Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium causing diseases in plants, animals, and humans, and its drug resistance is a major concern in medical care. Biofilms play an important role in P. aeruginosa drug resistance. Three factors are most important to induce biofilm: quorum sensing (QS), bis-(3′-5′)-cyclic diguanosine monophosphate (c-di-GMP), and small RNAs (sRNAs). P. aeruginosa has its own specific QS system (PQS) besides two common QS systems, LasI–LasR and RhlI–RhlR, in bacteria. PQS is interesting not only because there is a negative regulation from RhlR to pqsR but also because the null mutation in PQS leads to a reduced biofilm formation. Furthermore, P. aeruginosa dispersed cells have physiological features that are distinct between the planktonic cells and biofilm cells. In response to a low concentration of c-di-GMP, P. aeruginosa cells can disperse from the biofilms to become planktonic cells. These raise an interesting hypothesis of whether biofilm can be reversed through the QS mechanism in P. aeruginosa. Although a single factor is certainly not sufficient to prevent the biofilm formation, it necessarily explores such possibility. In this hypothesis, the literature is analyzed to determine the negative regulation pathways, and then the transcriptomic data are analyzed to determine whether this hypothesis is workable or not. Unexpectedly, the transcriptomic data reveal a negative regulation between lasI and psqR. Also, the individual cases from transcriptomic data demonstrate the negative regulations of PQS with laslI, laslR, rhlI, and rhlR under different experiments. Based on our analyses, possible strategies to reverse biofilm formation are proposed and their clinic implications are addressed.
Keywords: biofilm, P. aeruginosa, quorum sensing, transcriptome, positive feedback, negative feedback
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium living in soil and water. Being an opportunistic pathogen, P. aeruginosa can cause the bacterial soft rot in plants (Rahme et al., 2000; Walker et al., 2004), and diseases in animals (Ferris et al., 2017; Vingopoulou et al., 2018) and humans, including eye (Willcox, 2007), burn wound (Church et al., 2006), acute and chronic pulmonary infections, where cystic fibrosis is associated with substantial morbidity and mortality (Elborn, 2016; Klockgether and Tümmler, 2017).
Therefore, P. aeruginosa is a major concern in medical care because of its drug resistance against the traditional antibiotic therapy (Buhl et al., 2015; Oliver et al., 2015), that is particularly problematic for immunocompromised patients and the elderly in nosocomial environments (Xia et al., 2016). P. aeruginosa brings about its drug resistance through hydrolyzation of antibiotics with carbapenemases or extended-spectrum β-lactamases or ApmR (Vatcheva-Dobrevska et al., 2013; Fisher and Mobashery, 2014; Hakemi Vala et al., 2014), the low permeability of outer membrane (Eren et al., 2013; Zgurskaya et al., 2015), the multidrug efflux (Poole, 2004; Aghazadeh et al., 2014), etc. Also, the biofilm is an important player in P. aeruginosa drug resistance (Mah et al., 2003) because the dense extracellular matrix of biofilms reduces the efficacy of detergents and antibiotics (Mah et al., 2003). Such resistance could be increased a thousand times in some cases (Stewart and Costerton, 2001).
The dispersal of cells from the biofilm colony is a crucial and unique stage for biofilms to spread and colonize new surfaces (Monroe, 2007) and for the transition of dispersed cells from the biofilm to the planktonic growth phase. Could it be possible to stop the biofilm from happening, or reserve the biofilm back to the planktonic phenotype, or eradicate the biofilm in bacteria?
Theoretically, this hypothesis could be possible for P. aeruginosa, because its dispersed cells have physiological features that are distinct between the planktonic and the biofilm cells (Chua et al., 2014, 2015). In response to a low concentration of c-di-GMP, P. aeruginosa cells can disperse from the biofilm to become the planktonic cells. The drug resistance is not stronger in the biofilm cells than in the stationary-phase planktonic cells, but is stronger than in the logarithmic-phase planktonic cells (Spoering and Lewis, 2001). Additionally, P. aeruginosa produces cis-2-decenoic acid, which is a fatty acid messenger and induces dispersion and inhibits the growth of biofilm colonies (Davies and Marques, 2009). Furthermore, nitric oxide triggers the dispersal of biofilms in P. aeruginosa (Barraud et al., 2006), leading to the treatment of chronic infections in cystic fibrosis (Howlin et al., 2017).
The formation of biofilm is induced and regulated by numerous genes and environmental factors (Fazli et al., 2014), of which three are most important. The first one is the quorum sensing (QS), because QS controls about 10% genes in P. aeruginosa (Wagner et al., 2003), including many genes that are actively involved in the biofilm development and dispersal, although they are unlikely to be involved in the attachment and the initial of biofilm growth (Davies et al., 1998). The second one is the bis-(3′-5′)-cyclic diguanosine monophosphate (c-di-GMP), because its signaling network is the most complex secondary signaling system in bacteria (Hengge, 2009) and has the responsibility to decide whether bacteria adopt either planktonic or biofilm phenotype (Jenal and Malone, 2006). The third one is the small RNAs (sRNAs) although their role in biofilm is yet to be clear (Wolska et al., 2016).
Indeed, QS has a close relationship with biofilm (Wolska et al., 2016). It controls the synthesis of rhamnolipids that maintain the channels (Stoodley et al., 1994) for distributing nutrient and oxygen and removing waste products in mushroom-shaped structures (Davey and O’Toole, 2000). The channels can help in the release of a large amount of eDNA due to the autolysis of subpopulation of bacteria (Allesen-Holm et al., 2006) at the late stage of biofilm development. Various components of the biofilm matrix, such as extracellular DNA (eDNA), exopolysaccharides (EPS) and glucan, are closely related to biofilm matrix dynamics and bacterial virulence (Rainey et al., 2019). Also, there are other virulence factors, which play an important role in the QS regulation and biofilm formation. For example, pyocyanin promotes eDNA release and facilitates the biofilm formation (Klare et al., 2016).
