINGCBD complex (Figs. 7–10B).
Figure 7.
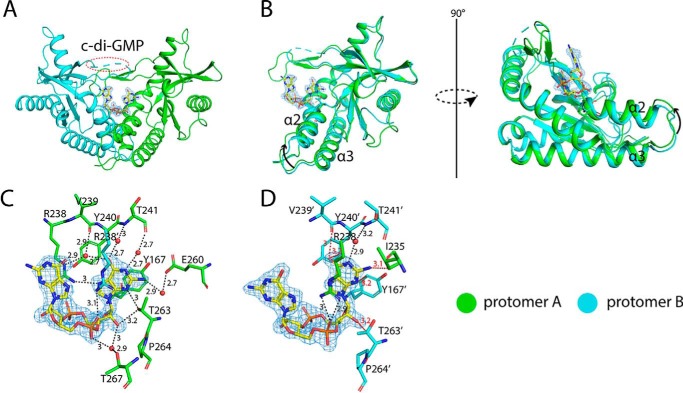
Structure of porcine STINGCBD in complex with c-di-GMP. The black dashed line represents the hydrogen bond formed between ligand and protein. The values indicate the length of hydrogen bond with the unit of Å. The red dashed line represents the repulsive van der Waals contact (bad contact or bump), and the values show the distance between nonhydrogen-bonding atoms. The waters are shown as red sphere. The ligand is shown in stick model colored in yellow. The simulated annealing omit Fo − Fc electron-density map for c-di-GMP (blue mesh) is contoured at 3 σ. A, overall structure of porcine STINGCBD–c-di-GMP complex (side view). The red dashed circle shows the disordered lid region in protomer B. B, structural comparison between two protomers in porcine STINGCBD–c-di-GMP complex. The arrow indicates the tilt of α2–α3 in protomer A relative to it in protomer B. C, detailed interactions between c-di-GMP and G site of STING protein. D, detailed interactions between c-di-GMP and A site of STING protein.
Figure 8.
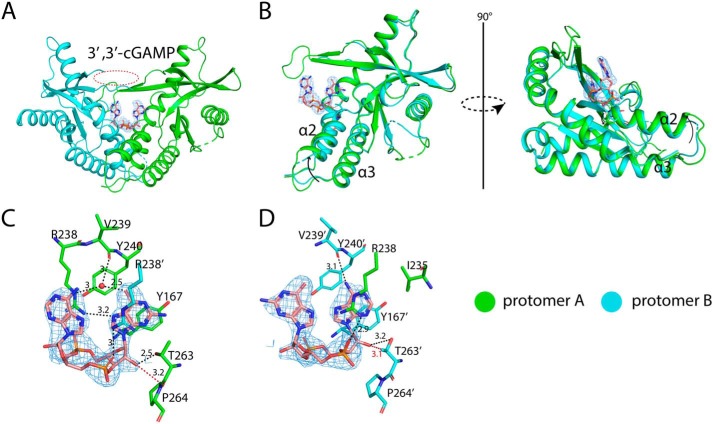
The structure of porcine STINGCBD in complex with 3′,3′-cGAMP. The black dashed line represents the hydrogen bond formed between ligand and protein. The values indicate the length of hydrogen bond with the unit of Å. The red dashed line represents the repulsive Van der Waals contact (bad contact or bump) and the values show the distance between nonhydrogen-bonding atoms. The waters are shown as red sphere. The ligand is shown as a stick model colored in salmon. The simulated annealing omit Fo − Fc electron-density map for 3′,3′-cGAMP (blue mesh) is contoured at 3 σ. A, overall structure of porcine STINGCBD-3′,3′-cGAMP complex (side view). The red dashed circle shows the disordered lid region in protomer B. B, structural comparison between two protomers in porcine STINGCBD-3′,3′-cGAMP complex. The arrow indicates the tilt of α2-α3 in protomer A relative to it in protomer B. C, detailed interactions between Guanosine moiety of 3′,3′-cGAMP and G site of STING protein. D, detailed interactions between Adenosine moiety of 3′,3′-cGAMP and A site of STING protein.
Figure 9.
