Abstract
A diverse collection of enzymes comprising the protocatechuate dioxygenases (PCADs) has been characterized in several extradiol aromatic compound degradation pathways. Structural studies have shown a relationship between PCADs and the more broadly-distributed, functionally enigmatic Memo domain linked to several human diseases. To better understand the evolution of this PCAD–Memo protein superfamily, we explored their structural and functional determinants to establish a unified evolutionary framework, identifying 15 clearly-delineable families, including a previously-underappreciated diversity in five Memo clade families. We place the superfamily's origin within the greater radiation of the nucleoside phosphorylase/hydrolase-peptide/amidohydrolase fold prior to the last universal common ancestor of all extant organisms. In addition to identifying active-site residues across the superfamily, we describe three distinct, structurally-variable regions emanating from the core scaffold often housing conserved residues specific to individual families. These were predicted to contribute to the active-site pocket, potentially in substrate specificity and allosteric regulation. We also identified several previously-undescribed conserved genome contexts, providing insight into potentially novel substrates in PCAD clade families. We extend known conserved contextual associations for the Memo clade beyond previously-described associations with the AMMECR1 domain and a radical S-adenosylmethionine family domain. These observations point to two distinct yet potentially overlapping contexts wherein the elusive molecular function of the Memo domain could be finally resolved, thereby linking it to nucleotide base and aliphatic isoprenoid modification. In total, this report throws light on the functions of large swaths of the experimentally-uncharacterized PCAD–Memo families.
Keywords: dioxygenase, isoprenoid, RNA modification, phosphorylase, hydrolase, AMMECR1, LigB, Memo, protocatechuate dioxygenase, YgiD
Introduction
The aromatic ring is an exceptionally stable molecular arrangement that is integral to a wide range of biomolecules. Its unique properties are central to the functions of essential biopolymers, like proteins (side chains of specific amino acids) and nucleic acids (the bases), as well as low-molecular-weight metabolites and signaling messengers. Their stability relative to other organic molecules also poses paradoxical challenges to the cell: catalyzing reactions that allow their utilization, appropriate modification, and proper incorporation into macromolecular building blocks on the one hand and the prevention of the accumulation of versions that can be toxic on the other hand. As part of this cellular management of compounds with aromatic rings, diverse molecular mechanisms have evolved for their opening and catabolism.
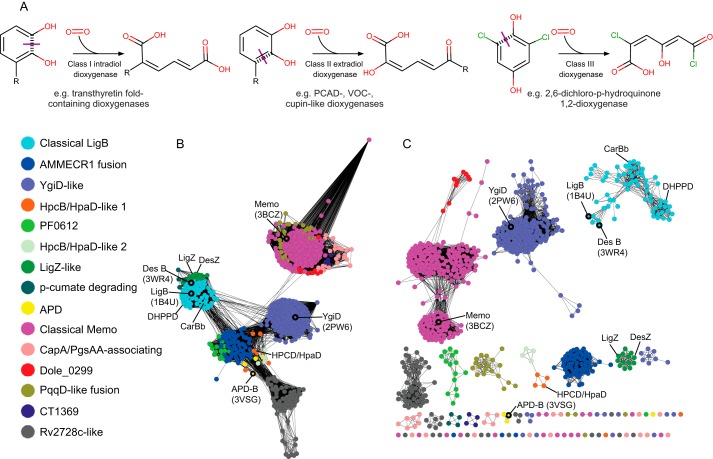
Enzymes involved in the breakage of aromatic rings act via oxidative ring activation, resulting in cleavage at an intradiol bond (class I) or an extradiol bond (class II), or in some cases independently of a diol (class III) (Fig. 1A) (1, 2). Extradiol bond-cleaving enzyme superfamilies catalyze ring breakage assisted by a coordinated metal ion and display at least three unrelated protein folds: 1) the vicinal oxygen chelate dioxygenase superfamily of the glyoxalase fold (type I extradiol dioxygenases); 2) the protocatechuate dioxygenase (PCAD)4 superfamily of the phosphorylase/peptidyl hydrolase fold (type II); and 3) the cupin family dioxygenases of the double-stranded β-helix fold (type III). Each of these extradiol dioxygenase “types” has been implicated in the catabolism of a wide range of aromatic ring-containing substrates in diverse pathways (1, 3–5).
Figure 1.
A, chemical reaction diagrams for the different classes of oxidative ring opening reactions involving diols. C–C bonds broken during ring opening are marked with purple lines. B and C, sequence similarity network of representative PCAD–Memo superfamily domains. Nodes represent groups of sequences that share <50% identity, and edges are shown if the pairwise BLAST e-value is <e −10 (A) or e −30 (B). Enzymatically and structurally characterized proteins are labeled, and nodes outlined in black include structurally characterized members. Nodes are colored according to family, as determined by BLASTCLUST analysis.
The type II extradiol dioxygenases of the PCAD superfamily share a common fold with catalytically unrelated clades of enzymes, namely the nucleosidase/nucleoside phosphorylases (PNP) and a diverse assemblage of peptidyl/amidohydrolases (6). The PCAD domains further show close structural and sequence affinities with the Memo family (7). The Memo protein was first identified as a mediator of ErbB2-induced cell motility in breast cancer cell lines (8). Subsequent studies have largely coalesced on the view that Memo acts as a general regulator of cell motility-related pathways with proposed involvement in actin reorganization (9), microtubule capture (10, 11), vascular development (12), and tumor migration (13). Although initial research pointed to a primary functional role in nonenzymatic phosphopeptide binding (7), subsequent research established metal ion-binding for Memo and thus pointed to a potential enzymatic role (13). The limited data collected on Memo enzyme activity to date do not implicate Memo in aromatic ring cleavage reactions (7, 13), instead implicating it in the reduction of molecular oxygen and generation of reactive oxygen species (13, 14). However, independently of this experimental data, comparative genome analyses identified conserved gene-neighborhoods that are again consistent with an enzymatic role in the modification of nucleic acid bases or lipids (15–18).
Although the reaction mechanisms, substrates, and family diversity of the PCAD domains have been studied to varying degrees in the past (19–21), an understanding of their total evolutionary history is generally lacking. To reconcile these findings with the role of the poorly-understood Memo and to better understand the internal relationships and the provenance of the PCAD–Memo superfamily, we initiated a comprehensive comparative genomic analysis. In our analysis, we sought to specifically address certain lacunae in the current literature, including 1) phyletic distributions of the PCAD clades based on complete genome sequences; 2) superfamily-wide substrate specialization along with prediction of novel pathways; 3) the extent of the dispersal of aromatic degradation pathways across organisms; 4) the origin, inter-family evolutionary relationships of the PCAD enzymes and the higher-order relationships with the Memo domain.
Through these analyses, we elucidated the evolutionary history of the unified superfamily, observing that the PCAD dioxygenases likely descended from the more ancient Memo-like clade, which in turn descends via a single circular permutation event of the protein fold from a PNP-peptidyl/amidohydrolase-like prototype with which they share a similarly located active-site pocket. This history establishes that the shift to a dedicated aromatic ring opening function likely happened only after the PCAD clade had diverged from the Memo-like clade. These observations open novel avenues of investigation into the precise molecular function of Memo, which remains poorly-understood despite increasing experimental evidence in the last decade linking it to various human diseases. Additionally, this analysis presents the first comprehensive comparative genomic account of the aromatic ring-opening dioxygenases of the PCAD superfamily, reporting a range of known and newly-predicted substrates as well as structural features influencing substrate recognition and likely processing efficiency. Finally, we report the discovery of a previously-unrecognized family in the PCAD superfamily present across the Actinobacteria, which, although likely an enzymatically inactive version of the superfamily domain, appears to function in the synthesis of a metabolite derived from a modified nucleotide.
Results
Assembly of the sequence and structure complement of the PCAD–Memo superfamily and its classification
To collect members of the PCAD–Memo superfamily, we initiated iterative sensitive sequence-based similarity searches using known members of the PCAD superfamily as seeds (see under “Experimental procedures”). Searches were run until convergence; sequences retrieved with borderline statistical significance (e-values between 0.01 and 0.001) were used as seeds in new searches to confirm membership in the superfamily. As an example, a search initiated with a sequence from the AMMECR1 fusion family (accession number: WP_006301058.1) returns a 2-aminophenol 1,6-dioxygenase (APD) family member from Cupriavidus (accession number: AEI74584.1; e-value: 2e-26; iteration: 2), a Rv2728c-like family member from Mycobacterium sp. 141 (accession number: WP_019969303.1; e-value: 9e-22; iteration: 2), a classical Memo family member from Arabidopsis thaliana (accession number: NP_565590.1; e-value 5e-04; iteration: 2), and a YgiD family member from Magnaporthe oryzae (accession number: ELQ40968.1; e-value: 3e-04; iteration: 3).
Similarly, we initiated structural similarity searches using known crystal structures as seeds (see under “Experimental procedures”). These searches retrieved increasingly distant members of the PCAD dioxygenase-Memo superfamily relative to the starting structure, followed by other structures belonging to the phosphorylase/peptidyl hydrolase fold, namely the purine nucleoside phosphorylases (PNP superfamily). For example, a structure-based homology search initiated with a crystal structure from the classical LigB family (PDB code 1B4U_B) first returns other PCAD structures (examples: 3VSH_D, z-score: 24.5; 2PW6_A, z-score: 19.2), then Memo family structures (example: 3BD0_A, z-score 15.3), and then PNP and peptidyl/amidohydrolase structures (example: 1TCV_A, z-score: 5.8). A search initiated with a Memo family member (PDB code 3BCZ_A) first detects other Memo structures and then detects various PCAD structures (examples: 5HEE_A, z-score: 19.0; 1B4U_B, z-score 15.3; 2PW6_A, z-score: 14.5), before finally detecting PNP and peptidyl/amidohydrolase structures (example: 1TCV_A, z-score 6.5).
The collected PCAD complement of 6619 sequences was subject to clustering analyses to gain insight into the distinct families composing the superfamily (see “Experimental procedures”) (22). In total, clustering by BLASTCLUST identified 15 clearly-delineable families in the PCAD–Memo superfamily, which, in accordance with the structure search results, revealed a fundamental divide between the PCAD and Memo clades of the superfamily. Five families were found to belong to the Memo clade, and nine clearly fell into the PCAD clade, and a single family falling into the PCAD clade in the sequence similarity network (Fig. 1, B and C), but not strongly associating with either clade in the BLASTCLUST analysis. The relationships among these families and the sparseness of experimentally-characterized versions are shown in a sequence similarity network depicted in Fig. 1.
Structure and sequence features of the PCAD–Memo superfamily
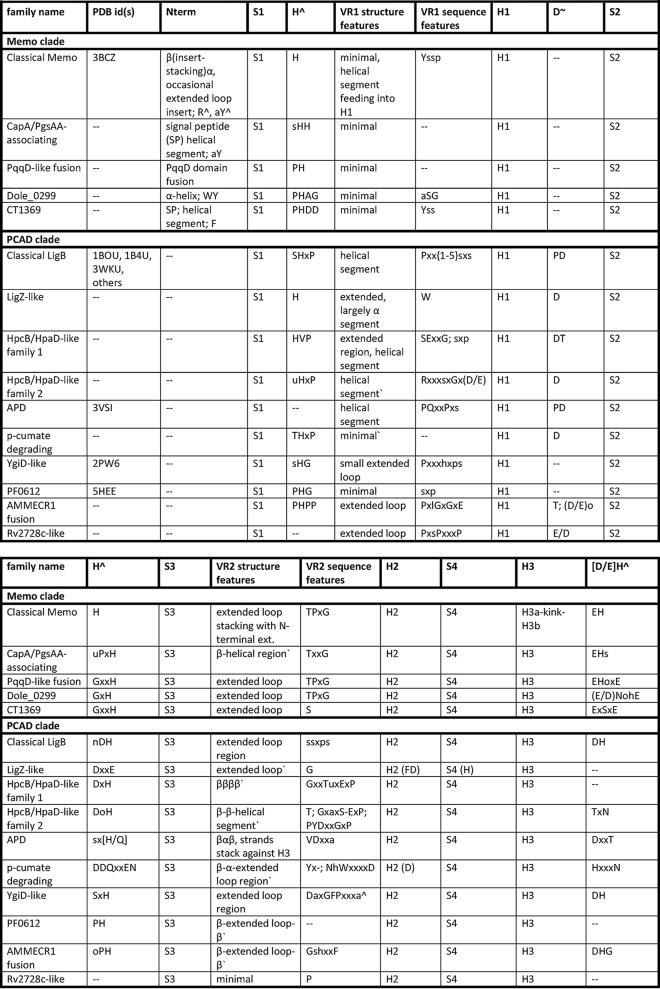
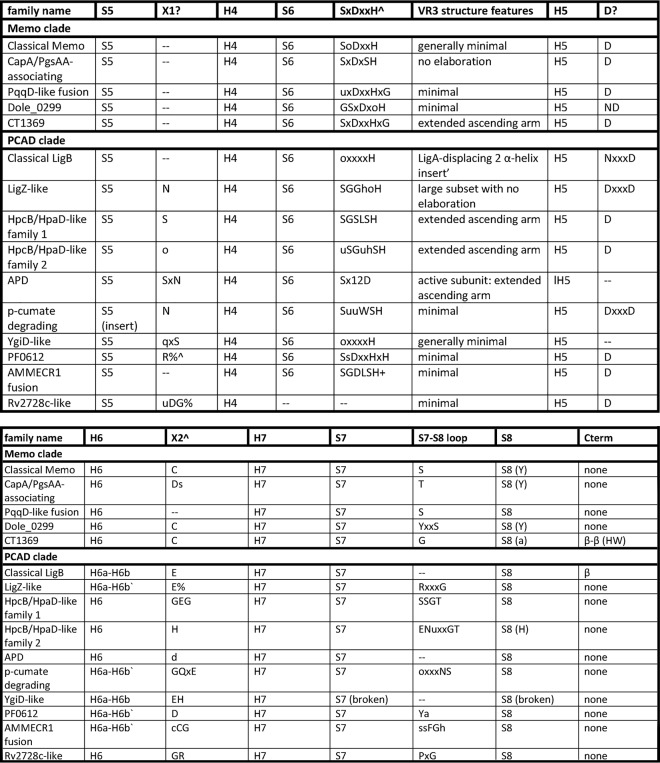
To provide context for a study of the natural history of the PCAD–Memo superfamily, we analyzed the above-collected complement of PCAD–Memo-solved structures and retrieved sequences to define the essential core of the domain and to catalog the complete range of structure and sequence variations. Table 1 presents the summary of the conserved and variable features identified across the superfamily, and these are further discussed below.
Table 1.
