Abstract
In matrilineal populations, the descent group affiliation is transmitted by women whereas the socio-political power frequently remains in the hands of men. This situation, named the ‘matrilineal puzzle’, is expected to promote local endogamy as a coping mechanism allowing men to maintain their decision-making power over their natal descent group. In this paper, we revisit this ‘matrilineal puzzle’ from a population genetics' point of view. Indeed, such tendency for local endogamy in matrilineal populations is expected to increase their genetic inbreeding and generate isolation-by-distance patterns between villages. To test this hypothesis, we collected ethno-demographic data for 3261 couples and high-density genetic data for 675 individuals from 11 Southeast Asian populations with a wide range of social organizations: matrilineal and matrilocal populations (M), patrilineal and patrilocal populations (P) or cognatic populations with predominant matrilocal residence (C). We observed that M and C populations have higher levels of village endogamy than P populations, and that such higher village endogamy leads to higher genetic inbreeding. M populations also exhibit isolation-by-distance patterns between villages. We interpret such genetic patterns as the signature of the ‘matrilineal puzzle’. Notably, our results suggest that any form of matrilocal marriage (whatever the descent rule is) increases village endogamy. These findings suggest that male dominance, when combined with matrilocality, constrains inter-village migrations, and constitutes an underexplored cultural process shaping genetic patterns in human populations.
This article is part of the theme issue ‘The evolution of female-biased kinship in humans and other mammals’.
Keywords: residence rule, matrilineal puzzle, inbreeding, human genetics, matrilocal, patrilocal
1. Introduction
In matrilineal populations (which represent about 12–17% of the world's populations), descent group affiliation is transmitted through the mother, whereas in patrilineal populations (about 45% of the populations), it is transmitted through the father [1,2]. These descent systems are not the symmetrical opposite of each other because, in both cases, authority and socio-political power (beyond the household) lie in the hands of men [3–6]. Indeed, while women play key roles within the domestic unit in terms of provisioning, childrearing and household organization, men are usually more empowered than women to control public sphere affairs [4,5,7,8]. Hence, it has been argued that matrilineal societies are caught in what has been named by A. Richards as a ‘matrilineal puzzle’ [5]. Indeed, in these populations, a man has to conciliate his loyalties to his conjugal and to his natal kin [8]: he is expected to exert, at the same time, his authority of husband and father over his spouse and children, and his authority of brother and uncle over his sisters and their children, who are members of his own clan and to whom material and immaterial forms of inheritance are transmitted. In addition, by following the matrilocal residence rule that exists in 70% of these matrilineal populations [2], men are supposed to move out to settle with their wife, possibly in a different village. The further away they marry, the more challenging it is for them to exert their authority over their sisters and their children. In addition, in these populations, the husband has to share authority with his brothers-in-law and other men in charge of his wife's lineage or clan, thus generating the problem of organizing relationships between the in-marrying husband and the male members of his wife's descent group [9].
The matrilineal puzzle has been discussed for over 60 years by different schools of anthropologists. Recent works by evolutionary anthropologists have outlined the evolutionary paradoxes of these matrilineal systems [10–12]. Indeed, in these societies, frequently men invest more in their sisters' children than in their own children, which violates the expectation of Hamilton's rule [13]. In addition, according to the Trivers–Willard hypothesis, it would be more beneficial for parents to transmit wealth to the sex that is most capable of converting it into large reproductive success, typically males [14]. The relative benefit of transmission to males over females depends on the nature of heritable wealth. For instance, livestocks and productive lands are both usually considered more beneficial to men than women because of their greater impact on male's capacity to acquire partners and to increase their reproductive success [10,12]. More recently, the matriliny as daughter-biased investment (MDBI) hypothesis proposes that daughter-biased investment could be an adaptive strategy if the risk of paternity uncertainty (usually high in matrilineal societies) outweighs the benefits of wealth transmission to sons [15]. On the other hand, the expendable male hypothesis suggests that the matrilineal puzzle may not be a puzzle in the evolutionary sense at all, and proposes that matriliny may emerge if females are capable of meeting the subsistence needs of their families while males invest little in children (their own, or their sisters'), this latter condition reconciling these systems with Hamilton's rule [7].
Here, we propose to come back to the original sense given to the matrilineal puzzle by A. Richards and other structural-functionalists [5,8], who were referring to the conflict in authority, and in particular to the constant ‘pull-father-pull-mother's brother’ stretch existing in these matrilineal populations. Interestingly, through the study of many matrilineal populations, these anthropologists have described several ‘solutions’ which may appease such tensions: (i) a handful of these matrilineal populations do not follow the matrilocal residence rule but follow a duolocal residence rule—the husband does not live with his wife but visits her regularly while staying with his sisters [8,16]; (ii) more often, the residence rule is avunculocal with, for example, fraternal extended families exerting full authority over the community, while men's sisters are loaned away to other communities and their children are reclaimed at puberty [5,17]; (iii) in the case of the matrilineal populations exhibiting a matrilocal residence rule, the eldest brother may be exempted from such a rule, thus exerting his authority over his sisters and their children [5,17]; (iv) in addition, matrilateral cross-cousin marriages are frequent in these populations, contributing to strengthen the authority of men who have contracted matrilocal marriages—by marrying their daughters to their sister's sons, they bring in their spouse's village, their nephews as sons-in-law, who come from their own descent group and natal village [5,18]; (v) finally, a very frequent ‘solution’ to the matrilineal puzzle is the strong preference for local endogamy observed in these populations—according to Murdock [19], 17 out of 24 matrilocal populations (70%) were found to be endogamous (as opposed to only 7 out of 101 patrilocal populations). This preference for marrying a woman from the same village, or from a nearby village, may allow men to stay close to the members of their maternal descent group and to exert their authority as brothers/uncles over them, as well as control their descent-group affairs.
