Abstract
Oncogenic microRNAs are essential components in regulating the gene expression of cancer cells. Especially miR21, which is a major player involved of tumor initiation, progression, invasion and metastasis in several cancers. The delivery of anti-miR21 sequences has significant potential for cancer treatment. Nevertheless, since anti-miR21 sequences are extremely unstable and they need to obtain certain concentration to function, it is intensely difficult to build an effective delivery system for them. The purpose of this work is to construct a self-assembled glutathione (GSH)-responsive system with tumor accumulation capacity for effective anti-miR21 delivery and cancer therapy. A novel drug delivery nanosphere carrying millions of anti-miR21 sequences was developed through the rolling circle transcription (RCT) method. GSH-responsive cationic polymer polyethyleneimine (pOEI) was synthesized to protect the nanosphere from degradation by Dicer or other RNase in normal cells and optimize the pompon-like nanoparticle to suitable size. Dehydroascorbic acid (DHA), a targeting molecule, which is a substrate of glucose transporter 1 (GLUT 1) and highly expressed on malignant tumor cells, was connected to pOEI through PEG, and then the polymer was used for contracting a RNA nanospheres into nanopompons. The anti-miR21 nanopompons showed its potential for effective cancer therapy.
KEY WORDS: MiRNA21, Dehydroascorbic acid, Rolling circle transcription, Self-assembly, RNAi nanoparticle, Tumor-targeting, Nanomedicine, Cancer treatment
Graphical abstract
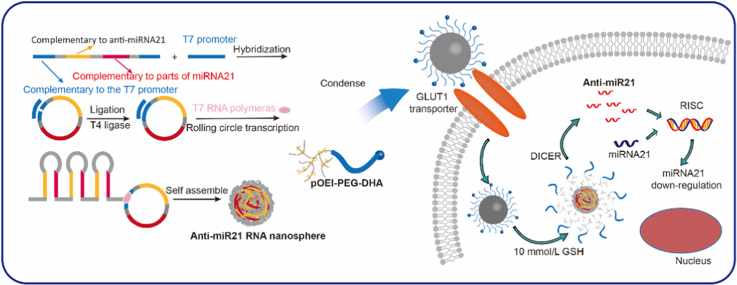
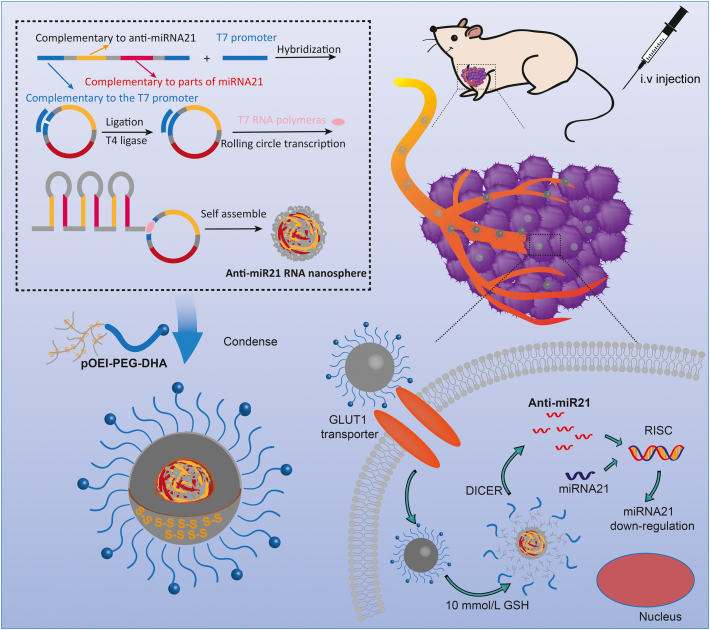
Preparation process, GLUT1-mediated transport and GSH-responsive drug release of DHA-targeting anti-miRNA21 nanopompons were reported. Nanopomponswere first obtained by rolling transformation transcription (RCT) and condensed by tumor-targteing pOEI-PEG-DHA.
1. Introduction
MicroRNAs (miRNAs) are regarded as post-transcriptional regulators of gene expression because they can degrade their target mRNAs or inhibit their translation1. Accumulating evidences have suggested that miRNAs have significant potentials for cancer treatment especially because one miRNA can regulate a multitude of target genes efficiently and synchronously, thus avoiding drug resistance and addressing the heterogeneous nature of cancer2., 3., 4.. miRNA21 is part and parcel of tumor initiation, progression, invasion and metastasis of various types of cancers and considered as a representative oncogenic miRNA. When the functions of endogenous miRNA21 were efficiently blocked, the tumor suppressor gene phosphatase and tensin homolog deleted on chromosome ten (PTEN) and programmed cell death protein 4 (PDCD4) could be up-regulated at the same time to suppress tumor development from different pathways5., 6., 7.. On the other hand, silencing miR21 can not only depress the invasion and proliferation but also promote apoptosis of the cancer cells8., 9.. With the rapid innovation of gene therapy, two strategies were developed to achieve tumor regression, including anti-miRNAs to knockdown tumorigenic miRNAs and imitator of miRNAs to upregulate antitumors miRNAs10., 11., 12., 13.. The delivery of miRNAs or anti-miRNAs with tumor suppressing capability holds great promise for cancer treatment, however, has been limited by the extreme instability and ineffective delivery14., 15., 16.. In spite of stability improvement of (anti)sense-miRNAs by modifying the nucleic acid structure, therapeutic miRNAs require effective delivery systems to become useful preparations. Scientists have designed and developed several delivery systems to protect nucleic acids from degradation of serum nucleases. Some well-designed nanoparticles could effectively inhibit endogenous miRNA function and significantly reduce the growth of tumor in vivo3., 4.. However, these nanoparticles showed slashed antitumor efficacy when the dose was reduced to around one half17. Consequently, it is essential to construct an efficient anti-miRNA delivery system for cancer treatment.
Rolling circle transcription (RCT) is a novel technology which could construct a nanoball (about 500 nm in diameter) containing plenty of RNAi sequences, and self-assembled with the carrier and cargo as a whole to delivery therapeutic RNA. Each nanosphere contains about a half million tandem copies of therapeutic RNA strands which are cleavable with dicer. Moreover, these RNA strands were layers of parcels with each other, so they have significantly better stability than single-stranded RNA18., 19.. RCT offered an option to delivery RNA drugs efficiently and therefore has been adopted to construct valid siRNA delivery system in general. But it has not been applied in (anti)miRNA delivery system yet.
