Abstract
Microtubule-associated proteins (MAPs) regulate microtubule polymerization, dynamics, and organization. In addition, MAPs alter the motility of kinesin and dynein to control trafficking along microtubules. MAP7 (ensconsin, E-MAP-115) is a ubiquitous MAP that organizes the microtubule cytoskeleton in mitosis and neuronal branching. MAP7 also recruits kinesin-1 to microtubules. To understand how the activation of kinesin-1 by MAP7 regulates the motility of organelles transported by ensembles of kinesin and dynein, we isolated organelles and reconstituted their motility in vitro. In the absence of MAP7, isolated phagosomes exhibit approximately equal fractions of plus- and minus-end–directed motility along microtubules. MAP7 causes a pronounced shift in motility; phagosomes move toward the plus-end ∼80% of the time, and kinesin teams generate more force. To dissect MAP7-mediated regulation of kinesin-driven transport, we examined its effects on the motility and force generation of single and teams of full-length kinesin-1 motors. We find that MAP7 does not alter the force exerted by a single kinesin-1 motor, but instead increases its binding rate to the microtubule. For ensembles of kinesin, a greater number of kinesin motors are simultaneously engaged and generating force to preferentially target organelles toward the microtubule plus-end.
Keywords: microtubule, microtubule-associated protein (MAP), intracellular trafficking, optical tweezers, kinesin, dynein, cytoskeleton, kinesin-1, kinesin-2, MAP7/ensconsin, intracellular transport
Introduction
Intracellular cargoes navigate a complex network of microtubule tracks to find their destination in the cell. Many cargoes (e.g. endosomes, mitochondria, peroxisomes, and mRNA) are transported by similar sets of kinesin and dynein motors, yet they exhibit different transport characteristics and localizations. What cues guide a specific cargo to its destination in the cell? Mounting evidence suggests a role for the microtubule cytoskeleton (1) through its organization (2–4), tubulin post-translational modifications (2, 5–7), and microtubule-associated proteins (MAPs)3 (8, 9).
MAPs regulate microtubule dynamics and organization (9–11). MAPs also control the interface between motor proteins and the microtubule surface to regulate motility. For example, the neuronal MAP tau reduces kinesin-1 processivity (12, 13). On cargoes transported bidirectionally by opposing kinesin and dynein motors, inhibition of kinesin-1 by tau biases transport toward the microtubule minus-end (14). MAP4 inhibits kinesin-1 motility in gliding assays (15) and regulates the aggregation and dispersion of pigment granules transported by kinesin-2 and dynein (16). Doublecortin and doublecortin-like kinase 1 promote the motility of kinesin-3 motors in neurons (17, 18). Thus, MAPs can act as both positive and negative regulators of motor protein activity to exert tight control on transport.
MAP7 is a microtubule-associated protein involved in diverse cellular processes. By stabilizing microtubules (19), MAP7 enhances neuronal branching (20, 21) and controls the length of microtubules in the mitotic spindle (22, 23). Further, MAP7 directly interacts with both the kinesin-1 stalk and microtubules (23, 24) to enhance kinesin-1 binding. Loss of MAP7 in oocytes results in impaired plus-ended motility (25) and defects in nuclear positioning in muscle cells (26). In single-molecule motility assays, MAP7 increases the frequency of processive events by kinesin-1 (24, 25, 27, 28). Whereas MAP7 is a positive regulator of kinesin-1, kinesin-3 (KIF1A) is inhibited by MAP7, and dynein is not affected (28).
The observation that MAP7 enhances the recruitment of kinesin-1 to microtubules suggests a potential mechanism for MAP7 to control the direction in which organelles move along microtubules by controlling the relative activity of kinesin and dynein motors. However, it is unclear how MAP7 might alter kinesin-1 under load or when functioning collectively and with other motors. To address these questions, we performed single-molecule motility and optical trapping assays on single full-length kinesin-1 motors, teams of kinesin-1, and native sets of kinesin and dynein motors on isolated organelles. Optical trapping assays indicate that MAP7 increases the binding rate of kinesin-1 to microtubules but does not affect the force generation or processivity of single kinesin motors. For teams of kinesin-1, this increased binding rate results in a greater number of motors being engaged with the microtubule at any given time, resulting in enhanced processivity and force generation. On native cargoes transported by teams of kinesin-1, kinesin-2, and dynein, kinesin motors produce higher forces in the presence of MAP7 to target transport toward the microtubule plus-end.
Results
MAP7 directs bidirectional cargoes to the microtubule plus-end
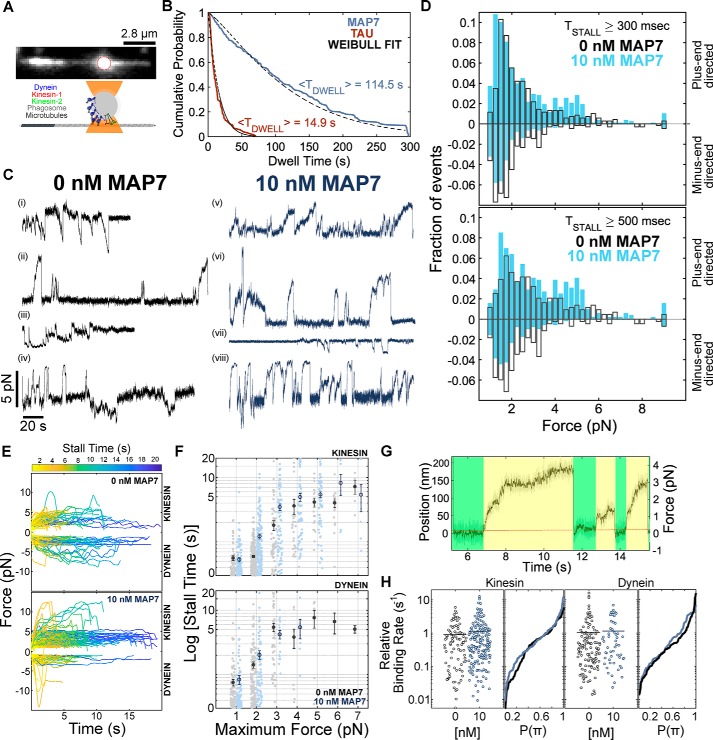
We isolated phagosomes along with their native transport machinery and reconstituted their motility along polarity-marked microtubules. Phagosomes are transported by teams of 1–3 kinesin-1, 2–6 kinesin-2, and 3–10 dynein motors (14, 29). Using an optical trap, we measured the forces exerted by teams of kinesin and dynein motors (Fig. 1A). In the absence of MAP7, isolated phagosomes spend an approximately equal fraction of time moving toward the microtubule plus- or minus-end (Fig. 1, C and D) (14, 30). MAP7 increases the frequency and magnitude of forces exerted by kinesin teams, whereas the frequency and magnitude of dynein-directed forces decrease (Fig. 1, D and E, and Fig. S1C).
Figure 1.