In this study, we propose to explore the potential impact of the matrilineal puzzle on the genetic evolution of these populations. A number of studies in the past 20 years have shown that social organizations shape the uniparental genetic diversities of human populations [20–29]. Fewer studies have explored the evolutionary implications of descent and residence rule on autosomal data [30–32]. Here, we propose to focus on the matrilineal puzzle, whose impact on human genetic diversity has been left untouched by population geneticists. Our working hypothesis is that the preference for local endogamy observed in matrilineal populations should increase the genetic inbreeding level in these matrilineal populations, in comparison to populations where such preference does not exist, in particular populations with patrilineal descent. Indeed, when local endogamy increases, we expect not only the proportion of consanguineous marriages to be higher (owing to the small size of the matrimonial market and its enrichment in relatives), but also the genetic drift to increase, both leading to higher genetic inbreeding [33]. In addition, we expect such preference for marrying within the same village, or in a nearby village, in matrilineal populations to generate isolation-by-distance patterns between villages [34]. Such isolation-by-distance patterns are less expected in patrilineal populations, because there is weaker pressure for local endogamy, leading to long distance gene flow.
To test such a hypothesis, we collected ethno-demographic data for 3261 couples as well as high density autosomal single nucleotide polymorphism (SNP) data for 675 individuals from 11 mainland Southeast Asian populations exhibiting a wide variety of social organizations, with different descent and residence rules, but living in similar tropical environments and having similar lifestyles based on rice farming. More precisely, we compared three populations (M) with matrilineal descent and matrilocal residence (Jarai, Tampuan and Kacho'), to four populations (P) with patrilineal descent and patrilocal residence (Khmu', Lamet, Ta-oih and Pacoh). This dataset has been completed by four populations (C) with cognatic descent and either matrilocal residence (Khmer and Bunong) or multilocal residence with final settlement in the wife's village (Brao and Kreung). We grouped these four cognatic populations into a single group of cognatic populations with predominant matrilocal residence. These populations were included to disentangle the effect of descent from the effect of residence on migrations of men. In particular, we investigated whether matrilocality by itself could generate a similar level of constraint on male migration as when it is associated with matrilineal descent. Indeed, it may be that under any form of matrilocal marriage, and independently of matrilineality, men find themselves, at least initially, in a position of subjection in their wife's village (while possibly losing a position of leadership in their village of origin) [5], a situation that can be lessened by marrying a woman from the same village [6]. Consequently, in populations following a matrilocal residence rule but with no matrilineal descent group (i.e. cognatic populations), we could expect a similar preference for endogamous marriages as in matrilineal populations. The populations under study are presented in table 1 (their geographical location is shown in electronic supplementary material, figure S1).
Table 1.
Description of the studied populations with sampling information.
| population | descent rule | residence rule | abbreviation (group) | sampled villages | DNA samples after QC | DNA samples after removing siblings | ethno-demographic interviews | effective population size (95% IC) |
|---|---|---|---|---|---|---|---|---|
| Tampuan | matrilineala | matrilocald | M | 8 | 101 | 96 | 65 | 13 052 (12 040–14 064) |
| Jarai | matrilineal | matrilocal | 6 | 92 | 89 | 56 | 13 641 (12 592–14 690) | |
| Kacho' | matrilineal | matrilocal | 3 | 32 | 30 | 27 | 13 407 (12 301–14 512) | |
| Bunong | cognaticb | matrilocal | C | 5 | 88 | 84 | 45 | 11 850 (10 895–12 804) |
| Khmer | cognatic | matrilocal | 5 | 92 | 92 | 44 | 15 503 (14 376–16 630) | |
| Brao | cognatic | multilocale | 6 | 49 | 49 | 39 | 12 023 (11 052–12 995) | |
| Kreung | cognatic | multilocal | 3 | 51 | 50 | 36 | 10 920 (9939–11 901) | |
| Khmu' | patrilinealc | patrilocalf | P | 8 | 72 | 68 | 65 | 13 868 (12 798–14 938) |
| Ramet | patrilineal | patrilocal | 4 | 26 | 26 | 40 | 15 051 (13 855–16 248) | |
| Ta-oih | patrilineal | patrilocal | 4 | 28 | 28 | 34 | 14 050 (12 922–15 179) | |
| Pacoh | patrilineal | patrilocal | 5 | 64 | 63 | 44 | 12 141 (11 140–13 143) | |
| total | 57 | 695 | 675 | 495 |
aMatrilineal descent: descent group affiliation is transmitted to the children through the mother.
bCognatic descent: recognition of descent from both sides of the family in the absence of any specified lines of descent.
cPatrilineal descent: descent group affiliation is transmitted to the children through the father.
dMatrilocal residence: the husband moves to his wife's natal village after marriage.
eMultilocal residence: the couple lives alternatively in the husband's and wife's natal villages before settling definitively in one place, which is most often, in the case of Brao and Kreung, the wife's village.
fPatrilocal residence: the wife moves to her husband's natal village after marriage.
2. Results
(a). Estimation of village endogamy
We estimated the village endogamy rate (as a proxy for local endogamy) for each population from ethno-demographic information collected by the research team for 3261 couples (figure 1). The village endogamy rate was defined as the proportion of couples for which both spouses were born in the same village. These rates were compared among social organizations using a generalized linear mixed model. As expected under the matrilineal puzzle hypothesis, village endogamy was significantly higher in M than P populations (0.87 versus 0.73 respectively, p-value = 0.029). In C populations, village endogamy was similar to M populations (0.84, p-value = 0.62) and higher than in P populations, although the difference was not statistically significant (p-value = 0.066). This suggests that the matrilocal residence rule alone (with no matrilineal descent groups but cognatic descent) may generate a similar level of constraints on migrations of men as when this residence rule is associated with matrilineal descent. In addition, social organization was estimated to explain 43% of the variance in village endogamy rate among populations.
Figure 1.
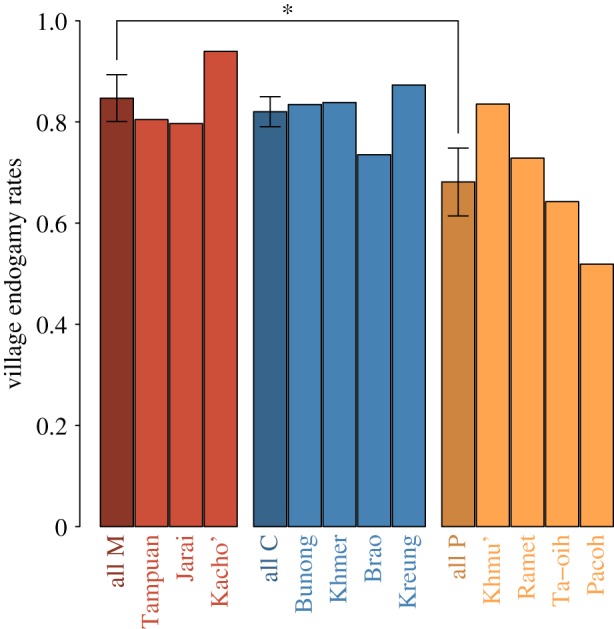
Mean village endogamy rate for each population. The first dark bar in each group represents the mean village endogamy rate in this group (with standard error). Asterisk indicates statistical significance (p-value < 0.05) assessed by generalized linear mixed model. (Online version in colour.)