Capital letters in column headers denote residues conserved across the superfamily, provided in linear order of appearance relative to core structural elements
The following abbreviations and symbols are used: S1–S8, strand 1–strand 8; H1–H7, helix 1–helix 7; VR1–VR3: variable region 1–variable region 3; Cterm, region C-terminal to domain core; Nterm, region N-terminal to domain core; ∧, likely involvement in metal coordination, catalytic mechanism; *, potential involvement in allosteric binding; ∼, potential involvement in dimerization, protein–protein contacts; ?, unknown function; ', based on secondary structure predictions; and %, conserved motif not positionally-equivalent.
Structural core of the superfamily
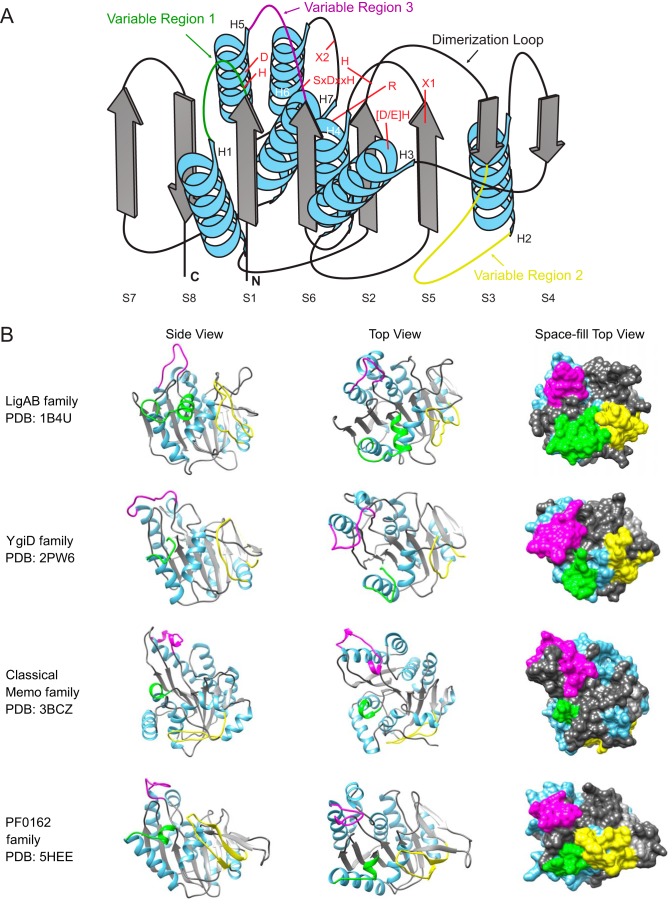
The core fold of the PCAD–Memo superfamily is a α/β three-layered sandwich with eight conserved strands forming a central β-sheet. There are seven conserved helices that sandwich the central sheet with two in one layer and five in the opposite layer (Fig. 2A). Comparison of the solved structures alongside multiple sequence alignments constructed for the families currently lacking any structure revealed three loop regions that consistently exhibit structural variation, and we hereafter refer to them as variable regions 1–3 (VR1–3; see below, Fig. 2A, and Table 1). Another region occasionally displaying variability across individual families is the “dimerization” loop between strand 2 (S2) and strand 3 (S3) and helix 6 (H6), which can sometimes “break” into two distinct elements (referred to as H6a and H6b in Table 1).
Figure 2.
A, topology diagram of core PCAD–Memo domain. α-Helices are depicted as blue coils and β-strands as gray boxed arrows. VR1–VR3 are placed on the core framework as colored loops. Conserved positions are identified with red letters at their approximate locations on the core. B, distinct structural views of PCAD–Memo families with representing solved crystal structures. Coloring is coordinated with core and variable regions in A. Proximity of variable regions to the active site is captured in the far-right space-fill column, with active-site residues colored red.
Previous research on the PCAD–Memo superfamily had reported extensive sequence diversity between and within individual families (23). Our analysis suggests that despite this diversity, several key sequence positions are conserved across the superfamily (see alignment columns shaded in black with white lettering, denoted by an asterisk in Fig. 3, Table 1, and Fig. S1). In total, eight positions are well-conserved across all enzymatically-active families in the superfamily; mapping of these positions onto available structures indicates that these are closely associated with the conserved active-site pocket (Fig. 2 and Table 1). Three of these positions (Fig. 2A and Fig. 3), namely the histidines after S1 and S2 and the acidic residue or cysteine between H6 and H7, are residues coordinating the active-site metal ion (7, 24–26). The histidine downstream of S6 is farther away from the coordinated metal but close to the substrate suggesting that it might contribute to aromatic substrate selectivity via π–π interactions (Figs. 2A and 3). Furthermore, the aromatic residue at the N terminus of H3 forms π–π interactions with both the above histidines coordinating the metal ion and may help maintain a delocalized π-cloud system for effective chelation (7, 24–26). The remaining conserved residues either help position the above residues or maintain the active-site conformation through hydrogen bonding. These observations also indicate that the PCAD–Memo superfamily falls entirely in the range of sequence conservation in the face of structural variation that has been described for several other families of enzymatic protein domains (27).
Figure 3.
Multiple sequence alignment of all defined families of the PCAD–Memo domain. Sequences are labeled at the left by gene name (where applicable), organism abbreviation, and accession number, separated by underscore. Top line provides consensus secondary structure, with α-helices and β-strands depicted as purple cylinders and orange arrows, respectively. Positions of conserved active-site residues are marked at top by * and position of conserved aspartate in H5 marked at top by ∧. Bottom line provides amino acid residue consensus conservation for individual positions in the alignment, abbreviations, and coloring as follows: p, polar shaded blue; c, charged shaded blue; s, small shaded green; u, tiny shaded green; h, hydrophobic shaded yellow; l, aliphatic shaded yellow; a, aromatic shaded yellow. Capitalized letters in consensus line represent conservation of specific amino acid residues. Residues involved or predicted to be involved in active-site catalysis or metal ion coordination are shaded in black and colored in white. Well-conserved aspartate identified in H5 is shaded gray and colored in white. Family names are provided at the right of alignment. Positions of VR1–VR3 are denoted by boxes and labeled at the top of the alignment.
The substrate is positioned within the active site via coordination interactions between the diol oxygens or a phenol oxygen and amino group nitrogen and the metal. This strongly suggests that the PCAD family is likely to be specific for substrates that contain an aromatic diol. The substrate is shielded from the solvent by elements of VR1 and sometimes VR3 or an additional subunit (e.g. LigA; see below) such that only the extradiol bond, which is cleaved, is exposed for attack by dioxygen. Beyond the above eight site residues closely associated with the active site, certain other well-conserved charged or polar positions were identified. Notable among these is a nearly absolutely-conserved aspartate residue in H5 (see column shaded in gray in Fig. 3, denoted by a caret symbol). While not extending into the active site itself, it appears to provide a stabilizing backbone contact that positions the loop between S6 and H5 as a “wall” delimiting one side of the active-site pocket (Fig. 3).
Variable regions and their relation to PCAD–Memo function and regulation
The above-mentioned three structurally variable regions in the superfamily (Fig. 2) occur between S1 and H1 (VR1), S3 and H2 (VR2), and S6 and H5 (VR3) (Fig. 2A and Table 1). VR1 tends to be the shortest and often takes the form of a small helical segment. The most minimal version of this represents a unifying feature across families of the Memo clade (Table 1 and Fig. 2B). More complex versions occur in clades like the LigZ-like family and take the form of longer predicted α-helical inserts. VR2 tends to be structurally more diverse than VR1 (Table 1). It might range from a complete lack of an insert (the Rv2728c-like family), through relatively simple forms consisting of predicted extended loops (e.g. the PqqD-like fusion and LigZ families), to large inserts composed of multiple delineable secondary structure elements in several families. For instance, in the Memo family VR2 contains elements stacking with core elements (7); in the APD family it contains a β–α–β unit, and in the HpcB/HpaD-like family-1 it is predicted to contain a four-stranded β-meander; several other families are predicted to form various α–β structures (Table 1). Solved structures for the APD and PF0612 (Table 1) provide an indication of the likely spatial positioning of the more elaborate VR2 inserts: in both cases, the VR2 inserts form a partial fourth layer to the core three-layered sandwich, stacking outside of the “forward” helical layer (Fig. 2B). The final variable region, VR3, typically forms or is predicted to form elongated loops in the proximity of the active-site pocket, likely acting as a cap, which at least in some families could participate directly in structural occlusion of the pocket. In some families, VR3 is predicted to house more structured elements, such as an insert with two α-helices in a subset of the classical LigB family (see below, Table 1, and Figs. 2 and 3).
These variable regions and their family-specific diversity (Table 1) are of note primarily for the following. 1) Their spatial positioning relative to the active-site pocket reveals that they contribute noticeably to the variability of the walls of the pocket (Fig. 2B). 2) VR1 and VR2 often contain family-specific conserved residues (Table 1), as noted previously for certain families in the PCAD–Memo superfamily (23). Mapping of the family-specific residues onto solved structures indicates that these generally fall into one of three categories: 1) residues that perform structural roles relating to formation of the active-site pocket (e.g. the conservation of prolines in VR1); 2) residues that contribute directly or indirectly to the active site (e.g. conserved aspartate residue Asp-73 in solved structure PDB code 3VSI of the APD family (Table 1, supporting data)); or 3) residues that are not directly involved in the active site but appear suitably positioned for allosteric regulation, as described previously in some families (e.g. conserved polar residue, often a glutamate, in the classical LigB family) (23). Residues from the final category often contribute to clearly-definable ancillary pockets in proximity to the active-site pocket. Using the experimentally-defined allosteric activating pocket for the LigB family (23), we found that four of the five families with solved crystal structures contain comparably-situated pockets. This suggests that allosteric regulation through these ancillary pockets could be a broader functional principle of the PCAD–Memo superfamily (Fig. 2B).
Multimerization
Prior experiments have shown that some members of the classical LigB family function as homodimers (24, 28). A range of other configurations have also been described: the gallate dioxygenase GalA, a member of the classical LigB family, functions as a homotrimer (29); the APD family forms a heterodimer between active and inactive paralogs (30); and the YgiD-like family has thus far been characterized as a functional monomer in bacteria (31) and plants (32). The significance of potential multimerization in the Memo clade remains unexplored. This variety in multimerization, including potentially differing modes even within the same family, could point to a role in fine-tuning substrate specificity and/or potentially influencing allosteric regulatory mechanisms in PCAD–Memo proteins.
The currently available structural data for the PCAD–Memo superfamily suggests a tendency for dimerization to occur end–to–end at the S4 edge of the core β-sheet, with the sheet in one monomer inverted in polarity relative to the opposing monomer sheet. The core β-sheets do not stack directly with each other, instead, the edge of one core sheet stacks with the S2–S3 loop (the “dimerization” loop noted above) and vice versa (Fig. 2A), comparable with what is seen in the classical LigB family. In the solved structure of the PF0612, this loop adopts a small β-strand conformation, which stacks alongside an extended S4 strand from the opposite monomer. This strand-stacking results in the formation of a barrel-like structure at the dimer interface. A third and considerably more elaborate dimerization mode is observed in the APD family, wherein the dimerization loop is captured by a “brace” of the opposing monomer.
Secondary structure prediction and sequence alignment of individual families without solved structures suggest that the dimerization modes are likely to more closely approximate PF0612 and classical LigB dimerization rather than some of the more aberrant modes listed above. Further experimental determination of the dimerization–pattern–space for the superfamily has the potential to throw light on any possible relationship between multimerization and substrate specificity/recognition, regulation of enzymatic activity, and the possible role of catalytic inactivation of one of the subunits in the heterodimeric forms.
Classification of the PCAD–Memo superfamily
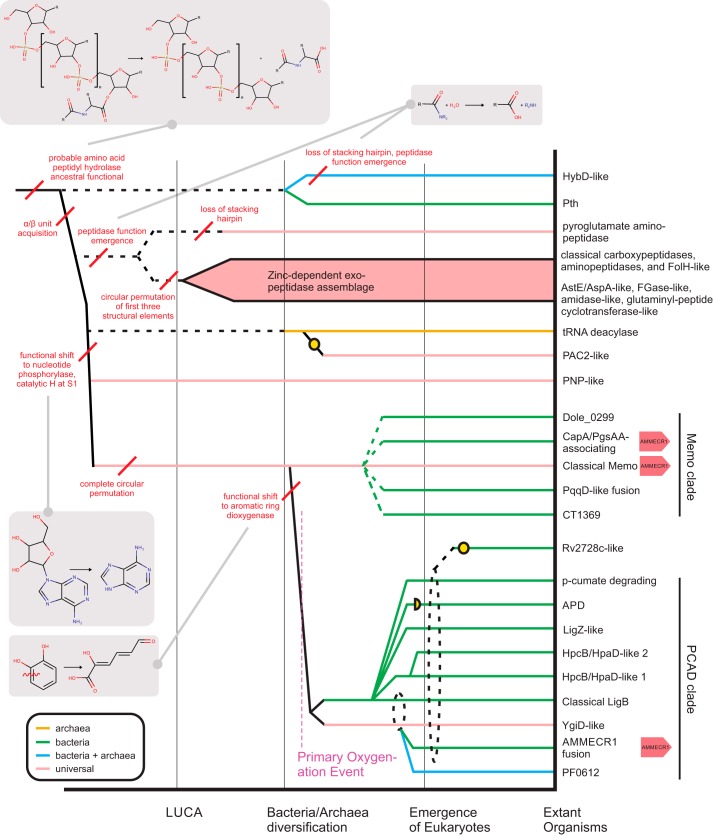
Reconstruction of the evolutionary history of protein domains allows the identification of shared and derived sequence and structure characters contributing to the functional diversification of families. This assists with the prediction of function in poorly-understood families within a superfamily. To this end, the families of the PCAD–Memo superfamily identified through collection and clustering were analyzed to establish a comprehensive natural history for the superfamily; these results are summarized in Table 1 and the supporting data. Additionally, internal relationships within individual families, where relevant, were established using conventional phylogenetic analyses (see “Experimental procedures”), and phyletic profiles were constructed based on these and clustering results. Furthermore, contextual information in the form of domain architectures and conserved gene neighborhoods were cataloged across the superfamily (see “Experimental procedures”). At this point, higher-order relationships between individual families were determined through comparison of shared structural features and other synapomorphies (shared derived characters). These relationships were then placed within the framework of their identified phyletic patterns to infer a relative temporal evolutionary scenario for the diversification of the PCAD–Memo superfamily. This is depicted as a temporal diagram (Fig. 4).
Figure 4.
Reconstructed evolutionary history of the PCAD–Memo domain and relation to other lineages in the PNP-peptide hydrolase fold. Major evolutionary transitions are marked by vertical black lines, distinguishing relative temporal eras. Individual lineages, labeled to the right, are traced to their maximal inferred evolutionary depth by horizontal lines, which are colored by extant phyletic distributions. Broken horizontal lines indicate a lineage cannot be traced beyond a certain point. Yellow circles denote loss of conserved residues needed for the enzymatic activities observed across fold lineages. Red lines mark key structural and/or functional transitions, further illustrated by chemical reaction diagrams in gray boxes. Recurrence of genomic AMMECR1 association across the PCAD–Memo domain are denoted at the right of the lineage names.
