Abstract
Similar to other prokaryotes, mycobacteria decorate their major cell envelope glycans with minor covalent substituents whose biological significance remains largely unknown. We report on the discovery of a mycobacterial enzyme, named here SucT, that adds succinyl groups to the arabinan domains of both arabinogalactan (AG) and lipoarabinomannan (LAM). Disruption of the SucT-encoding gene in Mycobacterium smegmatis abolished AG and LAM succinylation and altered the hydrophobicity and rigidity of the cell envelope of the bacilli without significantly altering AG and LAM biosynthesis. The changes in the cell surface properties of the mutant were consistent with earlier reports of transposon mutants of the closely related species Mycobacterium marinum and Mycobacterium avium harboring insertions in the orthologous gene whose ability to microaggregate and form biofilms were altered. Our findings point to an important role of SucT-mediated AG and LAM succinylation in modulating the cell surface properties of mycobacteria.
Keywords: mycobacteria, Mycobacterium smegmatis, polysaccharide, tuberculosis, cell surface, arabinogalactan, lipoarabinomannan, succinylation
Introduction
Covalent modification of cell envelope glycans with strategically placed discrete substituents such as various sugars, amino acids, phosphates, or acyl groups is a common strategy used by prokaryotes to adapt and survive under various stress conditions, including the environment of the infected host. Although not required for growth per se, these tailoring modifications may affect the biosynthesis, export, and/or biological activities of polysaccharides, thereby impacting the cell envelope composition and physical properties of the bacteria, their resistance to antimicrobials, and their interactions with host immune responses (1–9). A classic example of how such modifications may thwart host defenses is masking by Gram-negative and Gram-positive bacteria of the negative charges conferred by the phosphate groups of lipopolysaccharide, wall teichoic acids, and lipoteichoic acids with positively charged amines under the form of amino sugars, phosphoethanolamine, or D-alanyl esters to evade killing by positively charged antimicrobial peptides. Although devoid of the canonical lipopolysaccharide and (lipo)teichoic acids, mycobacteria produce polysaccharides and lipopolysaccharides with unique structures that play critical roles in their cell envelope integrity and pathogenicity (10–11). The finding of covalent substituents modifying the structures of two of the dominant heteropolysaccharides produced by all Mycobacterium species, arabinogalactan (AG)2 and lipoarabinomannan (LAM), suggests that mycobacteria have evolved similar strategies as other prokaryotes to promote their survival in different environments (12). To date, however, our understanding of the biological significance of these discrete substituents is, at best, rudimentary.
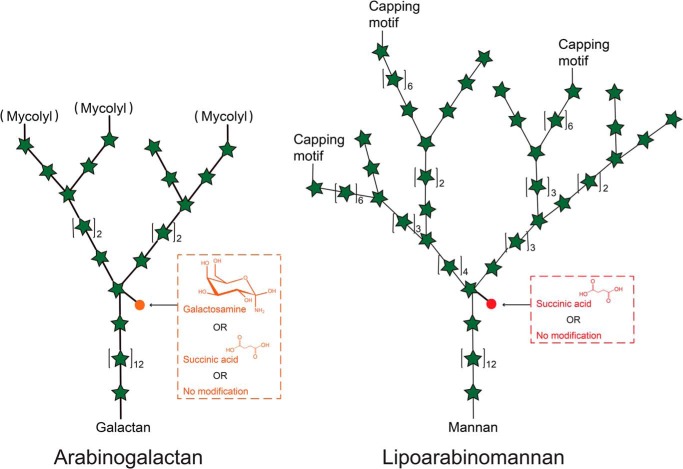
The arabinan domain of LAM is very similar to that of AG and made of stretches of α-1,5–linked arabinofuranosyl (Araf) residues with precisely positioned α-3,5 branch sites (Fig. 1). In Mycobacterium tuberculosis and a number of other slow- and fast-growing pathogenic mycobacteria (Mycobacterium avium, Mycobacterium kansasii, Mycobacterium bovis, Mycobacterium leprae, and Mycobacterium abscessus), the C2 position of a portion of the internal α-3,5–branched Araf residues of the arabinan domain of AG may be modified with galactosamine substituents, a motif not found in LAM (13) (Fig. 1). Studies from our laboratory have shed light on the biosynthetic origin of this substituent (13) and begun to elucidate its contribution to modulation of the host immune response (14). Interestingly, succinate groups were found to substitute the same positions of the arabinan domain as well as quantitatively minor α-1,5-Araf positions of AG from M. tuberculosis and Mycobacterium smegmatis (15), the internal α-3,5–branched Araf residues of the arabinan domain of LAM from M. bovis Bacillus Calmette-Guerin (BCG) (16), and