We observed variation in the village endogamy rate within groups (electronic supplementary material, table S1). In particular, the Kacho' population had a significantly higher village endogamy rate compared to the other M populations (Tampuan and Jarai). Among the P populations, the Khmu' had a significantly higher village endogamy rate than the Pacoh and Ta-oih. The Pacoh had significantly lower village endogamy rate than the Khmu' and Ramet.
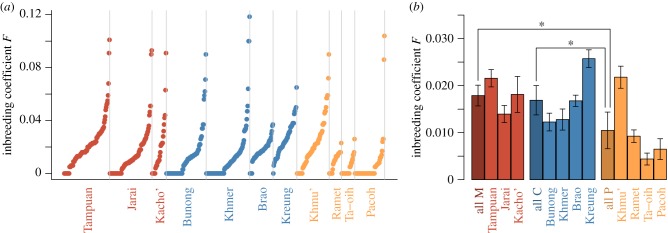
(b). Estimation of inbreeding coefficients
We tested whether M and C populations exhibited higher inbreeding levels in comparison to P populations as a result of their higher village endogamy rate. The FEstim software in the FSuite pipeline [35,36] was used to estimate the inbreeding coefficient of each individual (figure 2a) and to infer the genealogical relationship between the parents (parental mating type) of each individual (electronic supplementary material, table S2). M and C populations had similar mean inbreeding coefficients (0.018 and 0.017 respectively, p-value = 0.91, figure 2b), and both had higher mean inbreeding coefficient compared to P populations (0.011, both p-values < 0.05). In addition, social organization was estimated to explain 27% of the variance in inbreeding coefficients among populations.
Figure 2.
Genetic inbreeding coefficients. (a) Each point represents the inbreeding coefficient of an individual. Coefficients are sorted in ascending order in each population. (b) The first dark bar in each group represents the mean inbreeding level in this group. All values are represented with standard error. Asterisks indicate statistical significance (p-value < 0.05) assessed by linear mixed model. (Online version in colour.)
Mating type inference showed that M populations had a higher proportion of individuals whose parents were related (90.2%: 77.7% of second cousins, 12.1% of first cousins, and 0.5% of double first cousins) compared to C populations (81.8%: 70.6%, 9.8% and 1.5% for the same mating types, χ2 test p-value = 0.012) and compared to P populations (60.0%: 61.6% of second cousins, 7.6% of first cousins and 0.5% of avuncular relationship, p-value < 0.01, electronic supplementary material, table S2). In addition, C populations also had a higher proportion of individuals whose parents were related compared to P populations (p-value < 0.01).
We observed variation within groups in terms of inbreeding coefficient (figure 2 and electronic supplementary material, table S3). In particular, the Kreung population had a significantly higher inbreeding level than other C populations (F = 0.026 compared to 0.014 on average for the other C populations). This may relate to the fact that their effective size is lower (only significantly so compared to the Khmer) than the effective population size estimated for the other cognatic populations (see table 1 and electronic supplementary material , tables S4 and S5). In addition, the Khmu' population had a significantly higher inbreeding coefficient than other P populations (F = 0.022 compared to 0.0067 on average for the other P populations). Contrary to the case of the Kreung, this does not seem to be linked to differences in effective population size among P populations.
To further investigate the influence of village endogamy on inbreeding level and confirm that village endogamy is the social component that explains the differences in inbreeding level between social organizations, we measured the correlation coefficient between these two parameters at the population level (figure 3). The village endogamy rate was indeed significantly correlated with the inbreeding coefficient (Spearman's ρ = 0.73, p-value = 0.015).
Figure 3.
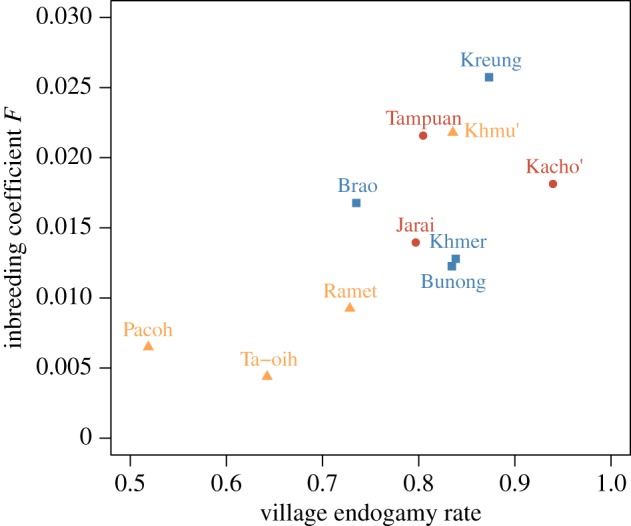
Relationship between village endogamy rate and mean genetic inbreeding coefficient by population. M populations are in red (circles), C populations in blue (squares) and P populations in yellow (triangles). (Online version in colour.)
