Abstract
The regulation of transcription in trypanosomes is unusual. To modulate protein synthesis during their complex developmental stages, these unicellular microorganisms rely largely on post-transcriptional gene expression pathways. These pathways include a plethora of RNA-binding proteins (RBPs) that modulate all steps of the mRNA life cycle in trypanosomes and help organize transcriptomes into clusters of post-transcriptional regulons. The aim of this work was to characterize an RNA regulon comprising numerous transcripts of trypomastigote-associated cell-surface glycoproteins that are preferentially expressed in the infective stages of the human parasite Trypanosoma cruzi. In vitro and in vivo RNA-binding assays disclosed that these glycoprotein mRNAs are targeted by the small trypanosomatid-exclusive RBP in T. cruzi, U-rich RBP 1 (TcUBP1). Overexpression of a GFP-tagged TcUBP1 in replicative parasites resulted in >10 times up-regulated expression of transcripts encoding surface proteins and in changes in their subcellular localization from the posterior region to the perinuclear region of the cytoplasm, as is typically observed in the infective parasite stages. Moreover, RT–quantitative PCR analysis of actively translated mRNAs by sucrose cushion fractionation revealed an increased abundance of these target transcripts in the polysome fraction of TcUBP1-induced samples. Because these surface proteins are involved in cell adherence or invasion during host infection, we also carried out in vitro infections with TcUBP1-transgenic trypomastigotes and observed that TcUBP1 overexpression significantly increases parasite infectivity. Our findings provide evidence for a role of TcUBP1 in trypomastigote stage-specific gene regulation important for T. cruzi virulence.
Keywords: RNA binding protein, RNA–protein interaction, RNA metabolism, gene regulation, trypanosome, post-transcriptional regulation, Chagas disease, RNA regulon, virulence
Introduction
Multiple events of spatial–temporal RNA metabolism occur during and after transcription to ultimately determine the proteomic content of each eukaryotic cell (1). A dynamic network of RNA–protein interactions is necessary to precisely direct the cytoplasmic fate of each transcript (2). The action of RNA-binding proteins (RBPs)4 is central, because these factors can accurately guide transcripts into distinct post-transcriptional regulatory processes (3, 4) and/or control checkpoints at various steps, including RNA processing, export, stabilization, localization, and translation (5–8). In recent years, researchers have observed an interesting strategy that some organisms use to manage the majority of their cellular transcripts (9). Different mRNAs that encode protein products necessary for a particular biological pathway are organized by one (or more) RBPs into specific messenger ribonucleoprotein (mRNP) complexes. Each member of a group of functionally related transcripts harbors a common regulatory sequence element within their noncoding region, identifying them as the target transcripts of a given trans-acting factor. The formed mRNPs might subsequently guide transcripts into the same post-transcriptional regulatory event (reviewed in Refs. 9 and 10). A group of functionally linked mRNAs together with the RBPs that coordinately modulate their expression is termed an RNA regulon (11), acknowledging their partial analogy to the polycistronic mRNAs produced by bacteria. The mRNP-driven organization of transcripts allows eukaryotic cells to manage products of genes that are dispersed throughout their genome but encoding proteins with similar functions. A complex but flexible level of regulation is achieved by this higher-order organization of transcripts, which also prompts a rapid readaptation of the cellular transcriptome in response to alterations in the environment (1). Post-transcriptional RNA regulons have been described in mammalian cells, fruit flies, and budding yeasts and are thought to be important for processes such as immune responses, oxidative metabolism, stress responses, the cell cycle, and disease (reviewed in Refs. 12 and 13).
Trypanosomes, which are protozoan parasites of the order Kinetoplastida, are responsible for diseases that affect humans and domestic animals worldwide (14). They are single-cell microorganisms with a complex life cycle that alternates between mammalian and insect hosts. These dixenic parasites must continuously adapt to environmental changes (15), and the adaptive processes can ultimately alter their transcriptomes (16–18). This is achieved by genetic expression programs that exhibit unique features (19, 20), including the nuclear compartmentalization of stage-specific transcripts (21). It currently appears that, in the absence of precise transcriptional control, gene expression is regulated mostly by post-transcriptional mechanisms (22–24). Nearly all trypanosomal coding genes are transcribed as part of polycistronic arrays that, with a few exceptions (25), do not exhibit any association of functionally related genes. Further RNA processing results in individual transcripts that are assembled into mRNP complexes, which channel them to their final fate (26). In this context, organized subsets of trypanosomal mRNP complexes must be post-transcriptionally co-regulated. The functionality of these RNA regulons has been extensively explored in trypanosomes (27–34). Trypanosoma cruzi is the etiologic agent of Chagas disease, which is a complex zoonosis that involves triatomine vectors and mammals that act as parasite reservoirs. T. cruzi infection is established in the mammal by the insect-derived metacyclic trypomastigote. It rapidly invades a wide variety of cells, in which it transforms into the intracellular replicative stage, named amastigote. After several binary divisions, amastigotes differentiate into the infective mammal form, trypomastigote, which is released into the bloodstream upon cell rupture. This form circulates in blood to infect other cells or undergo ingestion by the hematophagous vector. Within the insect, trypomastigotes differentiate to replicative epimastigotes, migrate along the digestive tract, and transform into infective metacyclic trypomastigotes. These forms are deposited on the mammalian host along with the insect feces during a blood meal and gain access to the bloodstream through a skin wound or the mucous membranes to complete the circle (14).
As mentioned, these transformation processes are mainly controlled by RBPs. T. cruzi U-rich RBP 1 (TcUBP1) is one of the best-studied RNA-recognition motif (RRM) proteins in trypanosomes (35). This single RRM domain cytoplasmic protein has a characteristic βαββαβ-fold and a C-terminal Gly–Gln–rich extension that is likely involved in protein–protein interactions (36). This RBP can form a complex with poly(A)-binding protein 1 on certain mRNAs and up- or down-regulate its target transcript in a stage-specific manner (35, 37). Although mRNAs from various functional categories (cell organization and division, RNA processing, stress and signaling, transcription, and transport) have been identified as being targets of TcUBP1, this RRM protein largely interacts with transcripts coding for surface glycoproteins and proteins involved in energy metabolism (36). Typically, these target transcripts harbor a 30-mer signature RNA motif with a stem–loop structure termed UBP1m that is frequently found within their 3′-UTR (34, 36).
The ability of T. cruzi to survive in the mammalian host is in part due to the expression of a plethora of surface proteins and signaling genes, which include members of the trans-sialidase and trans-sialidase–like (TcS) superfamily, mucins, and mucin-associated surface proteins, among others (38). Freitas et al. (38) reported that TcS is the largest gene family in T. cruzi and encompasses eight clusters (groups I–VIII). This superfamily is composed of more than 1,400 genes. Cluster I has been shown to contain the active trans-sialidase enzymes that are responsible for sialic acid metabolism, which is crucial for the biology of the parasite (39). The present work focused on TcS superfamily members belonging to groups II–VIII, which are preferentially expressed in the infective trypomastigote stage of the parasite, have a highly conserved sequence in their 3′-UTRs and are target transcripts of TcUBP1. We previously showed that overexpression of GFP-tagged TcUBP1 in epimastigote cells triggers their differentiation to the infective trypomastigote forms of the parasite (40). In this study, we further examined the biological role of this trypanosomal RBP by following the fate of a stage-specific group of surface glycoprotein TcUBP1 targets through analysis of their mRNA abundance, localization, and translation. Moreover, we examined the impact of TcUBP1 overexpression on the infective life stage of this parasite.
Results
TcS family mRNAs with a highly conserved element in their 3′-UTRs are more abundant in trypomastigotes
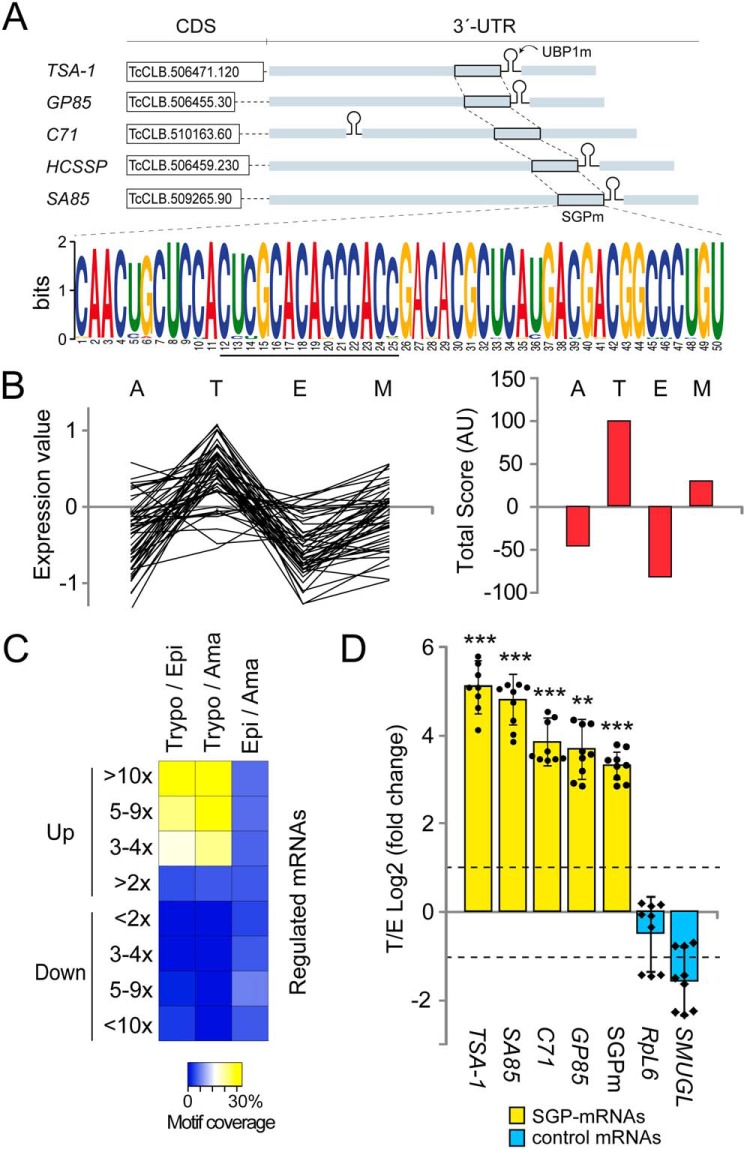
In previous studies on partners of the TcUBP1–mRNP complex by using in vivo RBP immunoprecipitation and sequencing (36), we described a sequence of ∼150 nt corresponding to a 3′-noncoding region highly conserved in transcripts encoding TcS proteins, all of them harboring UBP1m. Examples of these genes are: the 85-KDa surface antigen (GP85) (TcCLB.506455.30), the Trypomastigote surface glycoprotein (TSA-1) (TcCLB.506471.120), the amastigote cytoplasmic antigen Sx23 (TcCLB.511219.40), the C71 signaling surface protein (TcCLB.510163.60), the SA85 protein (TcCLB.508285.60), the host cell signaling surface protein HCSSP (TcCLB.506459.230), and the Tc13 surface antigen (TcCLB.510307.284) (Fig. 1A).
Figure 1.
