Abstract
Excitotoxic levels of glutamate represent a physiological stress that is strongly linked to amyotrophic lateral sclerosis (ALS) and other neurological disorders. Emerging evidence indicates a role for neurodegenerative disease–linked RNA-binding proteins (RBPs) in the cellular stress response. However, the relationships between excitotoxicity, RBP function, and disease have not been explored. Here, using primary cortical and motor neurons, we found that excitotoxicity induced the translocation of select ALS-linked RBPs from the nucleus to the cytoplasm within neurons. RBPs affected by excitotoxicity included TAR DNA–binding protein 43 (TDP-43) and, most robustly, fused in sarcoma/translocated in liposarcoma (FUS/TLS or FUS). We noted that FUS is translocated through a calcium-dependent mechanism and that its translocation coincides with striking alterations in nucleocytoplasmic transport. Furthermore, glutamate-induced up-regulation of glutamate ionotropic receptor α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)–type subunit 2 (GRIA2) in neurons depended on FUS expression, consistent with a functional role for FUS in excitotoxic stress. These findings reveal molecular links among prominent factors in neurodegenerative diseases, namely excitotoxicity, disease-associated RBPs, and nucleocytoplasmic transport.
Keywords: amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), excitotoxicity, RNA transport, neurodegeneration, glutamate, excitatory neurotransmission, fused in sarcoma/translocated in liposarcoma (FUS/TLS), glutamate ionotropic receptor AMPA type subunit 2 (Gria2), glutamate receptor 2 (GluA2), nucleocytoplasmic transport
Introduction
Glutamate is the major excitatory neurotransmitter in the central nervous system. Upon release from pre-synaptic terminals, relatively low levels of glutamate activate metabotropic glutamate receptors as well as the following ionotropic receptors: α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA),5 N-methyl-d-aspartate, and kainate, for normal neurotransmission. However, excessive glutamate exposure overstimulates neurons. This causes a massive influx of calcium, which triggers an excitotoxic cascade involving oxidative damage as well as mitochondrial and ER dysfunction (1). Excitotoxicity has been implicated in neuronal death and degeneration for various neurological conditions, including the fatal neurodegenerative disease amyotrophic lateral sclerosis (ALS). Pathological evidence for excitotoxicity includes elevated levels of glutamate in patient cerebrospinal fluid as well as aberrant processing of the AMPA subunit that controls calcium influx at both the transcript (Gria2) and protein (glutamate receptor 2 (GluA2)) levels in patient tissue and disease models (2). Furthermore, ALS-causing mutations are present in d-amino acid oxidase, an enzyme that regulates the degradation of the N-methyl-d-aspartate co-agonist, d-serine (3). Riluzole, the first Food and Drug Administration–approved treatment for ALS, is thought to reduce glutamate signaling through anti-excitotoxic effects (4). Despite this wealth of knowledge and profound disease relevance, the biological mechanisms underlying the cellular response to excitotoxicity have not been fully elucidated.
RNA-binding proteins (RBPs) have emerged as relevant factors in neurodegenerative disease pathogenesis, particularly in the context of ALS and the related disorder frontotemporal dementia (FTD) (5). RBPs belong to a unique class of biomolecules that undergo nucleocytoplasmic shuttling in response to various stimuli, including stress. For instance, the disease-linked RBPs fused in sarcoma/translocated in liposarcoma (FUS/TLS or FUS), TAR DNA-binding protein 43 (TPD-43), and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) all exhibit nuclear egress during hyperosmotic stress (6–8). The purpose of this translocation is unclear, and it may represent a functional response to cellular stress (6, 9). In support of this notion, cell viability under hyperosmotic stress is compromised when FUS expression is reduced (6). However, cell stress also represents a nongenetic factor that likely contributes to neurodegenerative disease pathogenesis (9). Indeed, chronic stress may contribute to the pathological accumulation of TDP-43 and FUS in some cases of ALS and FTD (10–14). For example, TDP-43 partitions into the insoluble fraction of cultured cells following oxidative stress or heat shock (15, 16), and disease-linked RBPs have been found to aggregate in vivo following cerebral ischemia (17). Intriguingly, the effects of stress on RBP translocation appear selective. Although ER stress, oxidative stress, and heat shock induce the cytoplasmic accumulation of TDP-43 and other RBPs (18, 19), these stressors fail to elicit a response of FUS (6, 20). Given the physiological relevance of excitotoxicity to neurodegenerative disease, an important but unanswered question is whether excitotoxic stress elicits a functional and/or pathological response from disease-associated RBPs.
Here, we demonstrate that excitotoxic levels of glutamate induce the nuclear egress of several ALS- and FTD-linked RBPs, including FUS, TDP-43, and hnRNPA1 into the cytoplasm of neurons. The nucleocytoplasmic equilibrium of FUS was especially sensitive to excitotoxic stress, as FUS was found to rapidly and robustly accumulate within soma and dendrites of cortical and motor neurons under stress. Furthermore, a glutamate-induced increase in dendritic Gria2 depended on FUS, consistent with a role for FUS in glutamatergic signaling during the cellular response to excitotoxic stress. Our results also revealed potentially adverse consequences of excitotoxicity, including the translocation of ALS-linked FUS variants and early signs of nucleocytoplasmic transport dysregulation. This study therefore demonstrates that excitotoxicity can trigger neurodegenerative disease-associated phenotypes, including cytoplasmic RBP accumulation and nucleocytoplasmic transport decline.
Results
Excitotoxic levels of glutamate shift the nucleocytoplasmic equilibrium of disease-linked RNA-binding proteins
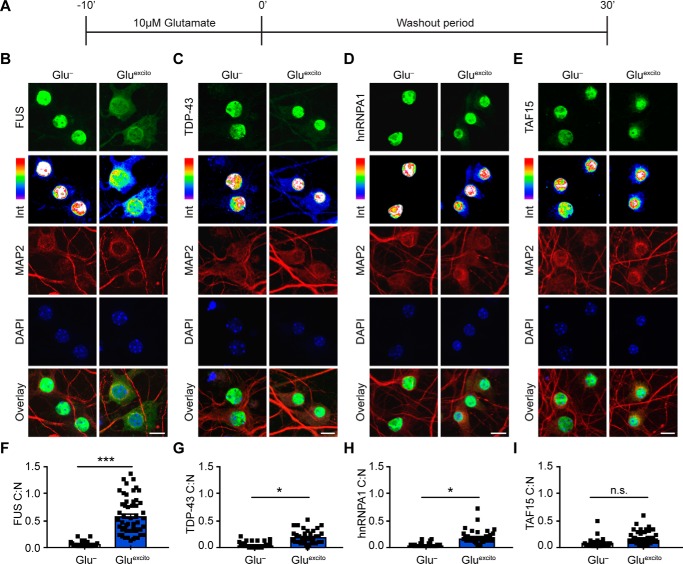
To investigate a potential relationship between excitotoxicity and neurodegenerative disease-linked RBPs, we first examined whether excitotoxicity affects the nucleocytoplasmic equilibrium of a panel of proteins, including FUS, TDP-43, hnRNPA1, and TATA-binding protein-associated factor 15 (TAF15). All four proteins have been linked to ALS (5), and FUS, TDP-43, and TAF15 are also associated with FTD (21). DIV 14–16 primary cortical neurons, the majority of which are excitatory, were bath-treated with excitotoxic and physiologically relevant levels of glutamate (22, 23) (10 μm; hereon referred to as Gluexcito) for 10 min followed by a 30-min washout period (Fig. 1A). Immunofluorescence was then used to assess the effect of Gluexcito on the cytoplasmic to nuclear ratio (C:N) of the endogenous RBPs (Fig. 1, B–I). Strikingly, the FUS C:N ratio significantly increased ∼15-fold from 0.04 ± 0.05 to 0.6 ± 0.3 in response to Gluexcito (Fig. 1, B and F). This increase is likely due to a rapid egress of FUS from the nucleus into the cytoplasm, as a Western blot analysis revealed total FUS protein levels are unchanged before and after stress (Fig. S1, A and B). Gluexcito likewise induced a significant increase in the C:N ratio of TDP-43 (Fig. 1, C and G) and hnRNPA1 (Fig. 1, D and H), although not to the same extent as FUS. Gluexcito did not significantly alter the C:N ratio of TAF15 (Fig. 1, E and I). As for FUS, protein expression levels of TDP-43, hnRNPA1, and TAF15 were unaffected by Gluexcito (Fig. S1, C–F).
Figure 1.
Endogenous FUS robustly translocates to the cytoplasm in response to excitotoxic stress. A, DIV 14–16 primary cortical neurons were bath-treated with 10 μm glutamate (Gluexcito) for 10 min, after which the glutamate-containing media was “washed out” and replaced with cultured neuronal media for an additional 30 min. B–E, immunofluorescence and confocal microscopy revealed the cellular localization of FUS, TDP-43, hnRNPA1, and TAF15 (green) in the absence and presence of Gluexcito. Endogenous RBP staining (green) visualized by a 16-color intensity map (Int) further demonstrates the cytoplasmic presence of these proteins. Neurons and dendrites were identified with anti-MAP2 staining (red), and nuclei with DAPI (blue). Scale bars = 10 μm. F–I, quantification of the cytoplasmic to nuclear ratio (C:N) from B–E was based on fluorescence intensities of the signal in each compartment as described under “Experimental procedures.” A significant nuclear egress of FUS (F), TDP-43 (G), and hnRNPA1 (H) but not TAF15 (I) was observed following Gluexcito treatment (n = 3–4 biological replicates). Black squares represent the C:N ratio of individual cells, and error bars correspond to S.E. Experimental means were calculated from the average C:N ratio across the individual biological replicates and significant comparisons were determined with a Student's t test (FUS: **, p = 0.0013; hnRNPA1: *, p = 0.0107; TDP-43: *, p = 0.0185; n.s., nonsignificant).
In light of the robust response of FUS to Gluexcito, we focused our attention on the properties of FUS translocation in more detail. First, endogenous FUS translocation in response to Gluexcito was confirmed using a panel of different anti-FUS antibodies (Fig. S2, A and B). We then examined the relationship between FUS translocation and glutamate concentration. With 10 μm glutamate, the vast majority of neurons (91.3 ± 11.5%) exhibited FUS egress (Fig. 2, A and B), whereas <5% of neurons exhibited translocation at ≤1 μm, revealing a dependence of FUS localization on glutamate concentration (Fig. 2B). Within the time course of our experiment (Fig. 1A), a significant accumulation of endogenous FUS was also detected throughout microtubule-associated protein 2 (MAP2)–positive dendrites (Fig. 2, C and D). The response of FUS to Gluexcito appears to be all–or–none, as lower concentrations of glutamate resulted in fewer neurons responding to but with similar degrees of FUS egress (Fig. S2C).
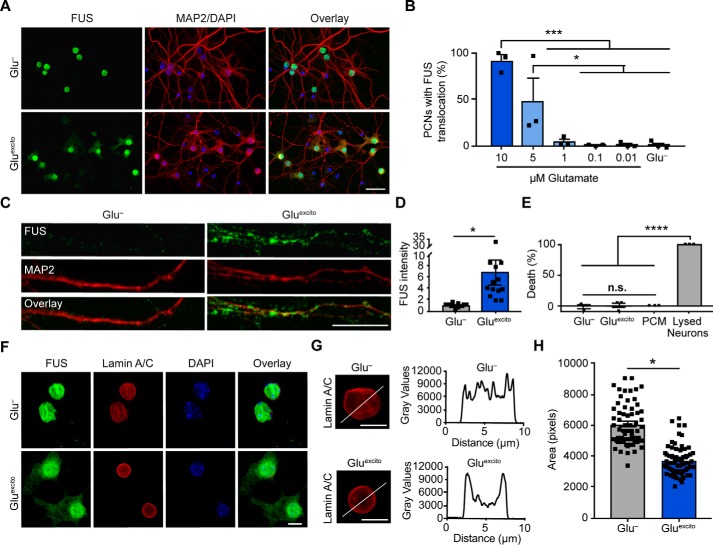
Figure 2.