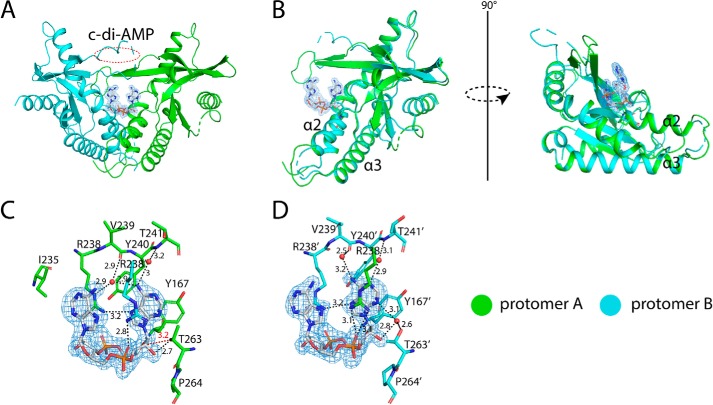
Structure of porcine STINGCBD in complex with c-di-AMP with incomplete lid region. The black dashed line represents the hydrogen bond formed between ligand and protein. The values indicate the length of hydrogen bond with the unit of Å. The red dashed line represents the repulsive van der Waals contact (bad contact or bump), and the values show the distance between nonhydrogen-bonding atoms. The waters are shown as a red sphere. The ligand is shown as a stick model colored in silver. The simulated annealing omit Fo − Fc electron-density map for c-di-GMP (blue mesh) is contoured at 3 σ. A, overall structure of porcine STINGCBD–c-di-AMP complex (side view). The red dashed circle shows the disordered lid region in protomer B. B, structural comparison between two protomers in porcine STINGCBD–c-di-AMP complex. C, detailed interactions between c-di-AMP and G site of STING protein. D, detailed interactions between c-di-AMP and A site of STING protein.
Figure 10.
Structure of porcine STINGCBD in complex with c-di-AMP with complete lid region. The black dashed line represents the hydrogen bond formed between ligand and protein. The values indicate the length of hydrogen bond with the unit of Å. The red dashed line represents the repulsive van der Waals contact (bad contact or bump), and the values show the distance between nonhydrogen-bonding atoms. The waters are shown as a red sphere. The ligand is shown as a stick model colored in slate blue. The simulated annealing omit Fo − Fc electron-density map for c-di-GMP (blue mesh) is contoured at 3 σ. A, overall structure of porcine STINGCBD–c-di-AMP complex (side view). B, structural comparison between two protomers in porcine STINGCBD–c-di-AMP complex. C, detailed interactions between c-di-AMP and G site of STING protein. D, detailed interactions between c-di-AMP and A site of STING protein.
In c-di-GMP–bound structure, the guanosine moiety in the A site shows unfavorable interactions with STING (Fig. 7D). For example, the extra-exocyclic-amino group at C2-position has bad contact with the aliphatic side chain of Ile-235; carbonyl oxygen in position-6 has bad contact with the carbonyl oxygen of Val-239; N3 nitrogen has bad contact with side chain of Tyr-167; N7 nitrogen and carbonyl oxygen in the C6-position have bad contacts with the side chain of Tyr-240; the free 2′-hydroxyl group in A site has the bad contact with the side chain of Thr-263 in protomer B. There is no hydrogen bond between Arg-238 and the N7 nitrogen in the guanine moiety within this binding pocket (supporting Table 1). These bad contacts can also be seen in the other CDN-porcine STING complexes (Figs. 8–10, C and D).