Our analyses confirm a fundamental divide in the superfamily between families of the “Memo-like” clade and those of the “PCAD-like” clade (Fig. 4). The Memo family itself, given its status as the only lineage conserved across all three Superkingdoms of Life, is the probable founder of the superfamily that diverged from the PNP-peptidyl/amidohydrolase clades, which share the same fold, sometime prior to the emergence of the Last Universal Common Ancestor (LUCA) (see below for further discussion). Barring the YgiD-like family, the other PCAD clade families are predominantly found in bacteria with only occasional occurrences outside, which can be attributed to secondary horizontal transfers (Fig. 4 and supporting data). Hence, we infer that the PCAD clade likely branched from the Memo family in bacteria after the time of the bacterial/archaeal split. We discuss further details of the classification below along with the conserved aspects of genome contextual information. Such information has previously been observed to be an effective means of predicting function of poorly-understood protein domain families (33, 34). However, as has been previously noted (20, 35), substrate promiscuity in the superfamily often complicates functional assignment/assessment at the family level. Particularly in the PCAD clade, functional partners are not always directly linked to each other on the genome (20, 21), precluding functional assignment by using conserved gene neighborhood analysis alone. However, several examples of previously-unrecognized associations point toward potential novel roles for several members of the superfamily.
PCAD clade
Conserved gene neighborhoods of interest for this clade are presented in Fig. 5, with known and predicted aromatic ring-opening reactions catalyzed by them provided in Fig. 6 for reference.
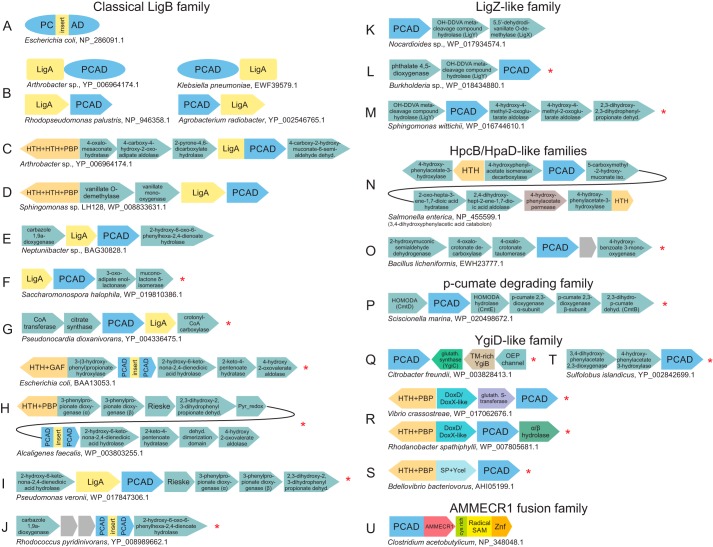
Figure 5.
Conserved genome associations of the PCAD clade. A–U, representative depictions of conserved domain architectures and gene neighborhoods containing PCAD domains (for comprehensive list of all genome associations, see supporting data). Domain architectures are depicted as adjacent shapes with a single shape corresponding to a single domain, not drawn to scale. Gene neighborhoods are depicted as boxed arrows. Each representative association is labeled below by organism name and accession number. PCAD domains are organized by family (labeled above) and colored blue. Domains known or predicted to participate in catabolic pathways are colored sea green. Associations identified for the first time in this study are marked to the left with a red asterisk. Genes observed in a neighborhood, which are not broadly conserved, are left unlabeled and shaded gray. Abbreviations used are: dehyd., dehydrogenase; OEP, outer membrane efflux protein; glutath., GSH; Cys-rich, cysteine-rich domain observed N-terminal to the radical SAM domain of the PCAD–Memo, AMMECR1, and radical SAM three-gene island.
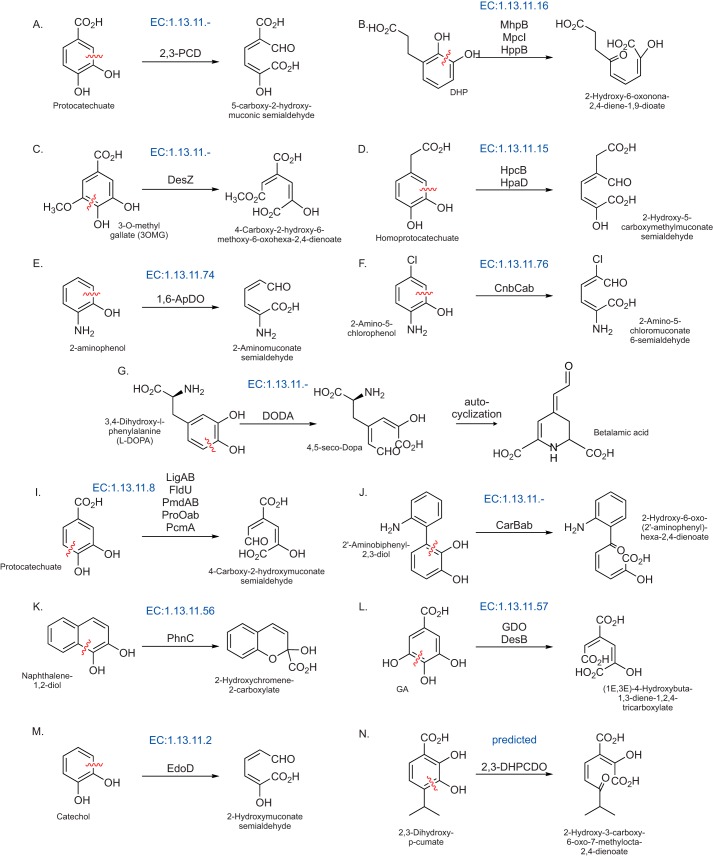
Figure 6.
Chemical reaction diagrams for known and predicted reactions catalyzed by members of the PCAD clade. Enzyme Commission (EC) numbers are provided in blue above the reaction, where applicable. Abbreviations used are: 1,6-ApDO, 2-aminophenol dioxygenase; GDO, gallate dioxygenase; DODA, 2,3-DHPCDO, 2,3-dihydroxy-p-cumate dioxygenase.
Classical LigB family
This family is extensively represented in α-, β-, and γ-proteobacteria and the actinobacteria, with only scattered representation in other prokaryotes (supporting data). Several of the currently biochemically characterized PCAD clade enzymes are members of this family (20, 21, 28, 35–38). These studies demonstrate their remarkable substrate diversity and promiscuity, sometimes through directly assaying catalytic efficiencies for enzymes against substrate panels, and suggest that the family is central to the degradation network of protocatechuate (PCA) and other phenolic compounds, particularly in the catabolism of lignins. The key to understanding the functional diversification of this family is a broad structural division that we identify for the first time in this study: the family can be divided into two subfamilies based on the presence or absence of a gene coding for LigA in the genome context. Versions lacking the LigA association, without exception, contain a large insert, which is unprecedented across the superfamily at the VR3 site (Figs. 2A, 3, and 5A, Table 1, and supporting data). Hereafter, we distinguish these subfamilies as the LigA-associating and insert-containing subfamilies.
The LigA protein is an all α-helical domain that forms a higher-order phylogenetic assemblage with several other helical domains, previously thought to have no relatives, including the Frankia-peptide domain (39).5 LigA itself is only observed in contexts with the classical LigB family. Previously solved structures have captured the LigA–LigB complex, which shows the LigA domain forming a helical cap-like structure over the LigB active site. In other enzyme superfamilies, such as the haloacid dehalogenase (HAD) superfamily of phosphoesterases (40), the cap modules help exclude water from the active site and “seal the reaction chamber” during catalysis, and LigA might play a comparable role for LigB. LigA can be fused directly to the PCAD domain of LigB at either the N or C termini or can be encoded by the gene immediately upstream or downstream of the LigB encoding in a conserved gene neighborhood (Fig. 5B and supporting data).
The VR3 insert of the insert-containing subfamily (Fig. 5A) is predicted to form at least two α-helices and occupy a spatially equivalent location to the LigA domain directly above the PCAD active-site pocket. The mutual exclusivity of the insert and LigA along with the predicted similarity in their positioning strongly suggests that the insert acts as a functional equivalent to the LigA domain, potentially occluding the active site in the course of the reaction. Given the respective phyletic distributions of these two subfamilies, it appears likely that the insert-containing clade emerged from a LigA-associating precursor through acquisition of the of insert followed by loss of LigA (supporting data).
We also observed a clear divide in the conserved gene-neighborhoods between the two subfamilies. Certain contexts for the LigA-associating subfamily have been reviewed previously (21) and are consistently observed as associating with genes encoding enzymes involved in PCA degradation, i.e. the 4,5-cleavage of PCA leading to oxaloacetate and pyruvate production (Fig. 6A). In contrast, the predominant genome contexts for the insert-containing subfamily point to a role in the degradation of trans-cinnamate (derived from benzoate) with all enzymes required to convert trans-cinnamate to acetyl-CoA being present in the neighborhood (see below for further discussion on substrate promiscuity in the classical LigB family, Figs. 5, C–J, and 6, and supporting data). This subfamily includes the well-studied MhpB and HppB enzymes (Fig. 6B) (38, 41–43), the former of which has been experimentally implicated in promiscuity by catalytic efficiency studies (38).
LigZ-like family
The LigZ-like family contains two previously-studied members, the titular LigZ and DesZ, both of which have been studied in distinct branches of the lignin degradation pathways (28, 44–46). Consistent with functions in lignin degradation, the LigZ family shares several sequence and structural features with the classical LigB family (Fig. 3 and Table 1). However, the LigZ family is distinguished from the other families of the superfamily by the presence of a predicted, likely α-helical insert with a highly-conserved tryptophan in VR1 (Table 1 and supporting data).
LigZ has been shown to catalyze the conversion of 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl to 4,11-dicarboxy-8-hydroxy-9-methoxy-2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate as part of the LigW–LigX–LigY–LigZ pathway that converts 5,5′-dehydrodivanillate to vanillate (44, 47). Gene neighborhoods of the LigZ-like family revealed the conserved presence of both LigY and LigX (although LigW is notably absent) across a range of proteobacteria and a few actinobacteria. This core is often joined by several additional enzymes suggesting potential linkages to other catabolic pathways (Fig. 5K and supporting data). For example, both the phthalate 4,5-dioxygenase and 4,5-dihydroxyphthalate decarboxylase are observed (Fig. 5L and supporting data), positing a linkage to phthalate degradation pathways. The 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase is also observed in some neighborhoods, suggesting an additional linkage to trans-cinnamate degradation (Fig. 5M and supporting data). Conspicuously, the DesZ enzyme itself, which catalyzes a 3-O-methyl gallate ring-opening reaction (45, 46) (Fig. 6C), lacks any conserved genome contexts. Taken together, these contexts suggest that like the related classical LigB family, the potential for substrate promiscuity in the LigZ-like family probably favored the colonization of similar biochemical niches.
HpcB/HpaD-like families
Two distinct families in the PCAD clade have been experimentally characterized as 3,4-dihydroxyphenylacetate 2,3-dioxygenases, which catalyze the degradation of 3,4-dihydroxyphenylacetate (also known as homoprotocatechuate) to 2-hydroxy-5-carboxymethylmuconate semialdehyde (Fig. 6D). The two families share several structural and sequence features and accordingly group together in clustering analysis, arguing for a higher-order relationship between the two families, including a predicted β-strand–rich region containing at least one shared sequence motif in VR2 (Table 1, Figs. 1 and 4, and supporting data). Most members of both families are part of tyrosine/tyramine/dopamine degradation pathways (48–51) and are almost always encoded as part of the 3,4-dihydroxyphenylacetic acid catabolon (Fig. 5N), which includes enzymes needed for processing of tyramine and related substrates to succinate (49). Consistent with previous reports, we observed that this regulon is found in several discrete units on the genome, and the composition of these units can vary between different species (supporting data). In a subset of firmicutes, the HpcB/HpaD-like family members, previously characterized in Paenibacillus sp. JJ-1b as a protocatechuate 2,3-dioxygenase (2,3-PCD), appear in a distinct 4-hydroxybenzoate degradation pathway context (Figs. 5O and 6A) (52, 53). This acquisition of a novel substrate again points to a certain degree of substrate promiscuity as noted for the above families, also supported by previous turnover number studies on 2,3-PCD (see further discussion below) (53).
APD family
This well-studied family further illustrates the versatility of the PCAD superfamily in utilizing diverse aromatic substrates; it contributes to the degradation of both 2-aminophenol (Fig. 6E, 1,6-ApDO) and 2-amino-5-chlorophenol (Fig. 6F, CnbCab) (30, 54–57). Although only sporadically observed in prokaryotes, the family is found across a diverse phyletic range, including several proteobacteria, actinobacteria, and on rare occasions in euryarchaeota. The family underwent an early duplication, spawning a closely-related yet inactive version that is tightly-coupled to the active version on the genome. The two then form a functional heterodimer with an unusual mode of dimerization (see above, Fig. 3, and supporting data). Phyletic pattern analysis suggests the gene pair has been disseminated via HGT, typically along with other downstream members of the nitrobenzene degradation pathway including 2-aminomuconic semialdehyde dehydrogenase and 2-aminomuconate deaminase (supporting data).
p-Cumate–degrading family
This family is restricted to a subset of actinobacteria and the deltaproteobacterium Candidatus Entotheonella. Although biochemically uncharacterized, its genomic contexts suggest that it is part of a pathway that catabolizes p-cumate to pyruvate (Fig. 5P and supporting data). A related pathway has been experimentally characterized in other organisms including Bacillus and Pseudomonas. In these organisms, the pathway proceeds through the unrelated glyoxylase fold extradiol dioxygenase (58), which appears to the be functional analog of this family suggesting potential analogous gene displacement between these families in the p-cumate utilization pathway (Fig. 6M).
YgiD-like family
This family is prototyped by the Escherichia coli YgiD protein and is the sole PCAD family to have widespread membership outside of the bacteria, with representatives found in eukaryotes such as fungi, plants, amoebozoans, kinetoplastids, certain chromalveolates, and few basal eukaryotes (Fig. 4 and supporting data). In addition, a sporadic presence is observed in Archaea, which similarly crosses diverse lineages, with at least one monophyletic clade containing representatives from Euryarchaea, Crenarchaea, and Asgardarchaea (supporting data).