Members of the TcS family containing a highly conserved sequence in their 3′-UTRs are over-represented in the infective trypomastigote stage of the parasite. A, a 50-nt-long conserved sequence within the 3′-UTR of TS-like subfamily members, called SGPm detected by the MEME tool. SGPm is present in 83% of the TS-like subfamily members (input: 59 sequences). B, summary of microarray analysis of 70 genes with the SGPm. The relative mRNA levels in the four developmental stages were obtained from TriTrypDB (left chart). The life cycle stage with the highest mRNA level was assigned score 2, the one with the second highest mRNA level was assigned score 1, and those with lower mRNA abundance were assigned scores −1 and −2. The total scores obtained for each stage were then plotted (right chart). M, metacyclic trypomastigotes. C, heat map representation of the percentages of genes having the SGPm in most up- or down-regulated transcripts in pair wise comparisons. Data of genes with >2, 3–4, 5–9, or >10-fold change differences were extracted from Li et al. (16). Trypo/Epi, trypomastigote versus epimastigote; Trypo/Ama, trypomastigote versus amastigote; Epi/Ama, epimastigote versus amastigote. The yellow/white color indicates a high correlation, whereas the blue color indicates a low correlation. D, results from RT-qPCR quantification assays showing the enrichment of surface glycoprotein transcripts in infective trypomastigotes over noninfective epimastigotes forms. TSA-1, GP85, SA85, C71, and SGPm are members of the TcS family (yellow bars). SMUGL is a target of TcUBP1 but not a member of TcS family. RPL6 is the ribosomal protein L6 mRNA that is not a TcUBP1 target. The values are expressed as the means ± S.D. of the fold changes in log2 scale from three independent experiments. The individual data points are shown by filled circles (SGP mRNAs) and diamonds (control). Dotted lines indicate a 2-fold change.
In silico analyses were performed to investigate whether other surface glycoprotein sequences harboring UBP1m also contained conserved elements in their 3′-UTRs. To address this, we analyzed the 3′-UTR of ∼60 TcUBP1 database target sequences annotated as trans-sialidases (Fig. S1). Using the motif discovery program MEME (41), we identified a 50-nt consensus element 5′-CAACUGCUCCACUCGCACACCCACCGACACGCUCAUGACGACGGCCCUGU-3′ (E -value 1.6E-1109). As a random control, the letters in the input sequences were shuffled, resulting in the lack of identification of statistically significant motifs. Subsequent BLASTN analyses indicated that the consensus element was widely distributed in more than 600 sequences in the genome of T. cruzi showing >95% identity and 100% query coverage (File S1). A comparison of these sequences revealed that they all have a high conservation degree of this shared untranslated sequence element, which we named surface glycoprotein motif (SGPm). Sequence logo graphics showed that all the motifs bear an AC-rich core sequence composed of CCACXCXCACACCCACC (Fig. 1A). A whole genome scan on all available sequences using the MAST program revealed that the cis-element is widely distributed in almost all chromosomes and that there are 329 copies of this sequence in the genome of T. cruzi when the cut off was set at an E value of E −1 (File S2). The SGP motif is widely distributed in the genome with >60 glycoprotein genes that are clustered within a 10-kbp genomic arrangement, including 18 genes separated by less than 2 kbp of intergenic regions (Table S1). Analysis of this gene cluster with the DAVID Functional Annotation Chart tool (NIAID, National Institutes of Health) revealed the enrichment of mRNAs encoding for trans-sialidase like proteins. Given that a large proportion of the TcUBP1 targets transcripts code for surface glycoproteins, we decided to examine how many of these 329 SGPm-mRNAs, listed in File S2, harbor the best RNA motif required for TcUBP1 binding. We found that 87% (287 genes) showed co-occurrence of both the UBP1m and SGPm in their 3′-UTRs (File S2). The data obtained revealed that the TcUBP1 target transcripts are enriched in the group of 50-nt SGP cis-element–containing genes, compared with the whole T. cruzi genome (χ2 test, p < 0.001). Strikingly, most of these genes (92.7%) have the SGPm located ∼200-nt downstream of the annotated stop codon and proximately upstream of the 30-mer regulatory element UBP1m, a configuration that we named as configuration b (Fig. S2). Configuration a refers to the minor fraction of the genes having both motifs separated by a dozen of nucleotides with the UBP1m located upstream, whereas configuration c refers to genes with UBP1m located downstream.
We also performed a bioinformatic analysis of previous microarray data (42) available in TriTrypDB.org to investigate whether there was a stage-specific pattern of expression of the genes containing the SGPm. For this cluster, we selected 70 genes for which there was transcriptome information available and plotted the relative mRNA levels and total scores in the four developmental forms. The analysis showed that the genes bearing the SGPm have higher mRNA levels in the trypomastigote stage (Fig. 1B). Additionally, we explored the T. cruzi transcriptome data reported by Li et al. (16) to evaluate the percentage of transcripts with the SGPm among the most up- or down-regulated genes in a pairwise comparison between the trypomastigote (T), epimastigote (E), and amastigote (A) stages. We searched the SGPm-containing genes in the transcripts sets with >2-, 3–4-, 5–9-, or >10-fold change differences between two stages (T versus E, T versus A, and E versus A). When analyzing the most up-regulated genes (>10-fold change), we found that the SGPm-containing genes represented the 25.1% in T/E comparison, 33.3% in the T/A comparison, and only 4.1% in the E/A comparison. No significant coverage was found for any of the down-regulated genes. This indicated that only the up-regulated genes in the trypomastigote stage were enriched in transcripts harboring the SGPm (ANOVA with post hoc Tukey test, p < 0.01) (see the significantly different clusters colored with yellow/white in the heat map depicted in Fig. 1C).
To validate the microarray data, we next selected four single-copy transcripts of the TcS family from the whole genome scan list (named TSA-1, GP85, SA85, and C71) and quantified the mRNA levels in infective trypomastigotes and noninfective epimastigotes by RT-qPCR. As experimental controls, we included Ribosomal Protein L6 (RPL6) and the TcSMUGL transcript (TcCLB.504539.20), a known target of TcUBP1 (43), which codes for a mucin-like glycoprotein anchored to and secreted from the surface of insect-dwelling epimastigotes but lacks the SGPm. The results showed a significant enrichment in these transcripts in the trypomastigote stage but not in the control mRNAs (Fig. 1D), with differences ranging from 15- (GP85) to 35-fold (TSA-1). We also measured the abundance of the conserved SGPm and found the same tendency: ∼10-fold increase in trypomastigotes over epimastigotes (log2 FC ∼3.3 in Fig. 1D). In contrast, no significant changes were observed in the controls TcSMUGL and RPL6. Thus, both the analysis of the microarray data and the results of the RT-qPCR indicated that the genes coding for this surface glycoprotein family are over-represented in the trypomastigote stage.
TcS transcripts are targets of a sequence-specific RBP
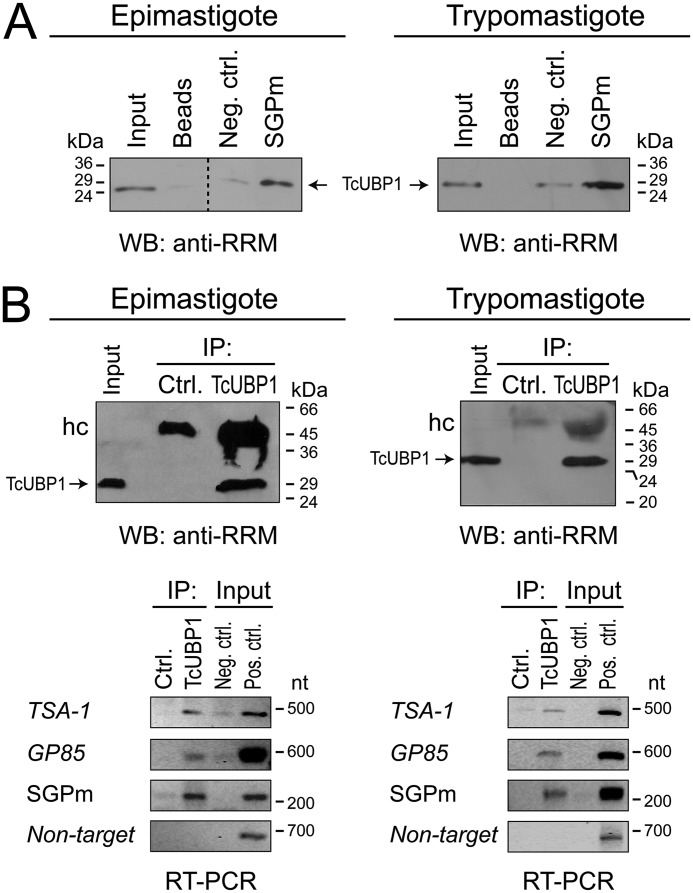
Next, we analyzed the possible interaction of sequence-specific RBPs with the 3′-UTR of surface glycoprotein-coding genes to verify whether the mRNA abundance of this group of genes is organized by an mRNP complex-driven mechanism. To this end, a region of the TSA-1 transcript containing the cis-acting SGPm and UBP1m was inserted into the pGEM-T Easy vector, transcribed in vitro in the presence of CTP-biotin, and incubated with recombinant GST-tagged TcUBP1, GST alone (as a control), or cell-free cytosolic extracts. A synthetic pGEM-T polylinker transcript without any insert was used as a negative control (Fig. 2A, Neg. ctrl.). In a first RNA-binding assay, we used recombinant proteins to test (qualitatively) whether these transcripts were bound by GST–TcUBP1 (Fig. S3A). The resulting RNA–protein complexes were pulled down, and the presence of GST–TcUBP1 in the sample obtained was revealed by Western blotting. The pGEM-T SGPm(+) construction effectively pulled down GST–TcUBP1 but failed to bind to the GST control protein (Fig. S3B). The control transcript (pGEM-T) failed to bind to any of the proteins. After confirming that recombinant GST–TcUBP1 effectively interacts in vitro with the region containing the SGPm and UBP1m, we repeated the same approach but now using parasite protein extracts (instead of a GST-tagged protein). This experiment corroborated that TcUBP1 was able to interact with the TcS transcripts bearing the SGPm in both epimastigote and trypomastigote cells but was not with the control transcript (Fig. 2A). Because this in vitro synthesized RNA sequence harbors the SGPm proximately upstream of the structural UBP1m, it will be interesting to dissect the RNA-binding activity of each of these motifs in the future (see “Discussion”).
Figure 2.
In vitro and in vivo TcUBP1 binding to the SGPm. A, in vitro binding assay between biotinylated transcripts corresponding to the pGEM-T polylinker fragment containing (or not) the SGPm and trypanosome cell-free cytosolic extracts. mRNP complexes were revealed by Western blotting (WB) with anti-TcUBP1 serum. B, in vivo RNA–protein interactions. The upper panels shows the immunoprecipitation assays with anti-TcUBP1 antibodies and preimmune serum (Ctrl.) using cell-free cytosolic extracts from epimastigotes (left panels) and trypomastigotes (right panels). The TcUBP1 RRM-type protein is indicated by arrows. The lower panels show the RT-PCR assays in mRNP complexes immunoprecipitated with anti-TcUBP1 or preimmune serum using specific primers for representative members of the RNA regulon. TSA-1, trypomastigote surface glycoprotein; GP85, 85-kDa surface antigen; SGPm, conserved element; Non-target, RNA-binding protein, putative (TcCLB.506649.80) (negative control). Input PCR controls: cDNA prepared from total RNA with (Pos. ctrl.) or without reverse transcriptase enzyme (Neg. ctrl.). IP, immunoprecipitation; hc, immunoglobulin heavy chain.