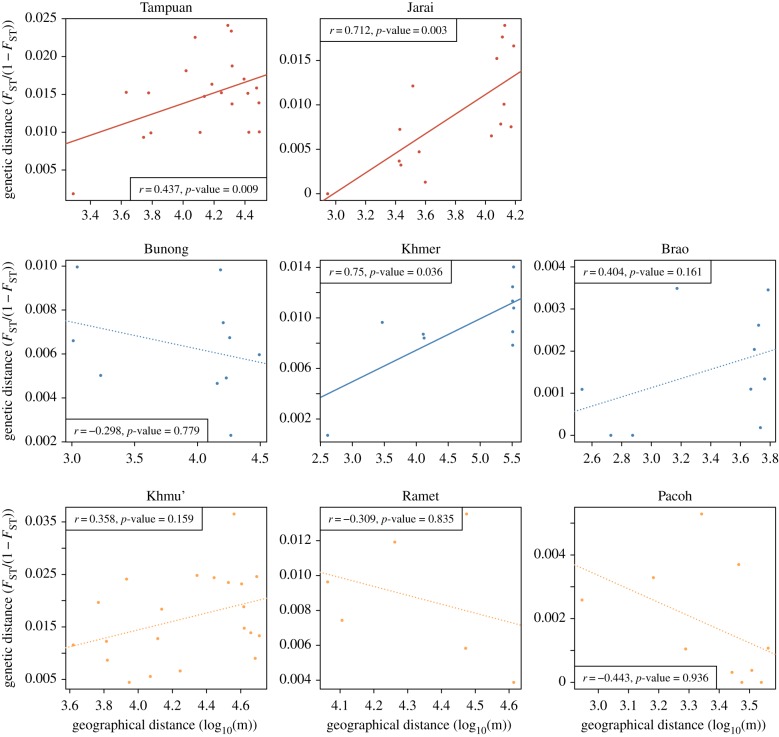
(c). Isolation-by-distance patterns
Finally, we explored the patterns of isolation-by-distance at the village level within each population (figure 4). We performed this analysis in populations with at least four sampled villages (after excluding villages with less than five sampled individuals). This filtering step excluded the Kacho', Kreung and Ta-oih from this analysis. We observed significant isolation-by-distance patterns in the two M populations included in this analysis (Tampuan and Jarai, both p-values < 0.05). Among the C populations, such isolation-by-distance pattern was found in the Khmer (p-value = 0.034) but not in the Bunong or Brao (both p-values > 0.05). P populations did not exhibit any significant isolation-by-distance pattern (all p-values > 0.05).
Figure 4.
Relationship between genetic distance and geographical distance between villages in each population. M populations are in red (top row), C populations in blue (middle row) and P populations in yellow (bottom row). The Pearson's correlation coefficient and the p-value estimated by Mantel test for each population are given. Continuous lines represent regression lines associated with a significant Mantel test (p-value < 0.05). Dotted lines represent regression lines associated with a non-significant Mantel test. (Online version in colour.)
3. Discussion
In this study, we observed that Southeast Asian matrilineal and matrilocal populations (M), but also cognatic populations with predominant matrilocal residence (C), have higher levels of genetic inbreeding than patrilineal and patrilocal populations (P). In addition, M populations exhibit isolation-by-distance patterns between villages. We hypothesize that such findings are the signature of the higher local endogamy resulting from what has been called the ‘matrilineal puzzle’, which takes root in the male dominance over socio-political power [5]: in matrilineal and matrilocal societies men are supposed to settle with their wife's family, possibly in a different village, while remaining actively involved in decision-making within their own descent groups. This becomes challenging as the geographical distance between the natal villages of the wife and the husband increases. Consequently, a preference for local marriages has been observed in these societies, as summarized by Murdock: ‘where residence is matrilocal, a man in marrying rarely settles in a new community. He merely takes his possessions from his parents' home and moves, so to speak, across the street’ [19, p. 214].
Several lines of evidence support the hypothesis that the matrilineal puzzle is responsible for the different genetic diversity patterns observed in the studied populations. First, we confirmed from our ethno-demographic dataset that M populations had higher village endogamy rates than P populations (on average 87% and 73% respectively). In addition, C populations had similar village endogamy (84%) to M populations. Previous ethnographic works on the matrilineal populations under study also confirmed the existence of a preference for local endogamy. Indeed, the Jarai marry according to a ‘the closest, the safest’ rule [18] and in the Tampuan population [37], the councils of elders are reluctant to integrate into their villages a man coming from a distant village, a preference still prevailing these days, that may contribute to increase the rate of village endogamy.
Secondly, the village endogamy rate was shown to be a good predictor of the genetic inbreeding levels in these populations. However, the preference for cousin marriages in these populations as reported by the ethnographic literature appears to be a poor predictor of their estimated genetic inbreeding levels: preferences for cousin marriages were reported for most M and P populations [18,37–40] but not for the C populations [38,41–43]. More generally, the percentages of populations with a preference for cousin marriages estimated in a worldwide population sample are higher for both matrilineal and patrilineal populations than for cognatic populations (42.3%, 40% and 19.7% respectively, [2]). These percentages do not fit with our observation that M and C populations have higher genetic inbreeding levels than P populations. Last but not least, a detailed ethnographic study in the Jarai population had shown that the preference for village endogamy was stronger than the preference for cousin marriages: the number of marriages within the same village exceeded the total number of preferential marriages, in particular between ego and mother's brother's daughter, and between ego and father's sister's daughter [18]. Consequently, the matrilineal puzzle, and its consequences in terms of endogamy, is a more likely candidate than the prevalence of cousin marriages to explain the observed differences in genetic patterns between M, C and P populations.
Note also that, despite the fact that M populations are famous for their high paternity uncertainty rate ([12] and references therein), we do not think this process could contribute to the observed genetic differences between M, C and P populations. Indeed, we would expect such paternity uncertainty to decrease, rather than increase, the genetic inbreeding level in these populations in comparison to patrilineal populations; for example, a child born from first cousins may have lower genetic inbreeding coefficient than expected because his parents may share the same grandmother but different grandfathers.
In patrilineal and patrilocal populations, the matrilineal puzzle does not occur as most men settle with their wife in their natal village whether they marry a woman from the same or from another village, with no risk of losing their position of influence or leadership, or their control over their descent-group affairs. This leads to comparatively lower local endogamy rates and lower inbreeding levels in these patrilineal populations, as well as an absence of isolation-by-distance patterns. In our dataset, we noticed one exception to this general observation: the Khmu', a patrilineal and patrilocal population, exhibits a matrilineal-like rate of village endogamy and genetic inbreeding level. The reasons for these differences from the other P populations remain to be investigated. One explanation could be that, although Khmu' follow a general patrilineal descent and patrilocal residence, there is in some families a period of matrilocal residence (up to three years) [40]. Despite the fact that this matrilocal residence is not permanent, this may generate a ‘nascent’ matrilineal puzzle, encouraging men to marry a woman from the same village.
As discussed in the introduction, local endogamy is probably not the only coping mechanism to the matrilineal puzzle; for example, one of the brothers could escape from the matrilocal rule, allowing him to stay in his natal village (with his wife coming from the same or from a different village) and deal with his descent-group matters [5]. However, our ethno-demographic data do not support such an alternative coping mechanism to the matrilineal puzzle in the Southeast Asian populations under study, since the matrilineal and matrilocal populations followed their residence rule more strictly than the patrilineal and patrilocal populations under study [26].