In this paper, we adopted RCT to delivery antisense of miRNA21 to prepare anti-miR21 nanospheres. Pompon-like nanoparticles were optimized to a diameter of 100 nm with the use of disulfide bonded polyethyleneimine (pOEI), which could be degraded by high concentration of glutathione in tumor cells. The pOEI molecule is much smaller than the nanosphere, therefore the tightly packed pOEI molecules could protect the nanosphere from degradation by dicer or other RNase in normal cells.
In order to accumulate more nanopompons in the tumor tissue, the nanopompons were modified with dehydroascorbic acid (DHA). As is well-known, cancer cells rewire their metabolism in distinct ways, such as aerobic glycolysis, also called as the “Warburg effect”20, which means that in order to satisfy the need of rapid proliferation, it is necessary to maintain high levels of glucose uptake and metabolism. Glucose transporter1 (GLUT1) is an important pathway for tumor glucose transport and therefore is highly expressed on the surface of most tumor cells20., 21., 22.. However, the transmembrane glucose transport of GLUT1 is bidirectional; when the intracellular glucose concentration reaches a certain level, the excess glucose will be excreted from the cell, so the concentration of intracellular glucose could maintain a certain level. DHA closely resembles the structure of glucose and also has been transported into the cell through GLUT123, but the transport mode is different from that of glucose. DHA is reduced to Vitamin C after being transported into the cell, so that it can be continuously transported into the cell. Due to changes in the structure of DHA after the transportation into cells, it cannot be recognized and effluxed by GLUT124. Therefore, the uptake of DHA by GLUT1 is a continuously enriched one-way transport process, and can be used to achieve the accumulation of nanopompons the tumor tissue. The targeting and GSH/dicer-responsive anti-miRNA21 delivery system was named as DHA-targeting anti-miRNA21 nanopompons (Fig. 1). The targeting delivery and GSH/dicer-responsive release are expected to guarantee sufficient accumulation of anti-miRNA21 sequences within tumor cells and can therefore ensure a safe and efficient cancer therapy.
Figure 1.
Preparation, GLUT1-mediated transport, and GSH-responsive drug release of DHA-targeting anti-miRNA21 nanopompons.
2. Materials and methods
OEI (ethylenediamine branched polyethylenimine, MW 800) was purchased from Sigma—Aldrich (St. Louis, MO, USA). Azide PEG succinimidyl carboxymethyl ester (AZIDE-PEG3500-SCM, MW 3500) was synthesized by Jenkem Technology (Beijing, China). 3,3′-Dithiobis(sulfosuccinimidylpropionate) (DTSSP) was obtained from ThermoFisher Scientific (Waltham, MA, USA). All others chemical were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). The T7promoter (TAATACGACTCACTATAGGGAT) and the linear DNA with anti-miR21 sequence (ATAGTGAGTCGTATTAACGTACCAACTAGCTTATCAGACTGATGTTGACTTGCAACATCAGTCCTCCCAGCTATAGAGGCATATCCCT) were obtained from Genscript Biotechnology Company (Nanjing, China). AmpliScribeTM T7 High Yield Transcription Kit was purchased from Epicenter Technologies (Madison, WI, USA). T4 DNA Ligase were obtained from Promega Corporation (Madison, WI, USA). BODIPY® FL NHS Ester (succinimidyl ester) was obtained from Life Technologies Corporation (Carlsbad, CA, USA). ECL Plus was pursed from Beyotime Biotechnology (Shanghai, China). Recombinant Human Dicer Enzyme Kit was acquired from the Genlantis (San Diego, USA). All antibodies were purchased from Abcam (Cambridge, England). OCT embedding medium was obtained from Sakura Tissue-Tek® (Torrance, CA, USA). All primers were synthesized by GenePharma Co., Ltd. (Shanghai, China).
2.1. Cell lines
MDA-MB-231-luci cells and 293 cells were kindly provided by Stem Cell Bank Chinese Academy of Sciences (China). All the cells were cultured in Dulbecco׳s modified Eagle׳s medium (DMEM) containing 10% fetal bovine serum, 1000 units/mL aqueous penicillin G, and 100 μg/mL streptomycin at 37 °C under 5% CO2 humidified atmosphere.
2.2. Animals
Balb/c nude mice (female, 20–25 g) were kindly provided by the Experimental Animals Department of Fudan University (China). All animal experiments were conducted in accordance with guidelines evaluated and approved by Fudan University Institutional Animal Care and Use Committee (IACUC, Shanghai, China). The xenograft tumor model was established by orthotopic injection of 1 × 106 MDA-MB-231-luci cells in 100 μL serum-free DMEM medium containing 5 mg/mL Matrigel into the mammary fat pad of mice.
2.3. Synthesis of DHA-PEG-pOEI
DHA (18 mg, 2 eq.), AZIDE-PEG3500-SCM (150 mg, 1 eq.), ascorbic acid (21 mg, 2.5 eq.) and PMDETA (44 μL, 5 eq.) were dissolved in 5 mL ultrapure water, and then the reaction system was protected by nitrogen. Finally add CuSO4 (5.4 mg, 0.5 eq.) into the mixture. The mixture was stirred at 40 °C for 12 h, after which the reaction was transferred to a dialysis membrane with a 1000 MWCO membrane and dialyzed in 10 mmol EDTA-2Na water solution (2 L) for 12 h. After which the dialysate was replaced with water, and then continued dialysis for 12 h. the crude product was lyophilized to obtain pure DHA-PEG-SCM. GSH-responsive pOEI can be easily synthesized by adding equimolar OEI and DTSSP into purified water, stirred at 35 °C for 48 h and purified by dialysis, and the specification of the dialysis-membrane was 7000. Then an equivalent amount of pOEI and DHA-PEG-SCM was added into PBS 8.0 and stirred at room temperature for 2 h. The reaction system was then purified by dialysis and lyophilization. Pure DHA-PEG-pOEI was dissolved in D2O and analyzed by 1H NMR spectroscopy. The molecular weight of pOEI and (DHA-)PEG-pOEI were measured by gel permeation chromatography (GPC). Molecular weight of pOEI, PEG-pOEI and DHA-PEG-pOEI were 9.8, 14.6 and 14.7 kDa, respectively. BODIPY-labeled OEI was synthesized by click reaction between the amino on OEI and BODIPY® FL NHS Ester (succinimidyl ester). All the BODIPY-labeled (DHA-)PEG-pOEI were prepared with BODIPY-labeled OEI.