As a second approach, we verified the ability of TcUBP1 to interact with these mRNA targets by immunoprecipitation assays (Fig. 2B). The presence of the transcripts TSA-1, C71, GP85, and the amplicon corresponding to the SGPm in the co-immunopurified RNA fractions was analyzed using specific primers (Table S2). The results confirmed that these mRNAs, containing the cis-acting sequences UBP1m and SGPm, are targets of TcUBP1 in epimastigotes, as well as in trypomastigotes (Fig. 2B), suggesting that TcUBP1 is involved as part of the SGPm regulon in the different developmental stages of the parasite.
Up-regulation of trypomastigote TcS transcripts in epimastigotes overexpressing TcUBP1–GFP
After showing that TcUBP1 interacts in vivo with TcS mRNAs, we next examined the biological role of this RNA-RBP interaction, because this RBP is expressed in both infective and noninfective forms of the parasite, whereas TcS mRNAs are preferentially expressed in infective forms. For this, we transfected epimastigotes with a DNA construction of TcUBP1–GFP (40) or GFP as a negative control, in a tetracycline-inducible system by using the pTcINDEX vector (44). Aliquots of stably transfected noninfective epimastigote cultures ectopically expressing TcUBP1 or GFP alone were analyzed after 96-h induction by fluorescence microscopy and indirect immunofluorescence (IIF) using specific anti-TcUBP1 serum (named anti-RRM) and noninduced transfected parasites as controls. In the induced populations transfected with the TcUBP1–GFP construct, co-localization of green (GFP) and red (corresponding to TcUBP1) signals was observed, confirming the cytosolic expression of this RBP (Fig. 3A). Next, RNA was purified from both GFP and TcUBP1–GFP parasite populations to evaluate by RT-qPCR the transcript levels for the four SGPm-containing mRNAs previously analyzed (TSA-1, SA85, C71, and GP85), the conserved SGPm, and a control mRNA. This allowed us to observe a significant enrichment for steady-state levels of all tested mRNAs in the parasites overexpressing TcUBP1–GFP (Tet+/−) compared with GFP transgenic parasites (Tet+/−) (p < 0.001, two-way ANOVA). In particular, we observe a 5–13-fold increase in SGPm-harboring mRNA levels in parasites overexpressing TcUBP1–GFP but not in the control SMUGL mRNA (Fig. 3B, depicted as log2 FC from ∼2 to ∼3.5). Also, the abundance of the target sequence SGPm, highly conserved in virtually all TcS family members, was also measured and showed similar results (>4-fold enrichment; Fig. 3B). Moreover, intra–TcUBP1–GFP analysis of induced over noninduced parasites revealed a clear significant enrichment of the abundances of SGP mRNAs compared with the TcSMUGL epimastigote control mRNA (p < 0.001, Bonferroni's t test post hoc). Finally, all the trypomastigote mRNAs tested: SA85, C71, TSA-1, GP85, and SGPm sequence displayed a significant (>2-fold) increase in steady-state mRNA levels in epimastigotes overexpressing TcUBP1–GFP (p < 0.05, Student's t test, two-tails, log2 FC compared with 1; upper dotted line in Fig. 3B).
Figure 3.
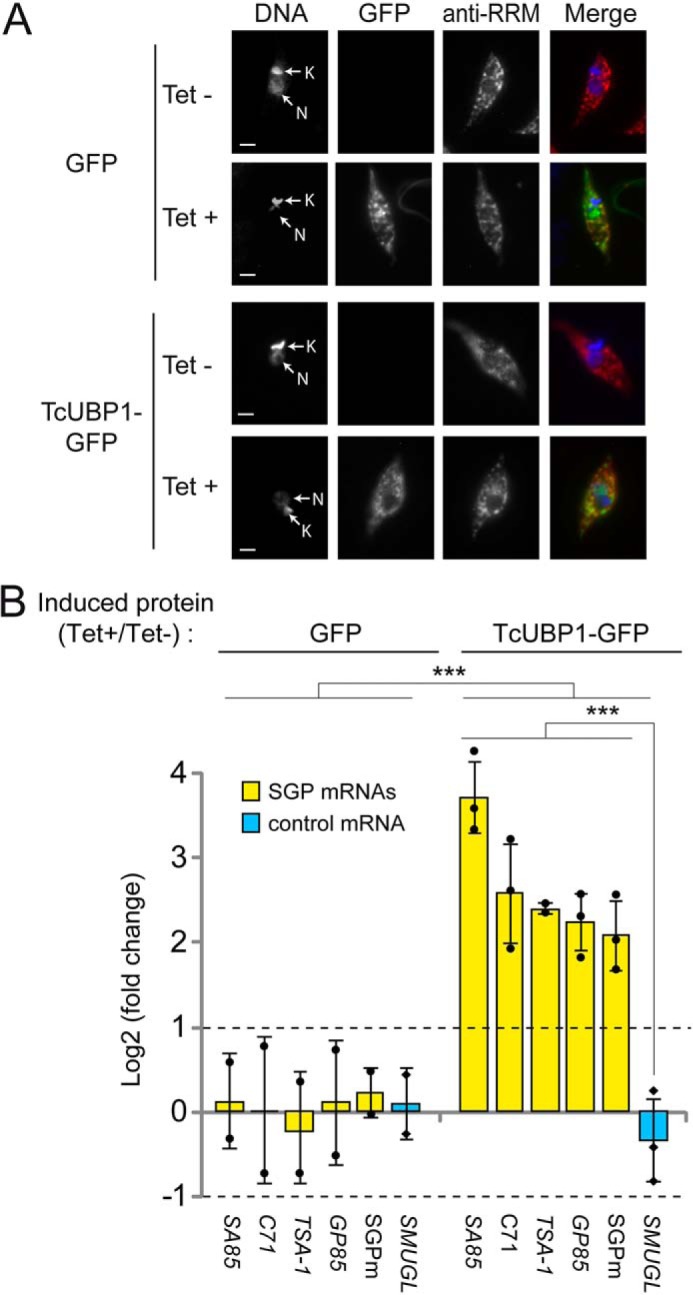
Levels of transcripts coding for trypomastigote surface glycoprotein in epimastigote parasites overexpressing TcUBP1–GFP. A, representative images of isolated epimastigotes transfected with GFP or TcUBP1–GFP and induced with tetracycline (Tet+) or not (Tet−). Grayscale images of each channel are shown on the left. Merged images showing expression of GFP (in green), TcUBP1 (anti-RRM signal, in red), and DAPI staining (in blue) for visualization of kinetoplast (K) and nuclear (N) DNA are shown on the right. B, RT-qPCR results for transcripts encoding trypomastigote surface glycoproteins (SA85, C71, TSA-1, and GP85), the SGPm element, and the SMUGL transcript coding for epimastigote surface glycoprotein as control. The values are expressed as the means ± S.D. of the fold changes in log2 scale from three independent experiments. The average data points are shown by filled circles (SGP mRNAs) and diamonds (control). ***, p < 0.001; two-way ANOVA with Bonferroni's post hoc test. Dotted lines indicate 2-fold enrichment (upper, log2 FC = 1) or depletion (lower, log2 FC = −1).
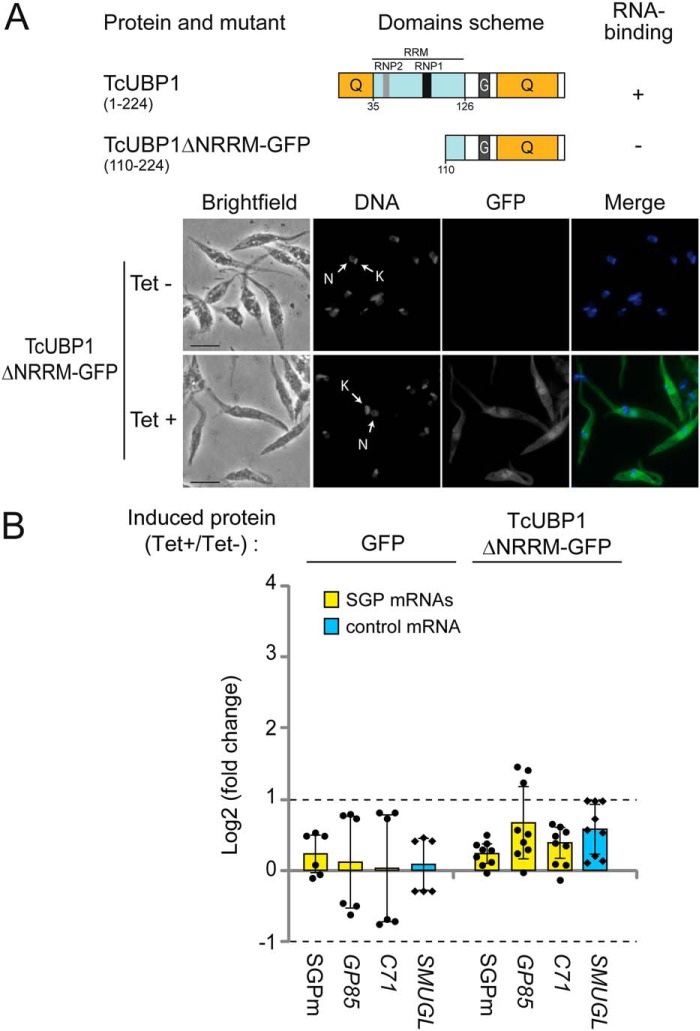
We then investigated whether the RNA-binding function of TcUBP1 is required to confer the effects observed. To answer this, we employed an RNA-binding defective variant of TcUBP1 as a control for the overexpression experiment (ΔNRRM construction) (Fig. 4A). Aliquots of stably transfected epimastigote cultures ectopically expressing this mutated version were analyzed after 96-h induction by fluorescence microscopy, confirming the proper expression of TcUBP1ΔNRRM-GFP (Fig. 4B). Then we quantified the mRNA levels in these parasites overexpressing TcUBP1ΔNRRM-GFP (Tet+/−) compared with GFP transgenic parasites (Tet+/−). The results showed no significant changes for any of the tested transcripts. We demonstrated that only the WT TcUBP1, but not an RNA-binding defective variant, can produce the up-regulation effect. Altogether, these results, obtained in transgenic noninfective epimastigote cells, suggest the presence of a post-transcriptional program coordinated by TcUBP1 for transcripts encoding trypomastigote surface glycoproteins.
Figure 4.
Levels of transcripts coding for trypomastigote surface glycoproteins in epimastigote parasites overexpressing TcUBP1ΔNRRM-GFP. A, scheme of TcUBP1 and mutant TcUBP1ΔNRRM showing the RRM (in light blue) and Q- and G-rich regions (in orange). Binding to RNA is based on previous evidence from our laboratory (40). Representative images of isolated epimastigotes transfected with GFP or TcUBP1ΔNRRM-GFP and induced (Tet+) or not (Tet−) with tetracycline. We obtained a fold change value of 9.3 ± 3.4 in induce samples. Grayscale images of each channel are shown on the left. Merged images showing expression of GFP (in green) and DAPI staining (in blue) for visualization of kinetoplast (K) and nuclear (N) DNA are shown on the right. B, RT-qPCR results for transcripts encoding trypomastigote surface glycoproteins (C71 and GP85), the SGPm, and the SMUGL transcript coding for epimastigotes surface glycoprotein as control. The values are expressed as the means ± S.D., of the fold changes in log2 scale from three independent experiments. In the GFP case, experimental data are the same as in Fig. 3B. The individual data points are shown by filled circles (SGP mRNAs) and diamonds (control). Dotted lines indicate 2-fold enrichment (upper, log2 FC = 1) or depletion (lower, log2 FC = −1).