Cell viability and nuclear membrane integrity are intact under conditions of Gluexcito that promote FUS translocation. A, following excitotoxic insult, FUS egress and cytoskeletal rearrangements were detected by anti-FUS (green) and anti-MAP2 (red) staining, respectively. Scale bar = 40 μm. B and C, quantification of FUS translocation revealed a dose-dependent response to glutamate in neurons using a one-way ANOVA and Tukey's post hoc test (10 μm compared with 1 μm, ***, p = 0.0002; compared with 0.1 μm, ***, p = 0.0001; compared with 0.01 μm, ***, p = 0.0002; and compared with Glu−, ***, p = 0.0002; 5 μm compared with 0.1 μm, *, p = 0.0404; compared with 0.01 μm, *, p = 0.0426; and compared with Glu−, *, p = 0.0451; n = 3 biological replicates). C, increased dendritic FUS staining (green) was observed by confocal microscopy following excitotoxic stress. Dendrites were labeled with anti-MAP2 (red). Scale bar = 10 μm. D, quantification of C. Black squares represent the intensity of dendritic FUS staining per cell. Means represent the average of n = 4 biological replicates (Student's t test; *, p = 0.0142) normalized to the control (Glu−). E, cytotoxicity induced by Gluexcito was assessed after the washout period (Fig. 1A) with the LDH assay. In contrast to the positive control (neurons treated with lysis buffer; lysed neurons), membrane permeabilization was not detected for neurons exposed to Gluexcito. Neurons cultured in the absence of Gluexcito (Glu−) served as a negative control. Wells containing only primary neuron–cultured medium (PCM) served as a background control. Results reflect n = 3 biological replicates analyzed with a one-way ANOVA and Tukey's post hoc test (for all statistical comparisons, ****, p < 0.0001, n.s. = nonsignificant). F, immunofluorescence with anti-lamin A/C staining (red) and confocal microscopy revealed the nuclear envelope was thickened yet still intact within neurons exhibiting translocated FUS (green) after Gluexcito exposure. The time point is the same as E. Scale bar = 25 μm. G, representative line scan analyses of lamin staining demonstrates enhanced lamin intensity at the nuclear periphery in neurons exposed to excitotoxic insult (scale bars = 10 μm). B, D, E, and H, error bars represent S.E. H, quantification of nuclear size using the nuclear counterstain, DAPI, revealed a significant decrease in nuclear area following excitotoxic insult. Black squares represent the area of individual neurons. Means represent the average of n = 4 biological replicates (Student's t test; *, p = 0.0154).
Given the toxicity of Gluexcito on neurons (22), we interrogated whether the rapid and robust accumulation of FUS outside the nucleus was simply a consequence of cell death and/or loss of nuclear envelope integrity. The extent of cell death was assessed using the LDH cytotoxicity assay, which detects the activity of LDH upon its release into the media from dead or dying cells. In contrast to neurons treated with lysis buffer, there was no evidence of cell death for neurons treated with Gluexcito (Fig. 2E). Furthermore, lamin A/C staining revealed an intact nuclear envelope in neurons exposed to Gluexcito (Fig. 2F). These observations support the premise that cytoplasmic FUS accumulation represents a cellular response to Gluexcito, rather than a nonspecific consequence of cell death. Moreover, RBP translocation appears selective, as TAF15 (Fig. 1, E and I) and the cytoplasmic protein, fragile X mental retardation protein (Fig. S3A), did not change localization following excitotoxic insult. It is noteworthy that Gluexcito affects neuron morphology at 30 min, potentially indicative of a stressed state. Anti-MAP2 staining revealed a rearrangement of the cytoskeleton; staining was more pronounced around the nucleus and indicative of dendritic fragmentation (Figs. 1 and 2). Likewise, the nuclear lamina appeared more concentrated around the nuclear envelope. This is illustrated by the line scan through the nucleus (Fig. 2G). Furthermore, nuclear size was significantly smaller in stressed neurons (Fig. 2H). As expected, neurons exposed to excitotoxic stimuli (10 μm, but not 1 μm glutamate) eventually undergo cell death within 24 h of the initial insult (Fig. S3, B and D) (22).
Excitotoxic stress induces egress predominately of nuclear ALS-linked FUS variants
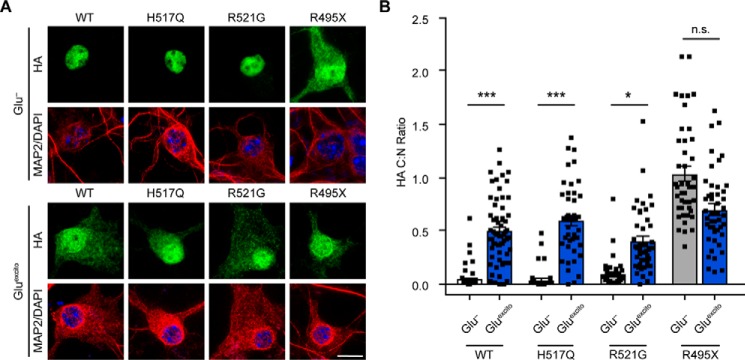
Next, we investigated the behavior of ALS-linked FUS variants under conditions of Gluexcito. To this end, a series of FLAG–HA-tagged FUS variants were transiently expressed in neurons, and the C:N ratio of exogenous FUS was determined in the absence and presence of Gluexcito (Fig. 3). In addition to wildtype (WT) FUS, we examined the following: H517Q, the only autosomal recessive FUS mutation associated with ALS; R521G, representing a mutational “hot spot” for ALS–FUS (24); and R495X, a particularly aggressive ALS-linked mutation (20). All of these variants have mutations within the nuclear localization sequence (NLS), yet the degree to which FUS H517Q and R521G mislocalize to the cytoplasm depends on their expression level. Near-endogenous levels of these variants exhibit a predominately nuclear localization in vitro (20) and in vivo (25), whereas overexpression leads to higher levels of cytoplasmic mutant FUS (10). In contrast, the entire NLS is omitted from the FUS R495X variant, which significantly mislocalizes to the cytoplasm regardless of expression level (20). Here, we observed minimal cytoplasmic mislocalization of mutant FUS H517Q and R521G without stress, indicating the expression levels of these exogenous proteins were below the threshold for significant cytoplasmic mislocalization in these cells. As expected, FUS R495X exhibited robust cytoplasmic mislocalization under basal conditions (Fig. 3). In response to Gluexcito, FLAG–HA–FUS WT exhibited a significant translocation to the cytoplasm as observed for endogenous FUS (Fig. 1, B and F). The C:N ratio of FLAG–HA–FUS H517Q and R521G also increased significantly with Gluexcito, providing evidence that the ALS-linked mutations did not interfere with the response of FUS to Gluexcito. In the case of FLAG–HA–FUS R495X, the C:N ratio was the same with and without Gluexcito, potentially indicative of a “ceiling effect” in that the normal nucleocytoplasmic distribution of R495X–FUS is equivalent to that of “maximally” redistributed endogenous FUS following excitotoxic stress.
Figure 3.
Effect of Gluexcito on ALS-linked FUS variants. A, cortical neurons transfected with the indicated FLAG–HA-tagged FUS variants were exposed to Gluexcito, and nuclear FLAG–HA–FUS egress was assessed by immunofluorescence. Exogenous FUS was detected using an anti-HA antibody (green) within MAP2-positive neurons (red). Nuclei were stained with DAPI (blue). Scale bar = 10 μm. B, quantification of the C:N ratio for FLAG–HA–FUS variants in A revealed a significant shift in equilibrium toward the cytoplasm for FLAG–HA–FUS WT, H517Q, and R521G, but not R495X (Student's t test; WT, ***, p = 0.0003; H517Q, ***, p = 0.0003; R521G, *, p = 0.0197, n.s. = not significant, n = 3–5 biological experiments). However, the C:N ratio of H517Q, R521G, and R495X was not significantly different from FLAG–HA–FUS WT following Gluexcito treatment (Glu−; one-way ANOVA and Dunnett's post hoc test; n.s. = nonsignificant, n = 3–5 biological replicates). Black squares represent individual, cellular C:N measurements. Error bars represent S.E.
Nucleocytoplasmic transport is disrupted in response to excitotoxic stress
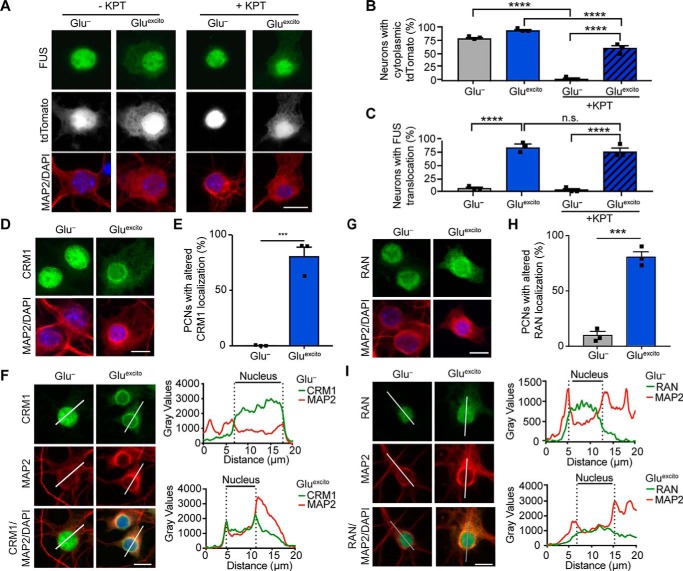
To understand the mechanism(s) underlying endogenous FUS egress in response to Gluexcito, we began with an examination of nucleocytoplasmic transport factors. FUS contains two predicted chromosome region maintenance 1 (CRM1)–dependent nuclear export sequences (NES) within the RNA-recognition motif (26). CRM1 is a major protein export factor, although whether this receptor controls nuclear FUS export is controversial (26, 27). To determine whether excitotoxic FUS egress is CRM1-dependent, we pretreated neurons with the CRM1 inhibitor, KPT-330, prior to treatment with Gluexcito (28). The CRM1-dependent NLS–tdTomato–NES shuttling reporter was used as a positive control (29). As expected, NLS–tdTomato–NES exhibited both a nuclear and cytoplasmic localization under basal conditions (Glu−, −KPT), whereas the localization of this reporter was significantly restricted to the nucleus in the presence of KPT-330 (Glu−, +KPT; Fig. 4, A and B). In contrast, KPT-330 had no effect on nuclear FUS egress in response to Gluexcito (Gluexcito ±KPT; Fig. 4, A and C). Surprisingly, KPT-330 also failed to fully restrict NLS–tdTomato–NES to the nucleus under conditions of Gluexcito (Fig. 4, A and B). Although there was a significant decrease in the percentage of cells with cytoplasmic NLS–tdTomato–NES in the presence of both KPT-330 and Gluexcito (60.1 ± 8.0%) compared with Gluexcito alone (98.3 ± 2.6, p = <0.0001), these results suggest that CRM1-mediated export is dysregulated under conditions of stress (Fig. 4B). Moreover, although endogenous CRM1 predominately localizes to the nucleus, significantly more cells exhibited altered CRM1 localization with Gluexcito. CRM1 alterations included enhanced cytoplasmic localization and a concentration of CRM1 signal around the nuclear membrane (Fig. 4, D–F). This finding prompted us to examine another critical nucleocytoplasmic transport factor, Ras-related nuclear protein (Ran). Ran is a GTPase that shuttles between the nucleus and cytoplasm and, depending on its nucleotide bound state, facilitates nuclear export and import (30). Indeed, Gluexcito also induced a significant change in the nucleocytoplasmic distribution of Ran in that a higher percentage of cells exhibited enhanced cytoplasmic Ran expression under stress (Fig. 4, G–I). These changes to CRM1 and Ran localization appear to be independent from changes in total protein expression as determined by a Western blot analysis of both proteins in the presence and absence of Gluexcito (Fig. S3, D–F). Taken together, Gluexcito caused the redistribution of critical transport factors and attenuated the effects of KPT-330 on CRM1 export.
Figure 4.