2.4. Preparation of naked anti-miR21 nanosphere
In order to establish circular DNA template, 1 mmol/L linear DNA and 1 mmol/L T7 were hybridized together by insulating at 95 °C for 2 min and gradually cooling down to 25 °C during 1 h. Then the loop was closed by T4 DNA ligase by adding 3 μL 10 × ligase buffer, 0.9 μL T4 DNA ligase solution and 5.1 μL RNase-free water into the circular DNA template solution and insulating the mixture at 25 °C for 3 h. Four kinds of ribonucleotide triphosphate (ATP, UTP, CTP and GTP) were then added into circular DNA template, each with a concentration of 7.5 mmol/L. The final spherical sponge-like structures were usually observed at about 16 to 20 h17. Meanwhile, excessive copy number may cause the self-assembly nanosphere to be too large, which is not conducive to subsequent compression into the appropriate particle size. Therefore, in order to generate a uniform formulation, the new mixture was incubated with 2 μL T7 RNA polymerase at 37 °C for 18 h in reaction buffer (total volume 20 μL). Afterwards, 1 MBU of RNase-Free DNase I was added into the reaction buffer and incubated for another 15 min at 37 °C to stop the transcription reaction. The anti-miR21 nanospheres were extracted with TE-saturated chloroform and pure chloroform, and then were precipitated by 5 mol/L ammonium acetate and washed with 70% ethanol and RNase-Free water for several times. They were finally redissolved in RNase-Free water with addition of RNase Inhibitor to maintain stability. We obtained the molecular weight of RNA nanospheres (2.01 × 1010 g/mol) from Zeta PALS software (Malvern Instruments Ltd., Malvern, Worcs, UK). The number of periodically repeated 88 base RNA strands in one RNA nanosphere was calculated as follows: molecular weight of 88 base RNA strand = 27,003 g/mol; number of cleavable RNA strands in one RNA nanosphere = 2.01 × 1010/27,002 = 7.44 × 105. So, 7,440,000 of siRNA can be maximally generated from one RNA nanosphere. The total RNA concentration of nanosphere was determined by UV–Vis spectrophotometry and calculated as follows to determine the amount of anti-miR21 therapeutic agent for drug-loading and animal test dose of nanopompons: RNA concentration (μg/μL) = OD260 × dilution ratio × 0.04, which must meet 1.7 < OD260/OD280 < 2.0.
2.5. Preparation of anti-miR21 nanopompons
The anti-miR21 nanospheres were prepared by sonication for 10 min. Afterwards, DHA-PEG-pOEI of gradient concentrations (1, 5, and 10 μg/μL) were added to nanospheres solutions, vortexed for 30 s and let it sit for another 30 min, forming three kinds of different anti-miR21 nanopompons.
2.6. Characterizations of anti-miR21 nanopompons
Different kinds of anti-miR21 nanopompons and naked anti-miR21 nanosphere were analyzed by dynamic light scattering to determine zeta potential and particle size. The morphologies were visualized using TEM (Tecnai G2 spirit Biotwin, Hillsboro, USA).
2.7. Agarose gel electrophoresis
Firstly, anti-miR21 nanopompons were incubated with 10 mmol/L GSH for 2 h at 37 °C, and then digested with 2 unit of recombinant dicer at 37 °C for 24 h, and then the reaction was stopped by adding 2 mL Dicer Stop Solution (according to the commercial specifications). Meanwhile, naked anti-miR21 nanospheres and anti-miR21 nanopompons were digested with recombinant dicer using the same method as mentioned above. Digested product were separated by 3% agarose gel (prestained with 0.5 mg/mL of ethidium bromide) electrophoresis in Tris-acetate-EDTA (TAE) buffer (20 mmol/L acetic acid, 1 mmol/L EDTA and 40 mmol/L Tris, pH 8.0) at 100 V for 60 min and visualized by UV light.
2.8. Cellular uptake
MDA-MB-231-luci cells were planked in 24-well culture plates at a density of 3 × 104 cells/well. When achieving 70%–80% cell density, the cells were incubated with Red BODIPY-labelled nanopompons (λex = 630 nm, λem = 650 nm) with different DHA modification degree at 0%, 10%, 20% or 50% with a concentration of 5 μg RNA in 200 μL/well of the glucose-free PBS under pH 7.4. Afterincubation for 30 min, cells were washed with PBS three times and then photographed under the fluorescent microscope (Leica DMI4000D, Germany).
2.9. Cellular uptake mechanism exploration
MDA-MB-231-luci cells were planked in 24-well culture plates at a density of 3 × 104 cells/well. The cells were pre-incubated with glucose-free PBS, when achieving 70%–80% cell density. After 15 min pre-incubation, cells were further incubated with Red BODIPY-labelled nanopompons at a concentration of 5 μg RNA in 200 μL/well for another 30 min. Other groups were incubated with Red BODIPY-labelled nanopompons at a concentration of 5 μg RNA in 200 μL/well as well and different inhibitors including 10 mmol/L d-glucose, 0.5 μg/mL filipin, 0.4 μg/mL phenylarsine oxide and 1 μg/mL colchicine, respectively. After incubation for 45 min, cells were washed with PBS for several times and then photographed under the confocal fluorescence microscope (IX2-RFACA, Olympus, Osaka, Japan).
2.10. Flow cytometry
After photographed under the confocal fluorescence microscope, pancreatin was added to separate cells. Then cells were diluted in Hank׳s solution and centrifuged at 3000 rpm (ThermoFisher Scientific, Waltham, MA, USA) for 5 min. After the supernate was discarded, the precipitates were redissolved with Hank׳s solution and analyzed by flow cytometry (cytoFLEX, Beckman Coulter, Brea, CA, USA).
2.11. Quantitative polymerase chain reaction (QPCR)
Cell samples after incubated with nanopompons for 2 days or freshly excised tumor tissues were prepared. Total RNA was isolated by TRIzol reagent. Possible DNA contaminant was removed with DNase I. The RNA product was re-purified by TRIzol reagent and the first strand cDNA was synthesized using reverse transcription kit. GADPH was also amplified to be an internal control.
2.12. In vitro cell viability test
In vitro cell viability was evaluated by MTT assay (n = 4). 293 cells were planked in 96-well plates at a density of 5 × 103 cells/well. When reaching 60%–70% confluence, cells were incubated with DHA-modified nanopompons, non-modified nanopompons, DHA-PEG-pOEI and PEG-pOEI at various concentrations in DMEM for 48 h at 37 °C. After incubation, the medium was removed and cells were washed by PBS for three times. Then 100 μL per well MTT solution with a concentration of 5 mg/mL was added and incubated with cells at 37 °C for 4 h. After incubation, the solution was removed and DMSO was added 100 μL per well. Then 96-well plates were shaken by the oscillating table for 10 min. The absorbance of formazan crystals was read at 590 nm using Multiskan MK3 microplate reader (Thermo Scientific, Waltham, MA, USA). Cells without treatment were considered as control.