TcS family transcripts harboring the SGPm change their subcellular distribution profile after TcUBP1–GFP overexpression
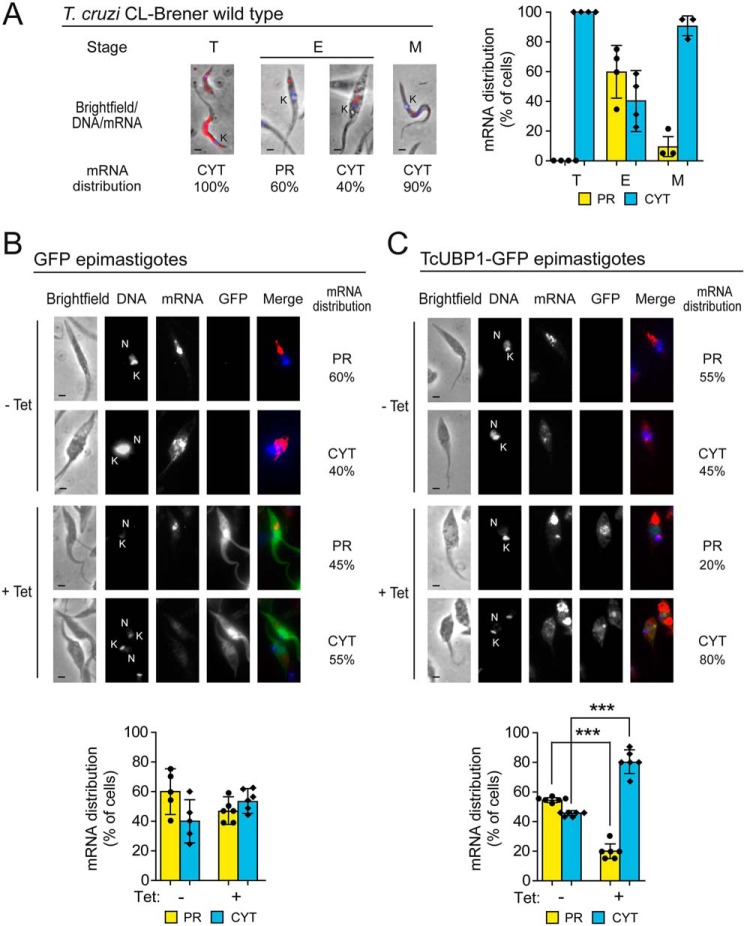
We next analyzed the subcellular distribution of transcripts encoding trypomastigote surface proteins during T. cruzi parasite development after inducing TcUBP1 overexpression. To this end, we synthesized a Cy3-conjugated probe complementary to the sequence encoding the SGPm (see “Experimental procedures”) and performed RNA hybridizations in WT infective and noninfective forms followed by microscopy analysis. Micrographs of infective trypomastigote forms of the parasite displayed a uniform cytosolic signal (CYT), whereas replicative epimastigotes showed an asymmetric RNA distribution (Fig. 5A, left panel). Quantification of these signals indicated that ∼60% of epimastigote cells displayed an RNA signal preferentially restricted to the posterior region of the cytoplasm (PR), whereas in cell-derived trypomastigotes and metacyclic trypomastigotes, the localization of the transcripts was uniformly distributed in the cytosol (90–100% of the cells) (Fig. 5A, right panel). To further investigate potential differences caused by a post-transcriptional program executed by TcUBP1, we evaluated the subcellular localization of SGPm-containing mRNAs after GFP or TcUBP1–GFP overexpression. In GFP epimastigotes, no differences were detected between induced and noninduced cells (Fig. 5B), showing intracellular mRNA localization similar to WT epimastigotes. Interestingly, epimastigotes overexpressing TcUBP1–GFP showed a statistically significant change of this localization toward the perinuclear and cytosolic regions, resembling what is observed in trypomastigote cells (Fig. 5C) (Student's t test, p < 0.001). Although our results do not show a direct association between mRNA localization and protein translation, the differential subcellular distribution of transcripts in areas previously described with high density of ribosomes suggests a putative change appropriate for RNA translation (45).
Figure 5.
Subcellular localization of TcS transcripts harboring SGPm in WT, GFP, and TcUBP1–GFP transgenic parasites. A, RNA localization of TcS family transcripts along the life cycle of T. cruzi. Left panel, representative merged images of cell-derived trypomastigotes (T), epimastigotes (E), and metacyclic trypomastigotes (M) parasites. The SGPm probe is indicated in red. DAPI staining (in blue) reveals nuclear (N) and kinetoplast (K) DNA. Right panel, percentage values are expressed as the means of three independent experiments (n = 30 cells). B, RNA localization in GFP transgenic epimastigotes in Tet-induced (+) and not induced (−) samples. Top, representative images of Tet-induced parasites with the percentage of these cells showing a distribution of SGP mRNAs preferentially located in the posterior cytosolic region (PR) or uniformly distributed in the cytosol (CYT). Grayscale images of each channel and merged images (DAPI in blue, mRNA in red, and GFP in green) are shown. Bottom, quantification of intracellular RNA localization of transcripts in Tet-induced (+) and not induced (−) cells. C, RNA localization in TcUBP1 transgenic epimastigotes in Tet-induced (+) and not induced (−) samples. The legend is as in B. The values are expressed as the means ± S.D. The individual data points are shown by filled circles (PR) and diamonds (CYT). ***, p < 0.001, Student's t test.
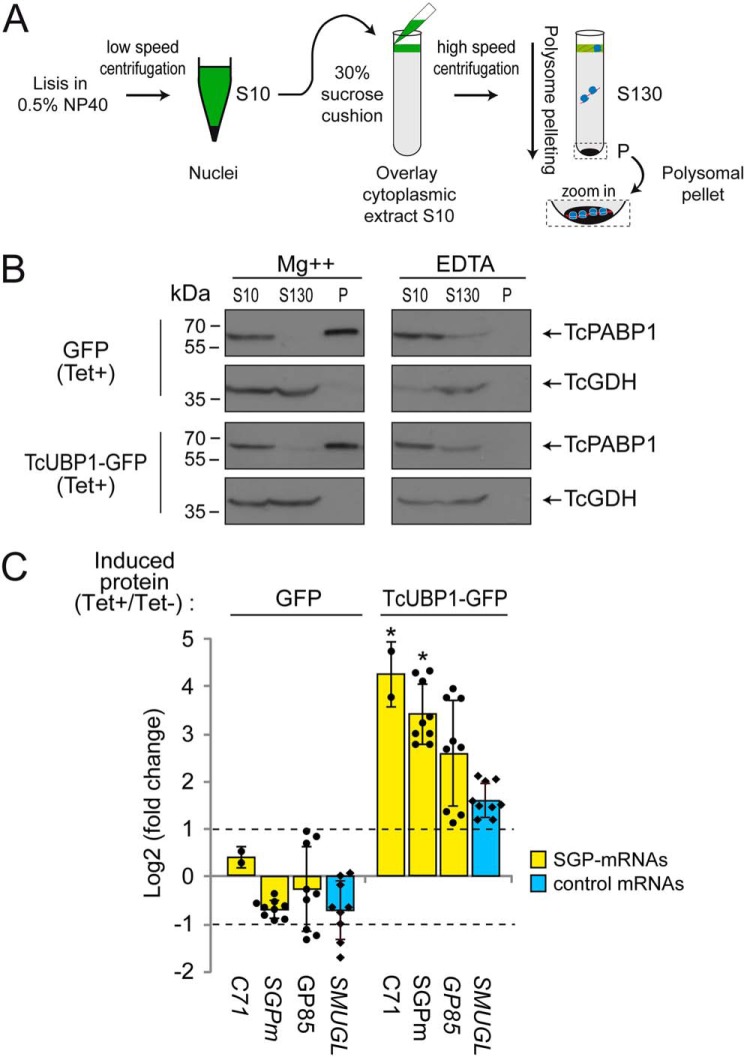
This result prompted us to investigate whether TcUBP1 can facilitate the remodeling of surface glycoproteins in T. cruzi by increasing the translation rate of TcS mRNAs. To this end, polysomal pellets were purified from each parasite population transfected with GFP or TcUBP1–GFP, using induced (Tet+) versus noninduced (Tet−) control cells (see “Experimental procedures” and Fig. 6A). Then we prepared a sucrose cushion in solutions containing either magnesium (required for ribosome stability) or EDTA (required for the dissociation of 40S and 60S ribosomal subunits) and analyzed the quality of the separation of both the prepolysomal S130 and polysomal P fractions by immunoblot with protein marker antibodies (Fig. 6B). As shown in the figure for both GFP and TcUBP1–GFP-induced parasites, in the presence of magnesium, PABP1 associated with polysomes, as indicated by the signal detected in the polysomal pellet (P), whereas the cytosolic glutamate dehydrogenase (GDH) showed a polysome-free localization indicated by the signal detected only in the S130 fraction. In the presence of EDTA, neither ribosomal nor ribosome-associated proteins were found at the bottom of the gradient. As expected, under this experimental condition, PABP1 and GDH migrated only near the top of the gradient (Fig. 6B). Two other trypanosomal marker antibodies from proteins not associated with polysomes (TcHSP70 (TcCLB.511211.170) and TcCruzipain (TcCLB.507603.260)) were used to check the specificity of the polysome purification (Fig. S4).
Figure 6.
TcUBP1–GFP overexpression enhances translation of trypomastigote SGPm-containing transcripts. A, scheme of polysome purification by high-speed centrifugation through a 30% sucrose cushion (for details of the procedure see “Experimental procedures”). B, Western blots of clarified trypanosome cell cytoplasmic extract (S10), prepolysomal (S130), and polysomal pellet (P) fractions from GFP and TcUBP1–GFP induced cultures obtained in the presence of magnesium (Mg2+) or EDTA. Aliquots of S10, S130, and 5-fold concentrated aliquots of the P fraction were probed with an anti-PABP1 or -GDH serum (indicated by arrows). C, RT-qPCR results in GFP and TcUBP1–GFP parasites for transcripts encoding trypomastigote surface glycoproteins (GP85 and C71), the SGPm, and the TcSMUGL and RpL6 transcripts as controls. The values are expressed as the means ± S.D. of the fold changes in log2 scale from induced (indicated as Tet+) to noninduced parasites (Tet−) obtained in three independent experiments. The individual data points are shown by filled circles (SGP-mRNAs) and diamonds (control). Dotted lines indicate 2-fold change. *, p < 0.05 using ANOVA.
We next performed an RT-qPCR analysis of polysomal pellets in GFP- or TcUBP1-overexpressing parasites. We tested two TcS transcripts (GP85 and C71) and the conserved SGPm and quantified the mRNA levels in induced versus noninduced control cells by RT-qPCR. For these quantifications, we also included the experimental control TcSMUGL transcript. The results showed a significant enrichment in TcUBP1 targets in TcUBP1-induced over noninduced cells but not in control GFP samples (ANOVA with post hoc Tukey test, p < 0.05; Fig. 6C). In TcUBP1-expressing parasites, this enrichment was more pronounced in the SGPm-containing mRNAs than on the control TcSMUGL, with differences ranging from 5-fold (GP85, log2 FC ∼2.5) to more than ∼10-fold enrichment (C71, SGPm) (log2 FC >3.3 in Fig. 6C). In contrast, GFP overexpression did not affect the abundance of any of these TcS or control mRNAs, because no significant changes were observed in these parasites (Fig. 6C). These results show that TcUBP1 overexpression trigger the mobilization of trypomastigote stage–specific transcripts to polysomes for active translation.