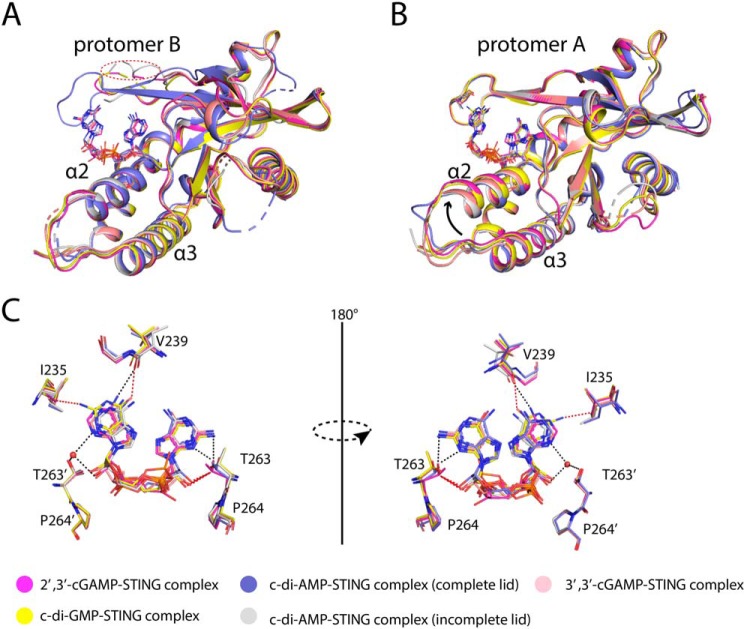
Superimposition of these five complex structures shows that overall these complexes look the same, especially for protomer B (Fig. 11A). The only difference in protomer A exists in α2 and α3 where it tilts more dramatically in the 2′,3′-cGAMP–STING complex than the other four CDN–STING complexes (Fig. 11B). As for the superimposed ligands, it clearly shows the preference for ligand binding. The unique sugar edge of GMP(2′,5′) moiety in 2′,3′-cGAMP is snugly engaged by the TP motif from protomer A in the G site, whereas the 3′,3′-CDNs are disfavored in this site because of steric clashes or the absence of hydrogen bonds (Fig. 11C). Similarly, the adenine-binding motif and Ile-235 selection lead to the preference of adenine in the A site. In other words, all the 3′,3′-CDNs adopting a symmetric conformation cannot fit them well in the asymmetric binding pocket resulting in a lower binding affinity complex.
Figure 11.
Structural comparison of porcine STINGCBD–CDNs complex. A, structural comparison of protomer B of porcine STINGCBD–CDNs complex. The lid regions of protomer B are incomplete except one of porcine STINGCBD–c-di-AMP complex. The red dashed circle shows the disordered lid region in protomer B. B, structural comparison of protomer A of porcine STINGCBD–CDN complex. The tilt of α2–α3 in 2′,3′-cGAMP is the most dramatic one. The black arrow indicates the upward movement of α2–α3. C, superimposition of CDNs shows the 2′,3′-cGAMP is the favorite ligand for STING. The black dashed line represents the hydrogen bond. The red dashed line represents the repulsive van der Waals contact (bad contact or bump).
Discussion
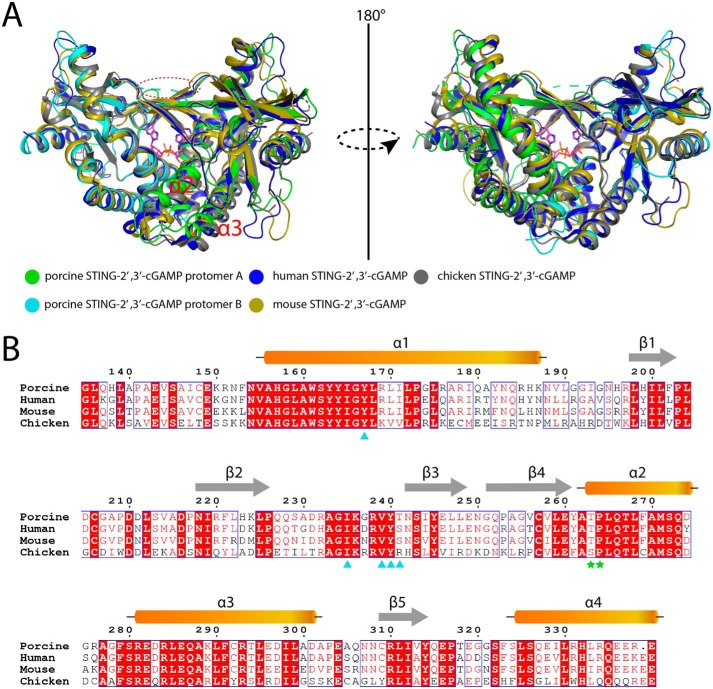
Since the identification of the CDN–STING pathway, many structural studies have revealed the binding and activation mechanism of STING by CDNs (22, 23). By superimposing porcine STING–ligand complex structure with other complex structures from human, mouse, and chicken, structural variations were observed among these STING proteins (Fig. 12A), although they share about 80% sequence identities (Fig. 12B). Except for its disordered lid region, protomer B from porcine STING is almost identical to the protomers from other species. However, protomer A from porcine STING cannot be superimposed well with its counterparts from other species. The striking difference is that the two helices (α2 and α3) in protomer A from porcine STING tilt upward, and in other species they adopt the same conformation as in protomer B. These differences make porcine STING exhibit an asymmetric pocket to engage asymmetric 2′,3′-cGAMP. There are several possible reasons why the STING-CDN complex structures from other species do not capture asymmetric conformation. As is known to all, the protein crystal structure is only a snapshot of various conformers in thermodynamic equilibrium. The crystals represent the state of the global minimum free energy taking into account both natural and unspecific interactions (36). Considering that the extensive interactions occurred between the N-terminal region and the CDN-binding domain of STING (23) and only the C terminus of STING itself is included in crystallization, the unspecific interactions observed in the crystalline state of STINGCBD may sacrifice the natural conformation and choose the lowest-energy one. Another point is that (e.g. PDB code 4KSY) the crystallographic 2-fold axis passes through the dimer interface, which can average the electron density resulting in two identical protomers. As for the chicken STING–2′,3′-cGAMP complex solved by the cryo-EM method, C2 symmetry was applied during 3D focus refinement, which will make the two protomers totally the same (23).