Nucleocytoplasmic transport is disrupted by Gluexcito. A–C, cortical neurons expressing the shuttling reporter, NLS–tdTomato–NES, were treated with or without 500 nm of the exportin 1 inhibitor KPT-330 (KPT) prior to Gluexcito exposure. Neurons were identified with anti-MAP2 staining (A; red). The percentage of MAP2-positive cells expressing cytoplasmic NLS–tdTomato–NES (A; white) or FUS (A; green) was quantified in B and C, respectively (n = 3 biological experiments). KPT-330 effectively prevents NLS–tdTomato–NES from localizing to the cytoplasm in the absence of stress (Glu−; two-way ANOVA and Tukey's post hoc test; for all statistical comparisons, ****, p < 0.0001), as expected. Conversely, in the presence of stress (Gluexcito), KPT-330 fails to restrict NLS–tdTomato–NES and FUS localization to the nucleus, indicative of dysregulated nucleocytoplasmic transport (B and C, compare Glu− to Gluexcito in the presence of KPT-330, two-way ANOVA, and Tukey's post hoc test; for all significant statistical comparisons, ****, p < 0.0001, n.s. = nonsignificant). The localization of nuclear transport factors CRM1 (D–F) and RAN (G–I) were significantly altered under conditions of Gluexcito in MAP2-positive neurons (red); cytoplasmic CRM1 and RAN (green in D and E, respectively) were more apparent in stressed cells. The percentage of neurons with CRM1 or RAN mislocalization was quantified in E and H, respectively (Student's t test; CRM1, ***, p = 0.0006; RAN, ***, p = 0.0003; n = 3 biological replicates). Error bars represent S.E. F–I, representative line scan analyses of CRM1 and RAN staining demonstrates an increase in the cytoplasmic presence of these proteins. Specific to CRM1, an increase in staining intensity at the nuclear periphery was observed in neurons exposed to excitotoxic insult. Scale bars = 10 μm.
Excitotoxic FUS egress is calcium-dependent
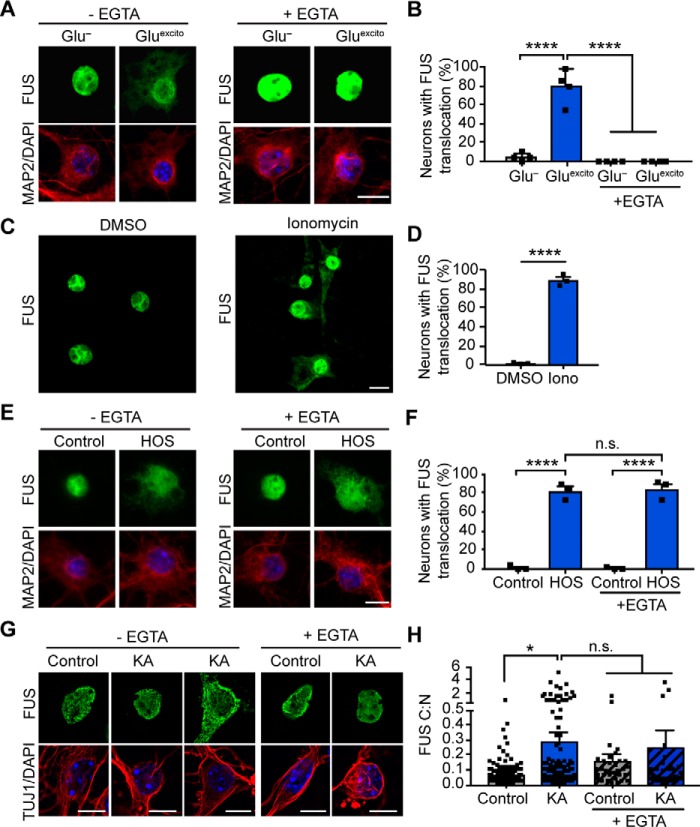
Knowing that calcium influx is a critical component of excitotoxicity (1), we sought to determine whether this signaling chemical is required for the response of FUS to excitotoxicity. To this end, the calcium chelator, EGTA, was included in the neuronal media during the experimental time course. Indeed, EGTA completely prevented Gluexcito-induced FUS egress in primary cortical neurons (Fig. 5, A and B). Furthermore, application of the calcium ionophore, ionomycin, was sufficient to induce FUS translocation in the vast majority (89.0 ± 5.6%) of neurons (Fig. 5, C and D). In light of our previous finding that hyperosmotic stress induces nuclear FUS egress (6), we wondered whether calcium also mediated this response. In contrast to Gluexcito, there was no effect of EGTA on FUS egress in neurons treated with hyperosmotic levels of sorbitol (Fig. 5, E and F), indicative of distinct mechanisms for FUS egress under these stress conditions.
Figure 5.
Calcium is necessary and sufficient for FUS translocation in primary cortical and motor neurons. A, reducing extracellular calcium levels with 2 mm EGTA attenuates FUS egress (green) in MAP2-positive neurons (red) following excitotoxic insult. Nuclei were stained with DAPI (blue). B, quantification of confocal microscopy findings in A confirmed the effect of EGTA treatment (two-way ANOVA and Tukey's post hoc test; for all statistical comparisons, ****, p < 0.0001; n = 4 biological replicates). C and D, application of 10 μm of the calcium ionophore, ionomycin (Iono), for 1 h induced FUS translocation relative to the DMSO control (Student's t test; ****, p < 0.0001; n = 3 biological replicates). E and F, FUS translocation induced by hyperosmotic stress (HOS) was not significantly attenuated by EGTA treatment (two-way ANOVA and Tukey's post hoc test; for all significant statistical comparisons, ****, p < 0.0001, n.s. = nonsignificant; n = 3 biological replicates). G and H, 10-min treatment of 300 μm kainic acid (KA) followed by a 1-h recovery induced FUS egress in primary motor neurons. Neurons were identified using the neuronal marker, TUJ1 (red), and nuclei were stained with DAPI (blue). H, kainic acid–induced FUS egress was statistically significant relative to the washout control; however, the addition of EGTA did not significantly restore FUS localization (two-way ANOVA and Tukey's post hoc test; *, p < 0.0190, n.s. = nonsignificant; control and KA, n = 8 biological replicates, EGTA and KA + EGTA n = 3 biological replicates). Black squares indicate individual cell measurements normalized to the average of the replicate control. Accordingly, means represent the normalized average of biological replicates. Error bars represent S.E. Scale bars = 10 μm.
Next, we investigated the effect of calcium on FUS localization in primary motor neurons, the neuronal cell type predominately affected in ALS. Consistent with cortical neurons, the application of ionomycin to DIV 6–8 motor neurons shifted the nucleocytoplasmic equilibrium of FUS toward the cytoplasm (Fig. S4). Application of the glutamatergic agonist, kainic acid, to motor neurons also induced a significant increase in the C:N ratio of FUS (Fig. 5, G and H). Kainic acid is known to induce motor neuron excitotoxicity (31) and was used here to avoid confounding effects of glutamate uptake by astroglia present in the motor neuron cultures (32). As observed for MAP2 staining in cortical neurons under Gluexcito (Fig. 2A), the cytoskeleton in motor neurons was affected by stress. For example, signals corresponding to neurofilaments (detected by SMI-32; Fig. S4A) and microtubules (detected by TUJ1; Fig. 5G and Fig. S4C) were altered and/or diminished under stress. Importantly, these treated motor neurons were viable as determined by the lack of pyknosis (Fig. 5G and Fig. S4). Furthermore, cleaved caspase-3 staining was not detected in cells with kainic acid–induced FUS translocation but was present in apoptotic cells treated with the proteasome inhibitor MG132 (Fig. S4C).
There was a relatively wide range in the C:N ratio of FUS in kainic acid–treated neurons. For example, a subpopulation of cells exhibited nearly-complete egress of nuclear FUS (Fig. 5, G and H), a result that was not observed in cortical neurons treated with glutamate. Efforts to characterize the properties of neurons that have different degrees of FUS translocation were hampered by an inability to perform live-cell imaging, as a GFP-tagged version of FUS did not undergo cytoplasmic translocation under excitotoxic stress (data not shown). We did notice a trend toward smaller nuclear areas for kainic acid–treated motor neurons, as observed for glutamate-treated cortical neurons (Fig. 2H); however, the former analysis did not reach statistical significance (Fig. S4D). Addition of EGTA to the media prior to kainic acid resulted in only a partial rescue of the FUS response; overall, there were fewer cells with high levels of cytoplasmic FUS (i.e. high C:N ratios), yet some cells were insensitive to EGTA under these conditions. Nevertheless, our collective findings are consistent with a role for calcium in modulating the cytoplasmic egress of FUS under conditions of excitotoxicity.
Excitotoxic stress represses translation independent of FUS expression and stress granule formation
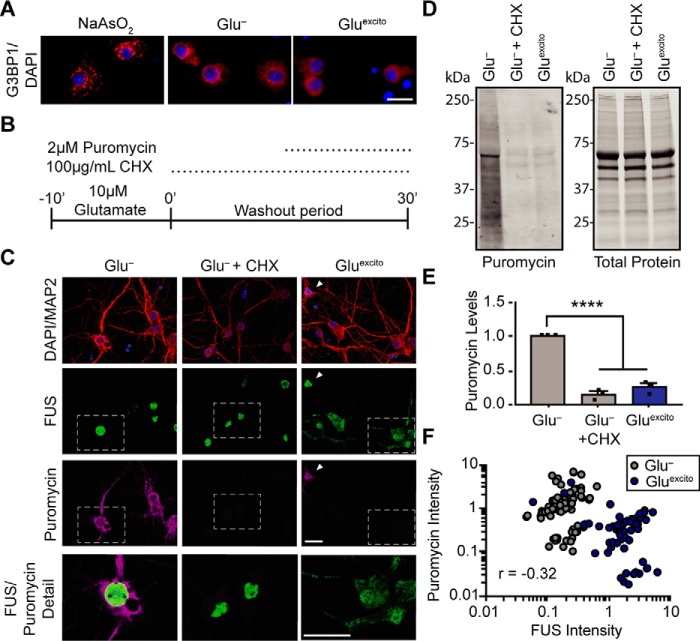
Translational repression and stress granule formation are common cellular responses to stress (33, 34). Given that cytoplasmic FUS has been linked to both translational regulation (35, 36) and stress granule formation (6, 20, 37, 38), we investigated both of these processes during excitotoxic stress. In contrast to neurons treated with sodium arsenite, a stressor known to induce the formation of stress granules (20), Gluexcito did not induce the formation of Ras GTPase-activating protein-binding protein 1 (G3BP1)–positive stress granules in neurons (Fig. 6A). Next, protein translation was assessed by pulse-labeling neurons with puromycin, a small molecule that incorporates into elongating peptides (Fig. 6B) (39). As observed by others, Gluexcito treatment inhibited global translation (40–42) and led to a reduction in puromycin incorporation (Fig. 6, C–E), likely through a mechanism involving inhibition of the translation elongation factor, eukaryotic elongation factor 2 (eEF2) (41, 43). Immunofluorescence detection of puromycin revealed a strong correlation between neurons exhibiting FUS translocation and translational repression; neurons with obvious translocated FUS were puromycin-reduced, and vice versa (Fig. 6, C and F). The degree of translational repression induced by Gluexcito was comparable with treatment of the translational inhibitor cycloheximide (Fig. 6, C–F), which did not promote FUS egress (Fig. 6C). Given the strong correlation between FUS localization and puromycin intensity, we investigated whether FUS expression was required for translational repression (Fig. 7). Knockdown of endogenous FUS with two different shRNAs (44) that target FUS (Fig. 7, A and B and Fig. S5) had no effect on puromycin intensity in the absence or presence of Gluexcito (Fig. 7, C–F). Furthermore, there was no net difference in puromycin intensity as a function of FUS expression between stressed and unstressed conditions (determined by comparing puromycin intensities between stressed and unstressed conditions, for cells with FUS expression or with FUS knockdown) (Fig. 7G). Following the same FUS knockdown protocol, we did not detect a change in cell viability after Gluexcito treatment as a function of FUS expression (data not shown); FUS knockdown was neither protective nor detrimental to the cell under these conditions.
Figure 6.
FUS translocation coincides with translational repression in neurons exposed to Gluexcito. A, immunofluorescence staining of stress granule marker, G3BP1 (red), shows neurons treated with sodium arsenite (NaAsO2) form stress granules unlike Gluexcito or Glu− conditions, where the G3BP1 signal remains diffuse. Scale bar = 20 μm. B, cellular translation in neurons was monitored by pulse treatment and incorporation of the small molecule, puromycin, into nascent peptides during excitotoxic and/or cycloheximide treatment (CHX; inhibitor of protein translation). C, localization of FUS (green) and incorporated puromycin (magenta) in MAP2-positive neurons (red) was assessed by immunofluorescence. Relative to Glu−, protein translation was reduced upon application of cycloheximide or Gluexcito; however, cycloheximide did not induce FUS egress from nuclei (DAPI; blue). The white arrowhead marks a neuron with predominately nuclear FUS and high puromycin staining under Gluexcito, whereas most neurons under this condition have cytoplasmic FUS and reduced puromycin staining. White boxes denote higher magnification details (below) to highlight neurons with representative levels of translation, as observed by anti-puromycin staining. Scale bars = 10 μm. D and E, Western and densitometry analysis of puromycin incorporation confirms a significant reduction in translation following cycloheximide or Gluexcito treatment relative to Glu− (one-way ANOVA and Tukey's post hoc test, for all statistical comparisons, ****, p < 0.0001, n = 3 biological replicates). Puromycin signal was normalized to total protein levels. Error bars represent S.E. F, plotting cytoplasmic FUS intensity (x axis) with puromycin intensity (y axis) for neurons treated with glutamate (blue) or control conditions (gray) revealed a significant, inverse correlation between cytoplasmic FUS and puromycin following excitotoxic insult. Statistical analysis was completed using data from n = 3 biological replicates (Pearson's correlation coefficient r = −0.3214, ****, p < 0.0001, n = 144 neurons).