It is worth reviewing literature to explore whether the biofilm is theoretically reversible through QS in P. aeruginosa, not only because P. aeruginosa is a causal organism of important health ailments but also because P. aeruginosa is a commonly used biofilm model organism (Rasamiravaka et al., 2015). More importantly, the synthesis of rhamnolipid in P. aeruginosa occurs at its late-exponential and stationary phases (Guerra-Santos et al., 1986). Rhamnolipid helps bacteria to utilize long-chain fatty acids as sources of carbon (Ochsner et al., 1994a) so it plays an important role in the biofilm formation (Stoodley et al., 1994; Davey and O’Toole, 2000; Allesen-Holm et al., 2006).
Reversing of biofilms could be plausible because QS is a target in many different circumstances such as attenuate virulence (Chan et al., 2015), bacterial metabolism (Goo et al., 2015), bacterial response to antibiotics (Rasamiravaka and El Jaziri, 2016), and therapy (LaSarre and Federle, 2013). Besides, the mechanism to form biofilms in P. aeruginosa is definitely different from other bacteria such as P. putida, P. fluorescens, Staphylococcus aureus, and Vibrio cholera (Wolska et al., 2016).
Needless to say, the reversing of biofilms is related to multiple factors, so a single factor such as QS could have very limited effects. However, we should theoretically explore those possibilities one by one at initial stage in view of the importance of biofilms in clinical meanings.
Positive and Negative Regulations in Qs
If we wish to reverse the biofilm through the QS, we need to find out whether the QS is reversible or not. So far overwhelmed evidence suggests that the QS is a positive feedback system, which implies that it is impossible to stop the QS once the QS is initiated. However, we have yet to know whether the ending point of QS is the biofilm formation? If this is the case, the stop of QS will either reverse the biofilm or stop the biofilm formation. To answer this issue, it is necessary to find out the negative regulation (feedback) in QS.
The QS is a cell-to-cell communication by means of production, detection, and response of chemical compounds, autoinducers, and thus the QS changes an individual or a population behavior upon the concentration of autoinducers, which are subject to the cell density (Fuqua et al., 1994).
Pseudomonas aeruginosa has three QS systems. (i) LasI–LasR that is related to the synthesis and the use of N-(3-oxo-dodecanoyl)-L-homoserine lactone (3OC12-HL) (Passador et al., 1993; Pearson et al., 1994), whose concentration is ranged from 1 to 5 μM (Pearson et al., 1994, 1995) (brown color items in Figure 1). (ii) RhlI-RhlR that is related to the synthesis and the use of N-(butyryl)-L-homoserine lactone (BHL) (Pearson et al., 1995), whose concentration is about 10 μM (Pearson et al., 1995) (yellow color items in Figure 1). (iii) Pseudomonas quinolone signal (PQS)-based QS, PqsABCDH-PqsR that is related to the synthesis and the use of 2-heptyl-3-hydroxy-4-quinolone (HHQ) (Mashburn-Warren et al., 2008; Kulkarni and Jagannadham, 2014), whose concentration is about 6 μM (Pesci et al., 1999) (green color items in Figure 1). The first two QS systems essentially are N-acylated homoserine lactone (AHL)-based QS systems (Pesci et al., 1997) and exist in many bacteria.
FIGURE 1.
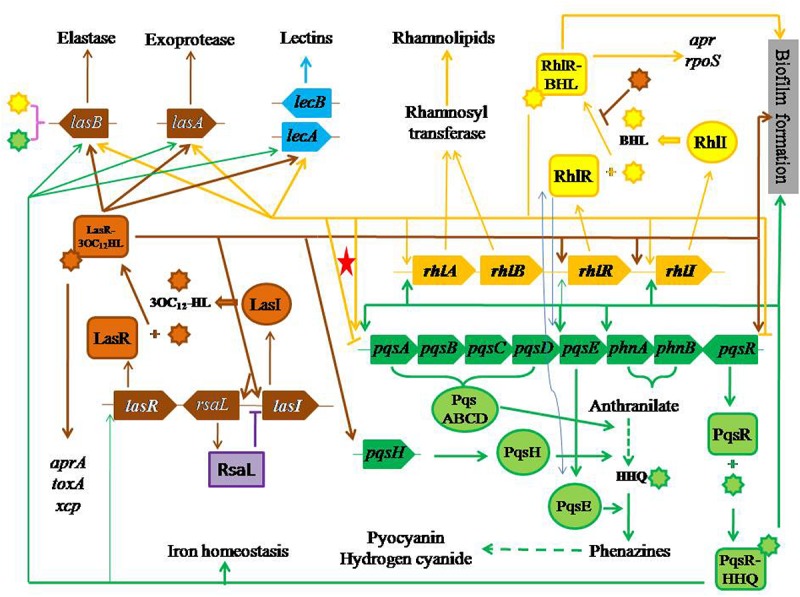
Three QS systems with their effects and regulatory pathways. The red star highlights bi-functional regulation of RhlI–RhlR system to PQS synthesis.