2.13. Western blot assay
Cell samples after incubated with nanopompons for 2 days or freshly excised tumor tissues were lysed with phenylmethanesulfonyl fluoride (1 mmol/L, RIPA lysis buffer). The protein concentration of cell sample was measured by BCA Protein Assay kit (Beyotime Biotechnology, Shanghai, China). Total proteins (50 μg/hole) were separated by 12% SDS—PAGE electrophoresis at 100 V for 1 h, and then transferred to PVDF membranes. After that, PVDF membranes were blocked with 5% fat-free milk for over 1 h, and incubated overnight with primary antibody (PTEN, Abcam, 1:1000; PDCD4, Abcam, 1:1000; actin, Beyotime, 1:100). After washed with TBST buffer 3 times for 10 min, the membranes were incubated with anti-rabbit or anti-mouse secondary antibodies (1:500) conjugated with horseradish peroxidase (HRP) for 1 h. Then second antibody solution was removed. The membranes were washed twice for 10 min with TBST buffer. The protein expression levels were detected by enhanced chemiluminescence autoradiography with the use of using ECL plus.
2.14. Real-time fluorescence imaging
Nude mice model of triple negative breast cancer (at the day 10 after implantation) were treated by tail vein injection with Red-BODIPY-labelled nanopompons (λex = 630 nm, λem = 650 nm) at the dose of 50 μg RNA per mouse. Normal saline was injected as control. At 4, 8 and 12 h after administration, the nude mice were visualized with the in vivo real-time fluorescence imaging system (IVIS Spectrum, Cailper PerkinElemer, Waltham, MA, USA). All operations were performed under brief anesthesia with inhalation of isoflurane. Then the excitation light was focused on the breast area to conduct 3D real-time image of DHA-targeting group 12 h after administration. Afterwards, mice were sacrificed, and tumors as well as other primary organs were excised carefully for comparing relative fluorescence accumulation.
2.15. Inspection of anti-tumor therapeutic effects on triple negative breast cancer (TNBC) model nude mice
At the day 7 after implantation, TNBC-bearing mice were randomly divided into three groups (n = 10 each group) according to the size of the tumor and body weight. One group was treated by tail vein injection with DHA-modified anti-miR21 nanopompons with a period of treatment of 5 injections every three days. The total RNA dose is 2.5 mg/kg. The other group was injected with non-modified anti-miR21 nanopompons through the same way. Normal saline-treated mice were served as control. Tumor volume (V) was calculated with the use of the Eq. (1)25:
| (1) |
where the shortest diameter (s) and the longest diameter (l) of tumors were measured by a digital caliper every other day. The body weight of mice was also measured every other day to evaluate the in vivo toxicity of nanopompons indirectly.
2.16. In vivo cell proliferation and apoptosis assay
Tumors excised from the TNBC model on day 18 were fixed with 4% paraformaldehyde for 24 h. Then tumors were dehydrated with sucrose solution, whose concentration was gradually increased from 15% to 30% for 24 h. The tumor tissues were then frozen in optimal cutting temperature compound (OCT) embedding medium at −80 °C and sliced with thickness of 10 µm. Tumor sections of control and (non-) targeting anti-miR21-nanopompons-treated group were de-paraffined by xylene and hydrated from 100% ethanol, 85% ethanol and 75% ethanol to pure water. Then antigens were retrieved by 10 mmol/L citric sodium buffer (pH 6.0) microwave antigen retrieval. Then sections were incubated with 3% H2O2 for 25 min to block endogenous peroxidase and washed by PBS. Afterwards, sections were blocked by 5% goat serum, and then were incubated with primary antibodies (cleaved caspase-3, Abcam, 1:1000; Ki67, Abcam, 1:1000) at 4 °C overnight, and then the sections were incubated with goat anti rabbit IgG conjugated with HRP at 25 °C for 60 min. The conjugated antibody was detected by diaminobenzidine. All sections were counterstained with hematoxylin, and then photographed under the fluorescent microscope (Leica, DMI4000D, Germany).
2.17. Statistical analysis
Analysis was performed using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA). Statistical comparisons were assessed by one-way ANOVA. Data are represented as mean±SD. Statistical significance was defined as P < 0.05.
3. Results
3.1. Physical characteristics of GSH/DICER-responsive anti-miR21 nanopompons
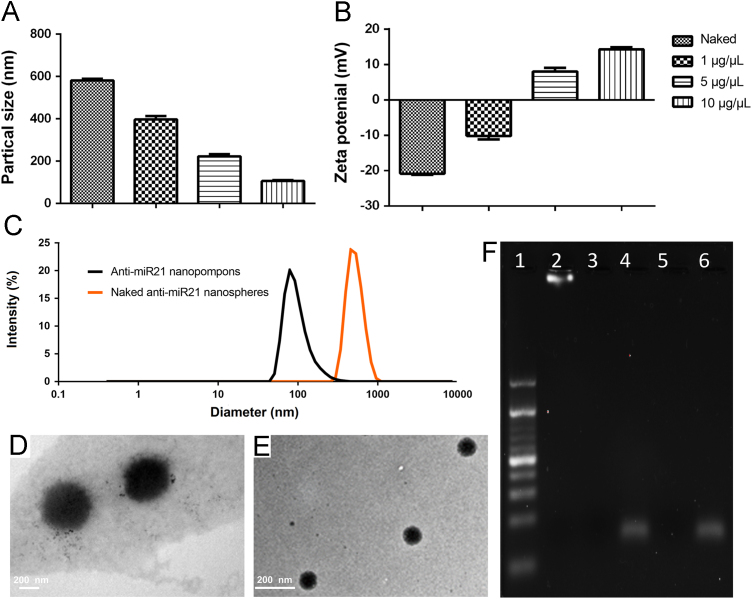
Naked anti-miR21 nanospheres were prepared by RCT and observed under transmission electron microscope (TEM) to confirm the successful preparation. Nanospheres shared a uniform particle size around 500 nm (Fig. 2C). Compared with the nanospheres carrying siRNA sequences18, stability of anti-miR21 nanospheres is so poor that degradation of edges can be observed under TEM (Fig. 2D). Stability experiment showed that naked nanospheres cannot be stable for more than 8 h (Supporting Information Fig. S1A). For greater stability and better particle size, after screening gradient pOEI-PEG (0, 1, 5 and 10 μg/μL), naked nanospheres were contracted by 10 μg/μL pOEI and PEG corona (Fig. 2A). The synthetic route of (DHA-)PEG-pOEI were shown in Supporting Information Scheme S1. 1H NMR spectrum of (DHA-)PEG-pOEI were shown in Supporting Information Fig. S2. Contracted nanopompons could be stable for more than 3 days (Fig. S1B). The mean diameter of the non-modified nanopompons are 107 nm (Fig. 2C), almost as same as DHA-modified nanopompons, because DHA molecule is too small to affect the particle size. Results of zeta potential showed that with the increase of pOEI concentration, zeta potential changed from negative to positive, and modification does not affect the potential. The final average zeta potential is +17 mV (Fig. 2B). Agarose gel electrophoresis showed that naked nanospheres had a sensitive dicer enzyme reactivity, and encapsulation can protect nanopompons from being degraded by the specific dicer enzyme in tumor cells, unless there is a high concentration of glutathione present, which has been proven to increase thousands of times in tumor cells26 (Fig. 2F).