possibly the same positions of LAM from M. leprae and M. tuberculosis (17–21). The presence of succinates has also been reported on the capsular polysaccharide d-arabino-d-mannan (AM) of M. tuberculosis, which shares with LAM a structurally identical arabinomannan domain (22). Succinates were finally found to substitute the C3 position of linear α-1,5–Araf residues of LAM in M. kansasii (23). Collectively, these observations are suggestive of the widespread distribution of succinate motifs in the AG and LAM of mycobacteria, even though their precise position on these glycans may be variable from species to species. Succinyl substituents were estimated to occur at the level of about one to three motifs per arabinan chain in AG, one to six motifs per LAM molecule, and two to three motifs per AM molecule (15–17, 22). Neither the enzyme(s) catalyzing the introduction of these motifs in the arabinan domain of AG, AM, and LAM nor the precise biological significance of polysaccharide succinylation in mycobacteria are currently known, even though an association was suggested between the charge of various LAM isoforms (in part driven by succinate content) and their ability to stimulate CD1b-restricted T cells (19, 21). It has further been proposed that the negatively charged succinyl residues interact with the protonated galactosamine substituents, leading to a more rigid and tightened AG structure (15). Assuming that succinates are introduced during elongation of the arabinan domains of AG and LAM, these motifs could also serve as molecular signatures regulating elongation and branching (12). Alternatively, the apparent lack of succinylation on the mycolylated arabinan chains of AG (Fig. 1) has led to the hypothesis that succinylation negatively controls mycolylation (15).
Figure 1.
Detail of the chemical modifications affecting the internal arabinan domains of AG and LAM in M. tuberculosis. Note that the succinylation of the mycolylated chains in arabinogalactan has been reported to be diminished or absent (15).
Here we report the identification of a conserved mycobacterial acyltransferase, hereafter referred to as SucT, responsible for the succinylation of AG and LAM and the significant impact of disruption of sucT on the physiology of M. smegmatis cells. The possibility of generating mycobacterial mutants deficient in succinylation of cell envelope polysaccharides paves the way for studies aiming to determine the contribution of this discrete substituent to the physiology of slow- and fast-growing mycobacteria, their adaptation to the environment, and immunopathogenesis.
Results
Identification of a candidate enzyme for succinylation of AG and LAM
A bioinformatics search for protein candidates with potential for catalyzing the succinylation of the arabinan domains of AG and LAM identified five integral membrane proteins (Rv0111, Rv0228, Rv0517, Rv1254, and Rv1565c) in M. tuberculosis and nine homologous proteins in M. smegmatis (MSMEG_0206, MSMEG_0319, MSMEG_0390, MSMEG_2021, MSMEG_3187, MSMEG_3490, MSMEG_5041, MSMEG_5537, and MSMEG_6230) showing sequence similarities with a family of trans-acylases (COG1835) involved in O-acylation of exported carbohydrate moieties in a variety of prokaryotes. Enzymes from this family include a number of O antigen, exopolysaccharide, and Nod factor acetyltransferases as well as macrolide acyltransferases (24). In line with the prototypical primary and secondary structures of these enzymes, all five M. tuberculosis candidate acyltransferases harbor nine to ten transmembrane segments and display conserved cytoplasmic and transmembrane amino acid residues reported to be critical for activity (24–26) (Fig. S1). All M. tuberculosis candidates are conserved in mycobacteria, with the exception of Rv0517, which is partially or completely deleted in some M. tuberculosis clinical isolates and is apparently missing from M. leprae. While TmaT (Rv0228 in M. tuberculosis and MSMEG_0319 in M. smegmatis) was recently shown to acetylate mycolic acids (27–28), other candidates have not yet been functionally characterized.