TcUBP1–GFP transgenic trypomastigotes have an enhanced capacity for infection
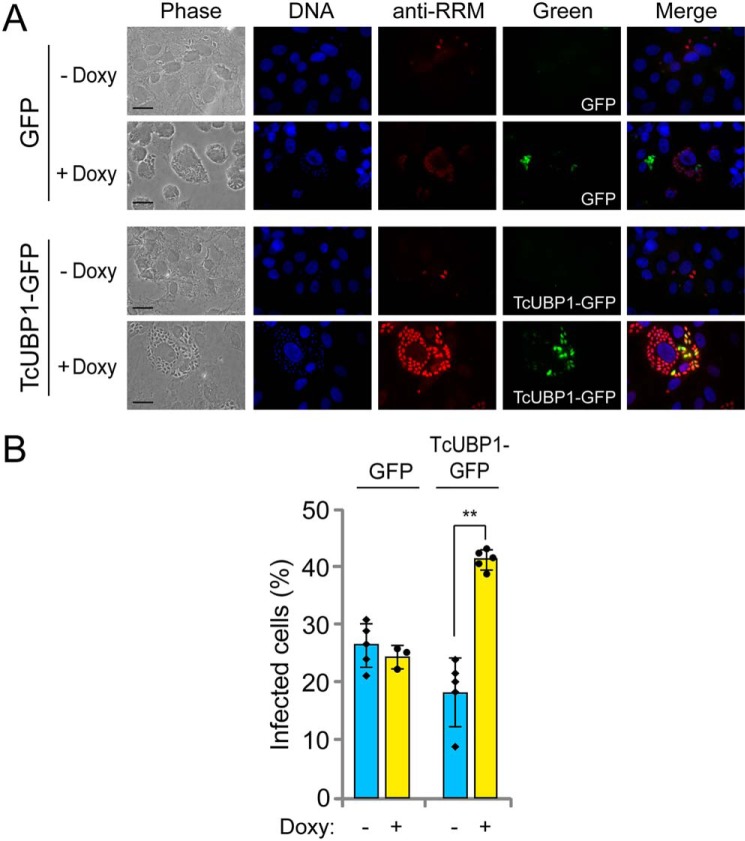
Previous studies describing the importance of the expression of trypomastigote surface glycoproteins during infection (16, 46) led us to evaluate whether the higher mRNA abundance of these cis-element–containing transcripts affected the infectivity of the trypomastigotes derived from transgenic epimastigotes. To test this possibility, epimastigotes transfected with pTcINDEX–GFP (used as control) or pTcINDEX–TcUBP1–GFP were starved and allowed to differentiate to metacyclic trypomastigotes. Both parasite populations were incubated with doxycycline and analyzed by fluorescence microscopy to detect induced GFP protein in these transfected cell lines. Trypomastigotes from the control populations showed a cytosolic green fluorescent signal, whereas TcUBP1–GFP trypomastigotes showed no fluorescent signal. Thus, the expression of TcUBP1–GFP was further analyzed by IIF, using antibodies against GFP (anti-GFP) and TcUBP1 (anti-RRM) (Fig. S5A). As expected, IIF analysis allowed us to detect GFP expression in control trypomastigotes with a consistent cytosolic signal (Fig. S5A). TcUBP1–GFP expression was also validated with both anti-GFP and anti-RRM antibodies, suggesting that these transgenic parasites were also able to ectopically express the fused protein in the cytosol (Fig. S5B). As a specificity control, no signal was detected in the microscopy samples performed with parasites without doxycycline incubation (Fig. S6). After confirming the expression of TcUBP1–GFP in the cytosol of transgenic trypomastigotes, we performed infection experiments and tested the infectivity of these parasites by inducing TcUBP1–GFP overexpression before each infection assay. By adding doxycycline during infections, we were able to visualize protein overexpression within replicating amastigotes. The representative images in Fig. 7A show TcUBP1 (anti-RRM) and GFP expression in parasites replicating within cells infected with transgenic parasites bearing the pTcINDEX–GFP (first row) and pTcINDEX–TcUBP1–GFP (second row) constructs. The evaluation of amastigogenesis by counting the mean number of intracellular parasites per infected cell showed nonsignificant statistical differences between cells infected with induced or noninduced TcUBP1–GFP or GFP (data not shown). In contrast, the percentage of infected cells was 2-fold higher for induced pTcINDEX–TcUBP1–GFP transfectants (+Doxy) than for both noninduced parasites (−Doxy) and parasites transfected with the pTcINDEX–GFP control (Fig. 7B). This indicates that overexpression of TcUBP1 enhanced the capacity of trypomastigotes for cell adherence and/or invasion during the infection process but did not affect differentiation to amastigotes or replication within the host cells.
Figure 7.
In vitro infections with transgenic trypomastigotes with pTcINDEX–GFP or pTcINDEX–TcUBP1–GFP. A, representative photographs of infected cells with induced (+Doxy) or not induced (−Doxy) trypomastigotes obtained from epimastigotes transfected with each construction. DAPI staining (in blue), TcUBP1 expression (anti-RRM signal, in red), and GFP expression (in green). Doxy, doxycycline. B, percentage of infected cells with cell-derived trypomastigotes transfected with GFP or TcUBP1–GFP with (+Doxy) or without (−Doxy) induction of protein expression with doxycycline. The values are expressed as the means of three independent experiments with the corresponding standard deviation bars. The individual data points are shown by filled circles (+Doxy) and diamonds (−Doxy). Student's t test, two tailed. **, p < 0.05.
Discussion
RBPs are able to mediate parasite differentiation in both T. cruzi and Trypanosoma brucei (40, 47–49) probably by coordinating a timely developmental program. This seems to be the case for TcUBP1, a protein that is expressed in all the developmental forms of the parasite. TcUBP1 leads to the stabilization/destabilization of several mRNAs, depending on the binding of other stage-specific components of mRNP complexes across the life cycle of the parasite (35, 43). In the present study, we showed the in vivo interaction of TcUBP1–SGPm RNAs in two different stages (Fig. 2). These mRNP complexes contain members of the surface glycoprotein TcS superfamily (harboring the 50-nt conserved sequence element, termed SGPm), usually expressed only in infective trypomastigotes (Fig. 1 and Table 1). Also, we demonstrated that TcUBP1 overexpression in epimastigote cells increased the abundance of these TcS transcripts (Fig. 3) and changed their subcellular localization to a perinuclear region (Fig. 5). This RNA distribution was usually observed in WT infective trypomastigotes (Fig. 5A) and has been previously reported for other trypomastigote-specific surface glycoproteins as the GP82 mRNA (50, 51). Together, these observations reflect a switch toward mRNA expression of infective trypomastigotes, in agreement with the fact that these TcUBP1-overexpressing parasites are committed to metacyclogenesis, being able to express the trans-sialidase enzyme that is usually expressed only in the infective trypomastigotes (40). These results are in agreement with previous reports showing that TcUBP1 protein levels are increased 3-fold during parasite differentiation to metacyclic trypomastigotes (52). Moreover, the translational status of TcUBP1-overexpressing parasites was reduced, although not inhibited (40). This is in line with previous studies revealing translation repression as a major regulatory mechanism in the infective form, a fact that could explain, at least partially, the proteome reduction reported for this stage (7, 53). Also, this differentiation process has been shown to be dependent on TcUBP1 binding to RNA (40). Thus, although the translation rate in epimastigotes overexpressing TcUBP1 seemed to be reduced, it is tempting to speculate that mobilization into polysomes and translation of stage-specific transcripts may contribute to triggering the differentiation to infective forms of the parasite and probably, but not exclusively, to the increased infectivity observed in the transgenic trypomastigotes (Fig. 7).
Table 1.
Number of members of each TcS group having SGPm cis-regulatory element
The numbers of transcripts for TcS group and total genes were extracted from Table S3 from Freitas et al. (38). The number of transcripts harboring the SGPm was obtained from File S1. Percentages of SGPm-containing targets within each TcS group are between parentheses.
| TcS group | Total genes | SGPm genes |
|---|---|---|
| I | 19 | 0 (0%) |
| II | 117 | 73 (62%) |
| III | 15 | 1 (7%) |
| IV | 25 | 22 (88%) |
| V | 227 | 213 (94%) |
| VI | 39 | 38 (97%) |
| VII | 17 | 4 (24%) |
| VIII | 46 | 31 (67%) |
RNA-sequencing analysis along an axenic epimastigote growth curve revealed that trans-sialidases are up-regulated during the stationary phase (days 7–10), a preadaptive stage for metacyclic trypomastigotes (54). A comparative transcriptome profiling revealed that the CL-14 T. cruzi clone, which shows reduced expression of gene families encoding surface proteins—such as trans-sialidases and mucins—is associated with a nonvirulent phenotype (46). Likewise, transfection of noninfective T. cruzi strains with virulence factors such as inactive trans-sialidases leads to the generation of infective trypomastigotes (55). These findings strongly suggest that RBPs are involved in the modulation of expression of virulence factors affecting T. cruzi infectivity. In this line of evidence, our results showing that trypomastigotes overexpressing TcUBP1 display an increased capacity for infection (Fig. 7) are in agreement with other findings showing a variation in T. cruzi infectivity after the overexpression of the trypanosomal-exclusive TcRBP19 (47, 56).
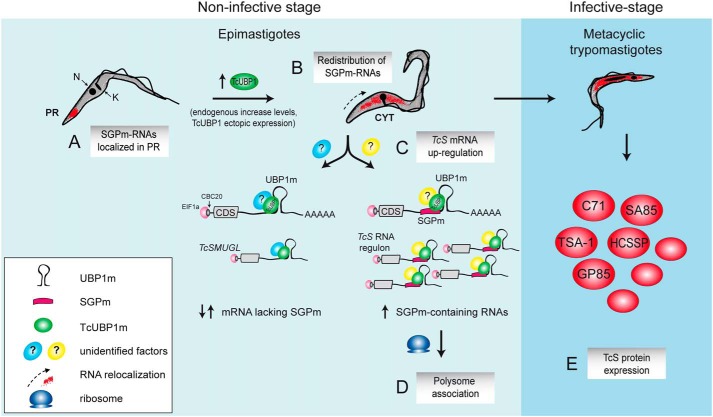
Our present results suggest that TcUBP1 can govern the final fate of the TcS mRNA members in parasites committed to differentiation to infective forms where the presence of the proteins they encode is relevant (50, 57). In our model, TcUBP1 interacts with infective stage–specific transcripts containing SGPm next to the RNA structural element UBP1m. In noninfective cells, these mRNAs are localized in the posterior region of the cell (Fig. 8A), being relocalized (Fig. 8B) and up-regulated (Fig. 8C) by TcUBP1 overexpression, resulting in an increased translatability (Fig. 8D). Although our results provide evidence for the role of TcUBP1 in the regulation of numerous trypomastigote TcS mRNAs (Fig. 8E), the differential behavior between its TcUBP1m targets (with or without the SGPm) require the detection of other yet-unidentified factors. In this context, it is not yet clear whether TcUBP1 directly binds the SGPm or whether the RNA is targeted by the UBP1m. Additional studies will be needed to dissect the functional relationship between these two motifs. Interestingly, we have reported in some mRNA target transcripts that the surrounding sequence of UBP1m harbors overlapping regulatory RNA motifs for other RBPs (such as TcRBP3), suggesting that it can adopt more than one structure, depending on the presence of other competing factors (36).
Figure 8.