The sophisticated QS systems in P. aeruginosa are described as follows. (i) LasI produces 3OC12-HL, which acts on LasR (Gambello and Iglewski, 1991; Pearson et al., 1994) (the upward brown arrow from lasR to LasR on the left side of Figure 1). (ii) LasR acts on aprA (Gambello et al., 1993), lasA (Toder et al., 1991) and toxA (Gambello and Iglewski, 1991; Gambello et al., 1993; Passador et al., 1993) (the downward brown arrow on the far left side in Figure 1). (iii) Both LasI and LasR act on lasB (Pearson et al., 1994, 1995) through 3OC12-HL, whose half-maximal expression needs 1.0 nM (Seed et al., 1995) (brown symbols on the left side of Figure 1). (iv) RhlI produces BHL, which acts on RhlR (Pearson et al., 1995, 1997) (the bright yellow arrow on the right side of Figure 1). (v) RhlR acts on pyocyanin synthesis (Meighen, 1991; Ochsner et al., 1994b; Brint and Ohman, 1995) (the long yellow arrow on the middle of Figure 1), lasA (Brint and Ohman, 1995) (the yellow arrow on the middle left of Figure 1), and rpoS (Latifi et al., 1996) (the yellow arrow on the upper right corner of Figure 1). (vi) Both RhlI and RhlR act on lasB through BHL (Brint and Ohman, 1995) (the yellow arrow on the upper right part of Figure 1), and rhlABR (Ochsner and Reiser, 1995) (the small yellow arrow on the middle of Figure 1), where rhlAB encodes rhamnosyltransferase (Ochsner et al., 1994a) (two yellow arrows on the upper middle part of Figure 1) together with rhlR positively regulate rhamnolipid synthesis (Ochsner et al., 1994b) (the yellow arrow on the middle upper part of Figure 1). (vii) LasR and RhlR positively regulate the synthesis of hydrogen cyanide (Pessi and Haas, 2000) (the downward yellow arrow on the lower middle part of Figure 1).
Still, Figure 1 displays the effects of PQS-based QS on their targets. (i) PqsABCDH produces HHQ requiring phnA and phnB through anthranilate (Gallagher et al., 2002) (the green curly lines on the middle right part of Figure 1), then HHQ acts on PqsR (Cao et al., 2001), regulating the production of elastase, PA-IL lectin, pyocyanin and rhamnolipid (Pesci et al., 1999; McKnight et al., 2000; Gallagher et al., 2002; Lee and Zhang, 2015) (the green lines from the lower right corner in Figure 1). (ii) PqsE positively acts on biosynthesis of various virulent factors, which is independent of HHQ or any compounds produced related to the function of pqsABCDE operon although the expression of pqsE and PqsE are controlled by HHQ and PqsR (Farrow, et al., 2008) (dashed green line on the lower right part of Figure 1). (iii) PqsR–HHQ is involved in iron homeostasis (Bredenbruch et al., 2006; Oglesby et al., 2008) (the lowest green line in Figure 1).
A positive feedback can be found in each of three QS systems. (i) The first positive feedback goes from LasR–3OC12-HL to LasI through lasI, whose half-maximal expression needs 0.1 nM 3OC12-HL (Seed et al., 1995) (light gray ellipse on the left part of Figure 1). (ii) The second positive feedback goes from RhlR–HL to RhlI through rhlI (Ochsner and Reiser, 1995) (light gray ellipse on the upper right part of Figure 1). (iii) The third positive feedback goes from PqsR–HHQ to pqsABCDE and phnAB operons (Cao et al., 2001; Gallagher et al., 2002; Wade et al., 2005) (light gray ellipse on the middle right part of Figure 1).
The relationship among three QS systems in P. aeruginosa is positive in the following regulations. (i) LasR positively regulates HHQ through the complex LasR–3OC12-HL on pqsH (Pesci et al., 1999; Schertzer et al., 2009) (brown arrow on the right middle part of Figure 1). (ii) RhlR positively regulates HHQ through PqsE (Pesci et al., 1999) (two arrow-blue lines on the middle right part of Figure 1). (iii) LasR positively regulates rhlR through the complex LasR–3OC12-HL (Latifi et al., 1996; Pesci et al., 1997) and rhlI (Latifi et al., 1996) (the brown horizontal line with two arrows in Figure 1). (iv) HHQ strongly acts on rhlI with BHL (two arrow-blue lines in middle right part of Figure 1) but weakly acts on lasR and rhlR (McKnight et al., 2000). (v) RhlR positively regulates PqsE, whose overexpression leads to a high rhamnolipid production (Farrow, et al., 2008) (the yellow arrow on the right middle part of Figure 1). (vi) PqsE changes the function of RhlR rather than that of BHL (Farrow, et al., 2008) (two arrow-blue lines on the middle right part of Figure 1). (vii) LasR/3OC12-HL controls pqsR (Camilli and Bassler, 2006) (the end arrow of brown horizontal line in Figure 1).
In fact, there is a negative regulation among QS systems, namely, RhlR negatively regulates pqsR in P. aeruginosa (Pesci et al., 1997; Wade et al., 2005), or RhlR and BHL together negatively affect the production of HHQ and other quinolones through pqsR to pqsABCDE operon transcription (McGrath et al., 2004; Jensen et al., 2006; Xiao et al., 2006b) (the yellow arrow from RhlR–BHL to the yellow horizontal line to the right end with downward dash end in Figure 1). On the other hand, a negatively regulatory pathway is not so sure (the yellow end line highlighted with a red star on the middle of Figure 1).
Can This Negative Regulation Work?
As PQS-based QS is so particularly relevant to Pseudomonas, its significance should not be ignored. This is because the null mutation in PQS leads to a reduced biofilm formation and decreased the productions of pyocyanin, elastase, PA-IL lectin and rhamnolipids (Rahme et al., 1997, 2000; Cao et al., 2001; Diggle et al., 2003). Indeed, PQS directly or indirectly controls 92 or 143 genes as shown in two transcriptomic analyses (Deziel et al., 2005, Bredenbruch et al., 2006). By contrast, the other two QS systems together influence the expression in 200-plus genes (Whiteley et al., 1999).