Given the presence of succinate motifs on the AG and LAM of M. smegmatis (15, 17, 20), individual KOs of each conserved candidate gene with clear orthologs in M. tuberculosis were generated by allelic replacement in this nonpathogenic, fast-growing model species (Fig. S2A), and their lipoglycans were analyzed by SDS-PAGE. As shown in Fig. 2 and Fig. S2B, the MSMEG_3187 mutant (orthologous to Rv1565c of M. tuberculosis) was the only one to present a lipoglycan profile significantly different from that of the WT parent strain. Although its lipomannan (LM) migrated similarly to that of the WT strain, its LAM migrated higher, suggestive of alterations in either size or charge (Fig. 2). Complementation of the MSMEG_3187 KO with a replicative plasmid expressing a WT copy of the MSMEG_3187 gene expressed from the hsp60 promoter reversed the slow migration of LAM on SDS-PAGE beyond WT levels. MSMEG_3187 shares 23% identity (40% similarity) on a 705-amino acid overlap with the O antigen acetyltransferase OafA from Salmonella (24). We renamed this protein SucT for the purpose of this study.
Figure 2.
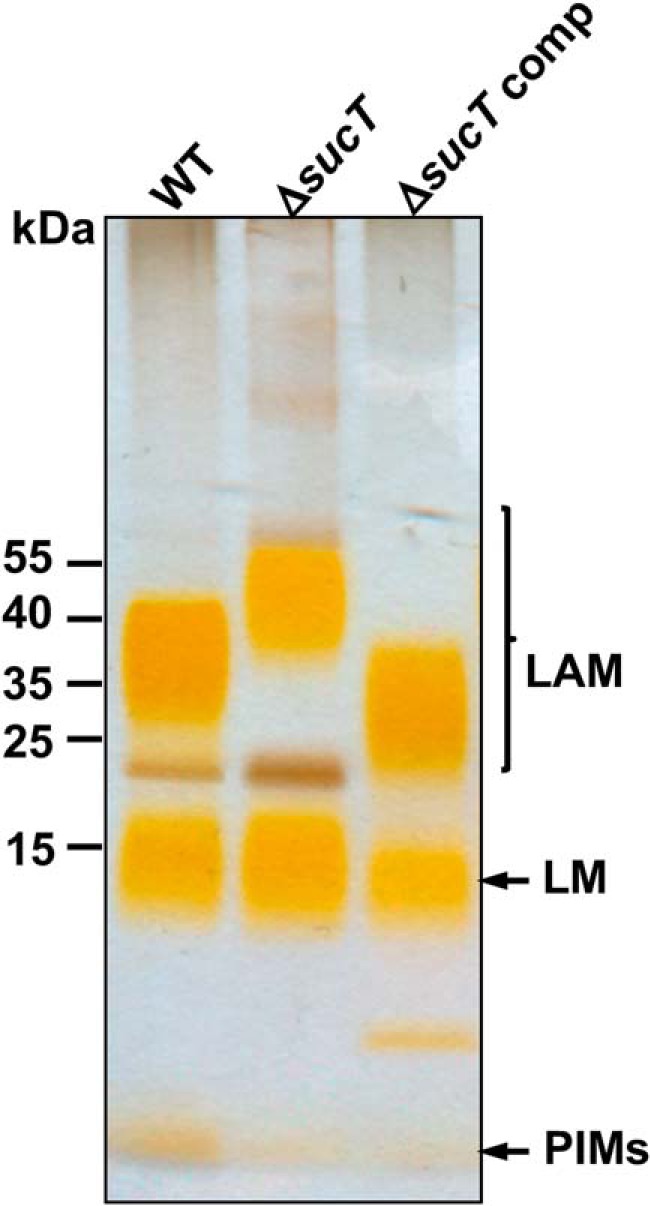
Electrophoretic mobility of lipoglycans from WT M. smegmatis mc2155, the sucT mutant, and the complemented sucT mutant. Lipoglycans extracted from WT mc2155, mc2ΔsucT, and mc2ΔsucT/pMVGH1-sucT (ΔsucT comp) were run on a 10–20% Tricine gel, followed by periodic acid–silver staining. PIMs, phosphatidylinositol mannosides.