Figure 12.
Structural comparison of STINGCBD–2′,3′-cGAMP complex from different species. A, protomer B except for the lid region in porcine STING is the same as protomers from other species. The red dashed circle shows the disordered lid region in protomer B. Protomer A in porcine STING cannot be superimposed well with protomers from other species. PDB code for human STINGCBD–2′,3′-cGAMP complex, 4LOH; PDB code for mouse STINGCBD–2′,3′-cGAMP complex, 4LOJ; PDB code for chicken STINGCBD–2′,3′-cGAMP complex, 6NT7. B, sequence alignment of human, mouse, chicken, and porcine STINGCBD domain. The residues in the adenine-binding motif are marked by cyan triangles. The residues in the TP motif are marked by green stars.
It is difficult to explain why STING prefers the asymmetric ligand by using the complex structures solved before. Shi et al. (32) reported that the 2′,3′-cGAMP does not undergo large conformational change when it binds to STING and has less entropy costs leading to the highest binding affinity for 2′,3′-cGAMP. However, they ignored the contribution of protein–ligand interaction, which plays a critical role in determining the binding affinity. Although all the CDNs can induce porcine STING to form an asymmetric binding pocket, only the asymmetric 2′,3′-cGAMP can fit into this pocket well, whereas other 3′,3′-CDNs can generate constraint and suboptimal contacts, thereby limiting the interaction.
All in all, our structural investigation of porcine STINGCBD–CDN complexes has provided novel insight into the structural basis for ligand binding and discrimination that has implications in activation and regulation of the STING pathway.
Experimental procedures
Protein expression and purification
The cDNA encoding the porcine STING CBD domain (from 152 to 379 amino acids) was amplified and inserted into the pET-22b(+) vector (Novagen) generating the pET-22b–STINGCBD expression vector. The BL21(DE3) cells carrying pET-22b–STINGCBD vector were grown in Luria broth (LB) medium. When the A600 reached 0.8, the protein expression was induced by using 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. After 14 h induction, the cells were collected by centrifugation (15 min, 12,000 × g). Then, the cell pellet was resuspended in ice-cold lysis buffer (25 mm Tris-HCl, pH 8.0, 500 mm NaCl) and lysed by sonication. The lysate was centrifuged at 30,000 × g for 45 min, and the supernatant was loaded onto a nickel-chelating Sepharose column (GE Healthcare) equilibrated with lysis buffer. After washing with wash buffer (25 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 mm imidazole), the bound protein was eluted by using elution buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 250 mm imidazole). Then, the protein was treated with the protease trypsin in a 1:10,000 mass ratio overnight at 4 °C to remove the flexible tail region. The protease inhibitor phenylmethylsulfonyl fluoride was added to the protein solution to stop the enzymatic reaction. The protein was then applied to the Q column and eluted using a linear 160-ml gradient of 0–0.5 m NaCl. Porcine STINGCBD was further purified by gel-filtration chromatography by using a Superdex-200 (GE Healthcare) column equilibrated with gel-filtration buffer (10 mm Tris-HCl, pH 8.0, 100 mm NaCl). The peak was pooled and concentrated to 20 mg ml−1 for crystallization.