Figure 2.
Characterization of anti-miR21 nanopompons. (A) Particle sizes and (B) zeta potentials after contraction by gradient pOEI (0, 1, 5 and 10 μg/μL). (C) Size distribution of naked anti-miR21 nanospheres and anti-miR21 nanopompons (pOEI 10 μg/μL) by DLS. (D)TEM images of naked anti-miR21 nanospheres. (E) TEM images of anti-miR21 nanopompons. (F) Enzyme electrophoresis of (contracted) anti-miR21 nanopompons (lane 1:10 bp RNA ladder; lane 2: naked anti-miR21 nanospheres lane 3: naked anti-miR21 nanospheres with incubation of 2.00 Unit dicer for 24 h; lane 4: condensed anti-miR21 nanoparticles without dicer; lane 5: condensed anti-miR21 nanoparticles with incubation of 2.00 Unit dicer for 24 h; lane 6: condensed anti-miR21 nanoparticles with incubation of 10 mmol/L GSH for 2 h and then 2.00 Unit dicer for 24 h).
3.2. Optimization of modification level and exploration of uptake mechanism
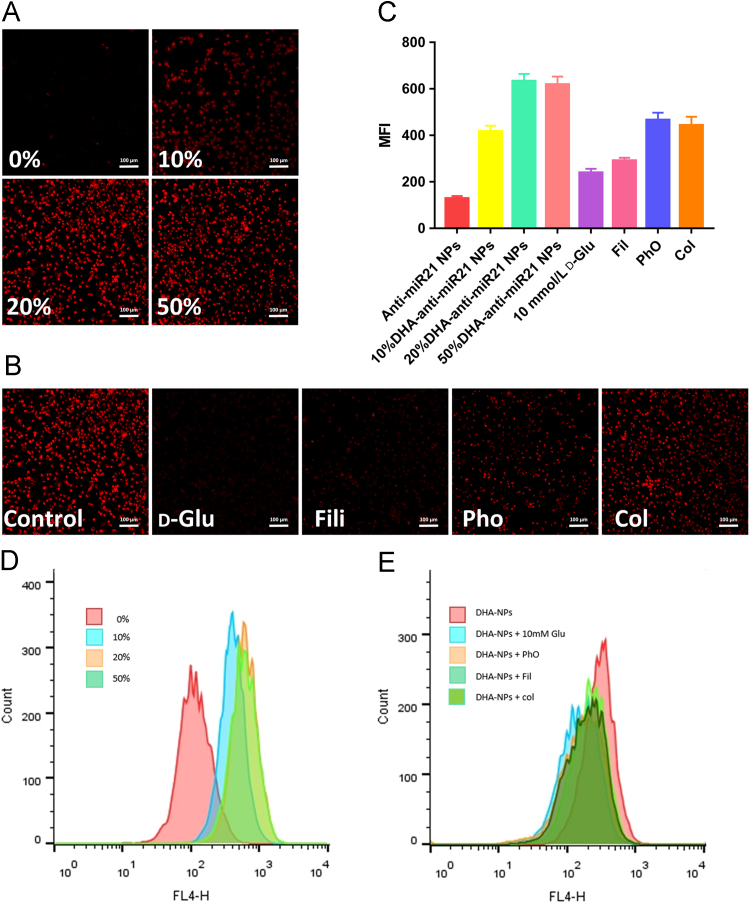
Red BODIPY-labelled nanopompons modified with different proportions of DHA were prepared for selection. Cellular uptake experiments of MDA-MB-231 Cell line was designed for screening a suitable final formulation. Cellular uptake experiments showed that with the increase of modification, the fluorescence intensity was significantly enhanced. This indicated an increase in cellular uptake, and reached a saturation point while the modification degree was 20% (Fig. 3A), which is probably due to the saturation of GLUT1. The results also proved that DHA modification has the ability to promote the accumulation of nanopompons in MB-MDA-231 cells. Same conclusion was obtained by flow cytometry quantitative test (Fig. 3D). In order to avoid the stability problems in vivo caused by excessive targeted molecular modification, 20% DHA modified nanopompons were selected as the final preparation.
Figure 3.
(A) The distribution of contracted anti-miR21 nanopompons in vitro. MBA-MD-231 cells were incubated with DHA modified anti-miR21 nanopompons and non-targeting nanopompons. Red: Red-BODIPY (λex = 630 nm, λem = 650 nm). Scale bars represent 100 µm. (B) Possible endocytosis pathway of anti-miR21 NPs study. Blocked by different inhibitors including: control; d-glucose (10 mmol/L); filipin (0.5 μg/mL); phenylarsine oxide (0.4 μg/mL); and colchicine (1 μg/mL). Scale bars represent 100 µm. (C) and (D) represent flow cytometry results of (A) and (B), respectively.
Then different kinds of cellular uptake inhibitors were used to further verify the role of DHA and explore the uptake pathway of nanopompons. Results showed that 10 mmol glucose can significantly inhibit the cellular uptake by saturating GLUT1 on the cell surface. Filipin has a comparable effect as glucose, which indicates that the uptake of nanopompons by MDA-MB-231 cells is mainly via a caveolin-mediated mechanism. Colchicine also has a certain inhibitory effect, indicating that some of the nanopompons can be taken up through pinocytosis. Phenylarsine oxide showed inhibitory effect to some extent27., 28. (Fig. 3B). The same conclusion was obtained by flow cytometry quantitative test (Fig. 3E).