Figure 7.
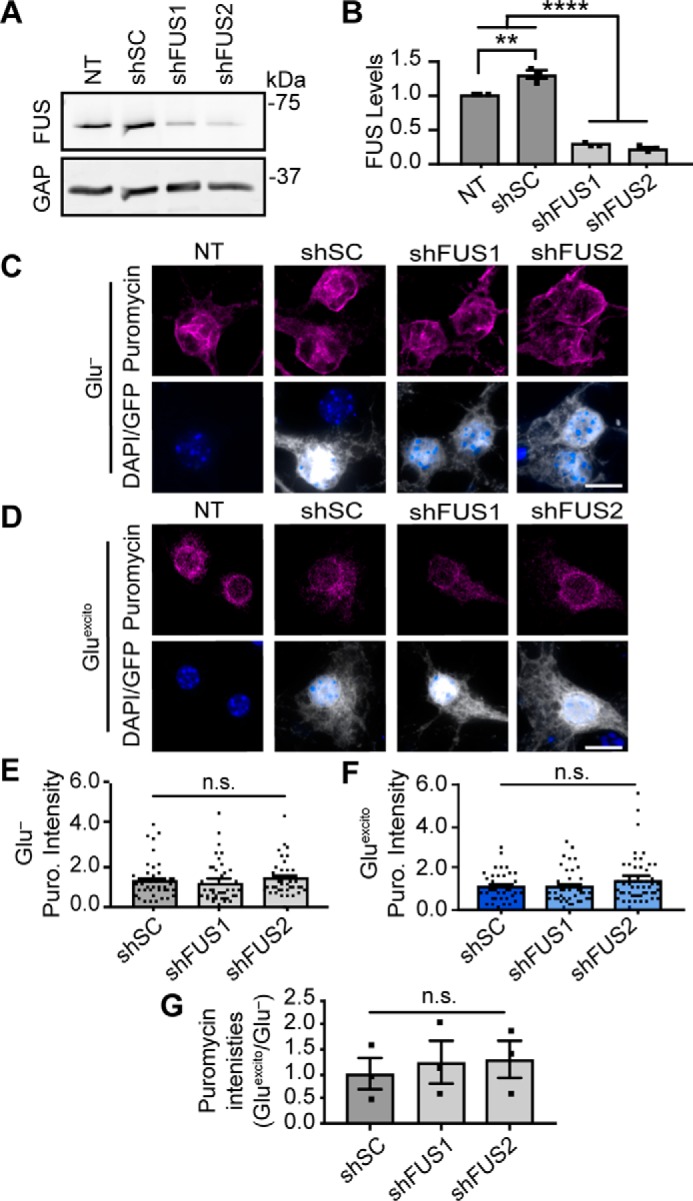
Reduced protein translation following excitotoxic stress is independent of FUS levels. Primary neurons were transduced with shRNAs against mouse FUS (shFUS1 and shFUS2) or a scrambled control (scrambled control) to induce FUS knockdown. A and B, Western and densitometry analysis confirms FUS knockdown relative to nontransduced (NT) and shSC conditions. A modest increase in FUS levels was observed upon expression of shSC relative the loading standard, GAPDH (GAP; n = 3; one-way ANOVA and Tukey's post hoc test, ****, p < 0.0001; **, p = 0.0020; n = 3 biological replicates). C and D, neurons were pulse-chased labeled with puromycin (Puro; magenta) to assess nascent protein translation in transduced cells (as in B–D). The intensity of puromycin (Puro) staining for each condition was normalized to the respective stressed or unstressed nontransduced (NT) control. Scale bar = 10 μm. E and F, quantification of puromycin (Puro) staining from C and D reveals no statistically significant difference in the somatic levels of translation following FUS knockdown (shFUS1 and shFUS2) relative to shSC (one-way ANOVA and Dunnett's post hoc test, n.s. = not significant, n = 3 biological replicates). G, furthermore, there was no significant difference in the relative amount of puromycin intensity between stress and nonstress conditions (Gluexcito/Glu−) as a result of FUS levels (shSC, shFUS1, and shFUS2). For stress and nonstress conditions, puromycin intensities were normalized to the respective, nontransduced control; black squares represent the means represent from n = 3 biological replicates (one-way ANOVA and Tukey's post hoc test, n.s. = nonsignificant). Error bars = S.E.
Gria2 mRNA is elevated in dendrites following excitotoxic insult in a FUS-dependent manner
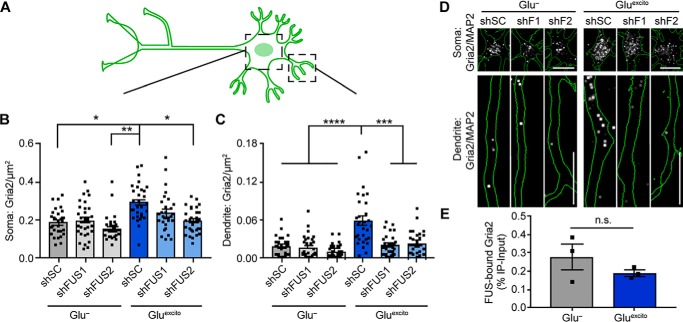
RBPs such as FUS play crucial roles in mRNA processing (9). Although FUS expression did not affect global protein synthesis (Fig. 7), this analysis would not necessarily detect differences in the translation of specific transcripts, especially those targeted to dendrites for local translation (45). Therefore, we investigated whether FUS modulates mRNA metabolism following excitotoxic insult and focused on Gria2, a transcript that is reportedly bound by FUS (46). Gria2 mRNA encodes the GluA2 protein subunit of the AMPA receptor and has been implicated in calcium dyshomeostasis in both ALS (1) and FTD (47). Following depolarization, dendritic GluA2 levels are enhanced (48). Under excitotoxic conditions, we uncovered a significant increase in Gria2 transcript density by FISH in both the soma and dendrites of cortical neurons (Fig. 8 and Fig. S6, A–C). To examine whether this increase in Gria2 mRNA density is FUS-dependent, endogenous FUS levels were knocked down using two shRNAs targeting distinct sequences within FUS as described for Fig. 7 prior to excitotoxic treatment. Consistent with previous findings (46), reduced FUS expression did not have a significant effect on the levels of Gria2 under basal conditions, as determined by FISH within the neuronal soma and dendrites (Fig. 8, B and C). In contrast, Gluexcito-induced changes to Gria2 were significantly attenuated upon FUS knockdown. Dendritic expression of Gria2 was particularly sensitive to FUS levels under Gluexcito, as knockdown of FUS restored dendritic Gria2 levels to baseline (Fig. 8, C and D). Within the time course of the analysis, we were unable to detect significant changes in GluA2 protein levels by Western blot analysis of whole-cell lysates (Fig. S6, D and E). Nevertheless, these data show that FUS expression is required for Gluexcito-induced changes to Gria2 localization in neuronal dendrites (Fig. 8). Indeed, Gria2 mRNA was found associated with FUS upon FUS immunoprecipitation from neurons under both unstressed (Glu−) and stressed (Gluexcito) conditions (Fig. 8E). Although FUS was known to interact with Gria2 under unstressed conditions, this interaction had not been investigated under conditions of stress. To investigate the effects of ALS-linked mutations on FUS-mediated Gria2 processing and localization, FLAG–HA–FUS constructs were transiently transfected into neurons as described for Fig. 3. However, in contrast to nontransfected neurons (Fig. 8), dendritic levels of Gria2 were unaffected by Gluexcito in FLAG–HA–FUS WT-expressing cells (data not shown), possibly a consequence of overexpressed FUS WT exerting a dominant-negative effect. Together, our data suggest that WT FUS directly affects the subcellular localization of Gria2 under Gluexcito and that the role of mutant FUS should be investigated in systems with endogenous levels of protein (25).
Figure 8.
Elevation of Gria2 mRNA in dendrites following Gluexcito requires FUS expression. A, lentivirus expressing a GFP reporter and scrambled control shRNA (shSC) or shRNA against FUS (shFUS1 and shFUS2) were used to reduce FUS levels in neurons to evaluate Gria2 mRNA distribution in soma and dendrites. B and C, following excitotoxic insult, the density of Gria2 was increased in both soma (B) and dendrites (C) of shSC-transduced neurons; upon FUS knockdown, dendritic Gria2 did not increase following treatment with Gluexcito using a two-way ANOVA and Dunnett's post hoc test (Soma: Glu− shSC versus Gluexcito shSC, *, p = 0368; Glu− shFUS2 versus Gluexcito shSC, **, p = 0.0050; Gluexcito shSC versus Gluexcito shFUS2, *, p = 0.0476; dendrites: ****, p < 0.0001; Gluexcito shSC versus Gluexcito shFUS1, ****, p = 0.0001; Gluexcito shSC versus Gluexcito shFUS2, ****, p = 0.0002; n = 3 biological replicates). Black squares indicate individual cell measurements. Error bars represent S.E. Scale bars = 10 μm. D, representative images of B and C. Gria2 mRNA was detected by FISH (white) in neurons outlined in green (raw images shown in Fig. 7-1B; image processing described in Fig. 7-1C). E, amount of Gria2 mRNA bound by immunoprecipitated neuronal FUS is not significantly different between stress and nonstress conditions (Student's t test, n.s. = nonsignificant; n = 3 biological replicates).
Discussion
This study uncovered an association between disease-linked RBPs and excitotoxicity, a stress that has particularly profound effects on the nucleocytoplasmic distribution of FUS in both cortical (Figs. 1 and 2) and motor neurons (Fig. 5). There is a compelling body of evidence linking glutamate-induced excitotoxicity to neurodegenerative diseases, including ALS (1, 2, 49). For instance, elevated levels of glutamate were detected in biological samples from ALS patients (50–52). Cell death caused by glutamate and calcium dysregulation has also been documented in multiple animal and cellular models (1, 2, 50, 53–55). The outcomes of this study shed new light on the excitotoxicity cascade and implicate a role for the ALS/FTD-linked protein FUS in this process.
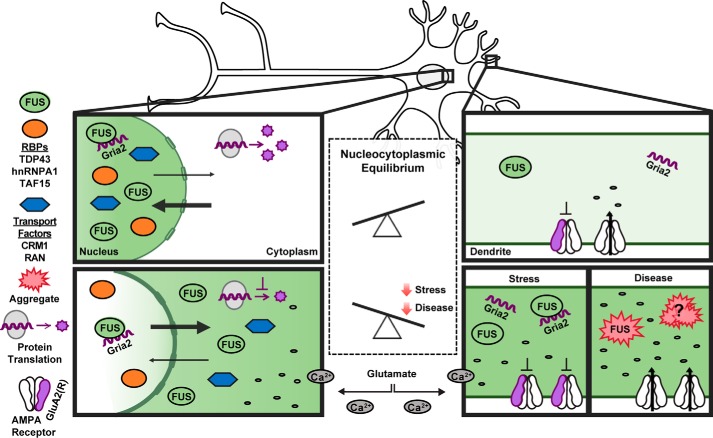
Our results are consistent with a functional role for FUS in response to glutamatergic signaling (56) rather than a nonspecific effect of cell death. First, FUS egress precedes cell death (Fig. 2 and Fig. S4C). Second, there is selectivity with respect to proteins that undergo a change in cellular localization; the response of FUS is particularly robust compared with the other proteins assessed in this study (Fig. 1). Third, the effects of excitotoxicity on Gria2 depend on FUS expression (Fig. 8). FUS binds Gria2 mRNA within introns and the 3′-untranslated region (UTR), and Gria2 splicing is affected by FUS expression under basal conditions (46). FUS binding to the 3′UTR of Gria2 is thought to occur synergistically with microRNAs that modulate Gria2 expression (57). Here, we confirmed that FUS binds Gria2 under unstressed conditions, and we show that FUS remains associated with Gria2 under conditions of Gluexcito (Fig. 8E). Subsequently, Gria2 density was enhanced in neuronal dendrites in a FUS-dependent manner under Gluexcito (Fig. 8, B and D). These data are consistent with a role for FUS in Gria2 nucleocytoplasmic transport. In the absence of stress, FUS binds Gria2 (Fig. 8E), and we suspect this interaction takes place predominately in the nucleus (Fig. 9). Upon exposure to glutamate stress, we posit that Gria2 is transported into the dendrites as part of a complex with FUS (Fig. 9). Gria2 encodes the GluA2 protein subunit of the AMPA receptor. Normally, GluA2 is post-transcriptionally edited, and GluA2-containing AMPA receptors are calcium-impermeable. As such, the calcium permeability of AMPA receptors and the susceptibility of neurons to excitotoxicity depend on GluA2 (1, 58). We speculate that the enhanced dendritic density of Gria2 may serve to increase the number of calcium-impermeable (GluA2-containing) AMPA receptors and thereby offset calcium influx caused by existing calcium-permeable (GluA2-lacking) receptors (Fig. 9). Accordingly, we expect FUS to serve a protective role under conditions of Gluexcito. We suspect that the acute toxicity imparted by Gluexcito was too strong to detect additional toxicity from FUS knockdown and that this relationship between FUS expression and cellular viability could be better addressed using in vivo models of chronic stress (59).