MAP7 targets bidirectional cargoes to the microtubule plus-end by increasing the number of engaged kinesin motors. A, isolated phagosomes containing fluorescent beads were positioned on polarity-marked microtubules, and the forces were measured using an optical trap. These isolated phagosomes are transported by teams of kinesin-1, kinesin-2, and dynein motors (14). B, MAP7 binds in static patches (Fig. S1J; Movie S6), whereas tau forms both static and diffusive patches along the microtubule lattice (Fig. S1K). The mean dwell time of MAP7 is 114.5 ± 6.16 s (n = 221 events from 11 recordings, two independent experiments), compared with 14.9 ± 1.19 s for tau (n = 128 events from 26 recordings, two independent experiments). C, force traces were acquired at 2 kHz and median-filtered at 20 Hz. We observe force events directed toward both the minus-end (i, iii, and vii) and plus-end (ii, v, and vi) and also bidirectional force events (iv and viii) (0 nm: n = 689 events from 38 recordings, 11 independent experiments; 10 nm MAP7: n = 787 events from 40 recordings, 11 independent experiments). D, maximum forces for all trap displacements greater than 300 ms (top) and 500 ms (bottom) in duration were included in the histogram. Consistent with previous results (14, 30), plus-end–directed force events consist of unitary stall forces of kinesin-1 and kinesin-2, events where the motors detach before reaching their stall force, and rare events driven by multiple kinesins. Minus-end–directed forces indicate events driven by teams of several dynein motors (Fig. S1C). The frequency and magnitude of kinesin-driven forces increase in the presence of 10 nm MAP7. In response, dynein-mediated forces are reduced. The Bayesian information criterion was used to determine the optimal number of components to describe the force histograms (Fig. S1F). Mean forces for plus-end directed motors are 1.57, 2.38, and 4.5 and for minus-ended motors are 1.14, 1.6, 2.8, and 5.8 pN. With MAP7, mean forces of the multicomponent fits for plus-end–directed forces are 1.4, 2.4, 4.5, and 7.3 pN, and for minus-end–directed forces, they are 1.4, 2.7, and ≥9 pN (p < 0.001; Kolmogorov–Smirnov test; Fig. S5A). E, teams of kinesin and dynein motors remain engaged to the microtubule under load for longer durations in the presence of MAP7. The color of the trajectory indicates the duration of a stall event. The duration of force events is significantly longer for kinesin-driven events with MAP7 (<T0 nM,+> = 1.5 s, <T10 nM,+> = 2.5 s, p < 0.01), whereas the duration of dynein-driven forces is slightly decreased (<T0 nM,−> = 2.26 s, <T10 nM,−> = 1.8 s, p = not significant) (Fig. S5C). F, comparing the duration of events of similar maximum force indicates that MAP7 does not alter the duration of events driven by similar numbers of motors. G, the relative binding rate was calculated as the reciprocal of the dwell time (green periods) preceding a force event (yellow). H, MAP7 increases the relative binding rate of plus-ended motors on MAP7 decorated microtubules by 28% (<π0 nM,+> = 0.89 s−1, <π10 nM,+> = 1.14 s−1; <π0 nM,−> = 1.05 s−1, <π10 nM,−> = 1.15 s−1). Error bars, S.E.
The amount of time that motors exert force before detaching provides a measurement of processivity under load. We observe that cargoes driven along MAP7-decorated microtubules stay engaged for longer periods of time before detaching. Kinesin teams remain engaged for ∼67% longer with MAP7, whereas the average stall time of minus-ended motors increases by only ∼18% (Fig. 1E). We examined the duration of events with similar maximum force as a way to compare events with similar numbers of engaged motors (Fig. 1F). Whereas high-force events are more frequent with MAP7 present, the time motors remain engaged at the same force is not affected, indicating that MAP7 does not alter the processivity of individual kinesin or dynein motors (Fig. 1F). We calculated the force-dependent unbinding rate using the method proposed by Berger et al. (31) and found that the unbinding rate for both kinesin and dynein is also unaffected by MAP7 (Fig. S1G). These analyses indicate that whereas MAP7 does not alter the processivity of individual motors, it enhances the processivity of kinesin teams by increasing the number of engaged motors.
To compare the effect of MAP7 on the rate at which kinesin and dynein motor teams bind to microtubules under similar conditions, we estimated the relative binding rate from the duration of the diffusive dwells preceding motor-driven events in the optical trap (Fig. 1G and supporting information). MAP7 increases the relative binding rate of plus-ended motors by ∼30%, whereas the binding rates of dynein teams increase by 10% (Fig. 1H). Accordingly, kinesin teams bind faster than dynein teams, and the fraction of kinesin-driven events increases, whereas dynein-driven events are less frequent in the presence of MAP7 (Fig. S1D). Taken together, these results suggest that MAP7 enhances kinesin binding, such that a larger fraction of the kinesin motors are engaged at any time and thus exert greater forces.
MAP7 recruits kinesin-1 to microtubules
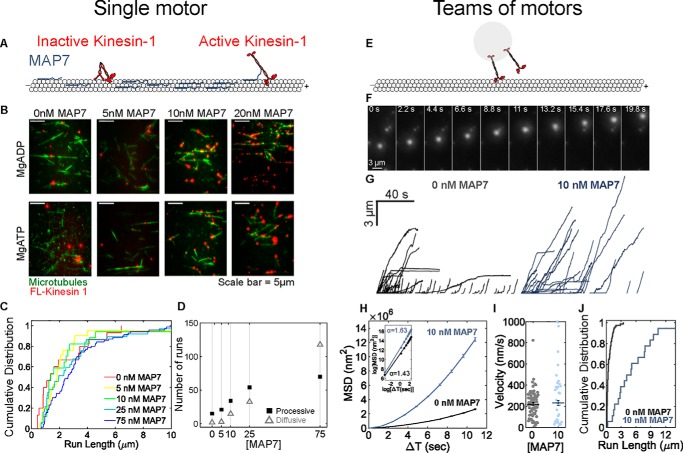
To examine the regulation of individual kinesin-1 motors by MAP7, we performed single-molecule motility assays with full-length kinesin-1 (Fig. 2A). MAP7 increases the binding rate of kinesin-1 motors on the microtubules in a dose-dependent manner (Fig. 2B). Whereas kinesin-1 run lengths increase (Fig. 2C), their velocities (Fig. S2A) are largely unchanged in the presence of MAP7. MAP7 binds to kinesin-1 on the stalk domain, near the hinge region that mediates autoinhibition (21, 24, 28, 32). Thus, we asked whether MAP7 acts both to recruit kinesin-1 to microtubules and relieve autoinhibition (33, 34). Full-length kinesin-1 exhibits both diffusive and processive events along the microtubule as it transitions between unfolded, active states and folded, autoinhibited states (35). MAP7 increases both the number of processive and diffusive events to a similar degree, suggesting that MAP7 recruits kinesin-1 to the microtubule but does not relieve autoinhibition. The fraction of diffusive events increases at high levels of MAP7 concentration (75 nm), indicating that MAP7 can act as an obstacle on the microtubule when present at high densities (Fig. 2D and Fig. S2B). Motility assays using Drosophila kinesin-1 and MAP7 (Fig. 2, A–D) or mammalian kinesin-1 and MAP7 (Fig. S2, C–H; Movies S1–S3) show similar results.
Figure 2.
MAP7 recruits kinesin-1 to microtubules. A and B, MAP7 recruits full-length kinesin-1 to microtubules in a dose-dependent manner (see also Fig. S2B). MgADP-bound kinesin-1 is in a weakly bound state. Kinesin-1 hydrolyzes MgATP to drive processive movement. C, when present at high levels, MAP7 increases kinesin-1 run lengths by up to ∼30%, possibly due to rapid re-attachment (average run length: 0 nm = 2033 ± 493.8 nm; 5 nm = 1640 ± 463 nm; 10 nm = 1800 ± 937 nm; 25 nm = 2227 ± 411.2 nm; 75 nm = 3002 ± 265.5 nm). D, the frequency of both processive and diffusive motility of kinesin-1 increases to a similar degree with increasing MAP7 concentration (0 nm MAP7: n = 15 events, three recordings; 5 nm MAP7: n = 17 events from three recordings; 10 nm MAP7: n = 21 events from three recordings; 25 nm MAP7: n = 37 events from three recordings; 75 nm MAP7: n = 70 events from three recordings). E and F, beads transported by ∼1–2 kinesin-1 motors were positioned on the microtubule using a weak optical trap (k ∼0.004 pN/nm) and imaged as they moved along the microtubule. G, long, directed events toward the microtubule plus-end indicate motility by multiple kinesin-1 motors. H, teams of kinesin-1 motors are more processive on MAP7-decorated microtubule (I and J). The average run length of kinesin-1 motors with MAP7 increases by ∼8-fold (average run length (0 nm) = 595 ± 60 nm; average run length (10 nm) = 5320 ± 890 nm), whereas the velocity is unaffected (average velocity (0 nm) = 220 ± 16 nm/s; average velocity (10 nm) = 292 ± 45 nm/s) (0 nm: n = 72 events from 21 trajectories, two independent experiments; 10 nm MAP7: n = 32 events from 18 trajectories, two independent experiments). Error bars, S.E.