Some eukaryotic YgiD-like sequences have been experimentally-characterized as 3,4-dihydroxy-l-phenylalanine (l-DOPA) dioxygenases (Fig. 6G) functioning in betalain biosynthesis pathways, for example in plants (32, 59, 60) and fungi (61, 62). Betalains are water-soluble, indole-derived pigments replacing anthocyanins in a limited set of plants, mostly of the order Caryophyllales and some fungi. Very recently, the E. coli YgiD was shown to exhibit DOPA dioxygenase activity in vitro (31). Given the limited distribution of betalain pigments and the much broader distribution of the YgiD-like family, it appears likely that betalain production is a specialized function of YgiD-like domains that is probably restricted to certain terminal eukaryotic lineages. The remaining members of the family, like the E. coli enzyme, could therefore be involved in channeling oxidized phenylalanine/tyrosine derivatives into other pathways or potentially be involved in the ring-opening of distinct substrates (see further discussion below and Fig. 5, Q–T). The lone YgiD crystal structure from E. coli (the Southeast Collaboratory for Structural Genomics (63)) presents a rather unusual active-site configuration for YgiD: the histidine at the N terminus of H3 is drawn away from its usual position to coordinate a second Zn2+ ion; similarly, the histidine downstream of S6 and the glutamate between H6 and H7 are drawn away from their usual positions to coordinate a 3rd Zn2+ ion. The three Zn2+ ions define a 120°–30°–30° triangle, and accordingly, the active-site pocket is unusually wide and “open” on the protein surface when compared with structures of other representatives in the superfamily (Fig. 2B). This feature suggests specialization to accommodate a larger substrate like l-DOPA with its carboxylate group coordinating with one of the additional Zn2+ ions. Furthermore, this active site could also favor the spontaneous cyclization of the oxidation product 4,5-seco-Dopa to betalamic acid that is observed downstream of the YgiD-catalyzed reaction (Fig. 6G). Alternatively, this active site might facilitate some substrate promiscuity.
PF0612 family
This family of PCAD domains, despite the recent publication of a solved structure (26), remains functionally enigmatic. No conserved genome context information was detectable for the family. It appears to have been transferred to certain archaeal lineages (Fig. 4 and supporting data).
AMMECR1 fusion family
This uncharacterized family is predominantly observed in the firmicutes; however, it appears to have been sporadically disseminated via HGT to several additional lineages, including proteobacteria, actinobacteria, synergistes, spirochaetes, and verrucomicrobia. This family is directly fused to a C-terminal AMMECR1 domain (15, 64) or in some cases contextually linked to the same in a conserved gene-neighborhood. These are further associated in a conserved gene-neighborhood with a radical SAM enzyme fused to a C-terminal zinc-ribbon domain (Fig. 5U and supporting data). Other associations reported for the AMMECR1 domain, coupled with previous experimental work on diverse radical SAM domain families, led to the prediction that this conserved gene neighborhood was likely catalyzing a nucleotide or nucleotide-base modification reaction (15, 16).
Strikingly, an association with orthologous radical SAM and AMMECR1 genes also occurs in the more ancient Memo clade of the superfamily (Fig. 4; see below), but Memo protein orthologs are largely absent in firmicutes. The mutual exclusivity of their respective phyletic patterns suggests this family of the PCAD clade displaced the Memo domain in this conserved three-gene island at or near the base of the firmicutes lineage (Fig. S2). Several other features are also shared between the Memo and AMMECR1 fusion family, including the notable conservation of a cysteine in place of an acidic residue in the loop between H6 and H7 (Fig. 3 and Table 1) seen in the other families. These potentially represent late, atavistic re-acquisition of Memo-like features in this family of the PCAD clade (Fig. 4). This observation carries several evolutionary and functional implications discussed below.
Memo clade
Contextual information in the form of conserved gene neighborhoods and domain architecture for this clade is provided in Fig. 7.
Figure 7.
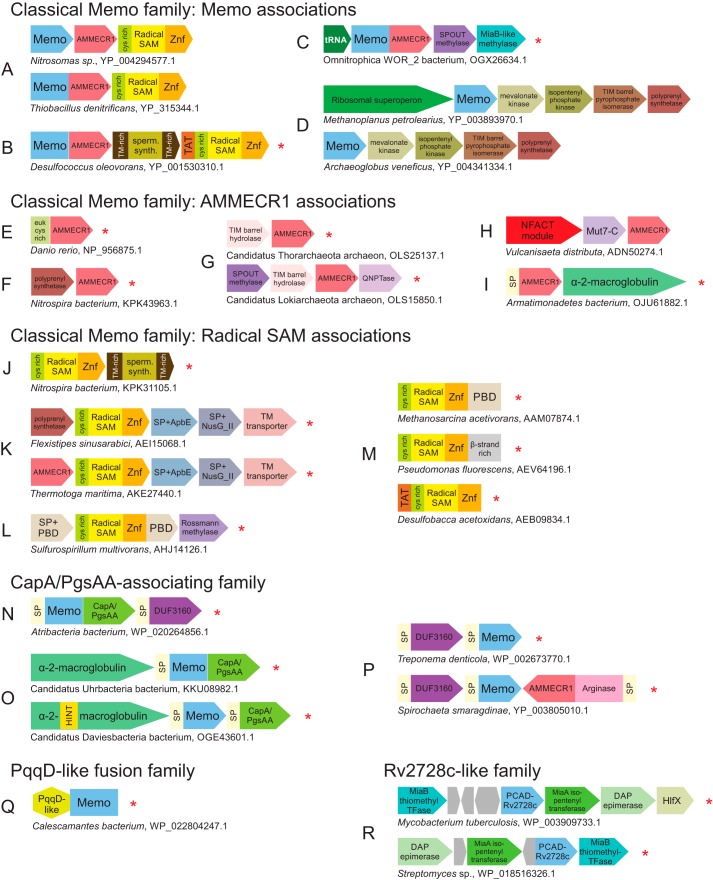
Conserved genome associations of the Memo clade and Rv2728c-like family. A–R, genome association depictions, labeling, and abbreviations are as described in Fig. 5 legend. Additional abbreviations used are: sperm. synth., spermidine synthase; euk cys rich, cysteine-rich N-terminal domain specifically fused to certain eukaryotic AMMECR1 domains; QNPTase, quinolinate/nicotinate phosphoribosyltransferase; PBD, periplasmic-binding protein/chelatase fold domain; TFase, transferase.
Classical Memo family
The classical Memo family is the only one in the PCAD–Memo superfamily clearly traceable to the LUCA. It is found across most major lineages of life, including a monophyletic branch of the family in the archaea that spans all known archaeal lineages. Against this backdrop, it is notable that certain terminal branches of life appear to have either lost the Memo clade or possess an alternative PCAD clade enzyme that appears to have displaced Memo (see the AMMECR1 fusion family above, and Fig. S2).
In prokaryotes, classical Memo associates broadly with the AMMECR1 domain either as a neighboring gene or in a direct fusion in the same polypeptide. Furthermore, these are also linked to a radical SAM family domain, which is fused to a C-terminal zinc-ribbon domain and a distinct N-terminal cysteine-rich domain (Figs. 5U and 7A). This three-gene neighborhood is broadly conserved across bacteria and some euryarchaea and thaumarchaea (supporting data). Although the core three-gene configuration is the overwhelmingly dominant association for the three components, each gene also has unique associations, wherein it may combine with one of the remaining two genes or occur in entirely independent contexts (Fig. 7, C–M, and see below). However, even in these cases, the three core components show a striking shared phyletic pattern, i.e. co-presence in the same genome (Fig. S2), suggesting that even when not linked, the three core components are likely to act in a certain common pathway in the cell (see below).
The FUNCOUP system (65) predicts a functional association between Memo and AMMECR1 in eukaryotes, predominantly stemming from shared mRNA co-expression patterns and high-throughput protein–protein interaction data along with support from shared transcription factor–binding sites across several genomes. We observe further that the radical SAM family enzyme of the three-gene prokaryotic island is present in some eukaryotes, including the diplomonad Giardia and the parabasalids Trichomonas and Tritrichomonas, as well as in Entamoeba and Blastocystis. However, the remaining eukaryotes have lost this enzyme (supporting data). We predict these detected eukaryotic radical SAM domain orthologs of the prokaryotic versions above are likely to interact with AMMECR1 and Memo in the limited set of eukaryotes that possess them. It is possible that the three-gene apparatus was inherited as a single unit in the last eukaryotic common ancestor with the radical SAM component displaced by an as-yet undetermined gene relatively early in eukaryote evolution. In this light, it is also of interest to note that in many, but not all, eukaryotes the AMMECR1 domain is fused to a novel N-terminal cysteine-rich domain that does not exist outside of this architectural context (Fig. 7E and supporting data). Its absence in the eukaryotes retaining the radical SAM family enzyme suggests a possible compensatory role for the domain.
Other families of the Memo clade
The remaining four families of the Memo clade, which remain uncharacterized in the current literature, share with the classical Memo family the conserved cysteine between H6 and H7 along with a minimal loop region in VR1 (Table 1). Their relatively restricted phyletic distributions (supporting data) indicate these are likely recent derivations from the ancestral classical Memo family. Two of these families, Dole_0299 and CT1369, have genome contextual associations that are minimally informative (supporting data).
The third of this set of families, the CapA/PgsAA domain fusion family, is characterized by a frequent direct C-terminal fusion to a calcineurin-like phosphoesterase domain specifically related to the version found in the CapA/PgsAA proteins (Fig. 7N and supporting data). The calcineurin-like phosphoesterases catalyze a broad range of activities, including nuclease, nucleotidase, phosphatase, sphingomyelin phosphodiesterase, and 2′,3′-cAMP phosphodiesterase activities (66, 67). CapA/PgsAA has been previously observed as part of a conserved four-gene operon that synthesizes poly-γ-glutamate (PGA). As part of this operon, CapB/PgsB is a well-characterized peptide-ligase that initiates and extends glutamate chains by activating them through the ATP-dependent formation of an acylphosphate (68, 69). Although CapA/PgsAA has been linked to increasing turnover of the peptide–ligase reaction (69, 70), its role is largely unclear. Given that it is a calcineurin phosphoesterase, we propose that it might function by hydrolysis of inappropriately phosphorylated carboxylates of glutamate during PGA chain elongation. Its association with the Memo clade is independent of the other genes involved in PGA synthesis. Across representatives of the recently-characterized radiation of novel bacteria (71), this family and the associated CapA/PgsAA phosphoesterase domain is also linked in a conserved neighborhood to bacterial α2-macroglobulin proteins (Fig. 7O and supporting data). The presence of signal peptides in representatives of this family as well as its diverse associating components suggests that together they form a periplasmic complex (see further discussion below, Fig. 7, N–P).
The fourth of these families is the PqqD-like fusion family, which features an N-terminal fusion to a PqqD-like winged helix–turn–helix (wHTH) domain (Fig. 7Q and supporting data). Although no other distinguishing structural, sequence, or genome contextual features were observed for the family, the PqqD-like domain is one member of a higher-order assemblage of peptide-binding wHTH domains (72, 73). The PqqD proper domain was initially characterized in the binding of the PqqA peptide and its presentation to an enzyme cascade that generates the bacterial cofactor pyrroloquinoline quinone (PQQ) (74–76). This family of Memo domains could therefore act on an unknown peptide bound by the fused PqqD-like domain. Although there is an as-yet unknown dioxygenase in the PQQ biosynthesis pathway (77), there is no clear contextual linkage between this version of the Memo domain and other enzymes for PQQ biosynthesis to suggest that this is the missing dioxygenase.
Rv2728c-like family
Members of this newly-identified and uncharacterized actinobacteria-specific family lack several of the standard active-site residues, including those involved in metal coordination (Fig. 3, Table 1, and supporting data), suggesting they are inactive. They are typically observed in conserved neighborhoods with the MiaA adenylate isopentenyltransferase (78), the MiaB isopentenyl adenylate thiomethyltransferase, a diaminopimelate epimerase-like protein, and occasionally a distal association with a GTPase of the HflX family (Fig. 7R). Although this version of the PCAD–Memo domain is likely incapable of coordinating a metal ion and therefore catalyzing the dioxygenase reaction, the presence of some conserved residues in the active-site pocket (Fig. 3 and Table 1) suggests it could still be involved in binding/recognition of a ligand, a trend commonly-observed in inactive members of several enzymatic families (79).
The other components encoded by this conserved gene neighborhood point to a function for this family in the synthesis or modification of an adenine nucleotide via the action of the MiaA- and MiaB-like enzymes. It is possible that the Rv2728c-like family domain plays a role in binding substrates for this reaction. The occasional association with HflX (Fig. 7R and supporting data), recently characterized as a 100S ribosome dissociation factor (80), could suggest a link to base modification of rRNA bases at the ribosome. Alternatively, we cannot rule out that this pathway could produce an actinobacterial secondary metabolite via modification of a nucleotide-derived, cytokinin-like molecule with an aromatic ring modification, potentially in conjunction with the action of a SLOG base-releasing enzyme (81, 82).
Functional analysis of the PCAD–MEMO superfamily
Certain enzyme superfamilies explore substrate space extensively, catalyzing essentially similar reactions on diverse substrates. Other enzyme superfamilies explore reaction space, catalyzing a diverse set of reactions on either an essentially similar set or a diverse set of substrates (83). The PCAD clade of the superfamily under consideration displays the former situation. However, shifts in reaction mechanism and substrate specificity might have happened earlier in the course of their divergence from the Memo clade and even earlier from the catalytically distinct PNP and peptidyl/amidohydrolase clades. We explore these aspects of the superfamily in greater detail below along with novel functional predictions inferred from genomic contexts.
Biochemical promiscuity in the classical LigB family
Within the PCAD clade, the classical LigB family displays extensive promiscuity in terms of substrates. Representatives catalyze ring-opening reactions on PCA and its precursors in-pathway to PCA generation (Fig. 6H) (21, 28, 36, 37). 1) The titular LigB/LigAB from Sphingobium sp. strain SYK-6 acts on PCA, gallate, and 3-O-methyl gallate (the latter being the apparent dedicated substrate of DesZ (see above and Fig. 6C)) (28, 35). 2) FldU from Sphingomonas sp. LB126 acts on PCA derived from fluorene (84). 3) PcmA acts on PCA derived from phthalate and phthalate-like precursors (85). 4) Enzymes like PmdAB from Comamonas testosteroni and ProOab from Pseudomonas ochraceae appear to act on PCA generated from several distinct pathways (36, 37).
The enzymes from this family have also been identified in the degradation of other aromatic compounds. 1) MhpB is a 2,3-dihydroxy-phenylpropionate 1,2-dioxygenase from E. coli K12 (41, 42), HppB from Rhodococcus globerulus PWD1 (38, 43), and MpcI from Alcaligenes eutrophus (41) catalyze the ring-opening of 3-(2,3-dihydroxyphenyl)propanoate or trans-2,3-dihydroxycinnamate in the benzoate degradation pathway proceeding through a trans-cinnamate intermediate (Fig. 6B). 2) The 2-aminobiphenyl-2,3-diol 1,2-dioxygenase CarBab from Pseudomonas sp. strain CA10 participates in carbazole degradation (Fig. 6I) (86). 3) PhnC, characterized as a 1,2-dihydroxynaphthalene dioxygenase, is implicated in naphthalene degradation by Burkholderia sp. RP007 (Fig. 6J) (87, 88). 4) Several enzymes operate on lignin-derived aromatic compounds: DesB from Sphingobium sp. strain SYK-6 and the GalA gallate dioxygenase from Pseudomonas putida KT2440 degrade gallate (Fig. 6K) (25, 28, 29, 89). 5) The native substrate of EdoD from Rhodococcus sp. I1 remains unknown (Fig. 6L) (90).