Disruption of sucT results in loss of succinyl substituents on the LAM of M. smegmatis
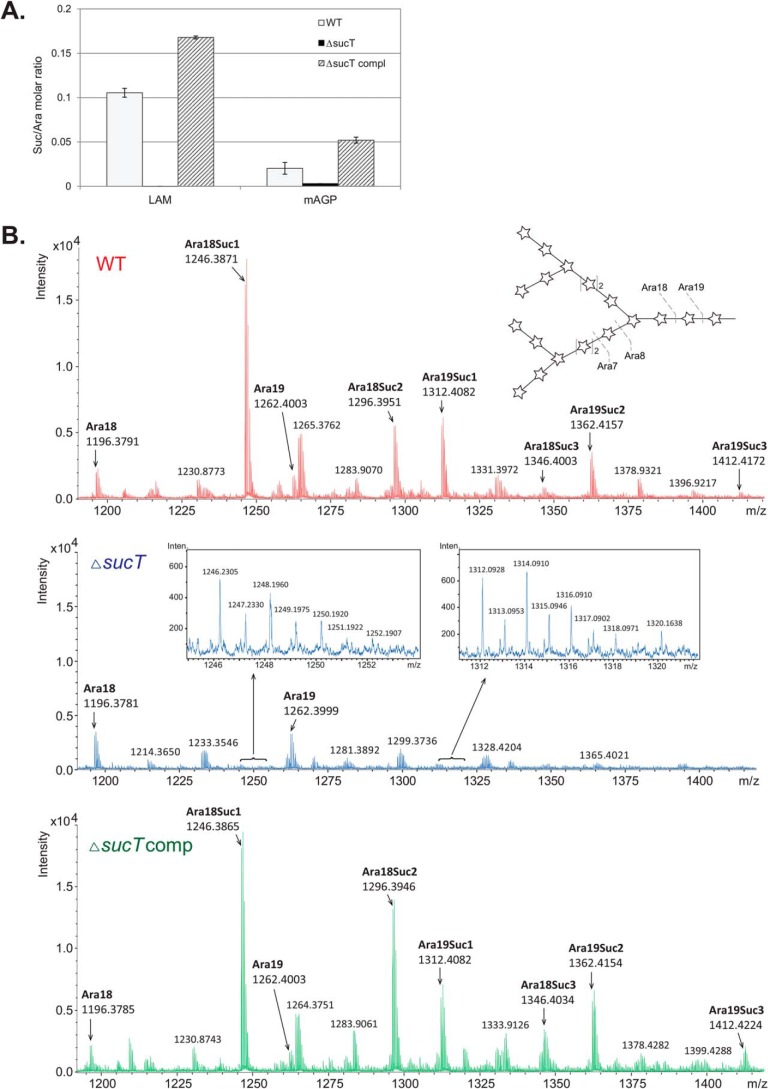
To determine whether a defect in succinylation and, thus, overall charge of the sucT mutant LAM might be responsible for its migration shift on SDS-PAGE, LAM was purified from the WT, sucT KO (MsmgΔsucT), and complemented mutant strain (MsmgΔsucT/pMVGH1-sucT) and submitted to a panel of analyses. Direct analysis of succinyl groups was performed by butanolysis of LAM prepared from the WT, mutant, and complemented mutant strains to yield dibutyl esters of any succinyl groups present (Supporting Methods). GC/MS analysis revealed a complete absence of succinates in the mutant LAM. LAM succinylation was restored beyond WT levels in the complemented mutant. Quantitation of the ratio of succinyl to arabinosyl groups showed a ratio of 1:9.5 (S.D. ± 0.4 for two determinations) for the WT strain and 1:6 (S.D. ± 0.1 for three determinations) following ectopic overexpression of MSMEG_3187 in MsmgΔsucT/pMVGH1-sucT (Fig. 3A).
Figure 3.