We next asked how enhanced recruitment by MAP7 might affect transport by teams of kinesin-1 motors. We prepared latex beads bound to mammalian full-length kinesin-1 motors and titrated the kinesin-1/bead concentration such that we observe ∼1–2 active kinesin-1 motors (Fig. 2, E and F; Movie S4; see “Experimental procedures”). The average run length for teams of ∼1–2 kinesin-1 motors is 594 nm (Fig. 2G), similar to the run length of a single kinesin-1 motor (36, 37). With MAP7, teams of kinesin-1 motors are more processive, as indicated by MSD analysis (Fig. 2, G and H; Movie S5), and there is an 8-fold increase in run length (Fig. 2J). Velocity is not affected (Fig. 2I). Our results, along with previous studies (25, 27, 28), show that MAP7 enhances the recruitment of kinesin-1 to microtubules. Further, for cargoes transported by multiple kinesins, MAP7 increases the number of motors engaged to increase cargo processivity.
Teams of kinesin-1 motors exert greater forces on MAP7-decorated microtubules
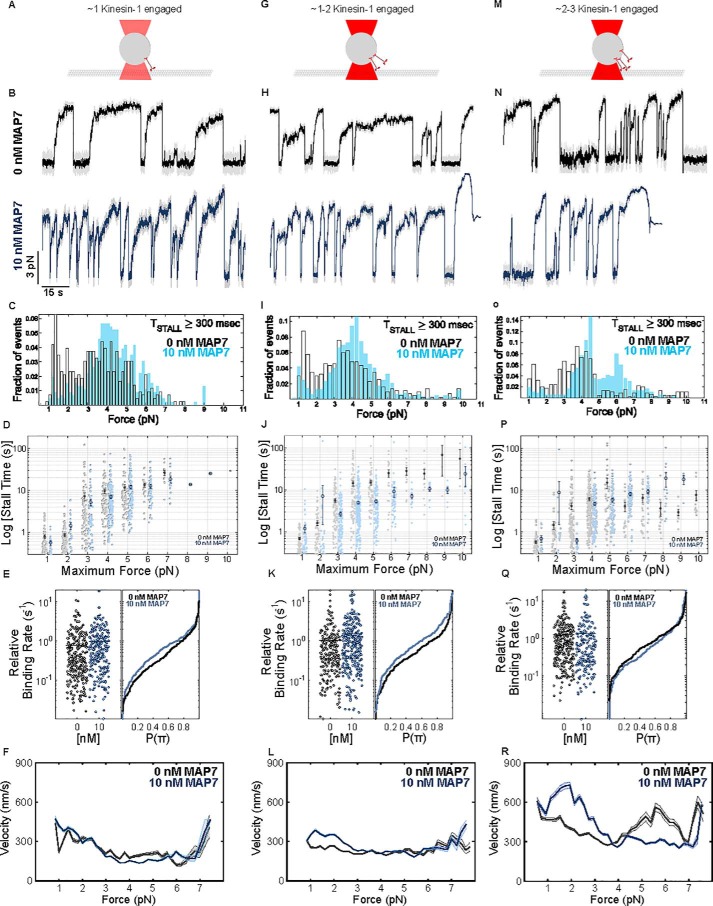
Evidence from multiple laboratories and experimental systems shows that MAP7 enhances kinesin-1 activity by recruiting it to microtubules (24, 25, 27, 28). However, it is not understood how enhanced binding influences force generation and cooperative transport by teams of kinesin-1s. We quantified the effect of MAP7 on the forces exerted by single motors and ensembles of kinesin-1 (Fig. 3). The force exerted by a single kinesin-1 motor is not affected by MAP7, although we do observe fewer substall detachments (Fig. 3, A–C). Force-dependent processivity is also not affected (Fig. 3D). However, the relative binding rate of kinesin-1 measured in the optical trap increases by ∼25% in the presence of MAP7 (Fig. 3E). When the density of kinesin-1 on the bead is increased by ∼2-fold, we observe frequent single-motor stall events (∼5 pN) and rare high-force events (>5 pN) driven by multiple engaged kinesins (Fig. 3, G–I). Similar to single kinesin-1 motors, MAP7 results in fewer substall detachments (Fig. 3I) and enhanced binding (Fig. 3K). At motor densities ∼4 times greater than single-molecule conditions, high-force events are still rare (Fig. 3, M–O), consistent with previous findings that multiple kinesin-1 motors rarely engage simultaneously (30, 37). Strikingly, at the same condition, high-force events are frequent on MAP7-decorated microtubules (Fig. 3O). Thus, MAP7 promotes the simultaneous engagement of multiple kinesin-1 motors.
Figure 3.
MAP7 increases the forces generated by teams of kinesin-1. We measured forces by single kinesin-1 motors and teams at three motor densities (ratio of motors to beads in the conjugation reaction). A, 3300 kinesin-1s/bead resulted in motility driven by a single kinesin-1, where <50% of beads interacted with the microtubule; G, 6600 kinesin-1s/bead resulted in motility due to 1–2 engaged kinesin-1 motors; M, 13,000 kinesin-1s/bead resulted in 2–3 kinesin-1 motors. B and C, on beads driven by single kinesin-1 motors, force distributions indicate unitary stall force of kinesin-1 and substall detachment forces (mean forces of the multicomponent fits Fcomp = 1.3 and 4.03 pN). With the addition of MAP7, substall detachment events decrease, and the frequency of unitary stall force events due to single kinesin-1 increases (Fcomp = 4.23 and ≥9 pN) (p < 0.001; Kolmogorov–Smirnov test; Fig. S5B) (0 nm: n = 444 events from 25 recordings, four independent experiments; 10 nm MAP7: n = 536 events from 25 recordings, four independent experiments). H and I, on beads driven by ∼1–2 kinesin-1s, force distributions show three distinct populations: forces at unitary stall force of kinesin-1, detachment forces, and rare multimotor events (Fcomp = 1.27, 3.47, and 7 pN). With the addition of MAP7, detachment and low force events decrease, whereas the frequency of unitary stall force events increases (Fcomp = 1.17, 4, and 7 pN) (p < 0.001; Kolmogorov–Smirnov test; Fig. S5B). The frequency and magnitude of multimotor force events remain unchanged (0 nm: n = 479 events from 26 recordings, four independent experiments; 10 nm MAP7: n = 623 events from 24 recordings, four independent experiments). N and O, on beads driven by ∼2–3 kinesin-1 motors, force distribution shows three distinct populations: forces at unitary stall force of kinesin-1, low-force events, and multimotor events (Fcomp = 1.24, 3.64, and 7.5 pN). With MAP7, we observe a shift toward frequent high-force events driven by multiple engaged kinesin-1s (Fcomp = 4.24, 5.97, and 7.29) (p < 0.001; Kolmogorov–Smirnov test; Fig. S5B) (0 nm: n = 485 events from 23 recordings, three independent experiments; 10 nm MAP7: n = 378 events from 22 recordings, three independent experiments). D, J, P, and Fig. S3, the duration of stall events at a given force is not strongly influenced by MAP7, indicating that MAP7 does not affect the processivity of single kinesin motors. Single kinesin-1 motors (E) and teams of 1–2 kinesin-1 motors (K) bind much faster to MAP7-decorated microtubules. Q, the increase in binding rate is not observed for larger teams of 2–3 kinesin-1s, likely because attachment is no longer limited by the single-motor binding rate when many motors are available for binding (single kinesin-1: 0 nm = 0.77 s−1 and 10 nm = 0.95 s−1; teams of ∼1–2 kinesin-1s: 0 nm = 1.04 s−1 and 10 nm = 1.55 s−1; ∼2–3 kinesin-1s: 0 nm = 1.24 s−1 and 10 nm = 1.35 s−1). F, L, R, the force–velocity curves indicate that more kinesin-1 motors are simultaneously engaged when MAP7 is present, as indicated by higher velocities at the same load. Error bars, S.E.
The enhanced kinesin-1 binding rate due to MAP7 alters transport by a single motor or teams differently. When a single kinesin-1 is available to engage with the microtubule (Fig. 3E), the association is slow (0.77 s−1) and is substantially accelerated by MAP7 (0.95 s−1). When many motors are present on the cargo and available to interact with the microtubule (Fig. 3Q), the attachment of the cargo to the microtubule is fast even in the absence of MAP7 (1.24 s−1) and is only increased slightly when MAP7 is present (1.35 s−1). Rather than accelerate association, the increased binding rate translates to a larger number of motors being engaged at the same time, resulting in higher forces (Fig. 3O). This idea is supported by our observations on isolated phagosomes, where MAP7 induces a subtle increase in the binding rate leading to plus-end–directed events (Fig. 1H), but plus-end directed forces increase substantially (Fig. 1D) due to a larger number of engaged kinesin motors. Further, kinesin-2 motors also contribute to phagosome motility (14) and are not likely to be affected by MAP7 (Fig. S4).