For PQS, it does not reach its maximal production until the late stationary phase of growth (McKnight et al., 2000). This implies that HHQ is not involved in sensing the cell density, so the observation that the QS response is not reversed for small decreases in population density in P. aeruginosa (Williams and Camara, 2009) is not the failure of PQS. An important time interval appears between QS systems, i.e., BHL is produced during the log phase of growth but HHQ is produced during late time in the stationary phase of growth (McKnight et al., 2000), so the positive regulation of HHQ on rhlI is more likely to be related to the second round of RhlI cycle. If HHQ would not function at this time interval, perhaps the QS would stop.
Another promising point is that the phenazine production requires HHQ in P. aeruginosa (McKnight et al., 2000; Mavrodi et al., 2001). In fact, phenazines may have a significant ecological impact on the biofilm formation in P. aeruginosa as well as other bacteria persisting in biofilms mixed with P. aeruginosa. Through affecting H2O2 generation, phenazines bring about the lysis of competing bacterial cells in mixed biofilms and the subsequent eDNA release (Das and Manefield, 2013).
Perhaps, one of the best ways to explore the possibility of whether the QS is reversible through PQS in P. aeruginosa is to analyze the transcriptomic data in order to find some common patterns. Accordingly, we analyzed the transcriptomic data on Affymetrix P. aeruginosa array with 5549 P. aeruginosa genes, platform GPL84, from Gene Expression Omnibus (GEO) (Edgar et al., 2002; Barrett et al., 2013), including all the data in 104 publications (Supplementary Information) with 274 datasets. Each dataset represents the response to a specifically experimental condition. With these all available transcriptomic data, we wish to determine if PQS could be depressed under different experimental conditions.
Table 1 shows correlation coefficients between any two genes of three QS systems. The rationale is that there are up-regulations and down-regulations in transcriptomic data. The correlation between two genes, which are both up-regulated or both down-regulated, would suggest a positive regulation with a positive correlation coefficient. By contrast, the correlation between two genes, which are regulated oppositely, would suggest a negative regulation with a negative correlation coefficient.
TABLE 1.
Correlation coefficient between any two genes of three QS systems using transcriptomic data.
| Gene | lasI | lasR | rhlI | rhlR | rhl | pqsA | pqsB | pqsC | pqsD | pqsE | pqsH | pqsR/mvfR | |
| PA1432 | PA1430 | PA3476 | PA3477 | PA3861 | PA0996 | PA0997 | PA0998 | PA0999 | PA1000 | PA2587 | PA1003 | ||
| lasI | PA1432 | 1 | 0.62 | 0.37 | 0.37 | 0.16 | 0.06 | 0.04 | −0.02 | −0.01 | 0.01 | 0.17 | −0.38 |
| lasR | PA1430 | 1 | 0.51 | 0.33 | 0.52 | 0.15 | 0.12 | 0.11 | 0.13 | 0.10 | 0.14 | −0.03 | |
| rhlI | PA3476 | 1 | 0.89 | 0.29 | 0.73 | 0.76 | 0.58 | 0.67 | 0.58 | 0.20 | 0.17 | ||
| rhlR | PA3477 | 1 | 0.13 | 0.84 | 0.88 | 0.66 | 0.76 | 0.68 | 0.05 | −0.04 | |||
| rhl | PA3861 | 1 | 0.15 | 0.14 | 0.19 | 0.22 | 0.14 | 0.16 | 0.24 | ||||
| pqsA | PA0996 | 1 | 0.97 | 0.85 | 0.95 | 0.82 | −0.05 | 0.25 | |||||
| pqsB | PA0997 | 1 | 0.89 | 0.96 | 0.84 | −0.03 | 0.24 | ||||||
| pqsC | PA0998 | 1 | 0.90 | 0.87 | −0.03 | 0.28 | |||||||
| pqsD | PA0999 | 1 | 0.81 | 0.00 | 0.30 | ||||||||
| pqsE | PA1000 | 1 | −0.07 | 0.23 | |||||||||
| pqsH | PA2587 | 1 | 0.27 | ||||||||||
| pqsR/mvfR | PA1003 | 1 |
Based upon the correlations within a single QS system in Table 1, the correlations between lasI and lasR, and between rhlI and rhlR confirm their auto-induction relationships (Gambello and Iglewski, 1991; Pearson et al., 1994) within each QS system. No negative correlation is found between the QS genes in the same QS system. As pqsR is named mvfR in gene bank, the auto-induction relationship with the rest of PQS genes are not very evident as the paired correlations between pdsA, pqsB, pqsC, pqsD, pqsE, but all paired correlations suggest a positive regulation within PQS system (Gallagher et al., 2002). Based upon the correlations between two QS systems in Table 1, the results conform there is a positive regulation between lasI–lasR and rhlI–rhlR, and between rhlI-rhlR and pqsABCDE. However, an undocumented negative regulation is revealed between lasI and pqsR/mvfR using these transcriptomic data. Could it be a potential pathway to reverse the biofilm formation?