Lastly, the design of this study, which includes cognatic populations with predominant matrilocal residence, allows us to disentangle the effect of descent from the effect of residence on the ‘matrilineal puzzle’. Indeed, as pointed out above, the cognatic populations (C) under study exhibit a similar rate of village endogamy and genetic inbreeding compared to the matrilineal populations (M). Altogether, these populations with matrilocal or predominant matrilocal residence exhibit higher village endogamy and higher genetic inbreeding than patrilocal populations (both p-values < 0.05, estimated by linear mixed model). Consequently, the matrilineal descent rule is probably not the main component exerting a constraint over male migrations. As pointed out by A. Richards, under any form of matrilocal marriage (whatever the descent rule is), men find themselves, at least initially, in a position of subjection in their wives' villages (while possibly losing a position of leadership in their natal village), an ‘irksome’ situation that can be avoided by marrying a woman from the same village [5]. As such, the ‘matrilineal puzzle’ could be renamed the ‘matrilocal puzzle’ in order to express the fact that it seems to affect not only matrilineal populations but also all matrilocal populations.
Our interdisciplinary study has a number of limitations. We could expect populations facing the matrilineal puzzle to exhibit not only a higher rate of village endogamy but also smaller distances between villages when couples are exogamous. However, our ethno-demographic dataset did not allow us to measure the geographical distance between spouses' natal villages. Consequently, we used the rate of village endogamy as a proxy for local endogamy in this study. In addition, our ethno-demographic dataset may suffer from certain sampling biases. For example, only individuals having their four grandparents from the same population were sampled, a criterion often used in population genetic studies, that may have biased our estimation of village endogamy (however, only slightly, as the proportion of interethnic marriages in these populations is low). There may be some other biases in such endogamy estimation; for example, the information regarding birth places as remembered by the interviewees for some of their relatives, especially their grandparents, may be erroneous, potentially biasing our estimation of village endogamy upwards (but equally so for M, C and P populations). Some matrilineal populations are famous for their duolocal residence mode, with husbands living with their sisters and visiting their wives [8]. However, such duolocal residence was not observed for any couple in our ethno-demographic survey, and was not reported in the ethnographic literature available on the populations under study [18,37–49], so we do not believe that the undetected occurrence of this residence mode could have biased our estimated endogamy rates. The cognatic populations included in this study were not fully matrilocal but include two multilocal populations with final settlement in the wife's village. Replication of this study in fully matrilocal populations is warranted.
Despite these limitations, our study not only suggests that the matrilineal puzzle is still in action in present-day Southeast Asia but also that such a puzzle shapes genetic diversity patterns in human populations, thus identifying a new cultural factor contributing to genetic diversity patterns among human populations. It has previously been shown that genetic inbreeding levels are greatly influenced by the prevalence of consanguineous marriages in human populations [33,50–55]. Our study shows that the association of matrilocality with local endogamy, which takes root in male dominance, may also contribute to some extent to higher inbreeding levels in human populations, thus revealing an additional layer of complexity to the interactions between socio-cultural factors and human genetic diversity patterns.
It remains to be investigated whether our result can be generalized to other matrilocal populations. It is likely to be so, as high endogamy has been reported by anthropologists as a general feature of matrilocal populations [6,19]. From our study, we can predict that other matrilocal populations will have higher genetic inbreeding levels than populations sharing the same environment, the same way of life, belonging to the same linguistic family (the criteria we used to select our populations to study) but having different social organizations. If such a prediction holds, high inbreeding could become an informative marker of matrilocality for ancient DNA studies trying to decipher the social organization of past human populations.
4. Methods
(a). Data collection
(i). Sampled populations
Twelve populations from Cambodia and Laos were sampled in 57 villages during three field missions carried out between 2011 and 2012: the Tampuan, Jarai, Kacho', Bunong, Khmer, Brao and Kreung from Cambodia and the Khmu', Ramet, Ta-oih, Pacoh and Prai from Laos (table 1). The populations were chosen for their differences in social organization. Most of them have been the focus of ethnographic works, describing in detail their social organization. The Tampuan, Jarai, Kacho' and Prai have matrilineal descent and matrilocal residence [18,37,38,48], the Khmu', Ramet, Ta-oih and Pacoh have patrilineal descent and patrilocal residence [38–40,46,47], the Bunong and the Khmer have cognatic descent and matrilocal residence [41,43,44,49], the Brao and Kreung have cognatic descent and multilocal residence [38,42,43,45]. Our previous analysis of ethno-demographic data collected in these populations [26] has shown that the four cognatic populations actually had comparable percentages of matrilocal couples (estimated to 43–48% of the exogamous couples), and that these matrilocal couples outnumbered the percentages of patrilocal couples (estimated to 13–38% of the exogamous couples). This shows that the final settlement of couples in the two multilocal populations is most often located in the wife's natal village. Consequently, we grouped these four populations into a single group of cognatic populations with predominant matrilocal residence. We refer in this paper to the matrilineal and matrilocal populations, to the cognatic populations with predominant matrilocal residence, and to the patrilineal and patrilocal populations respectively as M, C and P populations. All these populations belong to the Austro-Asiatic linguistic family, except the Jarai that speak an Austronesian language.
Note that in these Southeast Asian populations, the village is an important social unit [18,37,40]. In the case of the matrilineal and patrilineal populations under study, each village usually comprises families belonging to several clans. Some of the 57 villages that were integrated in this study were ancient and stable in time, with, for example, a great tree marking symbolically its location. Others had been moving, either in line with a traditional practice [37] or because of recent political changes in Cambodia and Laos in the past two generations [37,40]. The members of each village usually know the history of the village [37] and our ethno-demographic data collection allowed us, while in the field, to exclude from our sampling, villages for which social integrity had been lost due to political events in the past two generations.
(ii). Ethno-demographic interviews
We interviewed 532 individuals, conjointly with their spouse, and collected ethno-demographic information (place of birth, village of residence; see [26] for details) about them and their family members (parents, grandparents, siblings, children and their respective spouses). This procedure allowed us to gather ethno-demographic information for 3530 couples.