Scheme of TcS regulon model during parasite development. In the noninfective insect stage, epimastigotes have SGPm-containing RNAs localized in the posterior region of the cytoplasm (PR) (A), whereas TcUBP1 endogenous increase levels (40) or ectopic overexpression promotes redistribution of these transcripts to a uniform cytosolic localization (CYT) (B). The presence of TcUBP1, with other yet unidentified factors (?), might orchestrate the RNA regulon promoting mRNA TcS up-regulation (C). Also, after TcUBP1 overexpression, TcS transcripts are preferentially associated to polysomes, indicating a switch toward mRNA expression of infective trypomastigotes (D). In fact, TcUBP1 overexpressing parasites are committed to metacyclogenesis (40), being able to express the trypomastigote TcS proteins in the infective mammalian stage (E). N, nuclear DNA; K, kinetoplast DNA.
Some studies have indicated that RBP–mRNA protein complexes can act as RNA regulons in trypanosomes. In T. brucei, for example, Mayho et al. (58) found common elements within a group of stage-regulated nuclear-encoded mitochondrial proteins, whereas in Leishmania mexicana, Holzer et al. (59) found a family of promastigote-enriched mRNAs. Recently, Jha et al. (60) recently demonstrated the DRBD13-specific regulation of transcripts encoding cell surface coat proteins in T. brucei, whereas Estévez (61) and Das et al. (30) demonstrated that DRBD3 regulates the abundance of genes encoding membrane transporters and intermediate metabolism enzymes. Under oxidative conditions, DRBD3, which is a nucleocytoplasmic protein, changes its localization, but its target mRNAs remain bound, suggesting that it could be involved in transporting mRNAs within the cell (62). Many efforts have been made to elucidate the RNA motifs recognized by RBPs and mRNP composition (63, 64). To further explore RNA regulons, future challenges need to be directed to the identification of the signaling pathways that link mRNA turnover with environmental stimuli in trypanosomes and to the remodeling events that each mRNP complex might subsequently suffer. Furthermore, considering the combinatorial nature of RNA–protein interactions (65–67), the association of RBPs with more than one RNA element within the same transcript must finally determine post-transcriptional regulatory networks.
Experimental procedures
Plasmid constructions
A DNA fragment from TSA-1 gene (concerning 110–461 nt from the stop codon) containing the UBP1m motif was amplified by PCR using specific primers (Table S2) and cloned into pGEM-T Easy vector (Promega). The DNA constructs pTcINDEX–GFP, pTcINDEX–TcUBP1–GFP, and pTcINDEX–TcUBP1–ΔNRRM–GFP previously described by Romaniuk et al. (40) were used for parasite transfections. Protein expression values in Tet+-induced epimastigote samples after 96 h were determined relative to noninduced controls (Tet−) by Western blotting analysis of GFP levels normalized to total protein loading, as measured by Coomassie Blue staining. For TcUBP1–GFP, we obtained a value of 6.32 ± 2.08-fold change (n = 3).
Parasite cultures and transfections
T. cruzi epimastigotes from the CL Brener strain were cultured in BHT medium containing 10% heat-inactivated fetal calf serum (BHT 10%) at 28 °C. All parasite cultures were performed in plastic flasks without shaking unless otherwise stated. Parasites were transfected by electroporation subsequently with pLew vector and pTcINDEX constructions and selected with 500 μg/ml of G418 and 250 μg/ml hygromycin. For the induction of recombinant proteins from the pTcINDEX vector, the parasites were incubated in BHT 10% containing 0.5 μg/ml tetracycline for 96 h at 28 °C with shaking.
To obtain metacyclic trypomastigotes, epimastigotes cultures were grown until stationary phase (70 × 106 cells/ml) and starved until parasites attached to the bottom of the bottles. Then cultures were diluted until a parasite concentration of 20 × 106 cell/ml and maintained in BHT with 4% fetal bovine serum for 3 days at 28 °C. Then metacyclic trypomastigotes were collected by centrifugation at 5,000 rpm for 5 min and used for in vitro infections to obtain cell-derived trypomastigotes. For this, Vero cells were incubated with metacyclic trypomastigotes using an infection index of 100 for 24 h at 37 °C. After incubation, the parasite medium was changed every day until cell-derived trypomastigotes emerged from infected cells. One of the cells cultures infected with TcUBP1–GFP or GFP trypomastigotes were maintained with medium supplemented with 5 μg/ml doxycycline to induce protein overexpression in the emerging parasites. Cell-derived trypomastigotes were purified from infection supernatants by centrifugation at 5,200 × g for 10 min and allowing trypomastigotes to swim for 2 h at 37 °C. Then trypomastigotes from induced cell cultures were collected from the supernatant and incubated for additional 18 h in culture media containing tetracycline (5 μg/ml). As control, trypomastigotes from noninduced cell cultures were incubated with medium without tetracycline. The parasites were concentrated by centrifugation, resuspended in 4% minimum essential medium, and counted for the in vitro infection assays.
In vitro infections
Vero cells (20,000 in 0.5 ml of 4% minimum essential medium fetal bovine serum) were plated onto round coverslips 24 h before infection. Infections were performed for 4 h with 2 × 106 cell-derived trypomastigotes per coverslip. After infection, the cells were washed twice in 1× PBS and incubated in fresh medium (with the addition of 5 μg/ml of doxycycline for visualization of TcUBP1 overexpression of intracellular amastigotes in infections performed with induced parasites) for additional 48 h to allow amastigotes replication. Then coverslips were washed twice in 1× PBS, fixed with paraformaldehyde 4% for 20 min, washed again, and mounted in 5 μl of FluorSave reagent (Calbiochem) and 5 μl of DAPI (100 μg/ml final concentration) for nucleus and kinetoplastid staining and observed and photographed using a Nikon Y-FL fluorescence microscope. Each infection was performed in duplicate, and the percentage of infected cells was calculated using the cell counter plugin from ImageJ software. For this, 30 fields, each containing a mean of 20 cells, were photographed with the 100× magnification objective for each experiment.
Recombinant protein expression and antibodies
Recombinant GST and GST–TcUBP1 were synthesized and purified as previously described (43). The antibodies used in this work were polyclonal rabbit antibody reacting with RNA-binding domain of TcUBP1 (anti-RRM) (TcCLB.507093.220) (68–70), antibodies against GFP, anti-TcGDH (71), anti-TcPABP1 (TcCLB.53506885.70) (70), anti-TcHSP70 (72), and anti-Cruzipain (73).
Immunoprecipitation assay
The procedure for immunoprecipitation assay was performed as previously described (37) using epimastigote and trypomastigote protein extracts. Antibodies used were rabbit preimmune serum (as a control) or polyclonal rabbit anti-RRM. The presence of proteins in the immunoprecipitated material was revealed by Western blotting, and the rest of the sample was used for RNA extraction using TRIzol reagent (Invitrogen) following the manufacturer's instructions. RNA samples were resuspended in water and then used in RT-PCR using specific primers (Table S2).
In vitro biotin pulldown assay
RNA-binding assays using purified recombinant proteins or cytosolic extracts was performed as previously described (36). Briefly, pGEM-T Easy empty vector (Neg. ctrl.) or containing the SGP motif [SGPm(+)] were digested with SpeI restriction enzyme for in vitro transcription with T7 RNA polymerase (Ambion) and biotin-14-CTP (Invitrogen). When cytosolic cell-free extracts of parasites were used, the cell lysates (2 × 106 parasites/ml) were supplemented with 400 mg/ml of tRNA, 200 mg/ml BSA, and 10 units of RNase inhibitor. Epimastigote or trypomastigote protein extracts were separately incubated with biotin–RNA in binding buffer for 2 h at room temperature. RNA–protein complexes were recovered and the presence of RBPs revealed by Western blotting assay.
Western blotting
Protein samples were resolved by SDS-PAGE gels 10%, transferred onto Hybond C nitrocellulose membrane (GE Healthcare), probed with primary antibodies, and developed using horseradish peroxide–conjugated antibodies and Supersignal WestPico Chemiluminescent Substrate (Thermo Scientific).
Fluorescence in situ hybridization (FISH)
FISH assays were carried out 12 h postinduction of TcUBP1–GFP and GFP expression for both epimastigotes and metacyclic trypomastigotes. For this, parasites were allowed to adhere to poly-l-lysine–coated microscope slides for 20 min and fixed by incubation with 4% paraformaldehyde for 20 min at room temperature. After 1× PBS wash and incubation with 25 mm NH4Cl, parasites were washed with 1× PBS and incubated with a blocking buffer prepared in PBS containing 2% BSA, 5% normal goat serum, and 0.5% saponin. Prehybridization was performed in buffer containing 4× SSC, 5× Denhardt's solution, 10% BSA, 0.5 μg/μl tRNA wheat germ, dextran sulfate 5, and 5% deionized formamide for 2 h at room temperature. As a control, before incubation, parasites were treated with RNase I (1 unit/4 × 106 parasites) for 30 min at room temperature. We used a probe specific to the conserved region at the 3′-UTR of the TcS mRNAs (Table S2) that was conjugated with Cy-3 at the 5′ end. Hybridizations were performed in a prehybridization buffer with the probe at a concentration of 50 ng/μl. previously heated at 65 °C for 5 min. The cells were preincubated at 42 °C for 3 min and then allowed to hybridize to the probe at room temperature overnight. After incubation, the cells were washed five times with 1 ng/μl 4× SSC with 4% formamide, 4× SSC, or 2× SSC with DAPI (Sigma) and then washed twice with 1× SSC. Then coverslips were mounted in 5 μl of FluorSave reagent (Calbiochem), observed and photographed using a Nikon Y-FL fluorescence microscope, and visualized with a Nikon E600 microscope.
Indirect immunofluorescence
For immunofluorescence assays, coverslips were prepared following the FISH protocol and incubated 2 h at room temperature with rabbit antiserum directed toward the RRM of UBP1 (anti-RRM) in a dilution of 1:300 or anti-GFP rabbit antiserum in a dilution of 1/10,000 for detecting GFP expression in TcUBP1–GFP transfected trypomastigotes. Then after PBS washings, Alexa 568–conjugated goat anti-rabbit Igs G (H + L) (1:10,000, Molecular Probes) were added for 60 min at room temperature and washed as before. Coverslips were mounted in 5 μl of FluorSave reagent (Calbiochem) and 5 μl of DAPI (100 μg/ml final concentration) for the nucleus and kinetoplastid staining and observed and photographed using a Nikon Y-FL fluorescence microscope and photographed as described before. Fluorescence signals were quantified by image analysis using the ImageJ software. For this, 30 photographs, containing an average of 15 parasites each, were taken for each coverslip with the 100× magnification objective.
Reverse transcription and quantitative real-time PCR
Co-immunoprecipitated RNA or total RNA was extracted by TRIzol reagent (Invitrogen) from 150 × 106 parasites. Integrity and purity of RNA was verified by agarose gel and absorbance. cDNA was synthesized with the SuperScript II system (Invitrogen) and oligo(dT) (or random primers for WT parasites assays). Real-time PCR was carried out in a final volume of 10 μl of reaction mixture containing 0.1 μm of each primer (Table S2), 5 μl of SYBR Green reaction mix (SensiFAST SYBR qPCR kit, Bioline), and 4 μl of cDNA template. cDNA was quantified and analyzed using the 7500 software from Applied Biosystems. Program setup was as follows: initial denaturation at 95 °C for 5 min and 45 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 45 s. The quantification data were obtained using the LinReg PCR software and normalized by the levels detected for 18S rRNA, which bears a region containing 11 adenines (the region between positions 479–488 in GenBankTM accession no. 53917.1) that is recognized by the oligo(dT) primer, allowing the subsequent detection after retrotranscription assays. Quantifications were performed for three independent experiments. For comparison of transcripts levels between WT epimastigote and trypomastigote parasites, the data were normalized by the levels detected for the luciferase transcript because 18S rRNA levels are not steady between different life stage forms. Luciferase RNA was in vitro synthesized using the MEGAshortscript T7 transcription kit (Ambion) and the first strand cDNA retrotranscribed using random primers. Equal amounts of the luciferase RNA was added to samples before cDNA synthesis.