Kinesin-1 motors do not function effectively in ensembles. In contrast to dynein, only one member of the kinesin team is often engaged with the microtubule at any one time (30, 37, 38). One contributing factor is that teams of kinesin-1 motors do not share loads equally (37–39); the leading kinesin-1 carries a greater proportion of the load than the trailing kinesins. To analyze how MAP7 affects the ability of kinesin-1 teams to share loads, we estimated their force–velocity relationships from the stationary optical trap data (31) (see supporting information and Fig. S3T). Cargoes transported by kinesin-1 teams move faster at a given load in the presence of MAP7, indicating that by increasing the number of active kinesin-1 bound to the microtubule, MAP7 enables kinesins to distributed loads more effectively and increases their ability to carry opposing load (Fig. 3, F, L, and R). MAP7 does not alter the force–velocity curve for single kinesin-1 motors (Fig. 3F). For teams of kinesin-1, we observe higher velocities with MAP7 due to a larger number of engaged kinesins exerting force (Fig. 3, L and R). For cargoes that are driven by ∼2–3 kinesin-1 motors, velocities in the range of loads ∼5–6 pN are lower when MAP7 is present as high-force events are rare in the absence of MAP7 (Fig. 3O), and the high velocities observed are likely due to infrequent events where several motors are engaged. Collectively, these results demonstrate that by promoting recruitment of kinesin-1 to the microtubule, MAP7 results in a greater number of kinesins to engage simultaneously to exert higher forces (Fig. 3, C, I, and O) and move more processively (Fig. 2, G, H, and J).
MAP7 regulates bidirectional transport by tuning the kinesin-1 binding rate
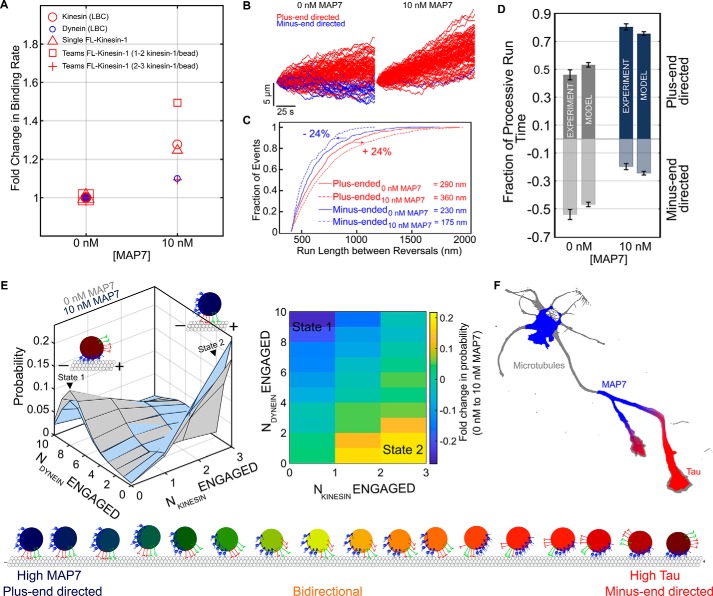
Bidirectional motility has been modeled as a stochastic tug-of-war, where force-dependent unbinding of opposing motor teams results in directional switches and pauses (40). We previously extended this model to describe the interactions between teams of three motors: dynein, kinesin-1, and kinesin-2 (14). To quantitatively relate our observations of the effect of MAP7 on single kinesin motors to organelles driven by teams of kinesin and dynein motors, we applied the increase in the binding rate of single kinesin-1 motors that we observed experimentally (Fig. 3E) to a simulation of transport by one kinesin-1, two kinesin-2, and 10 dynein motors (Fig. 4A; see “Experimental procedures”). Dynein motility is largely unaffected by MAP7 (28). To assess the possibility of an interaction between MAP7 and kinesin-2, we performed a global alignment of kinesin-1 (heavy chain) and kinesin-2 motors as MAP7 interacts with the stalk domain of the kinesin-1 heavy chain (Fig. S4A) (21). We observe no homology in the kinesin-1 and kinesin-2 stalk domains, indicating that MAP7 is unlikely to directly interact with kinesin-2.
Figure 4.
MAP7 directs transport toward the microtubule plus-end. A, we extended the mathematical model proposed by Muller et al. (40) to describe the interaction between teams of kinesin-1, kinesin-2, and dynein motors based on single-motor parameters including the motor stall force, detachment force, and unbinding and binding rates (14). We modeled the effect of MAP7 by increasing the binding rate of single kinesin-1 molecules by the same amount as we measured in optical trapping measurements. Kinesin-2 and dynein were assumed to be unaffected by MAP7. B, the increase in kinesin-1 binding rate biases simulated trajectories toward the plus-end (number of simulated trajectories, n = 100). C, we calculated run length as distance between reversals (29) and consider events >400 nm as processive. Plus-end–directed run lengths increase by ∼24%, whereas minus-end–directed run lengths decrease by ∼24%. D, the model results suggest that the shift in phagosome transport toward the microtubule plus-end that we observe can be described by simply increasing the kinesin-1 binding rate with no direct effect on kinesin-2 or dynein. E, the number of engaged kinesin and dynein motors changes in the presence of MAP7. A greater number of plus-ended motors are simultaneously engaged and exerting force, and as a result, minus-ended motors are under higher loads and are less processive. F, MAP7 and tau exhibit distinct localizations in neurons. Whereas MAP7 is enriched at axonal branches (21), tau is localized in a gradient along the axon. Thus, MAP7 might target cargoes to the microtubule plus-end at branch sites, whereas tau directs distal cargoes toward the cell body. Error bars, S.E.
When we apply the enhanced binding rate observed for kinesin-1 by MAP7 to the model of phagosome transport, motility dramatically shifts toward the microtubule plus-end (Fig. 4, B and D). The length of processive events of kinesin teams increases, whereas the processivity of dynein teams decreases (Fig. 4C). Next, we quantified the probability of each possible state in the model, where each state represents the number of kinesin and dynein motors that are engaged. The probability distribution indicates that the motility is driven by switching between two dominant states, one where only dynein motors are engaged and another where only kinesin motors are engaged. States where both kinesin and dynein motors are engaged are less frequent. In the absence of MAP7, the kinesin-driven and dynein-driven states have about equal probability, resulting in an equal fraction of plus- and minus-end–directed motility (Fig. 4). With MAP7, the probability of kinesin teams being engaged increases by 20%. In response, dynein teams are under greater loads and detach faster, resulting in a reciprocal decrease in dynein engagement (Fig. 4E). The model results suggest that the increase in kinesin-1 binding rate measured in single molecule experiments alone accounts for the shift in motility observed for isolated organelles transported by teams of kinesin and dynein.
Discussion
MAP7 promotes binding of kinesin-1 to microtubules to enable a range of kinesin-1–mediated roles, including intracellular transport, oocyte polarization, nuclear positioning, and centrosome separation (20, 21, 23–28). Given its central role in regulating kinesin-1 activity, we sought to uncover the mechanisms through which MAP7 controls organelle transport. We quantified the effect of MAP7 on the motility and force generation of single molecules of full-length kinesin-1, teams of kinesin-1, and organelles transported by kinesin-1, kinesin-2, and dynein motors. Our results suggest that MAP7 accelerates kinesin-1 binding to microtubules. On cargoes driven by single kinesin-1 motors, MAP7 increases the binding rate but does not alter processivity or force generation. For teams of kinesin-1, the increased binding rate results in a greater number of the motors associated with the cargo being engaged with the microtubule. This leads to enhanced processivity and higher forces exerted by teams of kinesin-1 motors. Vesicular cargoes and organelles are often transported by teams of opposing kinesin and dynein motors. Isolated phagosomes, driven by teams of kinesin and dynein motors, move with approximately equal fractions of plus- and minus-end directed motility. On MAP7-decorated microtubules, kinesin teams produce higher forces, such that ∼80% of motility is directed toward the microtubule plus-end.