Genome contexts for the LigA-associating subfamily, as mentioned above, primarily point to roles in terminal PCA degradation. This is supported by the associations with several other PCA degradation enzymes, including 4-carboxy-4-hydroxy-2-oxoadipate aldolase, 4-oxalomesaconate hydratase, 2-pyrone-4,6-dicarboxylate hydrolase, and 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase (Fig. 5C). However, this subfamily instead occasionally associates with components of pathways upstream of this core PCA meta-degradation pathway. For example, it might be linked with vanillate O-demethylase and vanillate monooxygenase enzymes, which catalyze the conversion of vanillate to PCA (Fig. 5D and supporting data). This either points to a direct linkage between lignin and PCA meta-degradation or it could point to this version of the classical LigB family being at the juncture of pathways processing multiple substrates. Additionally, the LigA-associating subfamily is also found in carbazole degradation pathway contexts, associating with carbazole 1,9a-dioxygenase and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (Figs. 5E and 6J and supporting data).
Beyond these, we also discovered gene-neighborhood associations pointing to a role for some LigA-associating subfamily members in previously-unrecognized pathways: 1) in degradation of catechol through a possible muconate intermediate. The specific genome context we identify here contains a muconolactone δ-isomerase enzyme and a likely 3-oxoadipate-enol-lactonase of the α/β-hydrolase fold (Fig. 5F), suggesting direct conversion of catechol to muconate by the PCAD dioxygenase before action by the isomerase enzyme. In previously-studied pathways, this dioxygenase reaction is catalyzed by catechol 1,2-dioxygenase, an unrelated transthyretin fold-containing dioxygenase (91). This suggests a functional equivalence between the two dioxygenases (Figs. 5B and supporting data). 2) We also observe a conserved linkage between primarily actinobacterial members of this subfamily and a citrate synthase-like enzyme, CoA transferase, and a Rossmann-like domain that is most closely related to crotonyl-CoA carboxylase/reductases (Fig. 5G). This could possibly represent an unexplored pathway that shunts an aromatic ring substrate, possibly through conversion to oxaloacetate or an upstream molecule, into the citrate metabolism pathway (supporting data).
In the insert containing subfamily of the classical LigB family, we occasionally observe variation in their standard gene neighborhoods. These suggested that the cis-3-(3-carboxyethenyl)-3,5-cyclohexadiene-1,2-diol intermediate is processed by the 3-phenylpropionate dioxygenase and 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase enzymes in lieu of the 3-(3-hydroxyphenyl)propionate hydroxylase (mphA) enzyme found in the more commonly observed configuration (Fig. 5H and supporting data). The PhnC enzyme, originally characterized as acting on bicyclic, i.e. naphthalene diols (87) (Fig. 6J), is found in this variant context, suggesting that it might also utilize the same benzoate derivative substrates as other well-studied members of this subfamily.
Analysis of phyletic patterns suggests that the helical insert emerged later and potentially physically displaced the LigA subunit. This probably went hand–in–hand with the emergence of the generally separate catalytic specialization within the two subfamilies. However, we observed several striking “crossovers” wherein gene neighborhoods typical of one subfamily contained a representative from the other (Fig. 5, I–J, and supporting data). For example, several predominantly actinobacterial LigA-associating subfamily members are found in association with components of the trans-cinnamate degradation pathways typical of the insert-containing subfamily (Fig. 5I). Conversely, certain insert-containing subfamily members associate with components involved in the carbazole degradation pathway otherwise typical of the LigA-associating subfamily (Fig. 5J). This suggests that despite the structural specialization within the two clades, the classical LigB family scaffold supports substrate promiscuity (20, 35, 38) allowing for occasional functional crossovers to have occurred. This experimentally-supported promiscuity probably relates to these enzymes being selected as part of “first-responder” pathways against atypical aromatic metabolites or xenobiotics in the environment.
Predicting functions for the YgiD family
Beyond the role in betalain synthesis (see above) (92, 93), the roles of the widespread YgiD family have remained unclear. Our analysis uncovered multiple conserved genome contexts that suggest that the YgiD-like family likely catalyzes aromatic diol oxidation in multiple pathways (Fig. 5, Q–T). First, across a range of γ-proteobacteria YgiD is found adjacent to the YgiC protein. YgiC contains an ATP-grasp fold amidoligase domain related to GSH synthetase (39, 94, 95). Several additional predicted membrane-spanning channel proteins are often encoded in the genomic vicinity of this YgiC–YgiD gene-dyad (Fig. 5Q and supporting data). Second, across several proteobacteria as well as sporadically in the actinobacteria, firmicutes, and planctomycetes, YgiD is linked to a protein containing the DoxD/DoxX-like transmembrane domain and a transcription factor that combines the N-terminal HTH domain (96) and a C-terminal ligand-binding PBP domain (97). This genome context might additionally feature either an α/β-hydrolase fold protein or a GSH S-transferase (Fig. 5R and supporting data). Notably, the DoxD/DoxX-like domain appears to be displaced in several proteobacteria as well as in some Bacteroidetes by a YceI-like protein, a β-barrel–forming member of the lipocalin fold with an N-terminal signal peptide (Fig. 5S).
Common to these themes is the association of YgiD-like proteins with diverse membrane or secreted proteins, which indicate that they likely function proximal to the membrane. Such associations might be interpreted as distinct mechanisms for the utilization of available aromatic compounds. Association with YgiC can be interpreted as the coupling of the dioxygenase activity to the modification of a metabolite with an aromatic ring by an amidoligase reaction catalyzed by the associated ATP-grasp domain (39). In contrast, DoxD/DoxX proteins found in the second major theme have been previously observed as a subunit of the DoxA–like p450 monooxygenase in the quinol:oxygen oxidoreductase complex (cytochrome aa3). The DoxA–DoxD pairing further works in concert with the DoxC transmembrane (TM) protein and the DoxB Rieske-type dioxygenase in the thiosulfate:quinone oxidoreductase complex, coupling reduction of dioxygen with thiosulfate oxidation with a decyl ubiquinone as an electron acceptor in the archaeon Acidianus ambivalens (98, 99). However, the systems with YgiD-like proteins differ from the above Dox systems in also being coupled to the α/β-hydrolase or GSH S-transferase (Fig. 5R). Several α/β-hydrolase fold proteins also catalyze a metal-independent dioxygenase reaction on oxoquinolines, and these could potentially catalyze a comparable reaction (100). Thus, the action of YgiD-like enzymes in these systems on aromatic diol-containing compounds could be coupled with either modification of the metabolite via glutathionylation or with further oxidation. This is consistent with the presence of a single-component HTH + PBP transcription factor, which suggests that the system is likely regulated by the direct sensing of such a metabolite by this protein (Fig. 5R). The YceI protein that displaces DoxD/DoxX in several gene neighborhoods (Fig. 5S) has been implicated in the trafficking of polyisoprenoid quinones, which are rife in bacteria as cofactors, most notably as part of respiratory electron transport at the membrane (101). Thus, the secreted YceI-like and DoxD/DoxX proteins might be functionally equivalent and help deliver polyisoprenoid quinones to the cell surface (Fig. 5, R–S, and supporting data), thereby coupling them to utilization of an aromatic compound oxidized by the YgiD-like family through a redox pathway.
In addition to these major associations, YgiD-like proteins are also observed in a few contextual themes that are much more restricted in their phyletic distribution. These themes are suggestive of the salvage/catabolic pathways typical of other PCAD families (supporting data). One such theme, exclusive to archaeal Sulfolobus species, combines the YgiD-like proteins with 3,4-dihydroxyphenylacetate 2,3-dioxygenase and 4-hydroxyphenylacetate 3-hydroxylase enzymes (Fig. 5A), pointing to a possible dioxygenase activity for these domains on a substrate like homoprotocatechuate.
Complex web of contextual connections characterizes Memo and its functional partners
Despite its widespread phyletic pattern and strong conservation, the Memo clade by far remains the most functionally obscure in the entire superfamily. Hence, we thoroughly analyzed the conserved gene-neighborhood and domain architecture associations for Memo and its two other functional partners AMMECR1 and the radical SAM enzyme beyond their linkage in the core three-gene system (see above and Figs. 5F and 7). We observed the following notable conserved associations for Memo.
1) Memo and AMMECR1, but not the radical SAM family domain, associate with a SPOUT domain methylase and a distinct radical SAM enzyme closely related to the MiaB family, often in conjunction with a tRNA gene (Fig. 7C and supporting data). This association is observed in the recently characterized radiation of bacteria termed the so-called “dark matter” of bacterial phylogeny (71).
2) In euryarchaea, crenarchaea, and thaumarchaea, the Memo gene by itself is located adjacent to either the ribosomal superoperon or another operon coding for mevalonate kinase, isopentenyl-diphosphate δ-isomerase, isopentenyl phosphate kinase, and polyprenyl synthetase or between both these conserved operons. The latter operon's products are components of a recently-characterized, archaea-specific alternative pathway for isopentenyl diphosphate (IPP) production (Fig. 7D and supporting data) (102–104). Based on the latter association, it was proposed that the Memo domain could act as a phosphomevalonate decarboxylase, an enzyme missing from this alternative pathway but essential for IPP production (102, 104). However, efforts to demonstrate this activity in Memo have not succeeded. Moreover, subsequent research has identified the decarboxylase for the alternative pathway in at least the euryarchaeon Haloferax volcanii (105). We identified a distinct, previously-uncharacterized member of the GHMP (Galacto-, Homoserine-, Mevalonate-, and Phosphomevalonate-) kinase fold in a subset of these neighborhoods from crenarchaea and predict these are the likely phosphomevalonate decarboxylases (106) at least in these organisms (supporting data).
3) Secreted versions of the Memo domain belonging to the CapA/PgsAA-domain fusion family show consistent associations with the so-called DUF3160 module (Fig. 7, N and P), which is also found in further genome associations with diverse membrane-associated domains, many of which are potentially involved in binding of peptidoglycan or lipid polysaccharides.5 In some spirochetes, an arginase domain fused to an AMMECR1 domain is further combined with the DUF3160 and Memo (Fig. 7P and supporting data). The DUF3160 module combines a helical N-terminal domain that features α-helices with a small, largely β-strand C-terminal domain. The long N-terminal α-helices of DUF3160 are suggestive of direct binding to a hydrophobic molecule like an isoprenoid derivative.
AMMECR1 is a member of the RAGNYA fold and features an absolutely-conserved cysteine residue predicted to be required for its catalytic activity (15). Even when occurring independently of Memo, it displays striking parallels to gene-neighborhood associations of Memo.
1) The AMMECR1 gene by itself is linked with the gene coding for a polyprenyl synthetase in several distinct prokaryotic lineages. Although this polyprenyl synthetase is specifically related to the above-mentioned versions from archaea, they do not occur with any other components of the IPP production pathway (Fig. 7F and supporting data).
2) In several archaea, AMMECR1 is combined with a gene encoding a metal-dependent TIM barrel hydrolase specifically related to the enzymes involved in base metabolism such as dihydropyrimidinase, dihydroorotase, and allantoinase (Fig. 7G). Often, these gene-neighborhoods also feature a SPOUT methylase, a Mut7-C-type PIN domain nuclease, and a quinolinate/nicotinate phosphoribosyltransferase (Fig. 7G), with the latter enzyme associating with AMMECR1 independently of the TIM barrel hydrolase in euryarchaea (supporting data).
3) We had earlier identified a conserved genome association between AMMECR1 and the DNA-glycosylase fold NFACT module together with the Mut7-C PIN endoRNase domain across a range of archaea (Fig. 7H) (16).
4) We also observed an association between a signal peptide-carrying AMMECR1 protein and an α2-macroglobulin domain protein (Fig. 7B and supporting data) (107). This notably parallels the comparable associations of the secreted versions of Memo (Fig. 7I).
The radical SAM enzyme associated with Memo and AMMECR1 is most closely related to the pyruvate formate lyase–activating enzyme. This and the related radical SAM families have been implicated in catalyzing the modifications of (poly)peptides, co-factors, and RNA (108) by means of the reactive radical generated by the cleavage of SAM (109). Its associations include the following.
1) When present with its usual partners Memo and AMMECR1, it might additionally associate with a spermine synthase domain (Fig. 7B) often embedded between the three core members of the neighborhood (Fig. 7B and supporting data). Additionally, it often associates with the same spermine synthase domain in gene-neighborhoods independently of Memo or AMMECR1 (Fig. 7J, and supporting data). In several cases, it can be fused to the twin-arginine translocation (TAT) signal peptide, which is required for translocation of fully folded proteins or oligomeric complexes across the membrane (Fig. 7B). This suggests that the TAT-associated versions are likely to function in the periplasm or extracellularly.
2) The radical SAM enzyme might associate with an ApbE-like flavin transferase, the predicted metal-binding NusG-II domain, and a predicted multi-TM transporter protein. The ApbE and NusG-II domain proteins bear signal peptides suggestive of being secreted. In certain bacterial lineages, this neighborhood appears to further include AMMECR1 or the polyprenyl-diphosphate synthase (Fig. 7K).
3) The radical SAM enzyme might be fused to the C-terminal periplasmic-binding protein/chelatase fold domain (PBD). It further associates in the same predicted operon with an additional PBD protein with a signal peptide, and a Rossmann fold methylase (Fig. 7L). Other than to the PBD, the radical SAM domain can also be directly fused at the C terminus to an uncharacterized β-strand–rich domain or the above-mentioned N-terminal TAT signal peptide (Fig. 7M).
Deciphering the functions of Memo from its contextual connections
The phyletic patterns of Memo, AMMECR1, and the radical SAM family together with their operonic and domain fusion associations suggest that all three of them were likely present in the LUCA. Such a phyletic pattern is strongly indicative of their combined involvement in universal processes of fundamental importance to the cell. The totality of the available genome contextual and experimental evidence point in two general directions: 1) involvement in a nucleotide or base modification reaction, as suggested previously (16–18), or 2) a role in a modification involving an aliphatic isoprenoid derivative. It is also possible that both roles are valid and that they come together in the context of the modification of a base by an isoprenoid derivative.