Succinate content of AG and LAM prepared from WT M. smegmatis mc2155, the ΔsucT mutant, and the complemented mutant strain. A, the amounts of succinates and arabinose residues in the same LAM and mAGP samples prepared from the WT, mutant, and complemented mutant (ΔsucT compl) strains were quantified as described in the Supporting Methods. Results are expressed as average ± S.D. succinate/arabinose molar ratios from three technical replicates. B, endogenous endoarabinanase digestion of mAGP from the WT, mutant, and complemented mutant strains. Analysis of the products of the reaction by QTOF LC/MS in the negative ion mode revealed characteristic [M-2H]−2 ions corresponding to oligoarabinosides with succinyl groups in the WT and complemented mutant that are not found in the sucT mutant mAGP.
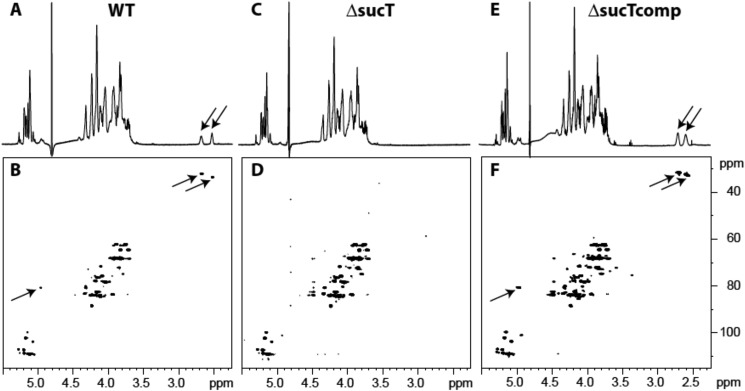
Next, LAM prepared from each of the three strains was analyzed for the presence of succinates by 1D and 2D NMR spectroscopy (16). The 1D 1H spectrum of LAM prepared from the WT strain showed the characteristic two pseudotriplets (J = 6.5 Hz) of similar intensities at 2.50 and 2.66 ppm (Fig. 4A) assigned to methylene groups of succinyl units. Their corresponding carbons were characterized at 34.7 and 33.3 ppm, respectively, on the 2D 1H-13C HMQC spectrum (Fig. 4B). The cross-peaks between 1H δ 4.96/13C δ 81.9 and 1H δ 4.94/13C δ 82.1 further typify the presence of succinyl residues on the C2 of α-3,5–branched Araf residues and, possibly, some adjacent α-1,5–Araf residues (16). These signals were absent from the 1D 1H and 2D HMQC spectra of the mutant LAM (Fig. 4, C and D) and restored upon genetic complementation Fig. 4, E and F). Collectively, our analyses support the conclusions that succinates substitute α-3,5–branched Araf residues of the arabinan domain of LAM in M. smegmatis, as reported previously in M. bovis BCG (16), and that the M. smegmatis sucT KO is devoid of any such substituents.
Figure 4.
NMR analysis of LAM prepared from the WT, mutant, and complemented mutant strains. Shown are 1D 1H (A, C, and E) and 2D 1H-13C (B, D, and F) HMQC NMR spectra. Arrows point to the signals typifying succinate detection and localization (see text for details).
Because of the report of succinates on the LM of an unspecified M. smegmatis strain (20), we next sought to verify whether the succinylation defect extended to the mutant's LM. Comparison of the WT and mutant LM by NMR spectroscopy, however, failed to identify any succinates on the LM of the M. smegmatis mc2155 parent strain used in this study (Fig. S3). Overall, no significant structural differences were noted between the LM prepared from MsmgΔsucT and the WT strain.