Computational analysis
The suite MEME (Multiple Em for Motif Elicitation) (41) was used to search for RNA motifs present in the noncoding regions of UBP1 database targets encoding trypomastigote glycoproteins. The parameters used in the MEME analysis were “−dna −mod zoops −nmotifs 1 −minw 3 −maxw 50 −evt 1e-5.” Subsequent motif discovery was implemented on the entire T. cruzi CL Brener genome using the MAST (Motif Alignment Search Tool) program. The parameters for MAST were “−mt 1e −10 −comp −text. The trypanosome database was obtained from the TcruziDB server (https://tritrypdb.org/tritrypdb/).5 3′ downstream genomic sequences were obtained using TcruziDB sequence retrieval tool. A length of 350-nt downstream to CDS was used to construct the 3′-UTR database, in agreement to previously reported data from trypanosomes (74). RNA motif UBP1m (36) was used to search against sequences using cmsearch algorithm from the Infernal program (75).
Polysome purification
Epimastigote cultures were treated with cycloheximide for 30 min to arrest protein synthesis. For EDTA treatment, samples were incubated with 20 mm EDTA (final concentration) for 30 min prior to cell lysis. The lysates (800 × 106 cells) were clarified by centrifugation at 4 °C for 15 min at 10,000 × g. The resulting supernatant (S10) was processed as described for the isolation of polysomes (76). Briefly, the S10 sample was layered onto a 30% sucrose cushion and centrifuged for 2.5 h at 4 °C at 130,000 × g (32,500 rpm in an SW41 rotor). The high-speed supernatant (S130) was harvested, and the polysomal pellet (P) was resuspended in a protease inhibitor buffer. To check the obtained fractions, samples were analyzed by Western blotting using sera toward typical protein markers: TcPABP1 for polysomes, and TcGDH, TcHSP70, and Cruzipain for polysome-free supernatants. RNA and protein samples were extracted and subjected to RT-qPCR and Western blotting analyses. All fractions were stored at −80 °C.
Author contributions
K. B. S., M. A. R., G. N., V. A. C., and J. G. D. G. methodology; K. B. S., V. A. C., and J. G. D. G. writing-original draft; V. A. C. and J. G. D. G. investigation; A. C., V. A. C., and J. G. D. G. formal analysis; V. A. C. and J. G. D. G. conceptualization; V. A. C. and J. G. D. G. funding acquisition; K. B. S., V. A. C., and J. G. D. G. visualization; V. A. C. and J. G. D. G. project administration; V. A. C. and J. G. D. G. writing-review and editing; J. G. D. G. supervision.
Supplementary Material
Acknowledgments
We are indebted to Agustina Chidichimo and Liliana Sferco for parasite cultures. We thank Dr. Graciela Boccaccio and Dr. María Gabriela Thomas at Fundación Instituto Leloir of the University of Buenos Aires for help with ultracentrifugation and Dr. Daniel Vigo for help with statistics.
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica Grants PICT 2016-0235 and PICT 2016-0465 (to J. G. D. G.) and PICT 2014-1798 (to V. A. C.); funds from the Research Career of CONICET (to M. A. R., A. C., V. A. C., and J. G. D. G.); and a CONICET research fellowship (to K. B. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Files S1 and S2, Tables S1 and S2, and Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RBP
- RNA-binding protein
- TcUBP1
- T. cruzi U-rich RBP 1
- RRM
- RNA-recognition motif
- SGP
- surface glycoprotein
- SGPm
- SGP motif
- qPCR
- quantitative PCR
- T
- trypomastigote stage
- E
- epimastigote stage
- A
- amastigote stage
- ANOVA
- analysis of variance
- IIF
- indirect immunofluorescence
- GDH
- glutamate dehydrogenase
- nt
- nucleotide(s)
- DAPI
- 4′,6′-diamino-2-phenylindole
- FISH
- fluorescence in situ hybridization.
References
- 1. Mansfield K. D., and Keene J. D. (2009) The ribonome: a dominant force in co-ordinating gene expression. Biol. Cell 101, 169–181 10.1042/BC20080055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore M. J., and Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 3. Kramer S., and Carrington M. (2011) trans-Acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 27, 23–30 10.1016/j.pt.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolev N. G., Ullu E., and Tschudi C. (2014) The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell. Microbiol. 16, 482–489 10.1111/cmi.12268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jensen B. C., Ramasamy G., Vasconcelos E. J., Ingolia N. T., Myler P. J., and Parsons M. (2014) Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics 15, 911 10.1186/1471-2164-15-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerreiro A., Deligianni E., Santos J. M., Silva P. A., Louis C., Pain A., Janse C. J., Franke-Fayard B., Carret C. K., Siden-Kiamos I., and Mair G. R. (2014) Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 15, 493 10.1186/s13059-014-0493-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smircich P., Eastman G., Bispo S., Duhagon M. A., Guerra-Slompo E. P., Garat B., Goldenberg S., Munroe D. J., Dallagiovanna B., Holetz F., and Sotelo-Silveira J. R. (2015) Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics 16, 443 10.1186/s12864-015-1563-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pérez-Díaz L., Silva T. C., and Teixeira S. M. (2017) Involvement of an RNA binding protein containing Alba domain in the stage-specific regulation of β-amastin expression in Trypanosoma cruzi. Mol. Biochem. Parasitol. 211, 1–8 10.1016/j.molbiopara.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 9. Keene J. D. (2014) The globalization of messenger RNA regulation. Nat. Sci. Rev. 1, 184–186 10.1093/nsr/nwu004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morris A. R., Mukherjee N., and Keene J. D. (2010) Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 162–180 10.1002/wsbm.54 [DOI] [PubMed] [Google Scholar]
- 11. Keene J. D., and Lager P. J. (2005) Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 13, 327–337 10.1007/s10577-005-0848-1 [DOI] [PubMed] [Google Scholar]
- 12. Keene J. D. (2007) RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- 13. Blackinton J. G., and Keene J. D. (2014) Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 34, 44–54 10.1016/j.semcdb.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., and Krishna S. (2003) The trypanosomiases. Lancet 362, 1469–1480 10.1016/S0140-6736(03)14694-6 [DOI] [PubMed] [Google Scholar]
- 15. Jackson A. P. (2015) Genome evolution in trypanosomatid parasites. Parasitology 142, S40–S56 10.1017/S0031182014000894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y., Shah-Simpson S., Okrah K., Belew A. T., Choi J., Caradonna K. L., Padmanabhan P., Ndegwa D. M., Temanni M. R., Corrada Bravo H., El-Sayed N. M., and Burleigh B. A. (2016) Transcriptome remodeling in Trypanosoma cruzi and human cells during intracellular infection. PLoS Pathog. 12, e1005511 10.1371/journal.ppat.1005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chávez S., Eastman G., Smircich P., Becco L. L., Oliveira-Rizzo C., Fort R., Potenza M., Garat B., Sotelo-Silveira J. R., and Duhagon M. A. (2017) Transcriptome-wide analysis of the Trypanosoma cruzi proliferative cycle identifies the periodically expressed mRNAs and their multiple levels of control. PLoS One 12, e0188441 10.1371/journal.pone.0188441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berná L., Chiribao M. L., Greif G., Rodriguez M., Alvarez-Valin F., and Robello C. (2017) Transcriptomic analysis reveals metabolic switches and surface remodeling as key processes for stage transition in Trypanosoma cruzi. Peer J. 5, e3017 10.7717/peerj.3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernández-Moya S. M., and Estévez A. M. (2010) Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev. RNA 1, 34–46 10.1002/wrna.6 [DOI] [PubMed] [Google Scholar]
- 20. Michaeli S. (2014) Non-coding RNA and the complex regulation of the trypanosome life cycle. Curr. Opin. Microbiol. 20, 146–152 10.1016/j.mib.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 21. Pastro L., Smircich P., Di Paolo A., Becco L., Duhagon M. A., Sotelo-Silveira J., and Garat B. (2017) Nuclear compartmentalization contributes to stage-specific gene expression control in Trypanosoma cruzi. Front. Cell Dev. Biol. 5, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rao S. J., Chatterjee S., and Pal J. K. (2017) Untranslated regions of mRNA and their role in regulation of gene expression in protozoan parasites. J. Biosci. 42, 189–207 10.1007/s12038-016-9660-7 [DOI] [PubMed] [Google Scholar]
- 23. Oliveira C., Faoro H., Alves L. R., and Goldenberg S. (2017) RNA-binding proteins and their role in the regulation of gene expression in Trypanosoma cruzi and Saccharomyces cerevisiae. Genet Mol. Biol. 40, 22–30 10.1590/1678-4685-gmb-2016-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pastro L., Smircich P., Pérez-Díaz L., Duhagon M. A., and Garat B. (2013) Implication of CA repeated tracts on post-transcriptional regulation in Trypanosoma cruzi. Exp. Parasitol. 134, 511–518 10.1016/j.exppara.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 25. De Gaudenzi J. G., Jäger A. V., Izcovich R., and Campo V. A. (2016) Insights into the regulation of mRNA processing of polycistronic transcripts mediated by DRBD4/PTB2, a trypanosome homolog of the polypyrimidine tract-binding protein. J. Eukaryot. Microbiol. 63, 440–452 10.1111/jeu.12288 [DOI] [PubMed] [Google Scholar]
- 26. Araújo P. R., and Teixeira S. M. (2011) Regulatory elements involved in the post-transcriptional control of stage-specific gene expression in Trypanosoma cruzi: a review. Mem. Inst. Oswaldo. Cruz. 106, 257–266 10.1590/S0074-02762011000300002 [DOI] [PubMed] [Google Scholar]
- 27. Guerra-Slompo E. P., Probst C. M., Pavoni D. P., Goldenberg S., Krieger M. A., and Dallagiovanna B. (2012) Molecular characterization of the Trypanosoma cruzi specific RNA binding protein TcRBP40 and its associated mRNAs. Biochem. Biophys. Res. Commun. 420, 302–307 10.1016/j.bbrc.2012.02.154 [DOI] [PubMed] [Google Scholar]
- 28. Das A., Morales R., Banday M., Garcia S., Hao L., Cross G. A., Estevez A. M., and Bellofatto V. (2012) The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA 18, 1968–1983 10.1261/rna.033829.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alves L. R., Oliveira C., Mörking P. A., Kessler R. L., Martins S. T., Romagnoli B. A., Marchini F. K., and Goldenberg S. (2014) The mRNAs associated to a zinc finger protein from Trypanosoma cruzi shift during stress conditions. RNA Biol. 11, 921–933 10.4161/rna.29622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das A., Bellofatto V., Rosenfeld J., Carrington M., Romero-Zaliz R., del Val C., and Estévez A. M. (2015) High throughput sequencing analysis of Trypanosoma brucei DRBD3/PTB1-bound mRNAs. Mol. Biochem. Parasitol. 199, 1–4 10.1016/j.molbiopara.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 31. Singh A., Minia I., Droll D., Fadda A., Clayton C., and Erben E. (2014) Trypanosome MKT1 and the RNA-binding protein ZC3H11: interactions and potential roles in post-transcriptional regulatory networks. Nucleic Acids Res. 42, 4652–4668 10.1093/nar/gkt1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Archer S. K., Inchaustegui D., Queiroz R., and Clayton C. (2011) The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One 6, e18425 10.1371/journal.pone.0018425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Freire E. R., Vashisht A. A., Malvezzi A. M., Zuberek J., Langousis G., Saada E. A., Nascimento Jde F., Stepinski J., Darzynkiewicz E., Hill K., De Melo Neto O. P., Wohlschlegel J. A., Sturm N. R., and Campbell D. A. (2014) eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA 20, 1272–1286 10.1261/rna.045534.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Gaudenzi J. G., Carmona S. J., Agüero F., and Frasch A. C. (2013) Genome-wide analysis of 3′-untranslated regions supports the existence of post-transcriptional regulons controlling gene expression in trypanosomes. Peer J. 1, e118 10.7717/peerj.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Gaudenzi J. G., Noé G., Campo V. A., Frasch A. C., and Cassola A. (2011) Gene expression regulation in trypanosomatids. Essays Biochem. 51, 31–46 10.1042/bse0510031 [DOI] [PubMed] [Google Scholar]