Figure 9.
Model depicting the impact of excitotoxic stress on neuronal homeostasis and disease pathogenesis. Under homeostatic conditions, shuttling RBPs such as FUS are predominately localized within the nucleus (top). Excitotoxic levels of glutamate (bottom) induce a massive influx of calcium, which is sufficient to induce the robust nuclear egress of FUS into the neuronal soma and dendrites. Excitotoxic stress also leads to translational repression, a re-distribution of nucleocytoplasmic transport factors, and increased levels of Gria2 transcript within dendrites. The expression of FUS is required for enhanced levels of dendritic Gria2 in response to excitotoxic stress, implicating an RNA-processing role for FUS under these conditions. Enhanced levels of edited Gria2 transcript may represent a mechanism to offset the toxic effects of calcium influx. We speculate that prolonged or severe stress could manifest in the pathological aggregation of RBPs, including FUS, in neurodegenerative diseases such as ALS and FTD. Aberrant processing of Gria2 and/or GluA2 can occur through several mechanisms (e.g. expression of mutant FUS in astrocytes, loss of FUS function due to aggregation, and other means as described in the text), and contributes to calcium dyshomeostasis during disease.
In ALS, a putative role for FUS under excitotoxic stress could be compromised as a result of dysregulated Gria2 editing and/or GluA2 expression (58, 60), particularly in motor neurons that rely heavily on AMPA receptor signaling (1, 2). Although we were unable to determine whether Gria2 processing is affected by exogenous expression of ALS-linked FUS, altered expression of genes involved in glutamate receptor signaling were recently found in humanized mutant FUS mouse models. Subsequent bioinformatics analyses predict an inhibition of this pathway within mutant FUS spinal cord tissues (25). A “loss of nuclear FUS function” may also impact gene expression under conditions of Gluexcito. For example, FUS egress under conditions of hyperosmotic stress coincided with altered splicing of a known FUS target, sodium voltage–gated channel α subunit 4 (SCN4A) (61). Furthermore, endogenous expression of a severely mislocalized FUS variant correlated with reduced levels of Gria2 in motor neurons, a phenotype that was attributed to both loss of nuclear and gain of cytoplasmic functions of the mutant protein (57). However, the relationship between FUS and Gria2 was not examined under conditions of stress in this prior study nor was the effect of FUS expression on Gria2 subcellular localization. Our data suggest that FUS directly influences the localization and expression of specific transcripts such as Gria2 but that FUS expression does not augment or attenuate global protein translation under either stressed or unstressed conditions (Fig. 7). It will be of interest to identify other genes that are influenced by FUS translocation under Gluexcito or hyperosmotic stress and as a function of disease (25, 35, 62).
While investigating the mechanism(s) underlying excitotoxic FUS egress, we uncovered changes to the CRM1 nuclear export pathway (Fig. 4). Inhibition of CRM1-mediated export by KPT-330 failed to restrict both NLS–tdTomato–NES (Fig. 4, A and B) and FUS (Fig. 4, A and C) within the nucleus under Gluexcito. Furthermore, CRM1 localization was significantly shifted toward the cytoplasm (Fig. 4, D and E). Despite these changes, nucleocytoplasmic transport was not completely dysregulated, as a partial inhibitory effect of KPT-330 on the shuttling reporter was observed (Fig. 4B). Our KPT-330 studies suggest that Gluexcito-induced FUS egress occurs through a mechanism other than active CRM1 export and could entail passive diffusion (26) or alternative transport factors (63). Selectivity of RBP egress following Gluexcito may stem from differences in nucleocytoplasmic shuttling dynamics, which are influenced by multiple factors, including binding interactions and post-translational modifications (64). An interesting area of future study could be to elucidate these factors and determine whether they are modulated by stress.
Alterations to CRM1 and Ran localization (Fig. 4) under Gluexcito may represent early signs of nucleocytoplasmic transport decline. Indeed, previous studies show that various forms of stress (e.g. excessive calcium influx, oxidative, and hyperosmotic stress) cause damage to nuclear pores and impair nucleocytoplasmic transport (65–69). Mice deficient in key astroglial glutamate transporters exhibited both nuclear pore degradation and motor neuron degeneration (59). Moreover, the nucleocytoplasmic transport pathway has been implicated in age-related neurodegeneration, particularly in the context of ALS and FTD (70). Whereas most ALS/FTD-associated studies have focused on the role of mutant proteins in dysregulating nucleocytoplasmic transport (30, 70), ALS/FTD-associated forms of cellular stress (e.g. excitotoxicity) may also contribute to nucleocytoplasmic transport defects in both inherited and sporadic forms of disease. In fact, nucleocytoplasmic transport is an emerging area of therapeutic development, and the CRM1 inhibitor KPT-350 is advancing toward ALS clinical trials. Partial inhibition of CRM1 is expected to offset defects in nuclear import. CRM1 inhibitors have had a therapeutic effect in some (70, 71), but not all (59, 63), models of neurodegeneration. Collectively, the available data support CRM1-mediated nucleocytoplasmic transport as a viable therapeutic target for neurodegenerative disorders. However, a combination therapy addressing additional effects of stress-induced nuclear pore degradation (i.e. calpain inhibitors (59)) may be required for a significant therapeutic outcome.
The calcium-mediated response of FUS to Gluexcito has additional implications for neurodegeneration, including cases of FUS-mediated ALS. For instance, motor neurons derived from human ALS–FUS-induced pluripotent stem cells are intrinsically hyperexcitable (72). Furthermore, the effects of ALS-linked FUS on calcium-mediated motor neuron toxicity is exacerbated by expression of the mutant protein in astrocytes (55, 58). Most ALS-linked FUS mutations are located within the NLS (24) and induce a shift in the nucleocytoplasmic equilibrium of the protein toward the cytoplasm, where it is believed to exert a gain of toxic function (20, 73). As ALS-linked variants R521G and H517Q translocate further into the cytoplasm under Gluexcito (Fig. 3), we predict these and other variants with impaired binding to nuclear import factors will accumulate in the cytoplasm under conditions of chronic stress in vivo (36, 65). Moreover, chronic stress may result in nuclear depletion and cytoplasmic aggregation of WT FUS and TDP-43 in sporadic cases as well (13, 14). We propose a model whereby FUS and related RBPs play a functional role in response to normal stimulation and moderate degrees of stress, but excessive or chronic stress severely disrupts their nucleocytoplasmic equilibrium and contributes to disease pathology (Fig. 9).
Experimental procedures
Cell culture and stress application
HEK293-T cells were cultured as described (6). All research involving animals for the primary neuron cultures described below was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Massachusetts Medical School. Dissociated primary cortical neuron cultures were prepared using cortices from C57BL/6 embryonic day 14–15 mice. Embryos were isolated in ice-cold Hanks' buffered saline solution (Corning, catalog no. 21-023-CV), and the meninges were removed. Cells were dissociated for 12 min in 0.05% trypsin (Invitrogen, catalog no. 25300-054) at 37 °C, diluted in Dulbecco's modified Eagle's medium (Invitrogen, catalog no. 11965118) containing 10% fetal bovine serum (MilliporeSigma, catalog no. F4135), and strained with a cell strainer before gently pelleting. Cells were then resuspended in Neurobasal media (Invitrogen, catalog no. 21103049), supplemented with 1% Glutamax (Invitrogen, catalog no. 35050-061), 1% penicillin/streptomycin (Invitrogen, catalog no. 15140122), and 2% B-27 (Invitrogen, catalog no. 0080085-SA), and plated at 1.8–2 × 105 cells/ml on polyornithine (final concentration of 1.5 μg/ml; MilliporeSigma; catalog no. P4957)-coated plates or coverslips. Neuronal cultures were grown under standard culture conditions (37 °C, 5% CO2, 95% air) and fed every 3–4 days by adding half-volumes of supplemented neurobasal media to each well/dish, with additional half-changes of media occurring every other feeding. Unless indicated, during the first feeding (DIV 2 or 3) neuron cultures were also treated with a final concentration of 0.5–1 μm cytosine β-d-arabinofuranoside hydrochloride (MilliporeSigma, catalog no. C6645) to inhibit non-neuronal cell growth. Experiments were performed days in vitro (DIV) 14–16.
Primary motor neurons were isolated from embryonic day 12.5 murine spinal cords as described (73). Briefly, after dissociation in 0.1% trypsin (Worthington, catalog no. LS003707) at 37 °C for 12 min, primary motor neurons were purified using a 6% Optiprep (MilliporeSigma, catalog no. D1556) density gradient and plated on glass coverslips coated with 0.5 g/liter polyornithine and natural mouse laminin (Thermo Fisher Scientific, catalog no. 23017015, Waltham, MA). Cells were grown in glia-conditioned Neurobasal medium and supplemented with 2% B27, 2% horse serum (MilliporeSigma, catalog no. H1270), and 10 ng/ml brain-derived neurotrophic factor (PeproTech Rocky Hill, NJ, catalog no. 450-02), glial cell line-derived neurotrophic factor (PeproTech catalog no., 450-44), and ciliary neurotrophic factor (PeproTech, catalog no. 450-50). Primary motor neurons were treated on DIV 6–8 with ionomycin or DMSO and on DIV 8 with kainic acid. For glutamate experiments, 100 mm glutamate (MilliporeSigma, catalog no. G5889) was freshly prepared in neurobasal media and diluted using primary neuron-cultured media to achieve 0.1–10 μm solutions. To apply stress, neuronal media were replaced with glutamate-containing primary cultured media or primary cultured media alone (glutamate-free control) for 10 min. After 10 min, treatment media were replaced with primary cultured media for 30 min or longer depending on the experiment prior to fixation or lysate collection. Kainic acid (Abcam, catalog no. ab144490) was diluted from 10 to 300 μm/ml in primary cultured media and added to motor neurons for 10 min followed by a replacement with glia-conditioned media for 1 h. Stock solutions of 5 mm ionomycin (MilliporeSigma, catalog no. I9657) or 1 m sodium arsenite (MilliporeSigma 71287) prepared in DMSO (Corning, catalog no. 25-950-CQC) or water were diluted to 10 μm or 1 mm in primary cultured media, respectively, before addition to neurons for 1 h. MG132 was prepared in DMSO and diluted to 1 μm in neuronal media prior to adding to primary motor neurons for 24 h. Sorbitol (MilliporeSigma, catalog no. S6021) was directly dissolved in primary cultured media to obtain a final concentration of 0.4 m and applied to cells for 1 h. For experiments in which EGTA (MilliporeSigma, catalog no. E3889) was added, a 100 mm stock was prepared in water, diluted to 2 mm in primary cultured media, and allowed to incubate for 30 min prior to use during the experimental time course. Translation was inhibited with 2 μm cycloheximide (MilliporeSigma, catalog no. C7698). Neurons were treated with 500 nm KPT-330 (Cayman Chemical, catalog no. 18127) dissolved in water on DIV 13 for 48 h prior to treatment with glutamate as well as during the experimental time course.