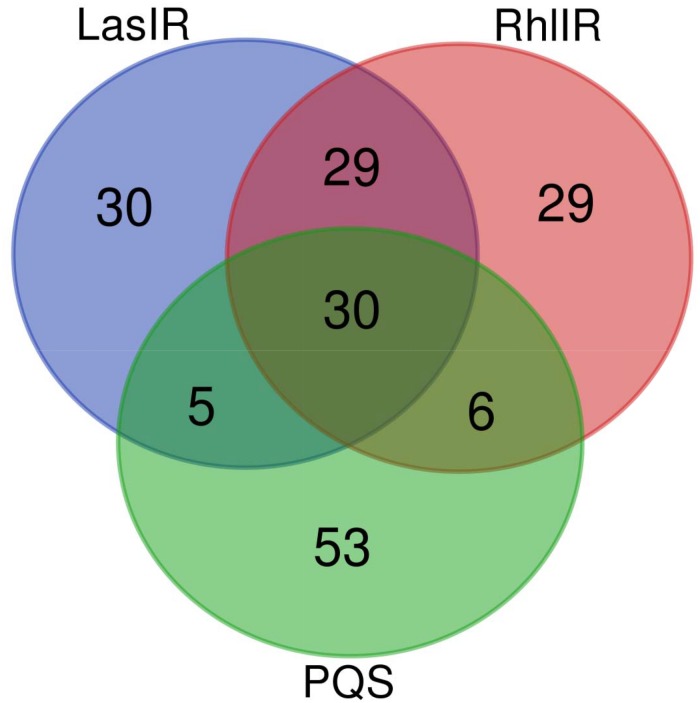
Furthermore, the responses of QS systems are analyzed under different transcriptomic experiments, and classified as down-regulation, down-regulation/no response, no response, no response/up-regulation, up-regulation and mixed responses. Figure 2 shows such analysis according to 94 transcriptomic experiments. No response on three QS systems was found in 30 transcriptomic experiments (the intersection of three circles in Figure 2). Both LasI-LasR and RhlI-RhlR have the same response in 29 transcriptomic experiments (the intersection of two upper circles in Figure 2), suggesting a good cooperation between them. By the contrast, only five and six transcriptomic experiments show the same response for PQS with LasI-LasR and RhlI-RhlR (the intersection of two upper and lower circles in Figure 2), respectively. The same response in both LasI-LasR and PQS systems includes no response in GSE24784, GSE26142, GSE35248, and GSE39044, and no response/up-regulation in GSE22684, indicating few positive impact of LasI-LasR on PQS. The same response in both RhlI-RhlR and PQS systems includes: down-regulation in GSE9255; down-regulation/no response in GSE5887; no response in GSE17179 and GSE61925; and no response/up-regulation in GSE65882 and GSE7402. Thus, the results from Venn diagram indicate that RhlI-RhlR has weak impacts on PQS. Figure 2 demonstrates the responses of 30, 29, and 53 transcriptomic experiments solely in LasI-LasR, RhlI-RhlR, and PQS, respectively, of which their response ranges from down-regulation to mixed response.
FIGURE 2.
Number of transcriptomic studies affecting QS in P. aeruginosa PAO1. The figure was made using online server at http://bioinformatics.psb. ugent.be/webtools/Venn/.
Finally, the negative regulation between different QS systems is found in four transcriptomic experiments (GSE4152, GSE8408, GSE6122, and GSE17296). In the study on Australian clonal strain (AES-1) in patients with cystic fibrosis in GSE6122 (Manos et al., 2009), lasI, rhlI, and rhlR were down-regulated while pqsA, pqsB, pqsC, pqsD, and pqsE were up-regulated. This highlights the PQS remarkable effect on the biofilm formation and enhanced infectivity. Another three transcriptomic experiments show that pqsA, pqsB, pqsC, pqsD, and pqsE were down-regulated whereas rhlI and rhlR were up-regulated (Teitzel et al., 2006; Tralau et al., 2007; Kawakami et al., 2010), of which lasI was up-regulated in copper-stressed (Teitzel et al., 2006), and both lasI and lasR were up-regulated in sulfate limitation (Tralau et al., 2007). Therefore, RhlI-RhlR does have a negative regulation on PQS (McGrath et al., 2004; Wade et al., 2005; Xiao et al., 2006a).
Conclusion
In this hypothesis, we apply the transcriptomic data to verify the hypothesis of whether the biofilm can be reversed in P. aeruginosa through QS because there are negative regulations between PQS and RhlI-RhlR. Interestingly, the transcriptomic data from 104 publications reveal a negative regulation between lasI and psqR, rendering a support to the hypothesis. Individual cases from transcriptomic data under different experiments demonstrate the negative regulations of PQS with laslI, laslR, rhlI, and rhlR.
In general, the relationships among different QS systems reveal positive regulations, which act together to promote the biofilm formation. However, the present analyses from literature and transcriptomic data provide the evidence that both LaslI-LaslR and RhlI-RhlR systems have negatively regulatory effects on PQS system. This is very important because these negative regulations lay the foundation for the biofilm reversion through QS. Although the exact pathways are still not fully discovered, the N-acylated homoserine lactone (AHL)-based QS systems can influence PQS-based QS system by inhibiting the expression of pqsABCDE operon and pqsR, resulting in the reduction of HHQ and PqsR synthesis. Consequently, the low concentration of PQS related products cannot maintain the biofilm, leading to its reversion. On the other hand, the down-regulated PQS-based QS system cannot perform well their function of positive regulations on LaslI-LaslR and RhlI-RhlR systems, which will further affect the biofilm formation, especially in the second round of RhlI cycle. Surely, there are other factors that play roles in the formation of drug-resistant multicellular biofilms, such as c-di-GMP. As mentioned in section “Introduction,” this signal can govern bacterial cells to adopt either planktonic phenotype or biofilm formation (Hengge, 2009). Recent study demonstrates that high levels of cAMP lead to the decrease of c-di-GMP content, which inhibits the biofilm formation in P. aeruginosa (Almblad et al., 2019).