Crystallization, data collection, structure determination, and refinement
Before crystallization screening, porcine STING was mixed with 2′,3′-cGAMP and three other 3′,3′-CDNs at a 1:1.2 and 1:5 molar ratio, respectively, and incubated for 2 h at 293 K. Sitting-drop vapor-diffusion method was used for the initial screening, and the hits were optimized by using hanging-drop vapor-diffusion method at 293 K. Porcine STINGCBD–c-di-AMP complex can be crystallized with 3.5 m sodium formate or 1.5 m Li2SO4 as precipitant at 0.1 m NaOAC, pH 4.6. Porcine STINGCBD–3′,3′-cGAMP, c-di-GMP, and 2′,3′-cGAMP complexes were crystallized in the same condition with 1.5 m Li2SO4 as precipitant at 0.1 m NaOAc, pH 4.6.
The crystals were cryoprotected by 20% (v/v) glycerol and flash-frozen in liquid nitrogen before data collection. The diffraction data were collected on beamline BL17U1 at Shanghai Synchrotron Radiation Facility (SSRF). The data collected were indexed, integrated, and scaled with HKL-2000 (16).
All five complex structures were solved by the molecular replacement method with the human STINGCBD structure (PDB code 4F5W) as the search model. Coot (17) and PHENIX (18) were used for manual model building and refinement, respectively. The final models were deposited in the PDB with entries 6A03, 6A04, 6A05, 6A06, and 6IYF. The statistics of data collection and structure refinement are listed in Table 1. All the structure figures were rendered with PyMOL.
Table 1.
Data collection and refinement statistics
Each dataset was collected from a single crystal.
| STINGCBD–c-di-GMP complex | STINGCBD–c-di-AMP complex (complete lid) | STINGCBD–c-di-AMP complex (incomplete lid) | STINGCBD–3′,3′-cGAMP complex | STINGCBD–2′,3′-cGAMP complex | |
|---|---|---|---|---|---|
| Protein Data Bank codes | 6A04 | 6A03 | 6IYF | 6A05 | 6A06 |
| Data collection | |||||
| Wavelength (Å) | 0.9798 | 0.9798 | 0.9798 | 0.9798 | 0.9798 |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | |||||
| a, b, c (Å) | 49.691, 63.949, 101.591 | 50.517, 80.459, 101.564 | 49.785, 65.896, 100.571 | 49.405, 64.741, 100.953 | 49.628, 63.311, 101.036 |
| α, β, γ | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00–1.90 (1.97–1.90)a | 50.00–2.60 (2.69–2.60) | 50.00–1.76 (1.79–1.76) | 50.00–2.20 (2.28–2.20) | 50.00–1.80 (1.86–1.80) |
| Rmerge(%) | 7.8 (41.3) | 13.7 (54.3) | 8.0 (71.1) | 8.7 (50.7) | 5.5 (44.9) |
| 〈I/σ(I)〉 | 42.20 (6.28) | 19.85 (3.87) | 21.37 (1.28) | 30.52 (4.59) | 47.45 (5.42) |
| Completeness (%) | 97.9 (100.0) | 100.0 (100.0) | 99.1 (84.1) | 100.0 (100.0) | 99.2 (99.0) |
| Redundancy | 6.6 (6.9) | 7.1 (7.3) | 7.1 (6.1) | 6.9 (6.9) | 7.0 (7.0) |
| Refinement | |||||
| No. of reflections | 25,597 | 13,319 | 33,277 | 16,950 | 30,287 |
| Rwork/Rfree (%) | 20.21/24.96 | 20.00/25.72 | 19.36/24.21 | 22.15/27.35 | 17.53/21.99 |
| No. of atoms | |||||
| Protein | 2910 | 2841 | 2795 | 2921 | 2929 |
| Ligand | 46 | 44 | 44 | 45 | 45 |
| Water | 151 | 76 | 136 | 37 | 214 |
| B-factors (Å2) | |||||
| Protein | 39.24 | 34.74 | 31.60 | 47.39 | 31.20 |
| Ligand | 33.53 | 28.79 | 30.05 | 41.84 | 22.92 |
| Water | 38.47 | 32.04 | 34.75 | 35.63 | 34.59 |
| Root mean square deviation | |||||
| Bond lengths (Å) | 0.007 | 0.003 | 0.011 | 0.008 | 0.006 |
| Bond angles (°) | 0.888 | 0.700 | 1.255 | 1.252 | 0.857 |
| Ramachandran plot | |||||
| Favored (%) | 96.97 | 94.99 | 97.46 | 94.6 | 96.69 |
| Allowed (%) | 3.03 | 4.72 | 2.54 | 5.4 | 3.31 |
| Disallowed (%) | 0 | 0.29 | 0 | 0 | 0 |
a Values in parentheses are for highest-resolution shell.