Immunofluorescence analysis
Primary cortical and motor neurons were fixed with 4% paraformaldehyde (Thermo Fisher Scientific, catalog no. AAA1131336) at room temperature for 15 min and permeabilized with 0.1–0.2% Triton X-100. Cortical neuron immunofluorescence experiments were conducted as described (6, 20) using antibodies described in Table 1. Primary motor neuron samples were blocked in 4% BSA for 45 min and hybridized overnight at 4 °C with primary antibodies (Table 1) and AlexaFluor-conjugated secondary antibodies (73).
Table 1.
Primary antibodies used for immunofluorescence and Western analyses
| Antibody | Speciesa | Company | Dilution/incubationb |
|---|---|---|---|
| FUS | R | Bethyl Laboratories A300–293A (Montgomery, TX) | IF: 1:1000 1 h or 1:500 O/N |
| FUS | M | Santa Cruz Biotechnology sc-47711 (clone 4H11, Dallas, TX) | IF: 1:200 1 h; WB: 1:1000 4 °C O/N |
| FUS | R | Genscript (Piscataway, NJ) (in-house) | IF: 1:200; WB: 1:1000 4 °C O/N |
| FUS | M | Santa Cruz Biotechnology sc-373698 (H-6) | IF: 1:200 1 h; WB: 1:1000 4 °C O/N |
| FUS | R | Bethyl Laboratories, A300–302A | IF: 1:1000 1 h or 1:500 O/N |
| MAP2 | M | MilliporeSigma M9942 (clone HM-2) | IF: 1:1000 1 h |
| MAP2-CY3 | M | MilliporeSigma MAB3418C3 | IF: 1:1000 1 h |
| NeuN | M | MilliporeSigma MAB377 | IF: 1:200 1 h |
| SMI-32 | M | Thermo Fisher Scientific | IF: 1:500 O/N |
| Alexa647-anti-TUBB3 (TUJ1) | M | Biolegend AA10 | IF: 1:500 1 h |
| Cleaved Caspase-3 | R | Cell Signaling Technology no. 9661S | IF: 1:400 4 °C O/N |
| GAPDH | R | MilliporeSigma G9545 | WB: 1:20,000 4 °C O/N |
| GAPDH | M | MilliporeSigma G8795 | WB: 1:2000 4 °C O/N |
| TAF15 | R | Abcam ab134916 (Cambridge, UK) | IF: 1:250 1 h, WB: 1:1000 4 °C O/N |
| hnRNPA1 | M | MilliporeSigma R4528 | IF: 1:1000 1 h, WB: 1:2000 4 °C O/N |
| TDP-43 | M | EnCor Biotechnology Inc. MCA-3H8 (Gainesville, FL) | IF: 1:250 1 h, WB: 1:500 4 °C O/N |
| Lamin A/C | M | MilliporeSigma SAB4200236 | IF: 1:500 1 h |
| HA | R | Cell Signaling Technology C29F4 (Danvers, MA) | IF: 1:500 1 h |
| RAN | R | Bethyl Laboratories A304–297A | IF: 1:250 1 h |
| CRM1 | R | Bethyl Laboratories A300–469A | IF: 1:250 1 h |
| Puromycin | R | Thermo Fisher Scientific, MABE343MI (clone 12D10) | IF: 1:250 1 h, WB: 1:1000 4 °C O/N |
| G3BP1 | R | Proteintech 13057–2-AP (Rosemont, IL) | IF: 1:2000 1 h |
| GFP | R | Invitrogen A-21311 | IF: 1:1000 1 h |
| GluA2 | M | MilliporeSigma MAB397 | WB: 1:1000 4 °C O/N |
a R is rabbit host; M is mouse host.
b IF is immunofluorescence; WB is Western blot; O/N is overnight.
Image acquisition and processing
Primary motor neuron images were imaged using a widefield fluorescence microscope (Nikon TiE, Melville, NY) equipped with a cooled CMOS camera (Andor, South Windsor, CT). Primary motor neuron images were acquired as Z-stacks (0.2-μm step size) using a ×60 lens. As indicated, fixed primary cortical neurons were imaged using a Leica TCS SP5 II laser-scanning confocal (Leica Microsystems, Buffalo Grove, IL) or Leica DMI6000B microscope as described (6). For confocal images of whole cells, 12-bit stacks (Δz = 0.25 μm steps, zoom = ×3, n = 23–30 planes) were acquired at ×40 with a pixel size of 126 nm (1024 × 1024 pixels; 1000 Hz). For dendrites, 12-bit stacks (Δz = 0.08 μm steps, zoom = ×3, n = 40–50 planes) were acquired at ×63 using a pixel size of 80 nm (1024 × 1024 pixels; 1000 Hz). For FISH, mFUS, and somatic puromycin analyses, widefield stacks of the entire cell were acquired (z = 0.2-.25 μm) and deconvolved using the LAS AF One Software Blind algorithm (10 iterations). All neuron images were analyzed using MetaMorph software (Molecular Devices Inc., San Jose, CA). The background and shading of stacks were corrected as described (20). Sum or maximum projections were created from corrected stacks for downstream analyses.
For the quantification of C:N ratios, a 20 × 20-pixel region was applied to the nucleus and perinuclear area in the soma for each cell (visualized by DAPI and MAP2, neuronal nuclei (NeuN), TUJ1, or SMI-32, respectively) as well as an area within each image that contained no cells (representing background fluorescence). Using MetaMorph or ImageJ, the integrated intensity for the signal of interest was obtained for each region, and a ratio of the cytoplasmic:nuclear signal was then generated following subtraction of background signal. For each experiment, the statistical comparison of C:N ratios with or without excitotoxic stress was completed using average C:N ratios collected from ≥3 independent, biological experiments. For the analysis of FUS levels in neuronal dendrites, microtubule-associated protein 2 (MAP2) was used to visualize neuronal dendrites and create a dendritic mask using MetaMorph. Using the MAP2-defined mask, the integrated intensity of FUS staining was obtained and used to quantify the relative amount of FUS staining in dendrites. For the quantification of nuclear area, DAPI images were thresholded using MetaMorph, and the area of individual nuclei per each cell was obtained. For the quantification of total neurons and neurons exhibiting FUS, CRM1, or RAN cytoplasmic translocation by immunofluorescence, ≥10 fields of view were imaged at ×40 for each condition tested. As indicated by MAP2 or NeuN staining, neurons were quantified from images with computer assistance from the “Cell Count” feature in MetaMorph. To assess the percent neurons with protein translocation, cells were scored for the presence of cytoplasmic protein divided by the total neuron number to generate the percent population exhibiting a response. All line scan analyses were completed using the Intensity Profile feature in ImageJ and plotted using GraphPad Prism.
Western blot analysis
Neurons were treated, washed twice with PBS, and lysed using RIPA buffer (Boston BioProducts, catalog no. BP-115-500, Ashland, MA) supplemented with protease (Roche Applied Science, catalog no. 11836170001, Basel, Switzerland) and phosphatase inhibitors (Roche Applied Science, catalog no. 4906837001). Lysates were centrifuged at 19,357 × g for 15 min, after which the supernatant was collected and its protein concentration determined using a bicinchoninic acid assay (Thermo Scientific Pierce catalog no. 23227, Rockford, IL). Lysates were subsequently used for Western and densitometry analysis as described (44). Gels were loaded with 8–20 μg of lysate, and GAPDH was used as a loading standard to determine relative protein levels. Primary antibodies used for analysis are described in Table 1; LI-COR (Lincoln, NE) secondary antibodies were used as described (44).
LDH analysis
Neuron toxicity to glutamate was analyzed by the LDH assay using the CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, catalog no. G1782, Madison, WI).
Plasmids and cloning
Human cDNA for FLAG–HA-tagged WT, H517Q, R521G, or R495X FUS were cloned into the lentiviral vector, pLenti-CMV-TO-Puro-DEST (Addgene, catalog no. 670-1, Cambridge, MA), using the In-Fusion HD Cloning Plus kit (Clontech catalog no. 638909). To achieve FUS knockdown, shRNA sequences (44) were packaged using In-Fusion HD cloning into the lentiviral backbone, CSCGW2 (a generous gift from Dr. Miguel Sena-Esteves, University of Massachusetts Medical School), which contains a green fluorescent protein (GFP)-reporter expressed under a separate CMV promoter. The shRNA targeting sequences were 5′-GCAACAAAGCTACGGACAA-3′ (shFUS1) and 5′-GAGTGGAGGTTATGGTCAA-3′ (shFUS2), as well as the scrambled control sequence 5′-AATTCTCCGAACGTGTCACGT-3′ (shSC). The shuttling reporter, NLS–tdTomato–NES (a generous gift from Dr. Martin Hetzer, Salk Institute (29)) was cloned into the pLenti-CMV-TO-Puro-DEST vector backbone (Addgene, catalog no. 670-1) using Gateway BP and LR Clonase reactions (Invitrogen, catalog nos. 1789020 and 11791020, respectively). The shuttling reporter contained an NLS sequence (PPKKKRKVQ) and NES sequence (LQLPPLERLTL) attached to tdTomato by a GGGG linker at the N and C termini, respectively.
Transient expression of ALS-mutant FUS
For transient transfection experiments, neurons were fed at DIV 6 and transfected with FLAG–HA–FUS constructs on DIV 7 using NeuroMag (Oz Biosciences, Marseille, France, catalog no. NM51000) transfection reagents. DNA (1.0 μg) and 1.75 μl of NeuroMag (for one 24-well plate with a 500-μl volume) were combined in an Eppendorf tube, and the volume was adjusted to 50 μl with neurobasal media. The DNA mixture was allowed to incubate for 20 min before addition to neurons. Neuron cultures were then placed on a NeuroMag magnet plate (Oz Biosciences, catalog no. MF10096) within the tissue culture incubator for 15 min to complete transfection. Transfected neurons were collected for experimental analyses on DIV 14–16.
Lentiviral production and application
High-titer lentivirus was prepared as described (74). Briefly, HEK-293T cells were individually transfected using calcium phosphate with the shRNA or NLS–tdTomato–NES constructs described along with the packing plasmids: CMVdR8.91 plasmid and VSV-G. DNA constructs were prepared using EndoFree Maxi Prep (Qiagen Germantown, MD, catalog no. 12362). Three hours after transfection, cell media were replaced with Opti-MEM (Invitrogen, catalog no. 31985070), and the virus was collected in open-top Beckman tubes (Beckman Coulter, Brea, CA, catalog no. 344058) by ultracentrifugation at a 133,907 relative centrifugal force in an SW32Ti rotor 72 h following transfection. Lentivirus titer was obtained by the transduction of HEK cells with serially diluted lentivirus. Upon titer determination, virus was added to DIV 6 noncytosine β-d-arabinofuranoside hydrochloride–treated neurons at an approximate titer of 1.2–1.810 transducing units/ml. For all transduction experiments except fluorescence in situ hybridization, neurons were cytosine β-d-arabinofuranoside hydrochloride–treated on DIV 7. Transduced neurons were collected 9 days post-transduction (DIV 15) for analysis.
Puromycin analysis
Based on previously described experiments (35), 4 mm stocks of puromycin (Invitrogen, catalog no. A11138-03) were prepared in water. Neurons were treated with glutamate as described, except that a final concentration of 2 μm puromycin was added to the primary cultured media during the last 15 min of the “washout” period. As a positive control of translational inhibition, 100 μg/ml cycloheximide (MilliporeSigma, catalog no. C7698) was included throughout the experimental time course. Neurons were then analyzed by Western or immunofluorescence using an anti-puromycin antibody (Table 1). For the analysis of puromycin immunostaining upon FUS knockdown, a 20 × 20-pixel region was placed in the soma of GFP-positive cells. Using MetaMorph, the integrated intensity of this region was obtained and used to quantify relative puromycin levels as described for the C:N signal intensity analyses above. Similarly, a 20 × 20-pixel region was applied to the neuronal soma to obtain integrated intensity values for cytoplasmic FUS and puromycin staining. Obtained values for a given cell were used for the correlation analysis of these variables.