Support for both these roles is observed in the persistent but more sporadic associations of the three core components of the Memo system with the other genes that we report above. For instance, the AMMECR1 association with the NFACT module, which has been implicated in the clearance of jammed ribosomes in eukaryotes via C-terminal alanine and threonine tail formation (110, 111) and the Mut7-C PIN domain RNase might support a role in RNA base modification coupled with endonucleolytic processing (16). This is echoed by the association of the Memo gene with the ribosomal superoperon (Fig. 7A). This is also consistent with the association we report with the SPOUT methylase and a MiaB-like radical SAM enzyme (Fig. 7A), both of which catalyze base modifications found in RNA (108, 112, 113). The association between AMMECR1 and the TIM barrel hydrolase, which belongs to a family involved in catalyzing base amidohydrolase reactions, again links it to base modification. In contrast, the persistent, independent, and combined connection of Memo, AMMECR1, and the radical SAM enzyme to polyprenyl synthetase enzymes is suggestive of modification of an isoprenoid or isoprenoid derivative (114, 115). For the more sporadically distributed paralogs of the three individual components, the associations point to a cell membrane or extracellular localization (Fig. 7, B–D, and supporting data). This observation could be consistent with the modification of membrane-linked isoprenoids (116, 117) or prenylated cell-surface proteins (114).
In conclusion, we posit that Memo together with its two partners catalyzes a conserved modification of aliphatic isoprenoids. The structure of the Memo protein reveals an active site with at least two pockets suggesting that it might interact with two distinct substrates (Fig. 2B). The active-site residues of Memo are congruent to the PCAD dioxygenases but have a distinguishing cysteine in place of the acidic residue between H6 and H7 (which convergently re-emerged in the PCAD clade AMMECR1 fusion family). It is conceivable that the isoprenoid substrate is cross-linked to this cysteine, whereas Memo catalyzes a reaction involving it and a second substrate. This is likely followed by a more drastic rearrangement mediated by the radical SAM enzyme. AMMECR1 probably mediates a transferase reaction via its proposed catalytic cysteine. However, the diversity of observed contexts suggests that the same reactions could be catalyzed in multiple distinct contexts on distinct substrates. A final functional consideration with the potential to bridge the two central themes would entail the isoprenoid modification of a base, consistent with the recent identification of widespread geranylation of RNA in bacteria (118) by the SelU transferase. This modification is vital for the subsequent selenylation of tRNA (118–121). The enzyme system described above could conceivably parallel SelU with the AMMECR1 conserved cysteine acting in a capacity similar to the conserved cysteine in the SelU rhodanese domain. As possible support for this proposal, we observe a degree of negative correlation in the phyletic profiles of the Memo-AMMECR1–radical SAM system and the SelU transferase (Fig. S2).
Discussion
Evolutionary themes in the PCAD–MEMO superfamily
Origin and early evolution of the superfamily
Profile–profile sequence similarity searches return hits of borderline significance between distant superfamilies across the PNP-peptidyl/amidohydrolase fold (see Fig. 4, Table S1, and “Experimental procedures”). However, structural similarity searches emphatically unify all versions of this fold. These also reveal a conserved core topology and several synapomorphies, most notably a shared active-site pocket and conserved catalytic residues projecting into the active site from the region downstream of strand S1 (Fig. 2A and Table S1). Through these comparisons, we also infer that the PCAD–Memo superfamily first emerged from an ancestral PNP-peptidyl/amidohydrolase domain through a circular permutation event. Analysis of the PNP-peptidyl hydrolase fold proteins reveals that they had already considerably diversified by the time of the LUCA. Reconstruction of the evolutionary history of the fold suggests a possible ancestral function in tRNA-peptidyl hydrolase activity, given the early-branching, catalytically-comparable lineages found in both bacteria and archaea (Fig. 4). Multiple independent emergences of peptidase/amidohydrolase activity are inferred to have occurred at different points in the evolution of the fold, with two other functional shifts predicted to have occurred prior to the LUCA: 1) emergence of a nucleotide phosphorylase function in the PNP lineage and 2) emergence of the PCAD–Memo lineage, which may have initially retained a nucleotide-acting role before shifting to dioxygenase activity (Fig. 4). The emergence of diverse functions in this fold may coincide with structural rearrangements arising from circular permutation events and loss or gain of specific elements associated with the edges of the central β-sheet (Fig. 4 and Table S1). The propensity for the domain to dimerize may have played a role in stabilizing and fixing circular permutations that followed duplication events. Distinct circular permutations are seen in the zinc-dependent exopeptidase assemblage and in the PCAD–Memo superfamily (Fig. 4).
In structure and sequence, the versions closest to the PCAD–Memo superfamily are as follows: 1) the d-amino acid peptidyl hydrolase domains that hydrolyze d-aminoacyl tRNAs; 2) the PAC2 proteasomal chaperones; 3) PNP domains that catalyze the phosphorolytic or hydrolytic cleavage of the base from the sugar of a nucleoside at the 1′ position. These share a core topology with the PCAD–Memo superfamily, featuring the strand-helix element in vicinity of VR2 (Figs. 2A and 4). Thus, the circularly permuted version of the fold found in the PCAD–Memo superfamily appears to be nested within a clearly-definable radiation in the PNP-peptidyl/amidohydrolase fold. Our analysis also showed that the Memo clade emerged prior to the LUCA. This finding therefore implies that the permutation resulting in the divergence of the PCAD–Memo superfamily from unpermuted versions of the PNP-peptidyl hydrolase fold happened prior to the LUCA (Fig. 4). Memo is linked via genome contexts to both base and aliphatic isoprenoid modification (Fig. 7, A–E).
Post-LUCA diversification and expansion of the superfamily
In bacteria, the ancient, widely-conserved Memo lineage underwent diversification yielding four distinct families (Fig. 4). Each of these four families was then subsequently dispersed across bacteria via HGT. At least one of these late-branching families appears to have been recruited to a clear role in isoprenoid modification at the membrane or the cell surface, potentially as an offshoot of a more generalized role for the ancient Memo lineage in this capacity. Furthermore, the largely nonoverlapping phyletic patterns for these four families raise the possibility of them performing comparable or equivalent functions in the respective lineages that possess them.
After the Memo clade, our analysis suggests that the YgiD-like family is likely the next earliest-emerging extant family and the potential progenitor of the rest of the PCAD families (Fig. 4). The observed monophyly of a small yet diverse set of archaeal YgiD-like proteins raises the possibility of its presence in the LUCA. Although some basal eukaryotes contain YgiD clade members, their sporadic distribution across the rest of the superkingdom, coupled with a lack of observed monophyly of the eukaryotic versions with each other or with the archaeal versions in phylogenetic analyses, suggests a later origin for the clade in bacteria with one or more subsequent transfer(s) from bacteria to archaea and eukaryotes. Such a scenario could be consistent with their specialized role in l-DOPA utilization and betalain biosynthesis in plants and fungi (32, 59, 61). Nevertheless, the l-DOPA ring-opening activity of the YgiD clade indicates that the emergence of this clade probably marked the origin of the classic aromatic diol/amino-phenol oxidizing capacity in the PCAD–Memo superfamily. In conclusion, the links to base modification for the Memo clade hinted in our analysis might mark the initial evolutionary transition from ancestral enzymes of PNP-peptidyl/amidohydrolase with potential roles in peptidyl hydrolysis during translation or removal of bases from nucleosides to enzymes that might operate in base modification. This transition was marked by the emergence of the multiple conserved histidines in the active site. The isoprenoid modification and membrane/cell-surface association for the Memo clade might have provided the ancestral adaptations for the emergence of the YgiD-like clade to catalyze the oxidative aromatic ring-opening proximal to the membrane as suggested by the above-reported contextual associations (Fig. 5A).
The emergence of the PCAD clade was accompanied by structural and sequence divergence associated with the variable regions in the core scaffold (Fig. 2). Family-specific sequence conservation patterns in these variable regions might have contributed to substrate versatility and/or opening avenues for allosteric regulation (Table 1, see above) (23). This structural diversification might have in turn been driven by selective forces coming from the availability of new environmental compounds rich in polyphenolic rings such as lignin, which emerged on multiple occasions in the larger plant lineage (122). The ability to utilize such compounds was then broadly disseminated via HGT. An alternative pathway for the diversification of these enzymes was their recruitment to novel functional niches through displacement of existing dioxygenases in the utilization of certain metabolites, such as in the p-cumate degradation pathway (Figs. 5H and 6N). This colonization of multiple aromatic compound utilization pathways in many of these families manifests a certain versatility in terms of their substrates. This versatility takes two forms: 1) intra-family substrate promiscuity, where a single family or even a single member of a family can process multiple substrates, sometimes in multiple distinct steps in the same pathway, and 2) the ability of the distinct families across the clade at large to operate on a wide range of distinct phenolic compounds.
Despite the functional and structural sequence diversification observed in the PCAD clade, one later-emerging family in the clade, as well as potentially the Rv2728c-like family, is predicted to perform functions comparable with the Memo clade (Figs. 4, 5, and 7). This indicates that despite the diversification of the PCAD clade, attained through relaxation of some structural constraints, certain intrinsic features of active-site architecture pre-dispose the domain toward participating in the ancestral Memo-like function.
Finally, even though the fold shared by the PCAD–Memo superfamily and the PNP-peptidyl/amidohydrolases emerged prior to the LUCA, the emergence and diversification of the PCAD clade proper happened much later, primarily in the bacteria. This temporal pattern of radiation is comparable with another major class of dioxygenases, namely the 2-oxoglutarate-dependent dioxygenases and the Jumonji-like domains, which emerged from within the more ancient double-stranded β-helix fold featuring several sugar-binding proteins and sugar isomerases (123). We had postulated that the widespread availability of molecular oxygen with the primary oxygenation event in earth's history might have favored the radiation of those dioxygenases. The phyletic patterns that we report here favor a similar driver for the radiation of PCAD clade concomitant with the emergence of active oxidative metabolism utilizing O2 (Fig. 4).
Conclusions
The results presented here provide the first complete evolutionary classification for the PCAD–Memo superfamily, resolving its origins and the relative temporal diversification of its major families. Despite increasing interest in members of the classical Memo family, particularly in light of its potential reported roles in cell motility and disease in human cells, its molecular function remains poorly understood. Herein, for the first time we propose unifying explanations for the uncharacterized Memo clade as well as the enigmatic members of the PCAD clade and expand the scope of the roles of representatives of the superfamily. We hope the predictions provided here regarding catalytic mechanisms and potential substrates will guide future experimental investigations.
Experimental procedures
Bona fide protein sequences of the PCAD–Memo superfamily were retrieved from the GenBank at the National Center for Biotechnology Information (NCBI) (124), and structures were retrieved from the PDB (125). These were used to seed iterative PSI-BLAST (126) and JackHMMER (127) searches against the nonredundant (nr) database using an expected (e)-value threshold of 0.01. New sequences detected in these searches were subject to reciprocal BLAST to confirm their inclusion in the superfamily. Profile–profile searches performed with the HHpred program (128) were also used to detect remote homologs, confirm membership of divergent members within the superfamily, and identify higher-order relationships between lineages in the PNP-peptidyl/amidohydrolase fold, searching against the PFAM (129) and PDB (125) databases. Structure similarity searches were performed using the DALI server (130). Structures were rendered, compared, and superimposed in the molecular visualization program PyMOL. Multiple sequence alignments were generated using the Kalign (131) and Muscle (132) programs with default parameters. These multiple sequence alignments were adjusted manually, guided by structure superimpositions, profile–profile alignments, and secondary structure prediction. The JPred (133) program was used to predict secondary structures.
An approximate maximum-likelihood method as implemented in the FastTree (134) program with default parameters was used to assess intra-family phylogenetic relationships. The FigTree program (http://tree.bio.ed.ac.uk/software/figtree/)6 was used to render phylogenetic trees. Co-occurring domain architectures and gene neighborhoods were retrieved through custom PERL scripts from the NCBI Genome database.
Clustering of protein sequences and the subsequent assignment of sequences to distinct families was performed through two methods. In the first method, the sequence similarity networks of PCAD–Memo domains were constructed using Pythoscape (135). Domains that are fused to PCAD–Memo domains were removed, because the added length artificially decreases e-values of multidomain proteins, making the single-domain proteins appear more divergent than multidomain proteins. Including all domains when calculating the network also introduced a spurious connection between the MEMO family and the PCAD AMMECR1 fusion family from the PCAD clade, because they share a common domain. In some cases, families identified by BLASTCLUST (below) dissociated into multiple clusters in the sequence similarity network, because sequence divergence at domain boundaries made it difficult to cleanly excise the additional domains, or because variation in sequence divergence within and between families prevented identification of a single BLAST e-value cutoff that recapitulated the BLASTCLUST family assignments, without either fragmenting some families or fusing others. After removing additional domains, the data set was filtered to 50% sequence identity. Representatives that share <50% identity were used to construct the networks by pairwise BLAST using an e-value cutoff of 10−10 or 10−30. The networks were visualized in Cytoscape using the Organic layout (136). In the second method, families were defined using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html), adjusting the length of aligned regions and bit-score density threshold empirically. Divergent sequences or small clusters of sequences typically belonging to the same phylogeny were added to a family if other lines of evidence, including shared sequence motifs, shared structural synapomorphies, reciprocal BLAST search results, and/or shared genome context associations supporting inclusion (Table 1 and supporting data). Divergent sequences for which no such evidence was observed were left without family assignment. The results from the two techniques displayed good concordance, both recovering the primary PCAD–Memo clade split and identifying roughly the same total number of families in the superfamily.
Author contributions
A. M. B., E. A. T., and L. A. conceptualization; A. M. B., E. A. T., and L. A. data curation; A. M. B., M. E. G., and L. A. formal analysis; A. M. B. and M. E. G. investigation; A. M. B. writing-original draft; A. M. B., M. E. G., K. P. B., E. A. T., and L. A. writing-review and editing; M. E. G., K. P. B., and E. A. T. visualization; E. A. T. and L. A. supervision; E. A. T. and L. A. methodology; E. A. T. and L.A. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Patricia Babbitt and David Mischel (University of California, San Francisco) for assistance with constructing sequence similarity networks.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S2, Table S1, and supporting data.
A. M. Burroughs and L. Aravind, personal observations.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PCAD
- protocatechuate dioxygenase
- SAM
- S-adenosylmethionine
- PNP
- purine nucleoside phosphorylase
- PBD
- periplasmic-binding protein/chelatase fold domain
- PDB
- Protein Data Bank
- APD
- 2-aminophenol 1,6-dioxygenase
- LUCA
- Last Universal Common Ancestor
- TM
- transmembrane
- l-DOPA
- 3,4-dihydroxy-l-phenylalanine
- HGT
- horizontal gene transfer
- PCA
- protocatechuate
- TAT
- twin-arginine translocation
- 2,3-PCD
- protocatechuate 2,3-dioxygenase
- PGA
- poly-γ-glutamate
- wHTH
- winged helix–turn–helix
- PQQ
- pyrroloquinoline quinone
- IPP
- isopentenyl diphosphate
- PBP
- periplasmic-binding protein.