In vitro motility assays (37, 41) and optical trapping in living cells (30, 38) indicate that even when multiple kinesins are bound to a cargo, only a single motor is often engaged with the microtubule at a time. In agreement, we observe that teams of motors rarely exert forces >5 pN (Fig. 3, C, I, and O). However, when MAP7 is present, the frequency of high-force events increases, and that of low-force events decreases. These results suggest that multiple kinesin-1 motors on a single cargo are able to engage simultaneously (Fig. S3, B and C), and negative interference between trailing and leading kinesin-1 motors can be overcome by increasing the binding rate. As a result, MAP7 reduces the effect of unequal load sharing among kinesin-1 teams, thus increasing their ability to share opposing load (Fig. 3, F, L, and R). Mathematical modeling suggests that the enhanced binding rate we observe for a single kinesin-1 motor (Fig. 4E) results in the ability of more motors to engage when operating as a team.
Previous studies have demonstrated that MAP7 is an essential regulator of kinesin-1 dependent transport in vivo (25, 27). We reconstituted the motility of isolated phagosomes driven by teams of kinesin-1, kinesin-2, and dynein motors. Phagosomes are transported by three motor proteins—kinesin-1, kinesin-2, and dynein—and fewer kinesin-1 motors (1–3 motors/phagosome) are present compared with kinesin-2 (2–6 motors) and dynein (3–10 motors) (14). Yet, MAP7 regulation is primarily mediated through controlling kinesin-1 activity. Mathematical modeling also indicates that regulation is mediated primarily through kinesin-1, as we applied the increase in the binding rate we observed for single kinesin-1 motors to the mathematical model while assuming that kinesin-2 and dynein are unaffected, and the resulting trajectories compare well with our experimental observations (Fig. 4). Tau uses an analogous mechanism to regulate bidirectional motility by inhibiting kinesin-1 processivity while having little direct effect on kinesin-2 and dynein (14). Scaffolding proteins like Miro/Milton and huntingtin that mediate interactions between motor proteins and cargoes have also been proposed to control kinesin-1 activity (42, 43). Together, these results suggest that 1) the net direction of transport of cargoes driven by multiple types of motor proteins can be regulated by controlling the activity of a single motor type, and 2) kinesin-1 activity is a common target for regulation.
Different MAPs have distinct effects on transport. In contrast to MAP7, tau inhibits kinesin-1 processivity to direct bidirectional cargoes toward the microtubule minus-end. Tau and MAP7 have distinct localizations in the axon. Tau localizes throughout the axon but is enriched in the distal axon (44). MAP7 localizes to axonal branches and contributes to branch formation (20, 21). Interestingly, MAP7 has been shown to compete with tau for binding along the microtubule lattice (28), such that tau and MAP7 may enrich on discrete regions of the microtubule cytoskeleton where they perform distinct functions. For example, tau may act to regulate the balance of plus- and minus-end–directed motility along the length of the axon (14). MAP7 may act either to promote movement through the branch site or potentially guide cargoes into a specific branch. Other MAPs are also likely involved. In the proximal axon and dendrites, MAP2 inhibits kinesin-1 (45) and dynein (46) but enhances the activity of kinesin-3 (47). Collectively, these results suggest that the regulation of organelle transport by MAPs is targeted to both the specific region of the cell, dictated by the localization of MAPs, and to specific motor proteins.
Experimental procedures
Cell culture and phagosome isolation
J774A.1 cells (ATCC) were plated in four 100-mm cell culture dishes and were grown in a 37 °C incubator at 5% CO2 to 80% confluence in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and 1% Glutamax (Thermo Fisher Scientific). Fluorescent latex beads (500-nm diameter, blue fluorescent, Thermo Fisher Scientific) were coated with BSA and incubated with the cells (48). After a 90-min chase, cells were washed with PBS, collected by a cell scraper, and washed again with cold PBS, where after each wash the cells were centrifuged at 2000 rpm in 4 °C for 5 min and resuspended in PBS. After the second wash, cells were resuspended in motility assay buffer (MAB; 10 mm PIPES, 50 mm potassium acetate, 4 mm MgCl2, 1 mm EGTA, pH 7.0) supplemented with protease inhibitors, 1 mm DTT, 1 mm ATP, and 8.5% (w/v) sucrose solution in MAB and lysed with a Dounce homogenizer. Following the lysis step, cell lysate was centrifuged at 2000 rpm in 4 °C for 10 min, and the supernatant (homogenate) was mixed with an equal volume of 62% sucrose. Sucrose solutions were supplemented with ATP and protease inhibitors and loaded in ultracentrifuge tubes (16 × 102 mm; Beckman catalog no. 344061) in the following order: 62% (3.61 ml), homogenate (∼1 ml), 35% (2.91 ml), 25% (2.91 ml), and 10% sucrose (2.91 ml). The sucrose gradient was centrifuged in a swinging bucket rotor (SW32.1; Beckman) at 24,000 rpm for 72 min. After the spin, the fraction containing phagosomes appears as a thin band at the interface between 25 and 10% sucrose.
Protein expression and purification
MAP7 expression and purification
MAP7 (accession number BC052637) was cloned from pEGFP-MAP7 (Addgene plasmid 46076) into the baculovirus expression vector pFastBac with N-terminal FLAG and SNAP tags. Recombinant baculovirus was prepared by standard protocols. Sf9 cells were infected with recombinant baculovirus for ∼72 h at 27 °C, harvested by centrifugation, and resuspended in lysis buffer (10 mm sodium phosphate, pH 7.5, 0.3 m NaCl, 7% sucrose, 0.5% glycerol, 2 mm DTT, 5 μg/ml leupeptin, 10 mm AEBSF, 10 mm phenylmethylsulfonyl fluoride, and 10 mm TLCK). Cells were lysed by sonication, and centrifuged at 257,000 × g for 35 min. The clarified lysate was added to 4 ml of FLAG affinity resin (Sigma-Aldrich) and incubated with rotation at 4 °C for 40 min. The resin was transferred to a column and washed with 200 ml of FLAG wash buffer (10 mm sodium phosphate, pH 7.5, 0.3 m NaCl, 0.5 mm DTT) and eluted with the same buffer containing 0.1 mg/ml FLAG peptide. Peak fractions were concentrated using an Amicon Ultra-15 centrifugal filter (Millipore) and dialyzed versus 10 mm HEPES, pH 7.5, 0.3 m NaCl, 1 mm DTT, 1 μg/ml leupeptin, 50% glycerol for storage at −20 °C. For some experiments, SNAP-MAP7 was labeled by incubating with a 2-fold excess of SNAP-Cell TMR substrate (S9105, New England Biolabs), supplemented with 1 mm DTT for 2 h at 4 °C. To remove the excessive unbound dye, the MAP7 solution was passed through a 10-kDa Centricon filter (MRCPRT010, Millipore Sigma).
Drosophila melanogaster ensconsin, isoform A (accession number NP_728941) with an N-terminal His6 tag in pET28a was a gift from Vladimir Gelfand (Northwestern University). Escherichia coli BL21 DE3 cells containing the plasmid were induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside and grown at 37 °C for 3 h for protein expression. Cells were pelleted and resuspended in lysis buffer (10 mm sodium phosphate, pH 7.5, 0.4 m NaCl, 0.5% glycerol, 7% sucrose, 7 mm β-mercaptoethanol, 5 μg/ml leupeptin, 10 mm AEBSF, 10 mm phenylmethylsulfonyl fluoride, and 10 mm TLCK) and sonicated. The clarified supernatant was applied to a HIS-Select nickel affinity column (Sigma-Aldrich) and washed with 10 mm sodium phosphate, pH 7.5, 0.4 m NaCl, 5 mm imidazole. Bound protein was eluted with the same buffer containing 0.2 m imidazole. Fractions were concentrated by dialysis against 50% glycerol, 10 mm imidazole, pH 7.5, 0.4 m NaCl, 1 mm DTT, and 1 μg/ml leupeptin and stored at −20 °C.