FISH analysis
Noncytosine β-d-arabinofuranoside hydrochloride–treated neurons were plated on coverslips and transduced with shFUS or shSC-expressing lentivirus on DIV 6 and harvested on DIV 15. Following stress application, neurons were fixed with fresh 4% paraformaldehyde (Thermo Fisher Scientific, catalog no. F79-500) diluted in RNase-free water (Corning, catalog no. 46-000-CM) for 30 min at ambient temperature. FISH labeling was completed using a QuantiGene ViewRNA ISH cell assay kit (Affymetrix, Santa Clara, CA, catalog no. QVC0001) according to the manufacturer's instructions. One exception to the protocol was that samples were dehydrated after fixation with 2-min incubations in 50, 70, and 100% ethanol at ambient temperature followed by a second addition of 100% ethanol and stored at −20 °C for 5 days before processing. The Gria2–Cy3 probe was designed and tested for specificity by Affymetrix. For post-FISH immunofluorescence staining, after probe labeling, coverslips were washed in PBS for 5 min and then blocked and processed for immunofluorescence as described (75). Coverslips were probed with MAP2 and GFP to visualize neurons and transduced cells, respectively. For analysis, neurons with at least two dendrites of >50 μm in length that did not excessively overlap with other cells were selected. Maximum projections of the imaged z-stacks were analyzed using MetaMorph software. For each neuron analyzed, 2–3 dendrites and the cell body were assessed for their area and the number of mRNA puncta present. Average dendrite data were reported for each cell, and 10 cells were analyzed per construct/condition. Images were prepared for visualization in the figures based upon methods previously described (76).
RNA-immunoprecipitation and quantitative PCR
Primary cortical neurons were plated on 1.5 μg/ml polyornithine-coated 10-cm plates and stressed as described above at DIV 13–15. Cells were harvested with polysome lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES, pH 7, 0.5% Nonidet P-40, freshly supplemented with protease inhibitors and 200 units of RNaseOUT, Invitrogen catalog no. 10777019). Lysates were cleared through centrifugation (16,000 × g) for 15 min at 4 °C. Protein concentrations were determined using the BCA protein assay kit (Thermo Fisher Scientific, catalog no. 23227). Lysates (1 mg of total protein) were subjected to immunoprecipitation (IP), and 100 μg (10%) of the lysates were reserved for “input” samples. Magnetic Surebeads protein A (100 μl/per condition; Bio-Rad, catalog no. 1614013) were washed with NT2 buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm MgCl2, 0.05% Nonidet P-40) and charged with 40 μg of rabbit anti-FUS antibody (Genscript) or Chrompure rabbit IgG whole molecule (Jackson ImmunoResearch, catalog no. 011-000-003) for 2 h at 4 °C by rotation. Charged beads were washed with NT2 buffer, and 1 mg of lysates were incubated with charged beads at 4 °C overnight by rotation. Supernatants were discarded, and beads were thoroughly washed six times with NT2 buffer. Beads were resuspended in 100 μl of NT2 buffer supplemented with 0.1% SDS and 30 μg of proteinase K (Invitrogen, catalog no. AM2546) and incubated for 30 min at 55 °C. RNA from both IP and input were extracted using TRIzol reagent (Invitrogen, catalog no. 15596026) following the manufacturer's protocol. Reverse transcription was performed using iScript Reverse Transcription Supermix (Bio-Rad, catalog no. 170-8841) as per the manufacturer's recommendation. Quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, catalog no. 1725122) with PrimePCR SYBR mouse GRIA2 probe (Bio-Rad, catalog no. qMmuCED0047098) on a CFX384 Real-Time PCR thermal cycler (Bio-Rad). Ct values were acquired through CFX ManagerTM software, and data were presented as % input, which is calculated as followed: ΔCt ((normalized IP) = Ct (IP) − (Ct (input) − log2 (input dilution factor)); % input = 2−ΔCt(normalized IP).
Experimental design and statistical analysis
At least n = 3 independent biological experiments were performed for every statistical analysis with GraphPad Prism software; this includes having an independent primary neuron preparation for each biological experiment. The number of independent biological experiments and the statistical test(s) performed are explicitly described in the figure legends as follows. A Student's t test at 95% confidence was used when comparing two conditions. For more than two conditions, either a one-way or two-way ANOVA was used followed by the appropriate post-hoc analyses (Tukey's or Dunnett's). A Pearson correlation coefficient was used for correlations analyses.
Author contributions
M. T., C. F., and D. A. B. conceptualization; M. T., D. M. B., Y.-C. L., K. V. G., C. F., and D. A. B. data curation; M. T., D. M. B., Y.-C. L., C. F., and D. A. B. formal analysis; M. T., D. M. B., Y.-C. L., K. V. G., J. E. L., C. F., and D. A. B. investigation; M. T., D. M. B., Y.-C. L., K. V. G., C. F., and D. A. B. methodology; M. T., C. F., and D. A. B. writing-original draft; M. T., C. F., and D. A. B. writing-review and editing; K. V. G., J. E. L., C. F., and D. A. B. resources; D. A. B. supervision; D. A. B. funding acquisition; D. A. B. project administration.
Supplementary Material
Acknowledgments
We thank Drs. Kensuke Futai (University of Massachusetts Medical School) and Miguel Sena-Esteves (University of Massachusetts Medical School) for sharing reagents and advice, Dr. Martin Hetzer (Salk Institute) for providing the NLS–tdTomato–NES construct, and all the members of the Bosco and Landers labs for their valuable input.
This work was supported by National Institutes of Health Grants R21 NS091860 (to D. A. B.) and R01 NS078145 (to D. A. B.) from NINDS; ALS Association Grant 18-IIA-418 (to C. F.), and a Zelda Haidek Memorial Scholarship from University of Massachusetts Medical School (to M. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ALS
- amyotrophic lateral sclerosis
- KA
- kainic acid
- ANOVA
- analysis of variance
- FUS
- fused in sarcoma
- TLS
- translocated in liposarcoma
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RBP
- RNA-binding protein
- ALS
- amyotrophic lateral sclerosis
- ER
- endoplasmic reticulum
- FTD
- frontotemporal dementia
- DIV
- day in vitro
- IP
- immunoprecipitation
- C:N
- cytoplasmic to nuclear ratio
- NES
- nuclear export sequence
- NLS
- nuclear localization sequence
- shSC
- scrambled control
- LDH
- lactate dehydrogenase
- DAPI
- 4′,6-diamidino-2-phenylindole
- CMV
- cytomegalovirus.
References
- 1. Van Dee Bosch L., Van Damme V. P., Bogaert E., and Robberecht W. (2006) The role of excitotoxicity in the pathogenesis of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1762, 1068–1082 10.1016/j.bbadis.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 2. Starr A., and Sattler R. (2018) Synaptic dysfunction and altered excitability in C9ORF72 ALS/FTD. Brain Res. 1693, 98–108 10.1016/j.brainres.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitchell J., Paul P., Chen H.-J., Morris A., Payling M., Falchi M., Habgood J., Panoutsou S., Winkler S., Tisato V., Hajitou A., Smith B., Vance C., Shaw C., Mazarakis N. D., and de Belleroche J. (2010) Familial amyotrophic lateral sclerosis is associated with a mutation in amino acid oxidase. Proc. Natl. Acad. Sci. U.S.A. 107, 7556–7561 10.1073/pnas.0914128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheah B. C., Vucic S., Krishnan A. V., and Kiernan M. C. (2010) Riluzole, neuroprotection and amyotrophic lateral sclerosis. Curr. Med. Chem. 17, 1942 10.2174/092986710791163939 [DOI] [PubMed] [Google Scholar]
- 5. Brown R. H., and Al-Chalabi A. (2017) Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 10.1056/NEJMra1603471 [DOI] [PubMed] [Google Scholar]
- 6. Sama R. R., Ward C. L., Kaushansky L. J., Lemay N., Ishigaki S., Urano F., and Bosco D. A. (2013) FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J. Cell. Physiol. 228, 2222–2231 10.1002/jcp.24395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dewey C. M., Cenik B., Sephton C. F., Dries D. R., Mayer P. 3rd., Good S. K., Johnson B. A., Herz J., and Yu G. (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31, 1098–1108 10.1128/MCB.01279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., and Cáceres J. F. (2000) The MKK3/6-p38–signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149, 307–316 10.1083/jcb.149.2.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sama R., Ward C., and Bosco D. (2014) Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro. 6, 1759091414544472 10.1177/1759091414544472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwiatkowski T. J. Jr., Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., Valdmanis P., Rouleau G. A., Hosler B. A., Cortelli P., de Jong P. J., et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- 11. Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H. A., and Mackenzie I. R. (2009) A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132, 2922–2931 10.1093/brain/awp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keller B. A., Volkening K., Droppelmann C. A., Ang L.-C., Rademakers R., and Strong M. J. (2012) Co-aggregation of RNA binding proteins in ALS spinal motor neurons: evidence of a common pathogenic mechanism. Acta Neuropathol. 124, 733–747 10.1007/s00401-012-1035-z [DOI] [PubMed] [Google Scholar]
- 14. Deng H.-X., Zhai H., Bigio E. H., Yan J., Fecto F., Ajroud K., Mishra M., Ajroud-Driss S., Heller S., Sufit R., Siddique N., Mugnaini E., and Siddique T. (2010) FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 67, 739–748 10.1002/ana.22051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyd J. D., Lee P., Feiler M. S., Zauur N., Liu M., Concannon J., Ebata A., Wolozin B., and Glicksman M. A. (2014) A high-content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J. Biomol. Screen. 19, 44–56 10.1177/1087057113501553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu G., Stevens S. M. Jr., Kobeissy F., Kobiessy F., Brown H., McClung S., Gold M. S., and Borchelt D. R. (2012) Identification of proteins sensitive to thermal stress in human neuroblastoma and glioma cell lines. PLoS ONE 7, e49021 10.1371/journal.pone.0049021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahl A., Blanco I., Jackman K., Baskar J., Milaganur Mohan H., Rodney-Sandy R., Zhang S., Iadecola C., and Hochrainer K. (2018) Cerebral ischemia induces the aggregation of proteins linked to neurodegenerative diseases. Sci. Rep. 8, 2701 10.1038/s41598-018-21063-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., and Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 10.1111/j.1471-4159.2009.06383.x [DOI] [PubMed] [Google Scholar]
- 19. McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., and Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 10.1093/hmg/ddr021 [DOI] [PubMed] [Google Scholar]
- 20. Bosco D. A., Lemay N., Ko H. K., Zhou H., Burke C., Kwiatkowski T. J. Jr., Sapp P., McKenna-Yasek D., Brown R. H. Jr., and Hayward L. J. (2010) Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19, 4160–4175 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito D., Hatano M., and Suzuki N. (2017) RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci. Transl. Med. 9, eaah5436 10.1126/scitranslmed.aah5436 [DOI] [PubMed] [Google Scholar]
- 22. Schubert D., and Piasecki D. (2001) Oxidative glutamate toxicity can be a component of the excitotoxicity cascade. J. Neurosci. 21, 7455–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiszman M. L., Ricart K. C., Latini A., Rodríguez G., and Sica R. E. (2010) In vitro neurotoxic properties and excitatory aminoacids concentration in the cerebrospinal fluid of amyotrophic lateral sclerosis patients. Relationship with the degree of certainty of disease diagnoses. Acta Neurol. Scand. 121, 120–126 10.1111/j.1600-0404.2009.01200.x [DOI] [PubMed] [Google Scholar]
- 24. Lattante S., Rouleau G. A., and Kabashi E. (2013) TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum. Mutat. 34, 812–826 10.1002/humu.22319 [DOI] [PubMed] [Google Scholar]