Interestingly, a screen for Mycobacterium marinum transposon mutants devoid of mannose caps on LAM yielded a strain harboring a transposon insertion in MMAR_2380, the ortholog of sucT in this species (75% identity/85% similarity at the protein level with MSMEG_3187) (29). Not only was this mutant confirmed to lack mannoside caps on LAM, it also displayed a higher degree of branching of both the mannan core and the arabinan domain compared with WT LAM and decreased acylation of the phosphatidylinositol anchor of both LM and LAM. In contrast to the M. marinum MMAR_2380 mutant, MsmgΔsucT failed to show any reduction in the incorporation of [1,2-14C]acetate into the acyl chains of LM and LAM (Fig. S4A). Likewise, quantitative analyses of the alditol acetate and per-O-methylated alditol acetate derivatives of the WT, mutant, and complemented mutant LAM did not point to any significant alterations in the Araf to Manp ratio of the mutant LAM (Table S1) or increase in the degree of branching of its mannan and arabinan domains (Table S2). The relative proportion of Ara4 to Ara6 arabinan termini of LAM was also similar in the WT and mutant strains (relative percentage of Ara2, Ara4, and Ara6 oligoarabinosides released upon Cellulomonas gelida endoarabinanase digestion: 58.7% ± 4.6%, 37.5% ± 5.2%, and 3.8% ± 0.7%, respectively, in the WT strain compared with 60.9% ± 3.4%, 34.3% ± 4.3%, and 4.8% ± 1.0% in the sucT mutant; average ± S.D. of three technical replicates). 31P NMR and LC/MS analysis of the oligoarabinosides released upon C. gelida endoarabinanase digestion of deacylated LAM further confirmed the presence of the expected phosphoinositol capping motifs of M. smegmatis LAM in the WT and mutant strains (Fig. S5, A and B). Metabolic labeling of WT and mutant cultures with [14C]glucose finally pointed to similar rates of de novo lipoglycan synthesis in both strains (Fig. S4B). Collectively, these results suggest that, besides abolishing succinylation, the disruption of sucT did not appreciably affect, either qualitatively or quantitatively, the biosynthesis of LAM in M. smegmatis.
Effect of inactivating sucT on the succinylation of M. smegmatis AG
Given the structural similarity of the arabinan domains of AG and LAM, we next sought to determine whether the disruption of sucT had any impact on the succinate content of AG from M. smegmatis. To this end, the mycolyl–AG–peptidoglycan (mAGP) complex prepared from the WT, mutant, and complemented mutant strains was subjected to the same butanolysis procedure as for LAM, and succinyl substituents as their dibutyl esters were analyzed by GC/MS. Compared with the WT parent, mAGP from the sucT KO showed an 84.7% decrease in succinate content. Quantitation of the ratio of succinyl to arabinosyl groups yielded a ratio of 1:52 (S.D. ± 14.3 for three determinations) for the WT strain, 1:350 (S.D. ± 20.9 for two determinations) for MsmgΔsucT, and 1:19 (S.D. ± 1.2 for three determinations) for MsmgΔsucT/pMVGH1-sucT (Fig. 3A). To gain further insight into the degree of succinylation of AG in the different strains, their mAGP was digested with endogenous M. smegmatis endoarabinanase, and the released oligoarabinosides were analyzed by LC/MS as described in the Supporting Methods. The mass spectra of the arabinans released from the WT and complemented mutant AG revealed nonsuccinylated and succinylated Ara18 and Ara19 oligoarabinosides, whereas only the nonsuccinylated variants were detected in MsmgΔsucT (Fig. 3B). Similar to the situation with M. tuberculosis (15), the fact that Ara7 fragments showed a significantly (∼ 3-fold) smaller degree of succinylation is interpreted as the succinyl residues being located on the interior branched α-3,5–Araf residue. This observation, combined with the fact that Ara18 may have up to four succinyl substituents, leads to the identification of quantitively minor positions of succinylation on adjacent linear α-1,5–branched Araf residues (Fig. 3B). The fact that MsmgΔsucT mAGP was not entirely devoid of succinates, according to the butanolysis analysis, could indicate that succinyl groups substitute, in a SucT-independent manner, other positions of AG not revealed by our LC/MS analysis of oligoarabinosides. Alternatively, because the GC/MS analysis of succinyl substituents was performed on mAGP rather than on purified AG, we cannot exclude that the remaining succinates arose from peptidoglycan and/or mycolic acids.
Analyses of the monosaccharide composition (Table S3) and glycosyl linkages (Table S4) of the WT, mutant, and complemented mutant AG otherwise failed to reveal any significant structural alterations in the galactan or arabinan domains of the mutant AG. The degree of mycolylation of the mutant AG was also similar to that measured in the WT strain (Table S3).