In clinic therapeutics for infectious diseases, antibiotic resistance has been spreading widely and rapidly, which becomes a major challenge for modern medicine. The strategy of interfering the biofilm formation is effective through bacterial cell-to-cell communication, especially with QS system (Soheili et al., 2019). More recently, QS inhibitors are drawing great attention in blocking the pathogenicity from P. aeruginosa (Calvert et al., 2018; Schütz and Empting, 2018). The transcriptional regulator PqsR becomes an attractive object and is considered to be one of the most appropriate targets. Currently, QS regulation mechanism in P. aeruginosa is mainly related to positive and negative regulation between QS systems. Clearly, exploration of regulation beyond QS should get attention in future.
Data Availability
All datasets generated or analyzed for this study are included in the manuscript and the Supplementary Files.
Author Contributions
GW designed the work. Both authors prepared and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was partly supported by the National Natural Science Foundation of China (Nos. 31460296 and 31560315) and the Key Project of Guangxi Scientific Research and Technology Development Plan (AB17190534).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01582/full#supplementary-material
References
- Aghazadeh M., Hojabri Z., Mahdian R., Nahaei M. R., Rahmati M., Hojabri T., et al. (2014). Role of efflux pumps: MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients. Infect. Genet. Evol. 24 187–192. 10.1016/j.meegid.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Allesen-Holm M., Barken K. B., Yang L., Klausen M., Webb J. S., Kjelleberg S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59 1114–1128. 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- Almblad H., Rybtke M., Hendiani S., Andersen J. B., Givskov M., Tolker-Nielsen T. (2019). High levels of cAMP inhibit Pseudomonas aeruginosa biofilm formation through reduction of the c-di-GMP content. Microbiology 165 324–333. 10.1099/mic.0.000772 [DOI] [PubMed] [Google Scholar]
- Barraud N., Hassett D. J., Hwang S. H., Rice S. A., Kjelleberg S., Webb J. S. (2006). Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188 7344–7353. 10.1128/jb.00779-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41 D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbruch F., Geffers R., Nimtz M., Buer J., Haussler S. (2006). The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 8 81318–81329. [DOI] [PubMed] [Google Scholar]
- Brint J. M., Ohman D. E. (1995). Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177 7155–7163. 10.1128/jb.177.24.7155-7163.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl M., Peter S., Willmann M. (2015). Prevalence and risk factors associated with colonization and infection of extensively drug-resistant Pseudomonas aeruginosa: a systematic review. Expert. Rev. Anti. Infect. Ther. 13 1159–1170. 10.1586/14787210.2015.1064310 [DOI] [PubMed] [Google Scholar]
- Calvert M. B., Jumde V. R., Titz A. (2018). Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J. Org. Chem. 14 2607–2617. 10.3762/bjoc.14.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A., Bassler B. L. (2006). Bacterial small-molecule signaling pathways. Science 311 1113–1116. 10.1126/science.1121357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Krishnan G., Goumnerov B., Tsongalis J., Tompkins R., Rahme L. G. (2001). A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U.S.A. 98 14613–14618. 10.1073/pnas.251465298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. G., Liu Y. C., Chang C. Y. (2015). Inhibiting N-acyl-homoserine lactone synthesis and quenching Pseudomonas quinolone quorum sensing to attenuate virulence. Front. Microbiol. 6:1173. 10.3389/fmicb.2015.01173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua S. L., Hultqvist L. D., Yuan M., Rybtke M., Nielsen T. E., Givskov M., et al. (2015). In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat. Protoc. 10 1165–1180. 10.1038/nprot.2015.067 [DOI] [PubMed] [Google Scholar]
- Chua S. L., Liu Y., Yam J. K., Chen Y., Vejborg R. M., Tan B. G., et al. (2014). Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5:4462. 10.1038/ncomms5462 [DOI] [PubMed] [Google Scholar]
- Church D., Elsayed S., Reid O., Winston B., Lindsay R. (2006). Burn wound infections. Clin. Microbiol. Rev. 19 403–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T., Manefield M. (2013). Phenazine production enhances extracellular DNA release via hydrogen peroxide generation in Pseudomonas aeruginosa. Commun. Integr. Biol. 6:e23570. 10.4161/cib.23570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E., O’Toole G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64 847–867. 10.1128/mmbr.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. G., Marques C. N. (2009). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191 1393–1403. 10.1128/JB.01214-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280 295–298. 10.1126/science.280.5361.295 [DOI] [PubMed] [Google Scholar]
- Deziel E., Gopalan S., Tampakaki A. P., Lepine F., Padfield K. E., Saucier M., et al. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol. Microbiol. 55 998–1014. 10.1111/j.1365-2958.2004.04448.x [DOI] [PubMed] [Google Scholar]
- Diggle S. P., Winzer K., Chhabra S. R., Worrall K. E., Camara M., Williams P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50 29–43. 10.1046/j.1365-2958.2003.03672.x [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elborn J. S. (2016). Cystic fibrosis. Lancet 388 2519–2531. 10.1016/S0140-6736(16)00576-6 [DOI] [PubMed] [Google Scholar]
- Eren E., Parkin J., Adelanwa A., Cheneke B., Movileanu L., Khalid S., et al. (2013). Toward understanding the outer membrane uptake of small molecules by Pseudomonas aeruginosa. J. Biol. Chem. 288 12042–12053. 10.1074/jbc.M113.463570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. M., III, Sund Z. M., Ellison M. L., Wade D. S., Coleman J. P., Pesci E. C. (2008). PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 190 7043–7051. 10.1128/JB.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M., Almblad H., Rybtke M. L., Givskov M., Eberl L., Tolker-Nielsen T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16 1961–1981. 10.1111/1462-2920.12448 [DOI] [PubMed] [Google Scholar]