3.3. Significant anti-miR21 knockdown at the molecular level and low toxicity of DHA-modified anti-miR21 nanopompons were proved in vitro
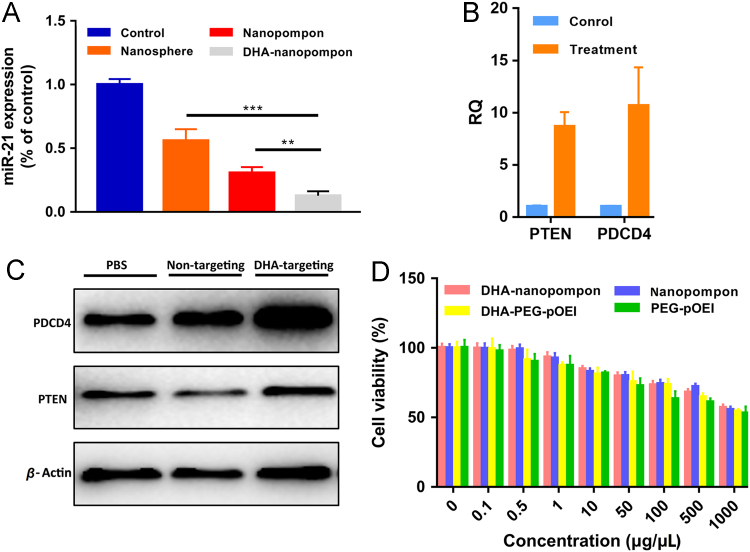
After incubation with nanopompons for 48 h, the level of miR21 was analyzed by real-time quantitative PCR. As shown in Fig. 4A, DHA-nanopompons have significantly reduced miR21 levels. Non-targeting preparation also down-regulated miR21 expression by nearly 50% probably because of the much longer incubation time compared with cellular uptake experiment in vitro. However, this effect was significantly dissipated in in vivo experiments (Fig. 6D). The expression of downstream target tumor suppressors of miR21–PTEN and PDCD proved the function of miR21. Increased expression of PTEN and PDCD4 at both mRNA and protein levels were correlated with the down-regulated miR21 expression level of the treatment group (Fig. 4B and C). Autophagy could sequester most nanomaterials and transport them to lysosomes for degradation26., 29., 30.. The relationship between nanopompons and autophagy was tested by acridine orange (AO) staining (Supporting Information Fig. S3). The maximum red autolysosome was caused by DHA-nanopompon, indicating that nanopompons could induce autophagy in cancer cells. Therefore, autophagy was demonstrated to be involved in the degradation of nanopompons31., 32., 33., 34..
Figure 4.
(A) miR21 gene down-regulation of anti-miR21 nanopompons in vitro. Asterisks (*) denote statistically significant differences (data represented as mean±SD, **P < 0.001, n = 3). (B) QRT-PCR and (C) Western blot result of the down-regulation at both mRNA and protein level of miR21 after treatment, bringing about up-regulation of two target genes PTEN and PDCD4 in vitro. (D) Cell viability test of DHA-nanopompon, nanopompon, DHA-PEG-pOEI and PEG-pOEI on 293 cells by MTT assay.
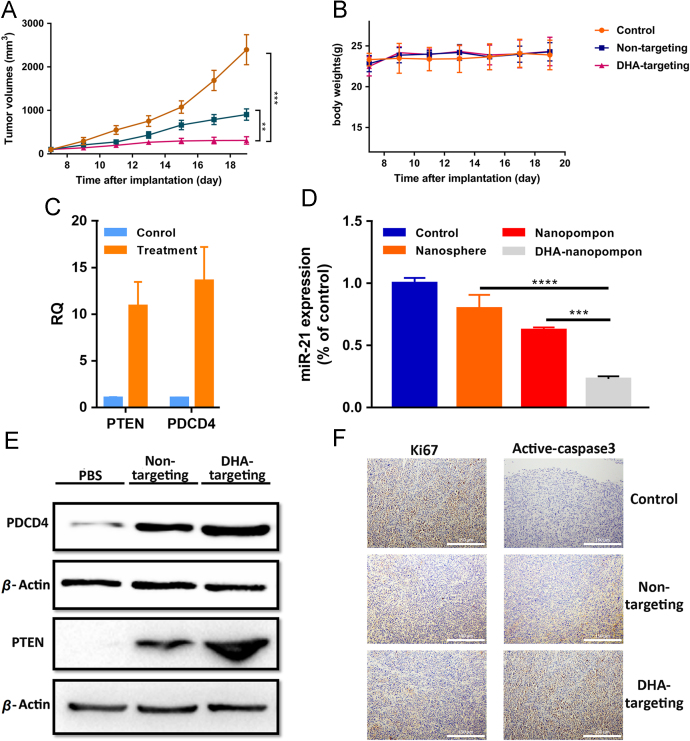
Figure 6.
(A) Tumor growth and (B) body weight curve over the course of 5 injections. (D) miR21 down-regulation of anti-miR21 nanopompons in vivo at miRNA level. Asterisks (*) denote statistically significant differences (data represented as mean±SD, ***P < 0.001, n = 3). (D) miR21 gene down-regulation of anti-miR21 nanopompons in vivo. Asterisks (*) denote statistically significant differences (data represented as mean±SD, **P < 0.001, n = 3). (C) QRT-PCR and (E) Western blot result of the down-regulation at both mRNA and protein level of miR21 after treatment, bringing about up-regulation of two target genes PTEN and PDCD4 in vivo. (F) Immunohistochemistry assay showed inhibition of tumor cell growth after treatment with the use of Ki67 as tumor proliferation indicator, and cleaved caspase-3 as tumor apoptosis indicator.
Cell viability test showed that both (DHA-)PEG-pOEI and nanopompons were proved to be low-cytotoxicity in high concentration and no cytotoxicity was observed in effective concentration (0.1 μg/μL). Nanopompons have lower cytotoxicity than (DHA-)PEG-pOEI because same concentration of the preparation contains less (DHA-)PEG-pOEI (Fig. 4D).