Alterations in the cell surface properties of the sucT mutant
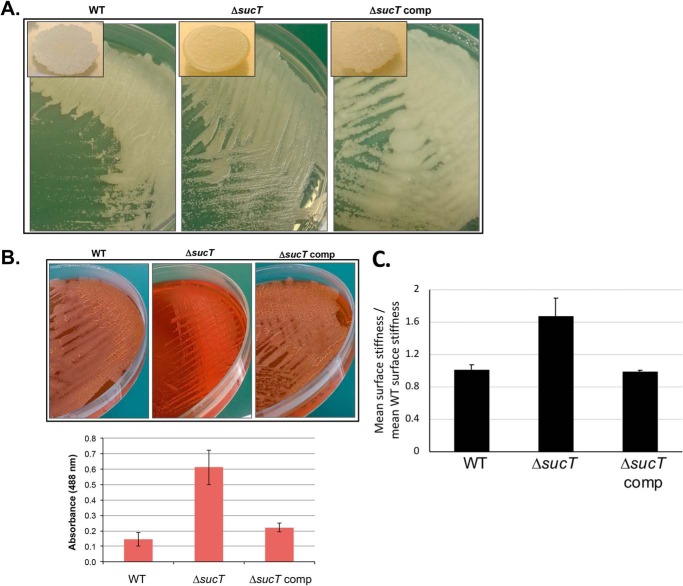
Two previous studies have reported on the isolation and partial characterization of Mycobacterium mutants carrying transposon insertions in sucT orthologs. First was the study by Driessen et al. (29), mentioned above, in which a M. marinum sucT KO mutant was identified as part of a screen for strains devoid of mannoside caps on LAM. Second was the screening by Yamazaki et al. (30) of an M. avium transposon mutant library for strains impaired in biofilm formation, which also yielded a mutant harboring an insertion in sucT. Although neither of these studies reported on the function of sucT and its impact on succinylation of AG and LAM, interesting observations were made regarding the growth and cell surface properties of the mutants. The M. marinum mutant, although displaying WT colony morphology on 7H10 agar plates, was reported to be hyperaggregative in 7H9-ADC-Tween 80 medium (29). The M. avium sucT mutant, on the other hand, was found to display an altered colony morphology on agar and to be impaired in its ability to form biofilms on polyvinyl chloride microplates (30). Our observations point to MsmgΔsucT sharing characteristics of both the M. marinum and M. avium sucT transposon mutants in that it aggregated in 7H9-ADC-Tween 80 medium and displayed an altered colony morphology on 7H11-OADC agar (Fig. 5A). Monitoring of the absorbance and colony-forming unit counts of MsmgΔsucT cultures over time in 7H9-ADC-tyloxapol medium (where satisfactory bacterial dispersion was achieved) also reproducibly pointed to a slight reduction in growth of the mutant compared with the WT strain, marked by an extended lag period (Fig. S6A). Despite having recovered the ability to succinylate both AG and LAM and a WT colonial morphology (Fig. 5A), the complemented mutant showed a growth rate more comparable with that of the sucT KO (Fig. S6A), a result that may tentatively be explained by the hypersuccinylation of AG and LAM in this strain. Further analysis revealed that MsmgΔsucT was slightly retarded in its ability to form surface biofilm pellicles in Sauton's medium, a phenotype probably resulting (at least in part) from its slower growth. Surface pellicle formation was partially restored upon genetic complementation (Fig. S6B).
Figure 5.
Alterations in the cell surface properties of the M. smegmatis sucT mutant. A, colony morphology of WT mc2155, mc2ΔsucT, and mc2ΔsucT/pMVGH1-sucT (ΔsucT comp) after 3 days of incubation on 7H11-OADC agar at 37 °C. B, Congo red binding on a TS agar plate and in TS liquid medium (graph). Shown on the graph are the average ± S.D. absorbances of acetone extracts measured for three biological replicates. C, analysis of the cell surface rigidity of the WT, mutant, and complemented mutant strains by correlated optical fluorescence and AFM. Two-sided rank sum test demonstrates a statistically significant difference in stiffness between the sucT mutant and both the WT (p = 0.0214) and the sucT complemented mutant (p = 0.0011).