- 36. Noé G., De Gaudenzi J. G., and Frasch A. C. (2008) Functionally related transcripts have common RNA motifs for specific RNA-binding proteins in trypanosomes. BMC Mol. Biol. 9, 107 10.1186/1471-2199-9-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z. H., De Gaudenzi J. G., Alvarez V. E., Mendiondo N., Wang H., Kissinger J. C., Frasch A. C., and Docampo R. (2012) A 43-nucleotide U-rich element in 3′-untranslated region of large number of Trypanosoma cruzi transcripts is important for mRNA abundance in intracellular amastigotes. J. Biol. Chem. 287, 19058–19069 10.1074/jbc.M111.338699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Freitas L. M., dos Santos S. L., Rodrigues-Luiz G. F., Mendes T. A., Rodrigues T. S., Gazzinelli R. T., Teixeira S. M., Fujiwara R. T., and Bartholomeu D. C. (2011) Genomic analyses, gene expression and antigenic profile of the trans-sialidase superfamily of Trypanosoma cruzi reveal an undetected level of complexity. PLoS One 6, e25914 10.1371/journal.pone.0025914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Callejas-Hernández F., Rastrojo A., Poveda C., Gironès N., and Fresno M. (2018) Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci. Rep. 8, 14631 10.1038/s41598-018-32877-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romaniuk M. A., Frasch A. C., and Cassola A. (2018) Translational repression by an RNA-binding protein promotes differentiation to infective forms in Trypanosoma cruzi. PLoS Pathog. 14, e1007059 10.1371/journal.ppat.1007059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bailey T. L., Williams N., Misleh C., and Li W. W. (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373 10.1093/nar/gkl198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minning T. A., Weatherly D. B., Atwood J. 3rd, Orlando R., and Tarleton R. L. (2009) The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10, 370 10.1186/1471-2164-10-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Gaudenzi J. G., D'Orso I., and Frasch A. C. (2003) RNA recognition motif-type RNA-binding proteins in Trypanosoma cruzi form a family involved in the interaction with specific transcripts in vivo. J. Biol. Chem. 278, 18884–18894 10.1074/jbc.M301756200 [DOI] [PubMed] [Google Scholar]
- 44. Taylor M. C., and Kelly J. M. (2006) pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnol. 6, 32 10.1186/1472-6750-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alves L. R., Guerra-Slompo E. P., de Oliveira A. V., Malgarin J. S., Goldenberg S., and Dallagiovanna B. (2013) mRNA localization mechanisms in Trypanosoma cruzi. PLoS One 8, e81375 10.1371/journal.pone.0081375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Belew A. T., Junqueira C., Rodrigues-Luiz G. F., Valente B. M., Oliveira A. E. R., Polidoro R. B., Zuccherato L. W., Bartholomeu D. C., Schenkman S., Gazzinelli R. T., Burleigh B. A., El-Sayed N. M., and Teixeira S. M. R. (2017) Comparative transcriptome profiling of virulent and non-virulent Trypanosoma cruzi underlines the role of surface proteins during infection. PLoS Pathog. 13, e1006767 10.1371/journal.ppat.1006767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pérez-Díaz L., Correa A., Moretão M. P., Goldenberg S., Dallagiovanna B., and Garat B. (2012) The overexpression of the trypanosomatid-exclusive TcRBP19 RNA-binding protein affects cellular infection by Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 107, 1076–1079 10.1590/S0074-02762012000800021 [DOI] [PubMed] [Google Scholar]
- 48. Kolev N. G., Ramey-Butler K., Cross G. A., Ullu E., and Tschudi C. (2012) Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science 338, 1352–1353 10.1126/science.1229641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alcantara M. V., Kessler R. L., Gonçalves R. E. G., Marliére N. P., Guarneri A. A., Picchi G. F. A., and Fragoso S. P. (2018) Knockout of the CCCH zinc finger protein TcZC3H31 blocks Trypanosoma cruzi differentiation into the infective metacyclic form. Mol. Biochem. Parasitol. 221, 1–9 10.1016/j.molbiopara.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 50. Bayer-Santos E., Gentil L. G., Cordero E. M., Corrêa P. R., and da Silveira J. F. (2012) Regulatory elements in the 3′ untranslated region of the GP82 glycoprotein are responsible for its stage-specific expression in Trypanosoma cruzi metacyclic trypomastigotes. Acta Trop. 123, 230–233 10.1016/j.actatropica.2012.03.014 [DOI] [PubMed] [Google Scholar]
- 51. Correa P. R., Cordero E. M., Gentil L. G., Bayer-Santos E., and da Silveira J. F. (2013) Genetic structure and expression of the surface glycoprotein GP82, the main adhesin of Trypanosoma cruzi metacyclic trypomastigotes. ScientificWorldJournal 2013, 156734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Godoy L. M., Marchini F. K., Pavoni D. P., Rampazzo Rde C., Probst C. M., Goldenberg S., and Krieger M. A. (2012) Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics 12, 2694–2703 10.1002/pmic.201200078 [DOI] [PubMed] [Google Scholar]
- 53. Tonelli R. R., Augusto Lda S., Castilho B. A., and Schenkman S. (2011) Protein synthesis attenuation by phosphorylation of eIF2α is required for the differentiation of Trypanosoma cruzi into infective forms. PLoS One 6, e27904 10.1371/journal.pone.0027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Santos C., Ludwig A., Kessler R. L., Rampazzo R. C. P., Inoue A. H., Krieger M. A., Pavoni D. P., and Probst C. M. (2018) Trypanosoma cruzi transcriptome during axenic epimastigote growth curve. Mem. Inst. Oswaldo Cruz 113, e170404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pascuale C. A., Burgos J. M., Postan M., Lantos A. B., Bertelli A., Campetella O., and Leguizamón M. S. (2017) Inactive trans-sialidase expression in iTS-null Trypanosoma cruzi generates virulent trypomastigotes. Front. Cell. Infect. Microbiol. 7, 430 10.3389/fcimb.2017.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pérez-Díaz L., Pastro L., Smircich P., Dallagiovanna B., and Garat B. (2013) Evidence for a negative feedback control mediated by the 3′ untranslated region assuring the low expression level of the RNA binding protein TcRBP19 in T. cruzi epimastigotes. Biochem. Biophys. Res. Commun. 436, 295–299 10.1016/j.bbrc.2013.05.096 [DOI] [PubMed] [Google Scholar]
- 57. Bayer-Santos E., Cunha-e-Silva N. L., Yoshida N., and Franco da Silveira J. (2013) Expression and cellular trafficking of GP82 and GP90 glycoproteins during Trypanosoma cruzi metacyclogenesis. Parasit. Vectors 6, 127 10.1186/1756-3305-6-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mayho M., Fenn K., Craddy P., Crosthwaite S., and Matthews K. (2006) Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 34, 5312–5324 10.1093/nar/gkl598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Holzer T. R., Mishra K. K., LeBowitz J. H., and Forney J. D. (2008) Coordinate regulation of a family of promastigote-enriched mRNAs by the 3′UTR PRE element in Leishmania mexicana. Mol. Biochem. Parasitol. 157, 54–64 10.1016/j.molbiopara.2007.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jha B. A., Gazestani V. H., Yip C. W., and Salavati R. (2015) The DRBD13 RNA binding protein is involved in the insect-stage differentiation process of Trypanosoma brucei. FEBS Lett. 589, 1966–1974 10.1016/j.febslet.2015.05.036 [DOI] [PubMed] [Google Scholar]
- 61. Estévez A. M. (2008) The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 36, 4573–4586 10.1093/nar/gkn406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fernández-Moya S. M., García-Pérez A., Kramer S., Carrington M., and Estévez A. M. (2012) Alterations in DRBD3 ribonucleoprotein complexes in response to stress in Trypanosoma brucei. PLoS One 7, e48870 10.1371/journal.pone.0048870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., Na H., Irimia M., Matzat L. H., Dale R. K., Smith S. A., et al. (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Inchaustegui Gil D. P., and Clayton C. (2016) Purification of messenger ribonucleoprotein particles via a tagged nascent polypeptide. PLoS One 11, e0148131 10.1371/journal.pone.0148131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., and Brown P. O. (2008) Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLos Biol. 6, e255 10.1371/journal.pbio.0060255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mittal N., Roy N., Babu M. M., and Janga S. C. (2009) Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc. Natl. Acad. Sci. U.S.A. 106, 20300–20305 10.1073/pnas.0906940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mittal N., Scherrer T., Gerber A. P., and Janga S. C. (2011) Interplay between posttranscriptional and posttranslational interactions of RNA-binding proteins. J. Mol. Biol. 409, 466–479 10.1016/j.jmb.2011.03.064 [DOI] [PubMed] [Google Scholar]
- 68. De Gaudenzi J., Frasch A. C., and Clayton C. (2005) RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryotic Cell 4, 2106–2114 10.1128/EC.4.12.2106-2114.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. D'Orso I., and Frasch A. C. (2001) TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J. Biol. Chem. 276, 34801–34809 10.1074/jbc.M102120200 [DOI] [PubMed] [Google Scholar]
- 70. D'Orso I., and Frasch A. C. (2002) TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and Poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 277, 50520–50528 10.1074/jbc.M209092200 [DOI] [PubMed] [Google Scholar]
- 71. Barderi P., Campetella O., Frasch A. C., Santomé J. A., Hellman U., Pettersson U., and Cazzulo J. J. (1998) The NADP+-linked glutamate dehydrogenase from Trypanosoma cruzi: sequence, genomic organization and expression. Biochem. J. 330, 951–958 10.1042/bj3300951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jäger A. V., De Gaudenzi J. G., Cassola A., D'Orso I., and Frasch A. C. (2007) mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 2035–2042 10.1073/pnas.0611125104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alvarez V. E., Niemirowicz G. T., and Cazzulo J. J. (2012) The peptidases of Trypanosoma cruzi: digestive enzymes, virulence factors, and mediators of autophagy and programmed cell death. Biochim. Biophys. Acta 1824, 195–206 10.1016/j.bbapap.2011.05.011 [DOI] [PubMed] [Google Scholar]
- 74. Campos P. C., Bartholomeu D. C., DaRocha W. D., Cerqueira G. C., and Teixeira S. M. (2008) Sequences involved in mRNA processing in Trypanosoma cruzi. Int. J. Parasitol. 38, 1383–1389 10.1016/j.ijpara.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 75. Nawrocki E. P., Kolbe D. L., and Eddy S. R. (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ji X., Kong J., and Liebhaber S. A. (2003) In vivo association of the stability control protein αCP with actively translating mRNAs. Mol. Cell Biol. 23, 899–907 10.1128/MCB.23.3.899-907.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.