Kinesin-1 expression and purification
D. melanogaster or mouse kinesin were cloned into the baculovirus transfer vector pAcSG2 (BD Biosciences). The mouse kinesin heavy chain (accession number BC090841) has a C-terminal biotin tag followed by a His6 tag. Its corresponding light chain (accession number BC014845) was cloned with a C-terminal YFP tag. The Drosophila kinesin heavy chain (accession number AF053733) was cloned with a C-terminal biotin tag followed by a FLAG tag. Its light chain (accession number AF055298) was cloned with a His6 tag at the C terminus. The biotin tag is an 88-amino acid sequence segment from the E. coli biotin carboxyl carrier protein, which is biotinylated at a single lysine during expression in Sf9 cells (49). This tag was used for attachment to streptavidin-conjugated Quantum dots (Qdots). Recombinant baculovirus was prepared by standard protocols.
Sf9 cells were co-infected with recombinant baculovirus coding for the kinesin heavy chain and light chain and grown in suspension for ∼72 h. The mouse kinesin (His6 tag) was purified by sonicating the infected Sf9 cells in buffer containing 10 mm sodium phosphate, pH 7.5, 0.3 m NaCl, 0.5% glycerol, 7% sucrose, 7 mm β-mercaptoethanol, 0.5 mm AEBSF, 0.5 mm TLCK, 5 μg/ml leupeptin, and 1 mm ATP. The cell lysate was clarified at 200,000 × g for 30 min, and the supernatant was applied to a HIS-Select nickel affinity column (Sigma-Aldrich). The resin was first washed with buffer A (10 mm sodium phosphate, 10 mm imidazole, pH 7.5, 0.3 m NaCl) and then with buffer A containing 30 mm imidazole. Kinesin was eluted from the column with buffer A containing 200 mm imidazole.
Drosophila kinesin was purified via its FLAG tag. Cells were sonicated in buffer containing 10 mm imidazole, pH 7.0, 0.3 m NaCl, 1 mm EGTA, 5 mm MgCl2, 7% sucrose, 2 mm DTT, 0.5 mm AEBSF, 0.5 mm TLCK, 5 μg/ml leupeptin, and 1 mm ATP. The cell lysate was clarified at 200,000 × g for 30 min, and the supernatant was applied to a FLAG affinity resin column (Sigma-Aldrich). The resin was washed with buffer containing 10 mm imidazole, pH 7.0, 0.3 m NaCl, and 1 mm EGTA. Kinesin was eluted from the column by using a 0.1 mg/ml solution of FLAG peptide in wash buffer.
For each purification, the fractions of interest were combined, and 1 mm DTT, 1 μg/ml leupeptin, and 10 μm ATP were added prior to concentrating with an Amicon centrifugal filter device (Millipore). Kinesin was dialyzed versus 10 mm imidizole, pH 7.4, 200 mm NaCl, 55% glycerol, 1 mm DTT, 10 μm MgATP, and 1 μg/ml leupeptin for storage at −80 °C in small aliquots.
Polarity-marked microtubules
Bright microtubule seeds were prepared by mixing 25% Alexa fluor 647–labeled tubulin and 75% unlabeled tubulin in BRB80 (80 mm K-PIPES, 1 mm MgCl2, 1 mm EGTA, pH 6.8) to a final concentration of 5 mg/ml supplemented with 1 mm GTP (Sigma-Aldrich) and polymerized at 37 °C for 20 min. After polymerization, bright microtubule seeds were dissolved in cold BRB80 supplemented with 1 mm GMPCPP (Jena Bioscience) and 2 mm MgCl2. This mixture was incubated at 37 °C for 30 min and then pelleted at 100,000 × g for 10 min at 4 °C in TLA100 rotor (Beckman). The pellet was resuspended in 70 μl of cold BRB80 and incubated on ice for 20 min for depolymerization. The solution was centrifuged at 100,000 × g for 10 min at 4 °C and resuspended with 2 mm MgCl2 and 1 mm GMPCPP. The pellet was then resuspended in BRB80 and aliquoted in 5-μl stocks and flash-frozen. To prepare polarity-marked microtubules, GMPCPP seeds were warmed at 37 °C for 1 min to which 8% labeled tubulin was added in 92% unlabeled tubulin in BRB80 and 1 mm GTP to a final concentration of 2 mg/ml. After a 25-min incubation, these microtubules were stabilized with 20 μm taxol (Cytoskeleton) and incubated for another 25 min at 37 °C.
Phagosome in vitro motility assays
Silanized coverslips were mounted on glass slides using vacuum grease and double-sided tape to form 20–25-μl flow chambers. After flowing 1 chamber volume of anti-β-tubulin (2.5:50 in BRB80), the chamber was surface-treated with F-127 (Sigma) to block any nonspecific binding. Diluted microtubules (2:50 in BRB80 supplemented with taxol) were flown through the chamber, following a wash with 2 chamber volumes of BRB80 supplemented with 20 μm taxol. Purified phagosomes were added to the chamber, supplemented with 0.2 mg/ml BSA, 10 mm DTT, 1 mm MgATP, 20 μm taxol, 15 mg/ml glucose, >2000 units/g glucose oxidase, >6 units/g catalase, and 1 mg/ml casein. For MAP7 experiments, microtubules were mixed with 10 nm MAP7 and incubated in the flow chamber for 15–20 min.
Kinesin-1/bead complex preparation
Anti-GFP–coated carboxylated polystyrene beads (500-nm diameter) were prepared as described (50). 3.9 μm full-length kinesin-1–YFP was titrated to three different concentrations (200, 400, and 800 nm) and incubated with 7.28 × 108 beads. This translated the motor/bead ratio to ∼3300 kinesin-1s/bead (for 200 nm), 6600 kinesin-1s/bead (for 400 nm), and 13,000 kinesin-1s/bead (for 800 nm). 0.25 μm ATP, 0.4 mg/ml BSA, and 2.5 μl of anti-GFP–coated beads were mixed, and the solution was then sonicated for 1 min. Afterward, full-length kinesin-1 (with the above-stated concentrations) was added to the solution and gently mixed (abrasive pipetting may damage the anti-GFP–coated beads). The final solution was then incubated for 20 min in 4 °C. Full-length kinesin-1–coated beads were added to the chamber with immobilized microtubules supplemented with 0.4 mg/ml BSA, 1 mm MgATP, 10 μm taxol, 15 mg/ml glucose, >2000 units/g glucose oxidase, and >5 units/g catalase.
Optical trapping
The optical trap was built on an inverted microscope (Eclipse Ti-E, Nikon) with a 1.49 numerical aperture oil-immersion objective. A 1064-nm laser beam (IPG Photonics) was expanded to overfill the back aperture of the objective. To measure the bead displacement and force, a quadrant photodiode was positioned conjugate to the back focal plane of the condenser. The optical trap stiffness (KTRAP) and position calibration (β) were determined by fitting a Lorenzian function to the power spectrum of the bead's thermal fluctuations. Force events are defined as displacement from the trap center where the force is greater than 0.5 pN. Short and low-force events are common, due to the detachment of motor teams at forces below their maximal force. To this end, we analyzed optical trap data at low (TSTALL ≥ 70 ms), intermediate (TSTALL ≥ 300 ms), and high (TSTALL ≥ 1000 ms) stall times. Low stall times indicate cases where teams of motors are engaged under load but rarely reach stall. On the other hand, a high stall time indicates cases where teams of motors are likely exerting their maximal forces (stall force). In the case of intermediate stall times, motors are expected to exert detachment and stall forces. Because our force data indicate a subpopulation of detachment and stall forces, we used a probabilistic model (Bayesian Gaussian mixture model) to analyze the expected values from the force histogram and used bootstrapping to determine the optimal number of components in our data set.