- 25. López-Erauskin J., Tadokoro T., Baughn M. W., Myers B., McAlonis-Downes M., Chillon-Marinas C., Asiaban J. N., Artates J., Bui A. T., Vetto A. P., Lee S. K., Le A. V., Sun Y., Jambeau M., Boubaker J., et al. (2018) ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron 100, 816–830.e7 10.1016/j.neuron.2018.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ederle H., Funk C., Abou-Ajram C., Hutten S., Funk E. B. E., Kehlenbach R. H., Bailer S. M., and Dormann D. (2018) Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci. Rep. 8, 7084 10.1038/s41598-018-25007-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kino Y., Washizu C., Aquilanti E., Okuno M., Kurosawa M., Yamada M., Doi H., and Nukina N. (2011) Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 39, 2781–2798 10.1093/nar/gkq1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grima J. C., Daigle J. G., Arbez N., Cunningham K. C., Zhang K., Ochaba J., Geater C., Morozko E., Stocksdale J., Glatzer J. C., Pham J. T., Ahmed I., Peng Q., Wadhwa H., Pletnikova O., et al. (2017) Mutant Huntingtin disrupts the nuclear pore complex. Neuron 94, 93–107.e6 10.1016/j.neuron.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hatch E. M., Fischer A. H., Deerinck T. J., and Hetzer M. W. (2013) Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim H. J., and Taylor J. P. (2017) Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 96, 285–297 10.1016/j.neuron.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fryer H. J., Knox R. J., Strittmatter S. M., and Kalb R. G. (1999) Excitotoxic death of a subset of embryonic rat motor neurons in vitro. J. Neurochem. 72, 500–513 10.1046/j.1471-4159.1999.0720500.x [DOI] [PubMed] [Google Scholar]
- 32. Rose C. R., Felix L., Zeug A., Dietrich D., Reiner A., and Henneberger C. (2017) Astroglial glutamate signaling and uptake in the hippocampus. Front. Mol. Neurosci. 10, 451 10.3389/fnmol.2017.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kedersha N., Ivanov P., and Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holcik M., and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- 35. Murakami T., Qamar S., Lin J. Q., Schierle G. S., Rees E., Miyashita A., Costa A. R., Dodd R. B., Chan F. T., Michel C. H., Kronenberg-Versteeg D., Li Y., Yang S.-P., Wakutani Y., Meadows W., et al. (2015) ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yasuda K., Zhang H., Loiselle D., Haystead T., Macara I. G., and Mili S. (2013) The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 203, 737–746 10.1083/jcb.201306058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M. E., Mackenzie I. R., Capell A., Schmid B., Neumann M., and Haass C. (2010) ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 10.1038/emboj.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gal J., Zhang J., Kwinter D. M., Zhai J., Jia H., Jia J., and Zhu H. (2011) Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging 32, 2323 10.1016/j.neurobiolaging.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidt E. K., Clavarino G., Ceppi M., and Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- 40. Orrego F., and Lipmann F. (1967) Protein synthesis in brain slices. Effects of electrical stimulation and acidic amino acids. J. Biol. Chem. 242, 665–671 [PubMed] [Google Scholar]
- 41. Marin P., Nastiuk K. L., Daniel N., Girault J. A., Czernik A., Glowinski J., Nairn A. C., and Prémont J. (1997) Glutamate-dependent phosphorylation of elongation factor-2 and inhibition of protein synthesis in neurons. J. Neurosci. 17, 3445–3454 10.1523/JNEUROSCI.17-10-03445.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barrera I., Hernández-Kelly L. C., Castelán F., and Ortega A. (2008) Glutamate-dependent elongation factor-2 phosphorylation in Bergmann glial cells. Neurochem. Int. 52, 1167–1175 10.1016/j.neuint.2007.12.006 [DOI] [PubMed] [Google Scholar]
- 43. Taha E., Gildish I., Gal-Ben-Ari S., and Rosenblum K. (2013) The role of eEF2 pathway in learning and synaptic plasticity. Neurobiol. Learn. Mem. 105, 100–106 10.1016/j.nlm.2013.04.015 [DOI] [PubMed] [Google Scholar]
- 44. Ward C. L., Boggio K. J., Johnson B. N., Boyd J. B., Douthwright S., Shaffer S. A., Landers J. E., Glicksman M. A., and Bosco D. A. (2014) A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 5, e1572 10.1038/cddis.2014.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holt C. E., and Schuman E. M. (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagier-Tourenne C., Polymenidou M., Hutt K. R., Vu A. Q., Baughn M., Huelga S. C., Clutario K. M., Ling S.-C., Liang T. Y., Mazur C., Wancewicz E., Kim A. S., Watt A., Freier S., Hicks G. G., et al. (2012) Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gascon E., Lynch K., Ruan H., Almeida S., Verheyden J. M., Seeley W. W., Dickson D. W., Petrucelli L., Sun D., Jiao J., Zhou H., Jakovcevski M., Akbarian S., Yao W.-D., and Gao F.-B. (2014) Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 20, 1444–1451 10.1038/nm.3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ju W., Morishita W., Tsui J., Gaietta G., Deerinck T. J., Adams S. R., Garner C. C., Tsien R. Y., Ellisman M. H., and Malenka R. C. (2004) Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat. Neurosci. 7, 244–253 10.1038/nn1189 [DOI] [PubMed] [Google Scholar]
- 49. Fogarty M. J. (2018) Driven to decay: excitability and synaptic abnormalities in amyotrophic lateral sclerosis. Brain Res. Bull. 140, 318–333 10.1016/j.brainresbull.2018.05.023 [DOI] [PubMed] [Google Scholar]
- 50. Spreux-Varoquaux O., Bensimon G., Lacomblez L., Salachas F., Pradat P. F., Le Forestier N., Marouan A., Dib M., and Meininger V. (2002) Glutamate levels in cerebrospinal fluid in amyotrophic lateral sclerosis: a reappraisal using a new HPLC method with coulometric detection in a large cohort of patients. J. Neurol. Sci. 193, 73–78 10.1016/S0022-510X(01)00661-X [DOI] [PubMed] [Google Scholar]
- 51. Plaitakis A., and Constantakakis E. (1993) Altered metabolism of excitatory amino acids, N-acetyl-aspartate and N-acetyl-aspartylglutamate in amyotrophic lateral sclerosis. Brain Res. Bull. 30, 381–386 10.1016/0361-9230(93)90269-H [DOI] [PubMed] [Google Scholar]
- 52. Rothstein J., Tsai G., Kuncl R. W., Clawson L., Cornblath D. R., Drachman D. B., Pestronk A., Stauch B. L., and Coyle J. T. (1990) Abnormal excitatory amino acid metabolism in amyotrophic lateral sclerosis. Ann. Neurol. 28, 18–25 10.1002/ana.410280106 [DOI] [PubMed] [Google Scholar]
- 53. Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., and Kwak S. (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427, 801–801 10.1038/427801a [DOI] [PubMed] [Google Scholar]
- 54. Hideyama T., Yamashita T., Aizawa H., Tsuji S., Kakita A., Takahashi H., and Kwak S. (2012) Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 45, 1121–1128 10.1016/j.nbd.2011.12.033 [DOI] [PubMed] [Google Scholar]
- 55. Kia A., McAvoy K., Krishnamurthy K., Trotti D., and Pasinelli P. (2018) Astrocytes expressing ALS-linked mutant FUS induce motor neuron death through release of tumor necrosis factor-α. Glia 66, 1016–1033 10.1002/glia.23298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G. G., and Takumi T. (2005) The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 15, 587–593 10.1016/j.cub.2005.01.058 [DOI] [PubMed] [Google Scholar]
- 57. Capauto D., Colantoni A., Lu L., Santini T., Peruzzi G., Biscarini S., Morlando M., Shneider N. A., Caffarelli E., Laneve P., and Bozzoni I. (2018) A regulatory circuitry between Gria2, miR-409, and miR-495 is affected by ALS FUS mutation in ESC-derived motor neurons. Mol. Neurobiol. 55, 7635–7651 10.1007/s12035-018-0884-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Damme P., Bogaert E., Dewil M., Hersmus N., Kiraly D., Scheveneels W., Bockx I., Braeken D., Verpoorten N., Verhoeven K., Timmerman V., Herijgers P., Callewaert G., Carmeliet P., Van Dee Bosch L., and Robberecht W. (2007) Astrocytes regulate GluR2 expression in motor neurons and their vulnerability to excitotoxicity. Proc. Natl. Acad. Sci. U.S.A. 104, 14825–14830 10.1073/pnas.0705046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sugiyama K., Aida T., Nomura M., Takayanagi R., Zeilhofer H. U., and Tanaka K. (2017) Calpain-dependent degradation of nucleoporins contributes to motor neuron death in a mouse model of chronic excitotoxicity. J. Neurosci. 37, 8830–8844 10.1523/JNEUROSCI.0730-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takuma H., Kwak S., Yoshizawa T., and Kanazawa I. (1999) Reduction of GluR2 RNA editing, a molecular change that increases calcium influx through AMPA receptors, selective in the spinal ventral gray of patients with amyotrophic lateral sclerosis. Ann. Neurol. 46, 806–815 [DOI] [PubMed] [Google Scholar]
- 61. Hock E.-M., Maniecka Z., Hruska-Plochan M., Reber S., Laferrière F., Sahadevan M. K. S., Ederle H., Gittings L., Pelkmans L., Dupuis L., Lashley T., Ruepp M. D., Dormann D., and Polymenidou M. (2018) Hypertonic stress causes cytoplasmic translocation of neuronal, but not astrocytic, FUS due to impaired transportin function. Cell Rep. 24, 987–1000.e7 10.1016/j.celrep.2018.06.094 [DOI] [PubMed] [Google Scholar]
- 62. Kamelgarn M., Chen J., Kuang L., Jin H., Kasarskis E. J., and Zhu H. (2018) ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl. Acad. Sci. U.S.A. 115, E11904–E11913 10.1073/pnas.1810413115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Archbold H. C., Jackson K. L., Arora A., Weskamp K., Tank E. M., Li X., Miguez R., Dayton R. D., Tamir S., Klein R. L., and Barmada S. J. (2018) TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci. Rep. 8, 4606 10.1038/s41598-018-22858-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rhoads S. N., Monahan Z. T., Yee D. S., and Shewmaker F. P. (2018) The role of post-translational modifications on prion-like aggregation and liquid-phase separation of FUS. Int. J. Mol. Sci. 19, E886 10.3390/ijms19030886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kodiha M., Chu A., Matusiewicz N., and Stochaj U. (2004) Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 11, 862–874 10.1038/sj.cdd.4401432 [DOI] [PubMed] [Google Scholar]
- 66. Bano D., Dinsdale D., Cabrera-Socorro A., Maida S., Lambacher N., McColl B., Ferrando-May E., Hengartner M. O., and Nicotera P. (2010) Alteration of the nuclear pore complex in Ca2+-mediated cell death. Cell Death Differ. 17, 119–133 10.1038/cdd.2009.112 [DOI] [PubMed] [Google Scholar]
- 67. Yasuda Y., Miyamoto Y., Saiwaki T., and Yoneda Y. (2006) Mechanism of the stress-induced collapse of the Ran distribution. Exp. Cell Res. 312, 512–520 10.1016/j.yexcr.2005.11.017 [DOI] [PubMed] [Google Scholar]
- 68. Zhang K., Daigle J. G., Cunningham K. M., Coyne A. N., Ruan K., Grima J. C., Bowen K. E., Wadhwa H., Yang P., Rigo F., Taylor J. P., Gitler A. D., Rothstein J. D., and Lloyd T. E. (2018) Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e17 10.1016/j.cell.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kelley J. B., and Paschal B. M. (2007) Hyperosmotic stress signaling to the nucleus disrupts the Ran gradient and the production of RanGTP. Mol. Biol. Cell 18, 4365–4376 10.1091/mbc.e07-01-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li N., and Lagier-Tourenne C. (2018) Nuclear pores: the gate to neurodegeneration. Nat. Neurosci. 21, 156–158 10.1038/s41593-017-0066-0 [DOI] [PubMed] [Google Scholar]
- 71. Haines J. D., Herbin O., de la Hera B., Vidaurre O. G., Moy G. A., Sun Q., Fung H. Y., Albrecht S., Alexandropoulos K., McCauley D., Chook Y. M., Kuhlmann T., Kidd G. J., Shacham S., and Casaccia P. (2015) Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nat. Neurosci. 18, 511–520 10.1038/nn.3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wainger B. J., Kiskinis E., Mellin C., Wiskow O., Han S. S., Sandoe J., Perez N. P., Williams L. A., Lee S., Boulting G., Berry J. D., Brown R. H. Jr., Cudkowicz M. E., Bean B. P., Eggan K., and Woolf C. J. (2014) Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 7, 1–11 10.1016/j.celrep.2014.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sama R. R, Fallini C., Gatto R., McKeon J. E., Song Y., Rotunno M. S., Penaranda S., Abdurakhmanov I., Landers J. E., Morfini G., Brady S. T., and Bosco D. A. (2017) ALS-linked FUS exerts a gain of toxic function involving aberrant p38 MAPK activation. Sci. Rep. 7, 115 10.1038/s41598-017-00091-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sena-Esteves M., Tebbets J. C., Steffens S., Crombleholme T., and Flake A. W. (2004) Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods 122, 131–139 10.1016/j.jviromet.2004.08.017 [DOI] [PubMed] [Google Scholar]
- 75. Baron D. M., Kaushansky L. J., Ward C. L., Sama R. R, Chian R.-J., Boggio K. J., Quaresma A. J., Nickerson J. A., and Bosco D. A. (2013) Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener. 8, 30 10.1186/1750-1326-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cajigas I. J., Tushev G., Will T. J., tom Dieck S., Fuerst N., and Schuman E. M. (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.