- Ferris R. A., McCue P. M., Borlee G. I., Glapa K. E., Martin K. H., Mangalea M. R., et al. (2017). Model of chronic equine endometritis involving a Pseudomonas aeruginosa biofilm. Infect. Immun. 85:e00332-17. 10.1128/IAI.00332-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. F., Mobashery S. (2014). The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg. Chem. 56 41–48. 10.1016/j.bioorg.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176 269–275. 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., McKnight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184 6472–6480. 10.1128/jb.184.23.6472-6480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello M. J., Iglewski B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173 3000–3009. 10.1128/jb.173.9.3000-3009.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello M. J., Kaye S., Iglewski B. H. (1993). LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61 1180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo E., An J. H., Kang Y., Hwang I. (2015). Control of bacterial metabolism by quorum sensing. Trends Microbiol. 23 567–576. 10.1016/j.tim.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Guerra-Santos L., Käppeli O., Fiechter A. (1986). Dependence of Pseudomonas aeruginosa continuous culture biosurfactant production on nutritional and environmental factors. Appl. Microbiol. Biotechnol. 24 443–448. [Google Scholar]
- Hakemi Vala M., Hallajzadeh M., Hashemi A., Goudarzi H., Tarhani M., Sattarzadeh Tabrizi M., et al. (2014). Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann. Burns Fire Disasters 27 8–13. [PMC free article] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Howlin R. P., Cathie K., Hall-Stoodley L., Cornelius V., Duignan C., Allan R. N., et al. (2017). Low-dose nitric oxide as targeted anti-biofilm adjunctive therapy to treat chronic Pseudomonas aeruginosa infection in cystic fibrosis. Mol. Ther. 25 2104–2116. 10.1016/j.ymthe.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Malone J. (2006). Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40 385–407. 10.1146/annurev.genet.40.110405.090423 [DOI] [PubMed] [Google Scholar]
- Jensen V., Lons D., Zaoui C., Bredenbruch F., Meissner A., Dieterich G., et al. (2006). RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 188 8601–8606. 10.1128/jb.01378-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Kuroki M., Ishii M., Igarashi Y., Arai H. (2010). Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12 1399–1412. 10.1111/j.1462-2920.2009.02109.x [DOI] [PubMed] [Google Scholar]
- Klare W., Das T., Ibugo A., Buckle E., Manefield M., Manos J. (2016). Glutathione-disrupted biofilms of clinical Pseudomonas aeruginosa strains exhibit an enhanced antibiotic effect and a novel biofilm transcriptome. Antimicrob. Agents Chemother. 60 4539–4551. 10.1128/AAC.02919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J., Tümmler B. (2017). Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Res. 6:1261. 10.12688/f1000research.10506.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni H. M., Jagannadham M. V. (2014). Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 160 2109–2121. 10.1099/mic.0.079400-0 [DOI] [PubMed] [Google Scholar]
- LaSarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77 73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Foglino M., Tanaka K., Williams P., Lazdunski A. (1996). A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21 1137–1146. 10.1046/j.1365-2958.1996.00063.x [DOI] [PubMed] [Google Scholar]
- Lee J., Zhang L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6 26–41. 10.1007/s13238-014-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T. F., Pitts B., Pellock B., Walker G. C., Stewart P. S., O’Toole G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426 306–310. 10.1038/nature02122 [DOI] [PubMed] [Google Scholar]
- Manos J., Arthur J., Rose B., Bell S., Tingpej P., Hu H., et al. (2009). Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol. Lett. 292 107–114. 10.1111/j.1574-6968.2008.01472.x [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L., Howe J., Garidel P., Richter W., Steiniger F., Roessle M., et al. (2008). Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69 491–502. 10.1111/j.1365-2958.2008.06302.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D. V., Bonsall R. F., Delaney S. M., Soule M. J., Phillips G., Thomashow L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183 6454–6465. 10.1128/jb.183.21.6454-6465.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath S., Wade D. S., Pesci E. C. (2004). Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230 27–34. 10.1016/s0378-1097(03)00849-8 [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Iglewski B. H., Pesci E. C. (2000). The Pseudomonas quinolone signal regulates rhl quorum-sensing in Pseudomonas aeruginosa. J. Bacteriol. 182 2702–2708. 10.1128/jb.182.10.2702-2708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighen E. A. (1991). Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55 123–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe D. (2007). Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 5:e307. 10.1371/journal.pbio.0050307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Fiechter A., Reiser J. (1994a). Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269 19787–19795. [PubMed] [Google Scholar]
- Ochsner U. A., Koch A. K., Fiechter A., Reiser J. (1994b). Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176 2044–2054. 10.1128/jb.176.7.2044-2054.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Reiser J. (1995). Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92 6424–6428. 10.1073/pnas.92.14.6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby A. G., Farrow J. M. I. I. I., Lee J. H., Tomaras A. P., Greenberg E. P., Pesci E. C., et al. (2008). The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283 28315558–28315567. 10.1074/jbc.M707840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Mulet X., López-Causapé C., Juan C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Update 2 41–59. 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Passador L., Cook J. M., Gambello M. J., Rust L., Iglewski B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260 1127–1130. 10.1126/science.8493556 [DOI] [PubMed] [Google Scholar]