(iii). DNA samples
Seven hundred and fifty-three individuals with all four grandparents from the same population were studied. We collected two saliva samples for each individual (4 ml each). Samples were kept in equivalent volume of lysis buffer with 800 µl of 10% SDS and 20 µl of proteinase K (20 mg ml−1). DNA was extracted from saliva samples using a standard ethanol precipitation protocol [56]. All participants provided written informed consents and the study was approved by the National Ethic Comities for Health Research in Cambodia and Laos as well as by the Comité Opérationnel pour l'Ethique (CNRS, France).
(iv). SNP genotyping
Samples were genotyped on Illumina Omni1 (529 individuals) and Omni2.5 SNP arrays (224 individuals). SNPs present on both chips were retained, leading to a dataset of 701 163 SNPs for 753 individuals. After quality control (electronic supplementary material, figure S2), the dataset contained 598 764 SNPs for 743 individuals.
We used the method described in Conomos et al. [57] in order to check if any siblings were present in the dataset. We removed 24 individuals in order to get a sibling-free dataset (which will be used when estimating the genetic inbreeding coefficients). This dataset contained 598 764 SNPs genotyped for 719 individuals.
In addition, we prepared a dataset excluding first and second-degree relationships in order to estimate effective sizes, FST, isolation-by-distance patterns, and allelic frequencies (necessary for the estimation of genetic inbreeding coefficients). To do so, first- and second-degree relationships were inferred using KING v. 2.1.6 [58]. Two hundred and thirty individuals were removed to generate this first- and second-degree relationships-free dataset, containing 598 764 SNPs genotyped for 489 individuals.
(b). Data analysis
(i). Selection of populations with similar effective population sizes
Firstly, we checked that all the studied populations have comparable effective sizes since this parameter is known to influence inbreeding levels [59], with smaller effective population sizes associated with higher inbreeding. We estimated the effective population size of each population using the method described in Auton & McVean [60] (electronic supplementary material, table S4). Effective population sizes were compared between populations by a Welch's t-test with a Bonferroni correction for multiple testing. Among all studied populations, Prai was the only population with a significantly lower effective population size compared to all other populations (7182 compared to 13 228 (s.d. ± 1403) on average for the other populations; p-values < 0.05; electronic supplementary material, tables S4 and S5). Consequently, the Prai population was not included in the analyses presented below (however, similar results and conclusions were reached when this population was included; see electronic supplementary material, figure S3 for a graphical summary of these results). The final dataset included 11 populations, with 598 764 SNPs genotyped for 675 individuals for the sibling-free dataset and 466 individuals for the first- and second-degree relationships-free dataset. The ethno-demographic dataset included 495 ethno-demographic interviews providing information for 3261 couples (table 1).
(ii). Village endogamy estimation
A full description of the post-marital residence patterns for each population under study is provided in our previous study [26]. Here, for each population, we estimated the proportion of couples for which both spouses were born in the same village (village endogamy rate). We then used logistic regression to assess the influence of social organization (M, P and C) on the probability that individuals marry partners from the same village with the ‘glmer’ function in ‘lme4’ package v. 1.1-9 in R [61]. We incorporated population as a fixed effect and village of residence and family as random effects in this model in order to account for potential sampling bias. P-values were estimated with the ‘lsmeans’ function in the ‘lsmeans’ package v. 2.27-2 in R.
(iii). Genetic inbreeding coefficients estimation
We used the FEstim software [36] integrated in the FSuite pipeline [35] to estimate the inbreeding coefficient and the parental mating types of each individual. Genetic maps were retrieved from the shapeit homepage [62]. The --hotspots option with hg19 build was used when creating the 100 submaps. Allele frequencies were estimated separately for each population (using the first- and second-degree relationships-free dataset).
Then, we used a mixed linear model to assess the influence of social organization (M, P and C) on inbreeding coefficients. We incorporated population as a fixed effect and village of residence and family as random effects in this model in order to take into account potential sampling bias. P-values were estimated with the ‘lsmeans’ function in the ‘lsmeans’ package v. 2.27-2 in R.
Spearman's correlation coefficient between village endogamy rate and mean inbreeding coefficient was estimated at the population level.
(iv). Isolation-by-distance pattern
Fixation indices (FST) between villages within each population were estimated using Genepop 4.7 [63]. Only villages with a minimum of five individuals were included in this analysis. Populations with less than four villages filling this condition were removed from the analysis. As such, Kacho', Kreung and Ta-oih were excluded from this analysis. The dataset was then pruned using Plink 1.9 [64] –indep-pairwise option with a window size of 50 SNPs, sliding by five SNPs and a pairwise r2 threshold of 0.5 to create a dataset of 252 680 SNPs in low linkage disequilibrium. Negative FST were changed to 0. A linear regression model was fitted with genetic distance between villages estimated by FST/(1 − FST) as the dependent variable and geographical distance in metre (decimal logarithmic value) as the explanatory variable for each population. Statistical significance of the correlation between genetic distances and geographical distances was evaluated using a Mantel test with 10 000 permutations.
All statistical analyses were performed in R v. 3.2.2 [61].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank all the volunteers for their participation, as well as Yves Buisson from the Institut de la Francophonie pour la Médecine Tropicale (IFMT) in Laos for their help in facilitating fieldwork, Hervé Pedry for his advice regarding regression models and Michael Houseman, Evelyne Heyer and Paul Verdu for helpful discussions.
Ethics
All participants provided written informed consents and the study was approved by the National Ethic Comities for Health Research in Cambodia and Laos as well as by the Comité Opérationnel pour l'Ethique (CNRS).
Data accessibility
Genotyping data has been deposited at the European Genome-phenome Archive (EGA, https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001003727.
Authors' contributions
R.C., S.P. and B.T. initiated the project; R.C., S.P., R.L., S.L., O.E., F.B., G.D. and C.M. contributed to the ethno-demographic and genetic sampling; S.L. and C.M. carried out the laboratory work; G.L. and R.L. analysed the data; G.L., R.C. and S.P. wrote the paper. All authors gave final approval for publication.
Competing interest
The authors declare no competing interests.
Funding
This work was supported by the ANR SoGen (JC09_441218) grant.