Single-molecule kinesin-1 motility assay
The procedure was essentially as described previously (35). Briefly, 1 μl of 0.2 μm streptavidin Quantum dot (Thermo Fisher Scientific) was mixed with 1 μl of 0.2 μm Drosophila kinesin (with C-terminal biotin tag) for 10 min at room temperature. The mixture was then diluted 200–400-fold in 80 mm PIPES, pH 6.9, 1 mm MgCl2, 1 mm EGTA, 1 mm MgATP, and an oxygen-scavenging system composed of 0.1 mg/ml glucose oxidase, 0.02–0.18 mg/ml catalase, and 3 mg/ml glucose) and flowed into a flow cell with microtubules attached to the surface via N-ethylmaleimide–modified myosin. The slide was mounted onto an inverted Nikon TE2000U microscope equipped with a Nikon objective lens (×100; numerical aperture 1.49) for through-the-objective total internal reflection fluorescence microscopy. Single molecule assays were performed at room temperature (25 ± 1 °C). Quantum dots and microtubules were excited with a 488-nm argon ion Spectra Physics laser. Fluorescence images of Quantum dots and microtubules were then split by color and projected side by side onto a Stanford Photonics Mega 10 camera. For typical experiments, 500–1000 images were captured at 10 frames/s with an exposure time of 100 ms. Using 2 × 2 binning, the pixel resolution is 117.6 nm/pixel. Images were then imported into ImageJ (National Institutes of Health, Bethesda, MD) for processing. Those events in which the kinesin moved on microtubules in both directions for at least 300 nm are classified as diffusive events. Those events in which kinesin only moved in one direction are classified as processive events. Run lengths were determined manually. Similar procedures were used to image full-length mammalian kinesin-1-YFP. In these experiments, exposure times were increased to 1 s.
Multiple-motor motility assay
Mammalian kinesin-1–coated beads with a titration of 10,000 kinesin-1s/bead held with a low stiffness optical trap were brought down to the microtubules and allowed to run out of the trap (13). Their motility was analyzed using FIESTA (Fluorescence Image Evaluation Software for Tracking and Analysis) (51). Position versus time trajectories from FIESTA tracking were compared with kymographs to validate the automated tracking. Run lengths were determined as net displacements from the trap center to the maximum position before the bead either detaches from the microtubule or slips back to the center of the trap. The one-dimensional mean squared displacement (MSD = 2Dtα) was calculated using internal averaging, where D is the diffusion coefficient, t is the time interval, and α is the scaling exponent corresponding to stationary (α = 0), diffusive (α = 1), and processive (α = 2) movement (52).
Mathematical modeling
Müller et al. (40) developed a mathematical model describing vesicle transport driven by two teams of opposing motor proteins. We extended the MKL model to describe transport by teams of kinesin-1, kinesin-2, and dynein motors (14). Directional switches are a result of stochastic binding and unbinding kinetics of the motors, governed by the force–velocity relation, v(NP,NM), unbinding rates, ϵ(NP,NM), and some constant binding rate, π(NP,NM). Here, NP are plus-ended kinesin-1 and kinesin-2 motors, whereas NM are minus-ended dynein motors. The force–velocity relationship is defined as follows,
| (Eq. 1) |
where v(NP,NM) is load-dependent motor velocity that linearly decreases from motor velocities at no load to motor velocities at stall or superstall force load. In the range of zero to stall force, motors walk in their respective direction with forward velocities, vf, whereas in the range of superstall forces, motors drastically slow down and in effect start walking backward with backward velocities, vb. The unbinding rate is as follows.
| (Eq. 2) |
Motors detach from the microtubules at some unbinding rate that exponentially increases with some force scale, α. Here, α is defined as F/Fd(NP,NM), where F is the motor force and Fd is the motor detachment force.
Statistical analysis
All of the data are presented as the mean ± S.E. The number of experiments and recordings (n) are mentioned in the figure legends. Statistical analysis was performed using MATLAB (Mathworks Inc.) Statistical analysis was performed using one-Factor analysis of variance, Student's t test, Efron's percentile bootstrapping method, the Kolmogorov–Smirnov test, and the Bayesian information criterion (Fig. S5). Key findings from this study are summarized in Tables S1 and S2.
Author contributions
A. R. C., K. M. T., and A. G. H. designed the research. A. R. C. performed optical trapping assays. H. L. performed single-molecule kinesin-1 motility assays. E. B. K. and C. S. B. expressed and purified kinesin-1 and MAP7. A. R. C. performed phagosome purifications. A. R. C., A. G. H., H. L., and K. M. T. analyzed the data and wrote the manuscript. A. G. H. and K. M. T. conceptualized the project and supervised the work.
Supplementary Material
Acknowledgments
We thank Gary Brouhard (McGill University, Montreal, Canada) for providing reagents, Christophe Leterrier (NeuroCyto, INP CNRS-Aix Marseille Université, Marseille, France) for providing the image of a neuron (Fig. 4F), Ricardo Henriques for providing the LaTeX template, and Linda Balabanian for preparing anti-GFP–coated carboxylated polystyrene beads. We are also grateful to Malina Iwanski, Loïc Chaubet, and Linda Balabanian for providing valuable input on the manuscript.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting information, including Figs. S1–S5, Tables S1 and S2, and Movies S1–S6.
- MAP
- microtubule-associated protein
- pN
- piconewtons
- AEBSF
- 4-benzenesulfonyl fluoride hydrochloride
- TLCK
- Nα-p-tosyl-l-lysine chloromethyl ketone
- YFP
- yellow fluorescent protein
- GMPCPP
- guanosine-5′-[(α,β)-methyleno]triphosphate, sodium salt.
References
- 1. Balabanian L., Chaudhary A. R., and Hendricks A. G. (2018) Traffic control inside the cell: microtubule-based regulation of intracellular transport. Biochemist 40, 14–17 [Google Scholar]
- 2. Tas R. P., Chazeau A., Cloin B. M. C., Lambers M. L. A., Hoogenraad C. C., and Kapitein L. C. (2017) Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271.e5 10.1016/j.neuron.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balabanian L., Berger C. L., and Hendricks A. G. (2017) Acetylated microtubules are preferentially bundled leading to enhanced kinesin-1 motility. Biophys. J. 113, 1551–1560 10.1016/j.bpj.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yogev S., Cooper R., Fetter R., Horowitz M., and Shen K. (2016) Microtubule organization determines axonal transport dynamics. Neuron 92, 449–460 10.1016/j.neuron.2016.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai D., McEwen D. P., Martens J. R., Meyhofer E., and Verhey K. J. (2009) Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 7, e1000216 10.1371/journal.pbio.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sirajuddin M., Rice L. M., and Vale R. D. (2014) Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKenney R. J., Huynh W., Vale R. D., and Sirajuddin M. (2016) Tyrosination of α-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 35, 1175–1185 10.15252/embj.201593071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atherton J., Houdusse A., and Moores C. (2013) MAPping out distribution routes for kinesin couriers. Biol. Cell 105, 465–487 10.1111/boc.201300012 [DOI] [PubMed] [Google Scholar]
- 9. Tortosa E., Kapitein L. C., and Hoogenraad C. C. (2016) Microtubule organization and microtubule-associated proteins (MAPs). In Dendrites: Development and Disease (Emoto K., Wong R., Huang E., and Hoogenraad C., eds) pp. 31–75, Springer Japan, Tokyo, Japan [Google Scholar]
- 10. Olmsted J. (1986) Microtubule-associated proteins. Annu. Rev. Cell Biol. 2, 421–457 10.1146/annurev.cb.02.110186.002225 [DOI] [PubMed] [Google Scholar]
- 11. Ramkumar A., Jong B. Y., and Ori-McKenney K. M. (2018) ReMAPping the microtubule landscape: how phosphorylation dictates the activities of microtubule-associated proteins. Dev. Dyn. 247, 138–155 10.1002/dvdy.24599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dixit R., Ross J. L., Goldman Y. E., and Holzbaur E. L. F. (2008) Differential regulation of dynein and kinesin motor proteins by Tau. Science 319, 1086–1089 10.1126/science.1152993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vershinin M., Carter B. C., Razafsky D. S., King S. J., and Gross S. P. (2007) Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. U.S.A. 104, 87–92 10.1073/pnas.0607919104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chaudhary A. R., Berger F., Berger C. L., and Hendricks A. G. (2018) Tau directs intracellular trafficking by regulating the forces exerted by kinesin and dynein teams. Traffic 19, 111–121 10.1111/tra.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tokuraku K., Noguchi T. Q., Nishie M., Matsushima K., and Kotani S. (2007) An isoform of microtubule-associated protein 4 inhibits kinesin-driven microtubule gliding. J. Biochem. 141, 585–591 10.1093/jb/mvm063 [DOI] [PubMed] [Google Scholar]