- Pearson J. P., Gray K. M., Passador L., Tucker K. D., Eberhard A., Iglewski B. H., et al. (1994). Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A. 91 197–201. 10.1073/pnas.91.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Passador L., Iglewski B. H., Greenberg E. P. (1995). A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92 1490–1494. 10.1073/pnas.92.5.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Pesci E. C., Iglewski B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179 5756–5767. 10.1128/jb.179.18.5756-5767.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Milbank J. B., Pearson J. P., McKnight S., Kende A. S., Greenberg E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96 11229–11234. 10.1073/pnas.96.20.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Pearson J. P., Seed P. C., Iglewski B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179 3127–3132. 10.1128/jb.179.10.3127-3132.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Haas D. (2000). Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182 6940–6949. 10.1128/jb.182.24.6940-6949.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10 12–26. 10.1111/j.1469-0691.2004.00763.x [DOI] [PubMed] [Google Scholar]
- Rahme L. G., Ausubel F. M., Cao H., Drenkard E., Goumnerov B. C., Lau G. W., et al. (2000). Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. U.S.A. 97 8815–8821. 10.1073/pnas.97.16.8815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L. G., Tan M. W., Le L., Wong S. M., Tompkins R. G., Calderwood S. B., et al. (1997). Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 94 13245–13250. 10.1073/pnas.94.24.13245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey K., Michalek S. M., Wen Z. T., Wu H. (2019). Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 85:e02247-18. 10.1128/AEM.02247-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasamiravaka T., El Jaziri M. (2016). Quorum-sensing mechanisms and bacterial response to antibiotics in P. aeruginosa. Curr. Microbiol. 73 747–753. 10.1007/s00284-016-1101-1 [DOI] [PubMed] [Google Scholar]
- Rasamiravaka T., Labtani Q., Duez P., El Jaziri M. (2015). The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015:759348. 10.1155/2015/759348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer J. W., Boulette M. L., Whiteley M. (2009). More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol. 17 189–195. 10.1016/j.tim.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Schütz C., Empting M. (2018). Targeting the Pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. Beilstein J. Org. Chem. 14 2627–2645. 10.3762/bjoc.14.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed P. C., Passador L., Iglewski B. H. (1995). Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177 654–659. 10.1128/jb.177.3.654-659.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soheili V., Tajani A. S., Ghodsi R., Bazzaz B. S. F. (2019). Anti-PqsR compounds as next-generation antibacterial agents against Pseudomonas aeruginosa: a review. Eur. J. Med. Chem. 172 26–35. 10.1016/j.ejmech.2019.03.049 [DOI] [PubMed] [Google Scholar]
- Spoering A. L., Lewis K. (2001). Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183 6746–6751. 10.1128/jb.183.23.6746-6751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P. S., Costerton J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358 135–138. 10.1016/s0140-6736(01)05321-1 [DOI] [PubMed] [Google Scholar]
- Stoodley P., Debeer D., Lewandowski Z. (1994). Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60 2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitzel G. M., Geddie A., De Long S. K., Kirisits M. J., Whiteley M., Parsek M. R. (2006). Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188 7242–7256. 10.1128/jb.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toder D. S., Gambello M. J., Iglewski B. H. (1991). Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol. 5 2003–2010. 10.1111/j.1365-2958.1991.tb00822.x [DOI] [PubMed] [Google Scholar]
- Tralau T., Vuilleumier S., Thibault C., Campbell B. J., Hart C. A., Kertesz M. A. (2007). Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J. Bacteriol. 189 6743–6750. 10.1128/jb.00889-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatcheva-Dobrevska R., Mulet X., Ivanov I., Zamorano L., Dobreva E., Velinov T., et al. (2013). Molecular epidemiology and multidrug resistance mechanisms of Pseudomonas aeruginosa isolates from Bulgarian hospitals. Microb. Drug Resist. 19 355–361. 10.1089/mdr.2013.0004 [DOI] [PubMed] [Google Scholar]
- Vingopoulou E. I., Delis G. A., Batzias G. C., Kaltsogianni F., Koutinas A., Kristo I., et al. (2018). Prevalence and mechanisms of resistance to fluoroquinolones in Pseudomonas aeruginosa and Escherichia coli isolates recovered from dogs suffering from otitis in Greece. Vet. Microbiol. 213 102–107. 10.1016/j.vetmic.2017.11.024 [DOI] [PubMed] [Google Scholar]
- Wade D. S., Calfee M. W., Rocha E. R., Ling E. A., Engstrom E., Coleman J. P., et al. (2005). Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187 4372–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V. E., Bushnell D., Passador L., Brooks A. I., Iglewski B. H. (2003). Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185 2080–2095. 10.1128/jb.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. S., Bais H. P., Déziel E., Schweizer H. P., Rahme L. G., Fall R., et al. (2004). Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134 320–331. 10.1104/pp.103.027888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Lee K. M., Greenberg E. P. (1999). Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox M. D. (2007). Pseudomonas aeruginosa infection and inflammation during contact lens wear: a review. Optom. Vis. Sci. 84 273–278. 10.1097/opx.0b013e3180439c3e [DOI] [PubMed] [Google Scholar]
- Williams P., Camara M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12 182–191. 10.1016/j.mib.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Wolska K. I., Grudniak A. M., Rudnicka Z., Markowska K. (2016). Genetic control of bacterial biofilms. J. Appl. Genet. 57 225–238. 10.1007/s13353-015-0309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Gao J., Tang W. (2016). Nosocomial infection and its molecular mechanisms of antibiotic resistance. Biosci. Trends 10 14–21. 10.5582/bst.2016.01020 [DOI] [PubMed] [Google Scholar]
- Xiao G., Deziel E., He J., Lepine F., Lesic B., Castonguay M. H., et al. (2006a). MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 62 1689–1699. 10.1111/j.1365-2958.2006.05462.x [DOI] [PubMed] [Google Scholar]
- Xiao G., He J., Rahme L. G. (2006b). Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152 1679–1686. 10.1099/mic.0.28605-0 [DOI] [PubMed] [Google Scholar]
- Zgurskaya H. I., Löpez C. A., Gnanakaran S. (2015). Permeability barrier of gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1 512–522. 10.1021/acsinfecdis.5b00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated or analyzed for this study are included in the manuscript and the Supplementary Files.