3.4. Real-time fluorescence imaging in animal models
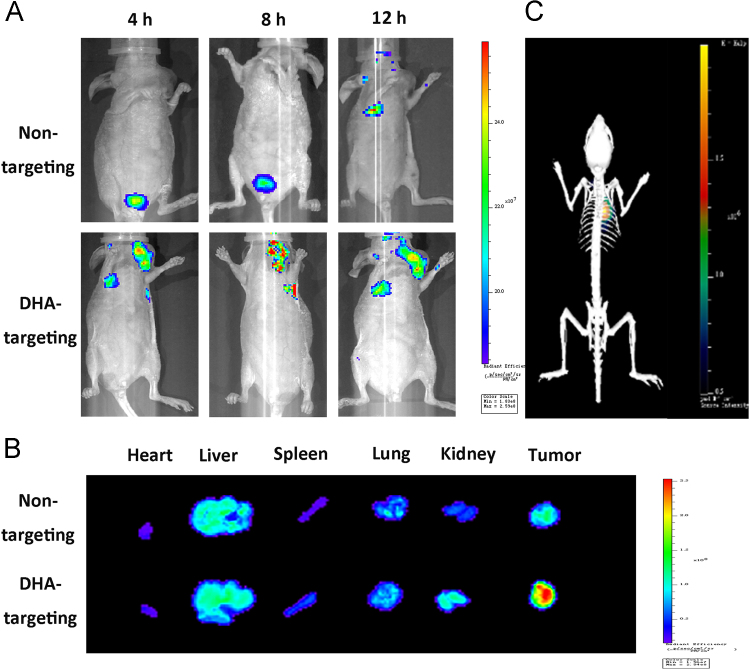
In order to evaluate tumor-targeting of anti-miR21 nanopompons, red BODIPY-labelled nanopompons were systemically administered into orthotopic TNBC-bearing mice via the tail vein. Images of normal tissues, organs, and tumors were taken after 4, 8 and 12 h (Fig. 5A), showing that DHA-targeting anti-miR21 nanopompons specifically accumulated in the tumors, and little accumulation was observed in healthy tissues and organs (Fig. 5A). Image of isolated organs (heart, liver, spleen, lung and kidney) and tumors was taken 12 h after administration, showing that DHA-targeting nanopompons can significantly improve tumor accumulation comparing with non-targeting nanopompons (Fig. 5B). Meanwhile, 3D real-time image of DHA-targeting group showed a significative signal at the tumor site (Fig. 5C).
Figure 5.
The distribution of anti-miR21 nanopompons in vivo. (A) Real-time fluorescence images of TNBC model nude mice injected with non-targeting anti-miR21 nanopompons, and DHA targeting anti-miR21 nanopompons at 4, 8, and 12 h after i.v. injection with BODIPY-anti-miR21 nanopompons, respectively. (B) The fluorescence images of excised tumors and organs at 12 h after i.v. injection. (C) In vivo 3D fluorescence imaging of model mouse at 12 h after i.v. injection.
3.5. Effective antitumor therapeutic efficacy of DHA-modified anti-miR21 nanopompons on TNBC model nude mice
Orthotopic TNBC-bearing nude mice model were constructed using MDA-MB-231 cells. When nanopompons were administered via the tail vein every 3 days, we measured the tumor size and assessed whether the systemically delivery of DHA-targeting anti-miR21 nanopompons can down-regulate miR21 and inhibit tumor growth (Fig. 6A). Tumor growth is noticeably inhibited after nanopompons injection compared with control group, and non-modified anti-miR21 nanopompons can also help tackling TNBC, but modification significantly enhanced the effect. After the TNBC model nude mice were treated with DHA-modified anti-miR21 nanopompons, body weight changed slightly (Fig. 6B). In order to confirm miR21 knockdown in vivo at the molecular level, tumor tissues were excised and lysed. The miR21 expression level was quantified by qRT-PCR assay (Fig. 6D), and the expression level of PTEN and PDCD4 was examined by qRT-PCR (Fig. 6C) and Western blot assay (Fig. 6E). Experimental evidence showed that tumors treated with DHA-targeting anti-miR21 nanopompons have reduced miR21 levels compared to control group. The knockdown of miR21 expression level was associated with increased expression of PTEN and PDCD4 at the mRNA and protein levels comparing with the control groups. In addition, DHA-targeting anti-miR21 nanopompons treatment could reduce cell proliferation in the tumor tissue (Fig. 6F), which could be visualized by decreased Ki67 staining. It could also induce tumor cell apoptosis, which could be visualized by increased cleaved caspase-3 level compared with the control groups. No significant systemic toxicity was observed according to H&E staining results (Supporting Information Fig. S4).
4. Discussion
In this research, MDA-MB-231 cell line, which comes from TNBC, a typical aggressive breast cancer type with the absence of progesterone receptor, estrogen receptor, and human epidermal growth factor receptor 2, was chosen as cell model. Patients with TNBC are poorly responsive to chemotherapy and easy to cause the tumor recurrence of the spread of cancer cells and metastasis, which leads to poor prognosis and short survival34., 35.. The great potential of gene therapy strategies for TNBC therapy tantalized scientists in the last few years, as for miR21, it is a widely recognized target gene for TNBC treatment3., 36.. In the previous studies of other investigators, naked anti-miR21 three-way junction (3WJ) motif3, nanocapsules carrying antisense of miR2136 and other subtle designs16 were established. However, most of these studies still face the poor delivery efficiency.
Antisense miR21 oligonucleotide with sequence-specific binding ability has been identified as an advanced inhibitor to silence miR218., 9.. Therefore, with the use of RCT, we designed a pompom-like nanoparticle carrying a half million tandem copies of therapeutic strands (antisense miR21 oligonucleotide) to improve delivery efficiency.
However, compared with previous work, in which siRNA sequences were transcribed using RCT18, the nanosphere containing anti-miRNA strands was much more unstable (Fig. 2C). In vitro stability test showed naked nanosphere could not remain stable when reaching tumor site (Fig. S1A), because arge particle size (about 500 nm) is also not conducive to its penetration in the tumor site. Therefore, it is essential to protect, compress and embellish the naked nanosphere.
For protection and compression, disulfide-bonded polyethyleneimine (pOEI) was designed. After nanospheres were wrapped and compressed, they become physiologically stable (Fig. S1B), and much smaller (about 100 nm) which are more suitable for tumor treatment. To further improve tumor accumulation, the nanopompons were modified with dehydroascorbic acid (DHA), which has been used by many drug delivery systems to improve tumor targeting37., 38.. From previous research, too many targeting small molecules once modified may affect stability of nanoparticles in vivo, and DHA is no exception. With comprehensive consideration of its ability to promote tumor cell accumulation, 20% DHA-modified nanopompons were selected as optimized preparation.
In this study, we exploited 3 components: inherent enzyme-responsive nanospheres, GSH-responsive outer-layer-releasing materials, and tumor-accumulating small molecules. Three components were combined organically and the formulation was optimized to achieve better drug efficacy. Compared with previous studies, nanoparticles accumulated much faster (in 2 h) in tumor accumulation, and achieved similar tumor accumulation in the case of halving RNA dose, which is related to RCT method. It is inseparable from the special properties of the nanospheres constructed by the RCT and the protection and modification of the materials.