- 16. Semenova I., Ikeda K., Resaul K., Kraikivski P., Aguiar M., Gygi S., Zaliapin I., Cowan A., and Rodionov V. (2014) Regulation of microtubule-based transport by MAP4. Mol. Biol. Cell 25, 3119–3132 10.1091/mbc.e14-01-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lipka J., Kapitein L. C., Jaworski J., and Hoogenraad C. C. (2016) Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 35, 302–318 10.15252/embj.201592929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J. S., Schubert C. R., Fu X., Fourniol F. J., Jaiswal J. K., Houdusse A., Stultz C. M., Moores C. A., and Walsh C. A. (2012) Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol. Cell 47, 707–721 10.1016/j.molcel.2012.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yadav S., Verma P. J., and Panda D. (2014) C-terminal region of MAP7 domain containing protein 3 (MAP7D3) promotes microtubule polymerization by binding at the C-terminal tail of tubulin. PLoS One 9, e99539 10.1371/journal.pone.0099539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tymanskyj S. R., Yang B., Falnikar A., Lepore A. C., and Ma L. (2017) MAP7 regulates axon collateral branch development in dorsal root ganglion neurons. J. Neurosci. 37, 1648–1661 10.1523/JNEUROSCI.3260-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tymanskyj S. R., Yang B. H., Verhey K. J., and Ma L. (2018) MAP7 regulates axon morphogenesis by recruiting kinesin-1 to microtubules and modulating organelle transport. Elife 7, e36374 10.7554/eLife.36374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallaud E., Caous R., Pascal A., Bazile F., Gagné J. P., Huet S., Poirier G. G., Chrétien D., Richard-Parpaillon L., and Giet R. (2014) Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J. Cell Biol. 204, 1111–1121 10.1083/jcb.201311094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Métivier M., Monroy B. Y., Gallaud E., Caous R., Pascal A., Richard-Parpaillon L., Guichet A., Ori-McKenney K. M., and Giet R. (2019) Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development 146, dev171579 10.1242/dev.171579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hooikaas P. J., Martin M., Mühlethaler T., Kuijntjes G. J., Peeters C. A., Katrukha E. A., Ferrari L., Stucchi R., Verhagen D. G. F., van Riel W. E., Grigoriev I., Altelaar A. F. M., Hoogenraad C. C., Rüdiger S. G. D., Steinmetz M. O., et al. (2019) MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol. 218, 1298–1318 10.1083/jcb.201808065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sung H. H., Telley I. A., Papadaki P., Ephrussi A., Surrey T., and Rørth P. (2008) Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev. Cell 15, 866–876 10.1016/j.devcel.2008.10.006 [DOI] [PubMed] [Google Scholar]
- 26. Metzger T., Gache V., Xu M., Cadot B., Folker E. S., Richardson B. E., Gomes E. R., and Baylies M. K. (2012) MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barlan K., Lu W., and Gelfand V. I. (2013) The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23, 317–322 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monroy B. Y., Sawyer D. L., Ackermann B. E., Borden M. M., Tan T. C., and Ori-McKenney K. M. (2018) Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 9, 1487 10.1038/s41467-018-03909-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W. 3rd, Tokito M., and Holzbaur E. L. F. (2010) Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20, 697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hendricks A. G., Holzbaur E. L. F., and Goldman Y. E. (2012) Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. U.S.A. 109, 18447–18452 10.1073/pnas.1215462109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berger F., Klumpp S., and Lipowsky R. (2019) Force-dependent unbinding rate of molecular motors from stationary optical trap data. Nano Letters 19, 2598–2602 10.1021/acs.nanolett.9b00417 [DOI] [PubMed] [Google Scholar]
- 32. Bathe F., Hahlen K., Dombi R., Driller L., Schliwa M., and Woehlke G. (2005) The complex interplay between the neck and hinge domains in kinesin-1 dimerization and motor activity. Mol. Biol. Cell 16, 3529–3537 10.1091/mbc.e04-11-0957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dietrich K. A., Sindelar C. V., Brewer P. D., Downing K. H., Cremo C. R., and Rice S. E. (2008) The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc. Natl. Acad. Sci. U.S.A. 105, 8938–8943 10.1073/pnas.0803575105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaan H. Y. K., Major J., Tkocz K., Kozielski F., and Rosenfeld S. S. (2013) “Snapshots” of ispinesib-induced conformational changes in the mitotic kinesin Eg5. J. Biol. Chem. 288, 18588–18598 10.1074/jbc.M113.462648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu H., Ali M. Y., Bookwalter C. S., Warshaw D. M., and Trybus K. M. (2009) Diffusive movement of processive kinesin-1 on microtubules. Traffic 10, 1429–1438 10.1111/j.1600-0854.2009.00964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klumpp S., and Lipowsky R. (2005) Cooperative cargo transport by several molecular motors. Proc. Natl. Acad. Sci. U.S.A. 102, 17284–17289 10.1073/pnas.0507363102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jamison D. K., Driver J. W., Rogers A. R., Constantinou P. E., and Diehl M. R. (2010) Two kinesins transport cargo primarily via the action of one motor: implications for intracellular transport. Biophys. J. 99, 2967–2977 10.1016/j.bpj.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rai A. K., Rai A., Ramaiya A. J., Jha R., and Mallik R. (2013) Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 10.1016/j.cell.2012.11.044 [DOI] [PubMed] [Google Scholar]
- 39. Wang Q., Diehl M. R., Jana B., Cheung M. S., Kolomeisky A. B., and Onuchic J. N. (2017) Molecular origin of the weak susceptibility of kinesin velocity to loads and its relation to the collective behavior of kinesins. Proc. Natl. Acad. Sci. U.S.A. 114, E8611–E8617 10.1073/pnas.1710328114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller M. J. I., Klumpp S., and Lipowsky R. (2008) Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. U.S.A. 105, 4609–4614 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Furuta K., Furuta A., Toyoshima Y. Y., Amino M., Oiwa K., and Kojima H. (2013) Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc. Natl. Acad. Sci. U.S.A. 110, 501–506 10.1073/pnas.1201390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X., and Schwarz T. L. (2009) The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163–174 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X. J., Saudou F., and Humbert S. (2008) Huntington phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27, 2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tint I., Jean D., Baas P. W., and Black M. M. (2009) Doublecortin associates with microtubules preferentially in regions of the axon displaying actin-rich protrusive structures. J. Neurosci. 29, 10995–11010 10.1523/JNEUROSCI.3399-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heins S., Song Y. H., Wille H., Mandelkow E., and Mandelkow E. M. (1991) Effect of MAP2, MAP2c, and tau on kinesin-dependent microtubule motility. J. Cell Sci. 1991, 121–124 10.1242/jcs.1991.Supplement_14.24 [DOI] [PubMed] [Google Scholar]
- 46. Lopez L. A., and Sheetz M. P. (1993) Steric inhibition of cytoplasmic dynein and kinesin motility by MAP2. Cell Motil. Cytoskeleton 24, 1–16 10.1002/cm.970240102 [DOI] [PubMed] [Google Scholar]
- 47. Gumy L. F., Katrukha E. A., Grigoriev I., Jaarsma D., Kapitein L. C., Akhmanova A., and Hoogenraad C. C. (2017) MAP2 defines a pre-axonal filtering zone to regulate KIF1-versus KIF5-dependent cargo transport in sensory neurons. Neuron 94, 347–362.e7 10.1016/j.neuron.2017.03.046 [DOI] [PubMed] [Google Scholar]
- 48. Hendricks A. G., Goldman Y. E., and Holzbaur E. L. F. (2014) Reconstituting the motility of isolated intracellular cargoes. Methods Enzymol. 540, 249–262 10.1016/B978-0-12-397924-7.00014-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cronan J. E. (1990) Biotination of proteins in vivo: a post-translational modification to label, purify, and study proteins. J. Biol. Chem. 265, 10327–10333 [PubMed] [Google Scholar]
- 50. Tomishige M., and Vale R. D. (2000) Controlling kinesin by reversible disulfide cross-linking: identifying the motility-producing conformational change. J. Cell Biol. 151, 1081–1092 10.1083/jcb.151.5.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ruhnow F., Zwicker D., and Diez S. (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100, 2820–2828 10.1016/j.bpj.2011.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nelson S. R., Ali M. Y., Trybus K. M., and Warshaw D. M. (2009) Random walk of processive, quantum dot-labeled myosin Va molecules within the actin cortex of COS-7 cells. Biophys. J. 97, 509–518 10.1016/j.bpj.2009.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.