References
- 1. Vaillancourt F. H., Bolin J. T., and Eltis L. D. (2006) The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 41, 241–267 10.1080/10409230600817422 [DOI] [PubMed] [Google Scholar]
- 2. Machonkin T. E., and Doerner A. E. (2011) Substrate specificity of Sphingobium chlorophenolicum 2,6-dichlorohydroquinone 1,2-dioxygenase. Biochemistry 50, 8899–8913 10.1021/bi200855m [DOI] [PubMed] [Google Scholar]
- 3. Fetzner S. (2012) Ring-cleaving dioxygenases with a cupin fold. Appl. Environ. Microbiol. 78, 2505–2514 10.1128/AEM.07651-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lipscomb J. D. (2008) Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr. Opin. Struct. Biol. 18, 644–649 10.1016/j.sbi.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He P., and Moran G. R. (2011) Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J. Inorg. Biochem. 105, 1259–1272 10.1016/j.jinorgbio.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 6. Murzin A. G., Brenner S. E., Hubbard T., and Chothia C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 10.1006/jmbi.1995.0159 [DOI] [PubMed] [Google Scholar]
- 7. Qiu C., Lienhard S., Hynes N. E., Badache A., and Leahy D. J. (2008) Memo is homologous to nonheme iron dioxygenases and binds an ErbB2-derived phosphopeptide in its vestigial active site. J. Biol. Chem. 283, 2734–2740 10.1074/jbc.M703523200 [DOI] [PubMed] [Google Scholar]
- 8. Marone R., Hess D., Dankort D., Muller W. J., Hynes N. E., and Badache A. (2004) Memo mediates ErbB2-driven cell motility. Nat. Cell Biol. 6, 515–522 10.1038/ncb1134 [DOI] [PubMed] [Google Scholar]
- 9. Meira M., Masson R., Stagljar I., Lienhard S., Maurer F., Boulay A., and Hynes N. E. (2009) Memo is a cofilin-interacting protein that influences PLCγ1 and cofilin activities, and is essential for maintaining directionality during ErbB2-induced tumor-cell migration. J. Cell Sci. 122, 787–797 10.1242/jcs.032094 [DOI] [PubMed] [Google Scholar]
- 10. Zaoui K., Benseddik K., Daou P., Salaün D., and Badache A. (2010) ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc. Natl. Acad. Sci. U.S.A. 107, 18517–18522 10.1073/pnas.1000975107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benseddik K., Sen Nkwe N., Daou P., Verdier-Pinard P., and Badache A. (2013) ErbB2-dependent chemotaxis requires microtubule capture and stabilization coordinated by distinct signaling pathways. PLoS ONE 8, e55211 10.1371/journal.pone.0055211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kondo S., Bottos A., Allegood J. C., Masson R., Maurer F. G., Genoud C., Kaeser P., Huwiler A., Murakami M., Spiegel S., and Hynes N. E. (2014) Memo has a novel role in S1P signaling and is [corrected] crucial for vascular development. PLoS ONE 9, e94114 10.1371/journal.pone.0094114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacDonald G., Nalvarte I., Smirnova T., Vecchi M., Aceto N., Dolemeyer A., Frei A., Lienhard S., Wyckoff J., Hess D., Seebacher J., Keusch J. J., Gut H., Salaun D., Mazzarol G., et al. (2014) Memo is a copper-dependent redox protein with an essential role in migration and metastasis. Sci. Signal. 7, ra56 10.1126/scisignal.2004870 [DOI] [PubMed] [Google Scholar]
- 14. Ewald C. Y., Hourihan J. M., Bland M. S., Obieglo C., Katic I., Moronetti Mazzeo L. E., Alcedo J., Blackwell T. K., and Hynes N. E. (2017) NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. Elife 6, e19493 10.7554/eLife.19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balaji S., and Aravind L. (2007) The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 35, 5658–5671 10.1093/nar/gkm558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burroughs A. M., and Aravind L. (2014) A highly conserved family of domains related to the DNA-glycosylase fold helps predict multiple novel pathways for RNA modifications. RNA Biol. 11, 360–372 10.4161/rna.28302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noma A., Kirino Y., Ikeuchi Y., and Suzuki T. (2006) Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154 10.1038/sj.emboj.7601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J., Xiao H., Bonanno J. B., Kalyanaraman C., Brown S., Tang X., Al-Obaidi N. F., Patskovsky Y., Babbitt P. C., Jacobson M. P., Lee Y. S., and Almo S. C. (2013) Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 498, 123–126 10.1038/nature12180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pornsuwan S., Maenpuen S., Kamutira P., Watthaisong P., Thotsaporn K., Tongsook C., Juttulapa M., Nijvipakul S., and Chaiyen P. (2017) 3,4-Dihydroxyphenylacetate 2,3-dioxygenase from Pseudomonas aeruginosa: an Fe(II)-containing enzyme with fast turnover. PLoS ONE 12, e0171135 10.1371/journal.pone.0171135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Masai E., Katayama Y., and Fukuda M. (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71, 1–15 10.1271/bbb.60437 [DOI] [PubMed] [Google Scholar]
- 21. Kamimura N., and Masai E. (2014) in Biodegradative Bacteria: How Bacteria Degrade, Survive, Adapt, and Evolve (Nojiri H., Tsuda M., Fukuda M., and Kamagata Y., eds) pp. 207–226, Springer, Tokyo [Google Scholar]
- 22. Heger A., and Holm L. (2000) Towards a covering set of protein family profiles. Prog. Biophys. Mol. Biol. 73, 321–337 10.1016/S0079-6107(00)00013-4 [DOI] [PubMed] [Google Scholar]
- 23. Barry K. P., Ngu A., Cohn E. F., Cote J. M., Burroughs A. M., Gerbino J. P., and Taylor E. A. (2015) Exploring allosteric activation of LigAB from Sphingobium sp. strain SYK-6 through kinetics, mutagenesis and computational studies. Arch. Biochem. Biophys. 567, 35–45 10.1016/j.abb.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 24. Sugimoto K., Senda T., Aoshima H., Masai E., Fukuda M., and Mitsui Y. (1999) Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions. Structure 7, 953–965 10.1016/S0969-2126(99)80122-1 [DOI] [PubMed] [Google Scholar]
- 25. Sugimoto K., Senda M., Kasai D., Fukuda M., Masai E., and Senda T. (2014) Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6. PLoS ONE 9, e92249 10.1371/journal.pone.0092249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nishitani Y., Simons J. R., Kanai T., Atomi H., and Miki K. (2016) Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase. Acta Crystallogr. F Struct. Biol. Commun. 72, 427–433 10.1107/S2053230X16006920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang D., Iyer L. M., Burroughs A. M., and Aravind L. (2014) Resilience of biochemical activity in protein domains in the face of structural divergence. Curr. Opin. Struct. Biol. 26, 92–103 10.1016/j.sbi.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kasai D., Masai E., Miyauchi K., Katayama Y., and Fukuda M. (2005) Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187, 5067–5074 10.1128/JB.187.15.5067-5074.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nogales J., Canales A., Jiménez-Barbero J., García J. L., and Díaz E. (2005) Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J. Biol. Chem. 280, 35382–35390 10.1074/jbc.M502585200 [DOI] [PubMed] [Google Scholar]
- 30. Li de F., Zhang J. Y., Hou Y. J., Liu L., Hu Y., Liu S. J., Wang da C., and Liu W. (2013) Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor. Acta Crystallogr. D Biol. Crystallogr. 69, 32–43 10.1107/S0907444912042072 [DOI] [PubMed] [Google Scholar]
- 31. Gandía-Herrero F., and García-Carmona F. (2014) Escherichia coli protein YgiD produces the structural unit of plant pigments betalains: characterization of a prokaryotic enzyme with DOPA-extradiol-dioxygenase activity. Appl. Microbiol. Biotechnol. 98, 1165–1174 10.1007/s00253-013-4961-3 [DOI] [PubMed] [Google Scholar]
- 32. Gandía-Herrero F., and García-Carmona F. (2012) Characterization of recombinant Beta vulgaris 4,5-DOPA-extradiol-dioxygenase active in the biosynthesis of betalains. Planta 236, 91–100 10.1007/s00425-012-1593-2 [DOI] [PubMed] [Google Scholar]
- 33. Huynen M., Snel B., Lathe W. 3rd, Bork P. (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 10, 1204–1210 10.1101/gr.10.8.1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aravind L. (2000) Guilt by association: contextual information in genome analysis. Genome Res. 10, 1074–1077 10.1101/gr.10.8.1074 [DOI] [PubMed] [Google Scholar]
- 35. Barry K. P., and Taylor E. A. (2013) Characterizing the promiscuity of LigAB, a lignin catabolite degrading extradiol dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry 52, 6724–6736 10.1021/bi400665t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Providenti M. A., Mampel J., MacSween S., Cook A. M., and Wyndham R. C. (2001) Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology 147, 2157–2167 10.1099/00221287-147-8-2157 [DOI] [PubMed] [Google Scholar]
- 37. Maruyama K., Shibayama T., Ichikawa A., Sakou Y., Yamada S., and Sugisaki H. (2004) Cloning and characterization of the genes encoding enzymes for the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci. Biotechnol. Biochem. 68, 1434–1441 10.1271/bbb.68.1434 [DOI] [PubMed] [Google Scholar]
- 38. Schlosrich J., Eley K. L., Crowley P. J., and Bugg T. D. (2006) Directed evolution of a non-heme-iron-dependent extradiol catechol dioxygenase: identification of mutants with intradiol oxidative cleavage activity. Chembiochem 7, 1899–1908 10.1002/cbic.200600296 [DOI] [PubMed] [Google Scholar]
- 39. Iyer L. M., Abhiman S., Maxwell Burroughs A., and Aravind L. (2009) Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. Biosyst. 5, 1636–1660 10.1039/b917682a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burroughs A. M., Allen K. N., Dunaway-Mariano D., and Aravind L. (2006) Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361, 1003–1034 10.1016/j.jmb.2006.06.049 [DOI] [PubMed] [Google Scholar]
- 41. Spence E. L., Kawamukai M., Sanvoisin J., Braven H., and Bugg T. D. (1996) Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and biochemical properties of a third family of extradiol dioxygenases. J. Bacteriol. 178, 5249–5256 10.1128/jb.178.17.5249-5256.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mendel S., Arndt A., and Bugg T. D. (2004) Acid-base catalysis in the extradiol catechol dioxygenase reaction mechanism: site-directed mutagenesis of His-115 and His-179 in Escherichia coli 2,3-dihydroxyphenylpropionate 1,2-dioxygenase (MhpB). Biochemistry 43, 13390–13396 10.1021/bi048518t [DOI] [PubMed] [Google Scholar]
- 43. Barnes M. R., Duetz W. A., and Williams P. A. (1997) A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: cloning and characterization of the hpp operon. J. Bacteriol. 179, 6145–6153 10.1128/jb.179.19.6145-6153.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuatsjah E., Chen H. M., Withers S. G., and Eltis L. D. (2017) Characterization of an extradiol dioxygenase involved in the catabolism of lignin-derived biphenyl. FEBS Lett. 591, 1001–1009 10.1002/1873-3468.12611 [DOI] [PubMed] [Google Scholar]
- 45. Kasai D., Masai E., Miyauchi K., Katayama Y., and Fukuda M. (2004) Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 186, 4951–4959 10.1128/JB.186.15.4951-4959.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kasai D., Masai E., Katayama Y., and Fukuda M. (2007) Degradation of 3-O-methylgallate in Sphingomonas paucimobilis SYK-6 by pathways involving protocatechuate 4,5-dioxygenase. FEMS Microbiol. Lett. 274, 323–328 10.1111/j.1574-6968.2007.00855.x [DOI] [PubMed] [Google Scholar]
- 47. Peng X., Egashira T., Hanashiro K., Masai E., Nishikawa S., Katayama Y., Kimbara K., and Fukuda M. (1998) Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64, 2520–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sparnins V. L., and Chapman P. J. (1976) Catabolism of l-tyrosine by the homoprotocatechuate pathway in Gram-positive bacteria. J. Bacteriol. 127, 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arcos M., Olivera E. R., Arias S., Naharro G., and Luengo J. M. (2010) The 3,4-dihydroxyphenylacetic acid catabolon, a catabolic unit for degradation of biogenic amines tyramine and dopamine in Pseudomonas putida U. Environ. Microbiol. 12, 1684–1704 10.1111/j.1462-2920.2010.02233.x [DOI] [PubMed] [Google Scholar]
- 50. Roper D. I., and Cooper R. A. (1990) Subcloning and nucleotide sequence of the 3,4-dihydroxyphenylacetate (homoprotocatechuate) 2,3-dioxygenase gene from Escherichia coli C. FEBS Lett. 275, 53–57 10.1016/0014-5793(90)81437-S [DOI] [PubMed] [Google Scholar]
- 51. Sparnins V. L., Chapman P. J., and Dagley S. (1974) Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J. Bacteriol. 120, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kasai D., Fujinami T., Abe T., Mase K., Katayama Y., Fukuda M., and Masai E. (2009) Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 191, 6758–6768 10.1128/JB.00840-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wolgel S. A., Dege J. E., Perkins-Olson P. E., Jaurez-García C. H., Crawford R. L., Münck E., and Lipscomb J. D. (1993) Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J. Bacteriol. 175, 4414–4426 10.1128/jb.175.14.4414-4426.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu J. F., Jiang C. Y., Wang B. J., Ma Y. F., Liu Z. P., and Liu S. J. (2006) Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl. Environ. Microbiol. 72, 1759–1765 10.1128/AEM.72.3.1759-1765.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu J. F., Sun C. W., Jiang C. Y., Liu Z. P., and Liu S. J. (2005) A novel 2-aminophenol 1,6-dioxygenase involved in the degradation of p-chloronitrobenzene by Comamonas strain CNB-1: purification, properties, genetic cloning and expression in Escherichia coli. Arch. Microbiol. 183, 1–8 10.1007/s00203-004-0738-5 [DOI] [PubMed] [Google Scholar]
- 56. Zhen D., Liu H., Wang S. J., Zhang J. J., Zhao F., and Zhou N. Y. (2006) Plasmid-mediated degradation of 4-chloronitrobenzene by newly isolated Pseudomonas putida strain ZWL73. Appl. Microbiol. Biotechnol. 72, 797–803 10.1007/s00253-006-0345-2 [DOI] [PubMed] [Google Scholar]
- 57. Lendenmann U., and Spain J. C. (1996) 2-Aminophenol 1,6-dioxygenase: a novel aromatic ring cleavage enzyme purified from Pseudomonas pseudoalcaligenes JS45. J. Bacteriol. 178, 6227–6232 10.1128/jb.178.21.6227-6232.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marín M., Plumeier I., and Pieper D. H. (2012) Degradation of 2,3-dihydroxybenzoate by a novel meta-cleavage pathway. J. Bacteriol. 194, 3851–3860 10.1128/JB.00430-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christinet L., Burdet F. X., Zaiko M., Hinz U., and Zrÿd J. P. (2004) Characterization and functional identification of a novel plant 4,5-extradiol dioxygenase involved in betalain pigment biosynthesis in Portulaca grandiflora. Plant Physiol. 134, 265–274 10.1104/pp.103.031914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sasaki N., Abe Y., Goda Y., Adachi T., Kasahara K., and Ozeki Y. (2009) Detection of DOPA 4,5-dioxygenase (DOD) activity using recombinant protein prepared from Escherichia coli cells harboring cDNA encoding DOD from Mirabilis jalapa. Plant Cell Physiol. 50, 1012–1016 10.1093/pcp/pcp053 [DOI] [PubMed] [Google Scholar]