References
- 1.Godelier M. 2004. Métamorphoses de la parenté. Paris, France: Fayard. [Google Scholar]
- 2.Murdock GP, White DR. 1969. Standard cross-cultural sample. Ethnology 8, 329–369. ( 10.2307/3772907) [DOI] [Google Scholar]
- 3.Fox R. 1972. Anthropologie de la parenté. Une analyse de la consanguinité et de l'alliance. Paris, France: Gallimard. [Google Scholar]
- 4.Mathieu N-C. 2007. Une maison sans fille est une maison morte. Paris, France: Maison des sciences de l'homme. [Google Scholar]
- 5.Richards AI. 1950. Some types of family structure amongst the Central Bantu. In African systems of kinship and marriage (eds Radcliffe-Brown A, Forde D), pp. 207–251. London, UK: Oxford University Press. [Google Scholar]
- 6.Kloos P. 1963. Matrilocal residence and local endogamy: environmental knowledge or leadership. Am. Anthropol. 65, 854–862. ( 10.1525/aa.1963.65.4.02a00050) [DOI] [Google Scholar]
- 7.Mattison SM, Quinlan RJ, Hare D. 2018. The expendable male hypothesis. bioRxiv ( 10.1101/473942) [DOI] [PMC free article] [PubMed]
- 8.Schneider DM, Gough K. 1961. Matrilineal kinship. Berkeley, CA: University of California Press. [Google Scholar]
- 9.Schneider DM. 1961. The distinctive features of matrilineal descent groups. In Matrilineal kinship (eds Schneider DM, Gough K), pp. 1–29. Berkeley, CA: University of California Press. [Google Scholar]
- 10.Mattison S. 2011. Evolutionary contributions to solving the ‘matrilineal puzzle’. Hum. Nat. 22, 64–88. ( 10.1007/s12110-011-9107-7) [DOI] [PubMed] [Google Scholar]
- 11.Fortunato L. 2012. The evolution of matrilineal kinship organization. Proc. R. Soc. B 279, 4939–4945. ( 10.1098/rspb.2012.1926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holden CJ, Sear R, Mace R. 2003. Matriliny as daughter-biased investment. Evol. Hum. Behav. 24, 99–112. ( 10.1016/S1090-5138(02)00122-8) [DOI] [Google Scholar]
- 13.Hamilton WD. 1964. The genetical evolution of social behaviour, I. J. Theor. Biol. 7, 1–16. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 14.Trivers RL, Willard DE. 1973. Natural selection of parental ability to vary the sex ratio of offspring. Science 179, 90–92. ( 10.1126/science.179.4068.90) [DOI] [PubMed] [Google Scholar]
- 15.Holden C, Mace R. 2003. Spread of cattle led to the loss of matrilineal descent in Africa: a coevolutionary analysis. Proc. R. Soc. Lond. B 270, 2425–2433. ( 10.1098/rspb.2003.2535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasternak B. 1976. Introduction to kinship and social organization. Englewood Cliffs, NJ: Prenctice-Hall. [Google Scholar]
- 17.Gough K. 1961. Variation in residence. In Matrilineal kinship (eds Schneider DM, Gough K), pp. 545–576. Berkeley, CA: University of California Press. [Google Scholar]
- 18.Dournes J. 1972. Coordonnées, structures jörai familiales et sociales. Paris, France: Muséum national d'Histoire naturelle; (Travaux et Mémoires de l'Institut d'Ethnologie n°77). [Google Scholar]
- 19.Murdock GP. 1949. Social structure. London, UK: Macmillan. [Google Scholar]
- 20.Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. 2001. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat. Genet. 29, 20–21. ( 10.1038/ng711) [DOI] [PubMed] [Google Scholar]
- 21.Hamilton G, Stoneking M, Excoffier L. 2005. Molecular analysis reveals tighter social regulation of immigration in patrilocal populations than in matrilocal populations. Proc. Natl Acad. Sci. USA 102, 7476–7480. ( 10.1073/pnas.0409253102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumonggor MK, Karafet TM, Downey S, Lansing JS, Norquest P, Sudoyo H, Hammer MF, Cox MP. 2014. Isolation, contact and social behavior shaped genetic diversity in West Timor. J. Hum. Genet. 59, 494–503. ( 10.1038/jhg.2014.62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besaggio D, Fuselli S, Srikummool M, Kampuansai J, Castrì L, Tyler-Smith C, Seielstad M, Kangwanpong D, Bertorelle G. 2007. Genetic variation in Northern Thailand hill tribes: origins and relationships with social structure and linguistic differences. BMC Evol. Biol. 7, 1–10. ( 10.1186/1471-2148-7-S2-S12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunnarsdóttir ED, Nandineni MR, Li M, Myles S, Gil D, Pakendorf B, Stoneking M. 2011. Larger mitochondrial DNA than Y-chromosome differences between matrilocal and patrilocal groups from Sumatra. Nat. Commun. 2, 226–228. ( 10.1038/ncomms1235) [DOI] [PubMed] [Google Scholar]
- 25.Kumar V, Langstieh BT, Madhavi KV, Naidu VM, Singh HP, Biswas S, Thangaraj K, Singh L, Reddy BM. 2006. Global patterns in human mitochondrial DNA and Y-chromosome variation caused by spatial instability of the local cultural processes. PLoS Genet. 2, 420–424. ( 10.1371/journal.pgen.0020053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ly G, et al. 2018. Residence rule flexibility and descent groups dynamics shape uniparental genetic diversities in South East Asia. Am. J. Phys. Anthropol. 165, 480–491. ( 10.1002/ajpa.23374) [DOI] [PubMed] [Google Scholar]
- 27.Chaix R, Quintana-Murci L, Hegay T, Hammer MF, Mobasher Z, Austerlitz F, Heyer E. 2007. From social to genetic structures in Central Asia. Curr. Biol. 17, 43–48. ( 10.1016/j.cub.2006.10.058) [DOI] [PubMed] [Google Scholar]
- 28.Heyer E, Chaix R, Pavard S, Austerlitz F. 2012. Sex-specific demographic behaviours that shape human genomic variation. Mol. Ecol. 21, 597–612. ( 10.1111/j.1365-294X.2011.05406.x) [DOI] [PubMed] [Google Scholar]
- 29.Verdu P, et al. 2013. Sociocultural behavior, sex-biased admixture, and effective population sizes in central African pygmies and non-pygmies. Mol. Biol. Evol. 30, 918–937. ( 10.1093/molbev/mss328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ségurel L, et al. 2008. Sex-specific genetic structure and social organization in Central Asia: insights from a multi-locus study. PLoS Genet. 4, e1000200 ( 10.1371/journal.pgen.1000200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillot EG, Hazelton ML, Karafet TM, Lansing JS, Sudoyo H, Cox MP. 2015. Relaxed observance of traditional marriage rules allows social connectivity without loss of genetic diversity. Mol. Biol. Evol. 32, 2254–2262. ( 10.1093/molbev/msv102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchi N, Mennecier P, Georges M, Lafosse S, Hegay T, Dorzhu C, Chichlo B, Ségurel L, Heyer E. 2018. Close inbreeding and low genetic diversity in Inner Asian human populations despite geographical exogamy. Sci. Rep. 8, 1–10. ( 10.1038/s41598-018-27047-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. 2012. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91, 275–292. ( 10.1016/j.ajhg.2012.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright S. 1943. Isolation by distance. Genetics 28, 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gazal S, Sahbatou M, Babron M-C, Génin E, Leutenegger A-L. 2014. FSuite: exploiting inbreeding in dense SNP chip and exome data. Bioinformatics 30, 1940–1941. ( 10.1093/bioinformatics/btu149) [DOI] [PubMed] [Google Scholar]
- 36.Leutenegger A-L, Prum B, Génin E, Verny C, Lemainque A, Clerget-Darpoux F, Thompson EA. 2003. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 73, 516–523. ( 10.1086/378207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourdier F. 2006. The mountain of precious stones: Ratanakiri, Cambodia: essays in social anthropology. Phnom Penh, Cambodia: Center for Khmer Studies.