MST assay
MST assay was performed to measure the affinity of the purified porcine STING for CDNs with Monolith NT.115 from Nanotemper Technologies. Proteins were fluorescently labeled according to the manufacturer's protocol, and the concentration of labeled protein used for each assay was about 200 nm. A solution of unlabeled CDNs was diluted for appropriate serial concentration gradients. The samples were loaded into silica capillaries (Polymicro Technologies) after incubation at room temperature for 30 min. Measurements were performed at 293 K in buffer containing 50 mm BisTris, pH 7.6, 150 mm NaCl, and 0.05% Tween 20, by using 12% LED power and 40% MST power. The assays were repeated three times for each affinity measurement. Data analyses were performed with Nanotemper Analysis software and OriginPro 8.0 software provided by the manufacturer.
Cell-based interferon-β luciferase reporter assay
PAM, RAW264.7 or HEK293 cells were seeded in 12-well plates and grown to 80% confluence, respectively. Cells were transfected with 0.5 μg of IFN-β promoter along with 0.25 μg of pRL-TK using Lipofectamine 3000 reagent (Invitrogen). At 18 h post-transfection, PAM and RAW264.7 cells were incubated with 60 μm c-di-AMP, c-di-GMP, 3′,3′-cGAMP, or 2′,3′-cGAMP for 19 h. For HEK293 cells, 60 μm c-di-AMP, c-di-GMP, 3′,3′-cGAMP, or 2′,3′-cGAMP were transfected into cells using Lipofectamine 3000 reagent. Luciferase activities were measured using a Dual-Luciferase reporter assay kit (Promega, Madison, WI) according to the manufacturer's protocol. The values were normalized with respect to Renilla luciferase activities. Then, the results were expressed as relative luciferase activities, which were shown as relative fold changes compared with the mock-treated control (untransfected cells). All assays were repeated at least three times, with each experiment performed in triplicate.
Author contributions
X. C., F. L., and G. S. formal analysis; X. C., Z. Y., Y. D., B. Wu, D. L., X. W., Y. Z., B. Wei, J. L., J. Wu, J. Wang, and J. Q. investigation; X. C., Z. Y., Y. D., B. Wu, D. L., X. W., Y. Z., J. Q., and G. S. methodology; X. C., Y. D., F. L., J. Q., and L. G. writing-review and editing; Z. Y. and G. S. writing-original draft; Y. D. and L. G. funding acquisition; S. X. visualization; J. Wang, J. Q., and L. G. supervision; J. Q. and G. S. conceptualization; G. S. data curation; G. S. validation.
Supplementary Material
Acknowledgments
We thank the staff of beamlines BL17U1 and BL19U at the Shanghai Synchrotron Radiation Facility (SSRF) for their support in the data collection. We also thank Dr. Xuewu Zhang for critical reading and editing of this manuscript.
This work was supported by National Natural Science Foundation of China Grants 31470732 (to L. G.), 31502051 (to X. C.), 31672609 (to J. Q.) and 21673244 (to Bo. W.) and the National Key Research and Development Program of China Grant 2016YFD0501505 (to Y. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
- PRR
- pattern recognition receptor
- PAMP
- pathogen-associated molecular pattern
- cGAS
- cyclic GMP-AMP synthase
- PDB
- Protein Data Bank
- MST
- microscale thermophoresis
- CDN
- cyclic dinucleotide
- CBD
- CDN-binding domain
- STING
- stimulator of interferon genes
- ER
- endoplasmic reticulum
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PAM
- Porcine alveolar macrophage
- IFN
- interferon.