- 61. Hinz U. G., Fivaz J., Girod P. A., and Zyrd J. P. (1997) The gene coding for the DOPA dioxygenase involved in betalain biosynthesis in Amanita muscaria and its regulation. Mol. Gen. Genet. 256, 1–6 10.1007/s004380050539 [DOI] [PubMed] [Google Scholar]
- 62. Girod P.-A., and Zryd J.-P. (1991) Biogenesis of betalains: purification and partial characterization of dopa 4,5-dioxygenase from Amanita muscaria. Phytochemistry 30, 169–174 10.1016/0031-9422(91)84119-D [DOI] [Google Scholar]
- 63. Liu Z. J., Tempel W., Ng J. D., Lin D., Shah A. K., Chen L., Horanyi P. S., Habel J. E., Kataeva I. A., Xu H., Yang H., Chang J. C., Huang L., Chang S. H., Zhou W., et al. (2005) The high-throughput protein-to-structure pipeline at SECSG. Acta Crystallogr. D Biol. Crystallogr. 61, 679–684 10.1107/S0907444905013132 [DOI] [PubMed] [Google Scholar]
- 64. Vitelli F., Piccini M., Caroli F., Franco B., Malandrini A., Pober B., Jonsson J., Sorrentino V., and Renieri A. (1999) Identification and characterization of a highly conserved protein absent in the Alport syndrome (A), mental retardation (M), midface hypoplasia (M), and elliptocytosis (E) contiguous gene deletion syndrome (AMME). Genomics 55, 335–340 10.1006/geno.1998.5666 [DOI] [PubMed] [Google Scholar]
- 65. Ogris C., Guala D., and Sonnhammer E. L. L. (2018) FunCoup 4: new species, data, and visualization. Nucleic Acids Res. 46, D601–D607 10.1093/nar/gkx1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sharples G. J., and Leach D. R. (1995) Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17, 1215–1217 10.1111/j.1365-2958.1995.mmi_17061215_1.x [DOI] [PubMed] [Google Scholar]
- 67. Aravind L., and Koonin E. V. (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26, 3746–3752 10.1093/nar/26.16.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Candela T., and Fouet A. (2006) Poly-γ-glutamate in bacteria. Mol. Microbiol. 60, 1091–1098 10.1111/j.1365-2958.2006.05179.x [DOI] [PubMed] [Google Scholar]
- 69. Ashiuchi M., Nawa C., Kamei T., Song J. J., Hong S. P., Sung M. H., Soda K., and Misono H. (2001) Physiological and biochemical characteristics of poly-γ-glutamate synthetase complex of Bacillus subtilis. Eur. J. Biochem. 268, 5321–5328 10.1046/j.0014-2956.2001.02475.x [DOI] [PubMed] [Google Scholar]
- 70. Ashiuchi M., and Misono H. (2002) Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl. Microbiol. Biotechnol. 59, 9–14 10.1007/s00253-002-0984-x [DOI] [PubMed] [Google Scholar]
- 71. Gasc C., Ribière C., Parisot N., Beugnot R., Defois C., Petit-Biderre C., Boucher D., Peyretaillade E., and Peyret P. (2015) Capturing prokaryotic dark matter genomes. Res. Microbiol. 166, 814–830 10.1016/j.resmic.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 72. Burkhart B. J., Hudson G. A., Dunbar K. L., and Mitchell D. A. (2015) A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 10.1038/nchembio.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Burroughs A. M., Iyer L. M., and Aravind L. (2009) Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins 75, 895–910 10.1002/prot.22298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Latham J. A., Iavarone A. T., Barr I., Juthani P. V., and Klinman J. P. (2015) PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. J. Biol. Chem. 290, 12908–12918 10.1074/jbc.M115.646521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tsai T. Y., Yang C. Y., Shih H. L., Wang A. H., and Chou S. H. (2009) Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins 76, 1042–1048 10.1002/prot.22461 [DOI] [PubMed] [Google Scholar]
- 76. Shen Y. Q., Bonnot F., Imsand E. M., RoseFigura J. M., Sjölander K., and Klinman J. P. (2012) Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51, 2265–2275 10.1021/bi201763d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Puehringer S., Metlitzky M., and Schwarzenbacher R. (2008) The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach. BMC Biochem. 9, 8 10.1186/1471-2091-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schweizer U., Bohleber S., and Fradejas-Villar N. (2017) The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 14, 1197–1208 10.1080/15476286.2017.1294309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. del Sol A., Fujihashi H., Amoros D., and Nussinov R. (2006) Residue centrality, functionally important residues, and active site shape: analysis of enzyme and non-enzyme families. Protein Sci. 15, 2120–2128 10.1110/ps.062249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Basu A., and Yap M. N. (2017) Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc. Natl. Acad. Sci. U.S.A. 114, E8165–E8173 10.1073/pnas.1709588114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Samanovic M. I., Tu S., Novák O., Iyer L. M., McAllister F. E., Aravind L., Gygi S. P., Hubbard S. R., Strnad M., and Darwin K. H. (2015) Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol. Cell 57, 984–994 10.1016/j.molcel.2015.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Burroughs A. M., Zhang D., Schäffer D. E., Iyer L. M., and Aravind L. (2015) Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654 10.1093/nar/gkv1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Anantharaman V., Aravind L., and Koonin E. V. (2003) Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 7, 12–20 10.1016/S1367-5931(02)00018-2 [DOI] [PubMed] [Google Scholar]
- 84. Wattiau P., Bastiaens L., van Herwijnen R., Daal L., Parsons J. R., Renard M. E., Springael D., and Cornelis G. R. (2001) Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152, 861–872 10.1016/S0923-2508(01)01269-4 [DOI] [PubMed] [Google Scholar]
- 85. Eaton R. W. (2001) Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183, 3689–3703 10.1128/JB.183.12.3689-3703.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sato S. I., Ouchiyama N., Kimura T., Nojiri H., Yamane H., and Omori T. (1997) Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J. Bacteriol. 179, 4841–4849 10.1128/jb.179.15.4841-4849.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Laurie A. D., and Lloyd-Jones G. (1999) The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Demanèche S., Meyer C., Micoud J., Louwagie M., Willison J. C., and Jouanneau Y. (2004) Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70, 6714–6725 10.1128/AEM.70.11.6714-6725.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tack B. F., Chapman P. J., and Dagley S. (1972) Metabolism of gallic acid and syringic acid by Pseudomonas putida. J. Biol. Chem. 247, 6438–6443 [PubMed] [Google Scholar]
- 90. Kulakov L. A., Delcroix V. A., Larkin M. J., Ksenzenko V. N., and Kulakova A. N. (1998) Cloning of new Rhodococcus extradiol dioxygenase genes and study of their distribution in different Rhodococcus strains. Microbiology 144, 955–963 10.1099/00221287-144-4-955 [DOI] [PubMed] [Google Scholar]
- 91. Vetting M. W., and Ohlendorf D. H. (2000) The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 8, 429–440 10.1016/S0969-2126(00)00122-2 [DOI] [PubMed] [Google Scholar]
- 92. Gandía-Herrero F., and García-Carmona F. (2013) Biosynthesis of betalains: yellow and violet plant pigments. Trends Plant Sci. 18, 334–343 10.1016/j.tplants.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 93. Khan M. I., and Giridhar P. (2015) Plant betalains: chemistry and biochemistry. Phytochemistry 117, 267–295 10.1016/j.phytochem.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 94. Burroughs A. M., Zhang D., and Aravind L. (2015) The eukaryotic translation initiation regulator CDC123 defines a divergent clade of ATP-grasp enzymes with a predicted role in novel protein modifications. Biol. Direct. 10, 21 10.1186/s13062-015-0053-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Galperin M. Y., and Koonin E. V. (1997) A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6, 2639–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Aravind L., Anantharaman V., Balaji S., Babu M. M., and Iyer L. M. (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262 10.1016/j.femsre.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 97. Tam R., and Saier M. H. Jr. (1993) Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57, 320–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Purschke W. G., Schmidt C. L., Petersen A., and Schäfer G. (1997) The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179, 1344–1353 10.1128/jb.179.4.1344-1353.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Müller F. H., Bandeiras T. M., Urich T., Teixeira M., Gomes C. M., and Kletzin A. (2004) Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160 10.1111/j.1365-2958.2004.04193.x [DOI] [PubMed] [Google Scholar]
- 100. Fischer F., Künne S., and Fetzner S. (1999) Bacterial 2,4-dioxygenases: new members of the α/β hydrolase-fold superfamily of enzymes functionally related to serine hydrolases. J. Bacteriol. 181, 5725–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Søballe B., and Poole R. K. (1999) Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145, 1817–1830 10.1099/13500872-145-8-1817 [DOI] [PubMed] [Google Scholar]
- 102. Grochowski L. L., Xu H., and White R. H. (2006) Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188, 3192–3198 10.1128/JB.188.9.3192-3198.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Matsumi R., Atomi H., Driessen A. J., and van der Oost J. (2011) Isoprenoid biosynthesis in Archaea–biochemical and evolutionary implications. Res. Microbiol. 162, 39–52 10.1016/j.resmic.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 104. Dellas N., Thomas S. T., Manning G., and Noel J. P. (2013) Discovery of a metabolic alternative to the classical mevalonate pathway. Elife 2, e00672 10.7554/eLife.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vannice J. C., Skaff D. A., Keightley A., Addo J. K., Wyckoff G. J., and Miziorko H. M. (2014) Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J. Bacteriol. 196, 1055–1063 10.1128/JB.01230-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vinokur J. M., Korman T. P., Sawaya M. R., Collazo M., Cascio D., and Bowie J. U. (2015) Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases. Protein Sci. 24, 212–220 10.1002/pro.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Budd A., Blandin S., Levashina E. A., and Gibson T. J. (2004) Bacterial α2-macroglobulins: colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 5, R38 10.1186/gb-2004-5-6-r38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Booker S. J., and Grove T. L. (2010) Mechanistic and functional versatility of radical SAM enzymes. F1000 Biol. Rep. 2, 52 10.3410/B2-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Buis J. M., and Broderick J. B. (2005) Pyruvate formate-lyase activating enzyme: elucidation of a novel mechanism for glycyl radical formation. Arch. Biochem. Biophys. 433, 288–296 10.1016/j.abb.2004.09.028 [DOI] [PubMed] [Google Scholar]
- 110. Shen P. S., Park J., Qin Y., Li X., Parsawar K., Larson M. H., Cox J., Cheng Y., Lambowitz A. M., Weissman J. S., Brandman O., and Frost A. (2015) Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 10.1126/science.1259724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kostova K. K., Hickey K. L., Osuna B. A., Hussmann J. A., Frost A., Weinberg D. E., and Weissman J. S. (2017) CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 10.1126/science.aam7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Anantharaman V., Koonin E. V., and Aravind L. (2002) SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4, 71–75 [PubMed] [Google Scholar]
- 113. Hori H. (2017) Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules 7, E23 10.3390/biom7010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 10.1038/nchembio834 [DOI] [PubMed] [Google Scholar]
- 115. Winkelblech J., Fan A., and Li S. M. (2015) Prenyltransferases as key enzymes in primary and secondary metabolism. Appl. Microbiol. Biotechnol. 99, 7379–7397 10.1007/s00253-015-6811-y [DOI] [PubMed] [Google Scholar]
- 116. Soderberg T., Chen A., and Poulter C. D. (2001) Geranylgeranylglyceryl phosphate synthase. Characterization of the recombinant enzyme from Methanobacterium thermoautotrophicum. Biochemistry 40, 14847–14854 10.1021/bi0111799 [DOI] [PubMed] [Google Scholar]
- 117. Nemoto N., Oshima T., and Yamagishi A. (2003) Purification and characterization of geranylgeranylglyceryl phosphate synthase from a thermoacidophilic archaeon, Thermoplasma acidophilum. J. Biochem. 133, 651–657 10.1093/jb/mvg083 [DOI] [PubMed] [Google Scholar]
- 118. Dumelin C. E., Chen Y., Leconte A. M., Chen Y. G., and Liu D. R. (2012) Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 8, 913–919 10.1038/nchembio.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang R., Ranganathan S. V., Basanta-Sanchez M., Shen F., Chen A., and Sheng J. (2015) Synthesis and base pairing studies of geranylated 2-thiothymidine, a natural variant of thymidine. Chem. Commun. 51, 16369–16372 10.1039/C5CC07479G [DOI] [PubMed] [Google Scholar]
- 120. Bartos P., Maciaszek A., Rosinska A., Sochacka E., and Nawrot B. (2014) Transformation of a wobble 2-thiouridine to 2-selenouridine via S-geranyl-2-thiouridine as a possible cellular pathway. Bioorg. Chem. 56, 49–53 10.1016/j.bioorg.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 121. Sierant M., Leszczynska G., Sadowska K., Komar P., Radzikowska-Cieciura E., Sochacka E., and Nawrot B. (2018) Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 592, 2248–2258 10.1002/1873-3468.13124 [DOI] [PubMed] [Google Scholar]
- 122. Martone P. T., Estevez J. M., Lu F., Ruel K., Denny M. W., Somerville C., and Ralph J. (2009) Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr. Biol. 19, 169–175 10.1016/j.cub.2008.12.031 [DOI] [PubMed] [Google Scholar]
- 123. Iyer L. M., Abhiman S., de Souza R. F., and Aravind L. (2010) Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 38, 5261–5279 10.1093/nar/gkq265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K. D., and Sayers E. W. (2018) GenBank. Nucleic Acids Res. 46, D41–D47 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., and Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 10.1093/nar/gkv397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Søding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G. A., Tate J., and Bateman A. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Holm L., and Sander C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 10.1016/S0968-0004(00)89105-7 [DOI] [PubMed] [Google Scholar]
- 131. Lassmann T., and Sonnhammer E. L. (2005) Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298 10.1186/1471-2105-6-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Drozdetskiy A., Cole C., Procter J., and Barton G. J. (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Price M. N., Dehal P. S., and Arkin A. P. (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Barber A. E. 2nd, and Babbitt P. C. (2012) Pythoscape: a framework for generation of large protein similarity networks. Bioinformatics 28, 2845–2846 10.1093/bioinformatics/bts532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.