5. Conclusions
Based on the overexpression of miR21 in tumor cells, nanopompons capable of down-regulating mir21 were designed to efficiently deliver a large number of anti-miR21 sequences and improve stability of RNA drug. Small molecule DHA with the capacity of tumor accumulation was designed based on the overexpression of GLUT1 modified on the surface of nanopompons in the tumor microenvironment. DHA-modified anti-miR21 nanopompons demonstrated outstanding TNBC accumulation, miR21-down-regulating and anti-TNBC therapeutic effect, which indicated the successful delivery of abundant therapeutic genes for an efficient TNBC-targeting RNAi therapy.
Acknowledgments
The work was supported by the grants from National Science Fund for Distinguished Young Scholars (81425023, China) and Program of Shanghai Academic Research Leader (18XD1400500, China).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data associated with this article can be found in the online version at doi:10.1016/j.apsb.2019.01.012.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Kota J., Chivukula R.R., O׳Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shu D., Li H., Shu Y., Xiong G., Carson W.E., III, Haque F. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piao L., Zhang M., Datta J., Xie X., Su T., Li H. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther. 2012;20:1261–1269. doi: 10.1038/mt.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 6.Qi L., Bart J., Tan L.P., Platteel I., van der Sluis T., Huitema S. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 8.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y., Cullen B.R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasinski A.L., Slack F.J. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry J.C., Azevedo-Pouly A.C., Schmittgen T.D. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28:3030–3042. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Tian Y, Tu MJ, Ho PY, Batra N, Yu AM. Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharm Sin B 2019;3:639–47 [DOI] [PMC free article] [PubMed]
- 14.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castanotto D., Rossi J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira D.M., Rodrigues P.M., Borralho P.M., Rodrigues C.M. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Babar I.A., Cheng C.J., Booth C.J., Liang X., Weidhaas J.B., Saltzman W.M. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.B., Hong J., Bonner D.K., Poon Z., Hammond P.T. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An S., Jiang X., Shi J., He X., Li J., Guo Y. Single-component self-assembled RNAi nanoparticles functionalized with tumor-targeting iNGR delivering abundant siRNA for efficient glioma therapy. Biomaterials. 2015;53:330–340. doi: 10.1016/j.biomaterials.2015.02.084. [DOI] [PubMed] [Google Scholar]
- 20.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 21.Amann T., Maegdefrau U., Hartmann A., Agaimy A., Marienhagen J., Weiss T.S. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol. 2009;174:1544–1552. doi: 10.2353/ajpath.2009.080596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amann T., Hellerbrand C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets. 2009;13:1411–1427. doi: 10.1517/14728220903307509. [DOI] [PubMed] [Google Scholar]
- 23.Rumsey S.C., Kwon O., Xu G.W., Burant C.F., Simpson I., Levine M. Glucose transporter isoforms GLUT1 and GLUT3 transport dehydroascorbic acid. J Biol Chem. 1997;272:18982–18989. doi: 10.1074/jbc.272.30.18982. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y., Zhang Y., Li J., Zhang Y., Lu Y., Jiang X. Cell microenvironment-controlled antitumor drug releasing-nanomicelles for GLUT1-targeting hepatocellular carcinoma therapy. ACS Appl Mater Interfaces. 2015;7:5444–5453. doi: 10.1021/am5091462. [DOI] [PubMed] [Google Scholar]
- 25.Naito S., von Eschenbach A.C., Giavazzi R., Fidler I.J. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 26.Tao W., Zhu X., Yu X., Zeng X., Xiao Q., Zhang X. Black phosphorus nanosheets as a robust delivery platform for cancer theranostics. Adv Mater. 2017;29:1603276. doi: 10.1002/adma.201603276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G., Fang Y.Z., Yang S., Lupton J.R., Turner N.D. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 28.Yameen B., Choi W.I., Vilos C., Swami A., Shi J., Farokhzad O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J Control Release. 2014;190:485–499. doi: 10.1016/j.jconrel.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C.N., Tian H.Y., Wang Y.B., Du Y., Chen J., Lin L. Anti-tumor effects of combined doxorubicin and siRNA for pulmonary delivery. Chin Chem Lett. 2017;28:807–812. [Google Scholar]
- 30.Cheng W., Nie J., Gao N., Liu G., Tao W., Xiao X. A multifunctional nanoplatform against multidrug resistant cancer: merging the best of targeted chemo/gene/photothermal therapy. Adv Funct Mater. 2017;27:1704135. [Google Scholar]
- 31.Zhang X., Dong Y., Zeng X., Liang X., Li X., Tao W. The effect of autophagy inhibitors on drug delivery using biodegradable polymer nanoparticles in cancer treatment. Biomaterials. 2014;35:1932–1943. doi: 10.1016/j.biomaterials.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 32.Chen S., Dong G., Wu S., Liu N., Zhang W., Sheng C. Novel fluorescent probes of 10-hydroxyevodiamine: autophagy and apoptosis-inducing anticancer mechanisms. Acta Pharm Sin B. 2019;9:144–156. doi: 10.1016/j.apsb.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J., Chang D., Yang Y., Zhang X., Tao W., Jiang L. Systematic investigation on the intracellular trafficking network of polymeric nanoparticles. Nanoscale. 2017;9:3269–3282. doi: 10.1039/c7nr00532f. [DOI] [PubMed] [Google Scholar]
- 34.Corti A., Casini A.F., Pompella A. Cellular pathways for transport and efflux of ascorbate and dehydroascorbate. Arch Biochem Biophys. 2010;500:107–115. doi: 10.1016/j.abb.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Brinkman A.M., Chen G., Wang Y., Hedman C.J., Sherer N.M., Havighurst T.C. Aminoflavone-loaded EGFR-targeted unimolecular micelle nanoparticles exhibit anti-cancer effects in triple negative breast cancer. Biomaterials. 2016;101:20–31. doi: 10.1016/j.biomaterials.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C., Wen J., Meng Y., Zhang K., Zhu J., Ren Y. Efficient delivery of therapeutic miRNA nanocapsules for tumor suppression. Adv Mater. 2015;27:292–297. doi: 10.1002/adma.201403387. [DOI] [PubMed] [Google Scholar]
- 37.Shao K., Zhang Y., Ding N., Huang S., Wu J., Li J. Functionalized nanoscale micelles with brain targeting ability and intercellular microenvironment biosensitivity for anti-intracranial infection applications. Adv Healthc Mater. 2015;4:291–300. doi: 10.1002/adhm.201400214. [DOI] [PubMed] [Google Scholar]
- 38.Shao K., Ding N., Huang S., Ren S., Zhang Y., Kuang Y. Smart nanodevice combined tumor-specific vector with cellular microenvironment-triggered property for highly effective antiglioma therapy. ACS Nano. 2014;8:1191–1203. doi: 10.1021/nn406285x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material