References
- 1. Pandey S., Kawai T., and Akira S. (2014) Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 7, a016246 10.1101/cshperspect.a016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar H., Kawai T., and Akira S. (2011) Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- 3. Walsh D., McCarthy J., O'Driscoll C., and Melgar S. (2013) Pattern recognition receptors–molecular orchestrators of inflammation in inflammatory bowel disease. Cytokine Growth Factor Rev. 24, 91–104 10.1016/j.cytogfr.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 4. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 5. Barbalat R., Ewald S. E., Mouchess M. L., and Barton G. M. (2011) Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 10.1146/annurev-immunol-031210-101340 [DOI] [PubMed] [Google Scholar]
- 6. Wu J., and Chen Z. J. (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- 7. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 8. Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., Wang Y. Y., Zhang X. L., and Shu H. B. (2009) The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30, 397–407 10.1016/j.immuni.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 9. Jin L., Waterman P. M., Jonscher K. R., Short C. M., Reisdorph N. A., and Cambier J. C. (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 10.1128/MCB.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T., Yamamoto N., Kawai T., Ishii K., Takeuchi O., Yoshimori T., and Akira S. (2009) Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 20842–20846 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishikawa H., Ma Z., and Barber G. N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobbs N., Burnaevskiy N., Chen D., Gonugunta V. K., Alto N. M., and Yan N. (2015) STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 10.1016/j.chom.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanaka Y., and Chen Z. J. (2012) STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y. T., Grishin N. V., and Chen Z. J. (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 17. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenal U., Reinders A., and Lori C. (2017) Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- 19. Davies B. W., Bogard R. W., Young T. S., and Mekalanos J. J. (2012) Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370 10.1016/j.cell.2012.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A. A., Gaffney B. L., Shuman S., Jones R. A., Deng L., Hartmann G., Barchet W., Tuschl T., and Patel D. J. (2013) Structure–function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 10.1016/j.cell.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., and Chen Z. J. (2013) Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 10.1016/j.molcel.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato K., Omura H., Ishitani R., and Nureki O. (2017) Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 86, 541–566 10.1146/annurev-biochem-061516-044813 [DOI] [PubMed] [Google Scholar]
- 23. Shang G., Zhang C., Chen Z. J., Bai X. C., and Zhang X. (2019) Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393 10.1038/s41586-019-0998-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ouyang S., Song X., Wang Y., Ru H., Shaw N., Jiang Y., Niu F., Zhu Y., Qiu W., Parvatiyar K., Li Y., Zhang R., Cheng G., and Liu Z. J. (2012) Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 10.1016/j.immuni.2012.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shu C., Yi G., Watts T., Kao C. C., and Li P. (2012) Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19, 722–724 10.1038/nsmb.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y. H., Liu X. Y., Du X. X., Jiang Z. F., and Su X. D. (2012) The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 19, 728–730 10.1038/nsmb.2333 [DOI] [PubMed] [Google Scholar]
- 27. Shang G., Zhu D., Li N., Zhang J., Zhu C., Lu D., Liu C., Yu Q., Zhao Y., Xu S., and Gu L. (2012) Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19, 725–727 10.1038/nsmb.2332 [DOI] [PubMed] [Google Scholar]
- 28. Yin Q., Tian Y., Kabaleeswaran V., Jiang X., Tu D., Eck M. J., Chen Z. J., and Wu H. (2012) Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 10.1016/j.molcel.2012.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chin K. H., Tu Z. L., Su Y. C., Yu Y. J., Chen H. C., Lo Y. C., Chen C. P., Barber G. N., Chuah M. L., Liang Z. X., and Chou S. H. (2013) Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta Crystallogr. D Biol. Crystallogr. 69, 352–366 10.1107/S0907444912047269 [DOI] [PubMed] [Google Scholar]
- 30. Zhang H., Han M. J., Tao J., Ye Z. Y., Du X. X., Deng M. J., Zhang X. Y., Li L. F., Jiang Z. F., and Su X. D. (2015) Rat and human STINGs profile similarly towards anticancer/antiviral compounds. Sci. Rep. 5, 18035 10.1038/srep18035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kranzusch P. J., Wilson S. C., Lee A. S., Berger J. M., Doudna J. A., and Vance R. E. (2015) Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′-cGAMP signaling. Mol. Cell 59, 891–903 10.1016/j.molcel.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi H., Wu J., Chen Z. J., and Chen C. (2015) Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc. Natl. Acad. Sci. U.S.A. 112, 8947–8952 10.1073/pnas.1507317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denessiouk K. A., Rantanen V. V., and Johnson M. S. (2001) Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins 44, 282–291 10.1002/prot.1093 [DOI] [PubMed] [Google Scholar]
- 34. Stefl R., Oberstrass F. C., Hood J. L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R. B., and Allain F. H. (2010) The solution structure of the ADAR2 dsRBM–RNA complex reveals a sequence-specific readout of the minor groove. Cell 143, 225–237 10.1016/j.cell.2010.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masliah G., Barraud P., and Allain F. H. (2013) RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell. Mol. Life Sci. 70, 1875–1895 10.1007/s00018-012-1119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krissinel E. (2010) Crystal contacts as nature's docking solutions. J. Comput. Chem. 31, 133–143 10.1002/jcc.21303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.