- 38.LeBar FM, Hickey GC, Musgrave JK. 1964. Ethnic groups of mainland Southeast Asia. New Haven, CT: Human Relations Area Files Press. [Google Scholar]
- 39.Izikowitz KG. 1951. Lamet hill peasants in French Indochina. Göteborg, Sweden: Etnografiska Museet. [Google Scholar]
- 40.Evrard O. 2006. Chroniques des cendres. Paris, France: IRD Edition. [Google Scholar]
- 41.Ebihara M. 1977. Residence patterns in a Khmer peasant village. Ann. N. Y. Acad. Sci. 293, 51–68. ( 10.1111/j.1749-6632.1977.tb41805.x) [DOI] [Google Scholar]
- 42.Baird IG. 2008. Various forms of colonialism : the social and spatial reorganisation of the Brao in southern Laos and northeastern Cambodia. PhD thesis, University of British Columbia.
- 43.UNDP Cambodia. 2010. Kreung ethnicity documentation of customary rules indigenous people in Pu-Trou village. Phnom Penh, Cambodia: UN Development Programme.
- 44.Martel G. 1975. Lovea. Village des environs d'Angkor. Aspects démographiques, économiques et sociologiques du monde rural cambodgien dans la province de Siem-Réap. Paris, France: Ecole française d'Extrême-Orient. [Google Scholar]
- 45.Matras-Troubetzkoy J. 1983. Un village en foret. L'essartage chez les brou du Cambodge. Paris, France: Peeters. [Google Scholar]
- 46.Lindell K, Samuelsson R, Tayanin D. 1979. Kinship and marriage in northern Kammu villages: the kinship model. Sociologus 29, 60–84. [Google Scholar]
- 47.Schmutz J. 2013. The Ta'oi language and people. MonKhmer Stud. 42, i–xiii. [Google Scholar]
- 48.Dessaint WY. 1981. The T'in (Mal), dry rice cultivators of Northern Thailand. J. Siam Soc. 69, 107–137. [Google Scholar]
- 49.Ledgerwood JL. 1995. Khmer kinship—the matriliny matriarchy myth. J. Anthropol. Res. 51, 247–261. ( 10.1086/jar.51.3.3630360) [DOI] [Google Scholar]
- 50.Bittles AH, Hamamy H. 2010. Endogamy and consanguineous marriage in Arab populations. In Genetic disorders among Arab populations (ed. Teebi AS.), pp. 85–108. Berlin, Germany: Springer. [Google Scholar]
- 51.Gazal S, Sahbatou M, Babron M-C, Génin E, Leutenegger A-L. 2015. High level of inbreeding in final phase of 1000 Genomes Project. Sci. Rep. 5, 17453 ( 10.1038/srep17453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF. 2010. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 5, e13996 ( 10.1371/journal.pone.0013996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McQuillan R, et al. 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83, 359–372. ( 10.1016/j.ajhg.2008.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Al-Bustan S, Feng Q, Guo W, Ma Z, Marafie M, Jacob S, Al-Mulla F, Xu S. 2014. The influence of admixture and consanguinity on population genetic diversity in Middle East. J. Hum. Genet. 59, 615–622. ( 10.1038/jhg.2014.81) [DOI] [PubMed] [Google Scholar]
- 55.Leutenegger A-L, Sahbatou M, Gazal S, Cann H, Génin E. 2011. Consanguinity around the world: what do the genomic data of the HGDP-CEPH diversity panel tell us? Eur. J. Hum. Genet. 19, 583–587. ( 10.1038/ejhg.2010.205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. 2006. Evaluation of saliva as a source of human DNA for population and association studies. Anal. Biochem. 353, 272–277. ( 10.1016/j.ab.2006.03.021) [DOI] [PubMed] [Google Scholar]
- 57.Conomos MP, Reiner AP, Weir BS, Thornton TA. 2016. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 98, 127–148. ( 10.1016/j.ajhg.2015.11.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873. ( 10.1093/bioinformatics/btq559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacquard A. 1968. Evolution des populations d'effectif limité. Popul. (French Edn) 23, 279–300. ( 10.2307/1527488) [DOI] [Google Scholar]
- 60.Auton A, McVean G. 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 17, 1219–1227. ( 10.1101/gr.6386707.scheme) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at https://www.R-project.org/.
- 62.Delaneau O, Marchini J, Zagury J-F. 2012. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181. ( 10.1038/nmeth.1785) [DOI] [PubMed] [Google Scholar]
- 63.Rousset F. 2008. Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. ( 10.1111/j.1471-8286.2007.01931.x) [DOI] [PubMed] [Google Scholar]
- 64.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 ( 10.1186/s13742-015-0047-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotyping data has been deposited at the European Genome-phenome Archive (EGA, https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001003727.