Because the hyperaggregative properties and biofilm defect of the M. smegmatis sucT KO were suggestive of changes in cell surface hydrophobicity, the WT, mutant, and complemented mutant were next compared for their ability to bind the lipophilic dye Congo red (31). In liquid TS broth, MsmgΔsucT cells bound more than four times more Congo red than the WT and complemented strains, pointing to a clear increase in cell surface hydrophobicity (Fig. 5B). This phenotype also reflected on TS agar supplemented with Congo red, where the WT and complemented mutant strains grew as dry white colonies, whereas the sucT KO grew as glossy red colonies (Fig. 5B). Drug susceptibility testing further indicated that the sucT mutant was eight times more susceptible to rifampicin, a drug thought to penetrate the M. smegmatis outer membrane through passive diffusion (Table 1).
Table 1.
Susceptibility of the M. smegmatis sucT knockout mutant to antibiotics
MIC were determined in 7H9-ADC-tyloxapol and MIC values are in micrograms per milliliter. MIC determinations were performed two to three times on independent culture batches. HYG, hygromycin; KAN, kanamycin; GEN, gentamycin; CIP, ciprofloxacin; INH, isoniazid; EMB, ethambutol; RIF, rifampicin; ND, not determined.
| Strains | HYG | KAN | GEN | RIF | INH | CIP | EMB |
|---|---|---|---|---|---|---|---|
| WT | 10 | 1.2 | 2.5 | 50 | 12.5 | 0.3 | 0.8 |
| mc2ΔsucT | 5–10 | 1.2 | 2.5 | 6.2 | 12.5 | 0.15 | 0.8–1.6 |
| mc2ΔsucT/pMVGH1-sucT | ND | 0.6 | ND | 50 | 25 | 0.3 | 0.8 |
To gain further insight into the cell envelope properties of the sucT mutant, the surface rigidity of the WT, mutant, and complemented mutant cells in relation to their height was measured using correlated optical fluorescence microscopy and atomic force microscopy. Results pointed to an ∼ 1.7-fold increase in the mean surface rigidity of the mutant compared with the WT and complemented mutant strains (Fig. 5C).
To investigate the possible reasons underlying the increased hydrophobicity and other surface alterations of the M. smegmatis sucT mutant, the surface lipids from this strain were finally extracted and compared with the cell surface lipid contents of the WT and complemented mutant. TLC analyses pointed to the reproducible buildup of three compounds in the sucT mutant that were either not detected or present in much lesser quantities at the surface of the WT and complemented strains (Fig. S7A). The nature of these products is at present not known, but staining with α-naphthol suggests that they may be glycolipids. TLC analyses of total lipids from the same M. smegmatis strains upon metabolic labeling with [1,2-14C]acetate did not reveal any other notable differences between the WT and sucT KO (Fig. S7B). Likewise, comparative analysis by LC/MS of the contents of the WT and sucT mutant in another succinylated lipopolysaccharide, known as the methylglucose lipopolysaccharide, failed to reveal any significant differences between the two strains (Fig. S8).
Discussion
Similar to other mycobacterial species, M. smegmatis has been known to acylate its two major cell envelope glycans, AG and LAM, with succinyl groups even though their biological significance and biosynthetic origin have remained elusive. The data presented here show that succinyl substituents precisely modify the C2 position of α-3,5-branched Araf residues of the arabinan domain of LAM in M. smegmatis and possibly the same position of the arabinan domain of AG, with the possibility for adjacent linear α-1,5-branched Araf residues to be succinylated as well. Data further show that SucT (MSMEG_3187) is the sole enzyme responsible for this modification in both LAM and AG.
The identification of SucT as the enzyme responsible for the modification of AG and LAM with succinyl groups raises questions regarding where and when the introduction of these substituents occurs. Both the arabinan domains of AG and LAM are synthesized on the periplasmic face of the plasma membrane through what appears to be sequential addition of Araf residues by polyprenyl-phosphate-arabinose–dependent arabinosyltransferases (10). It is thus reasonable to assume that the succinylation of AG and LAM takes place on the periplasmic side of the membrane. The mechanism of acyl transfer catalyzed by the family of integral membrane trans-acylases to which SucT belongs, however, has to date not been fully been resolved. The fact that the repeating unit of O-antigen from Salmonella Typhimurium is synthesized intracellularly originally led Slauch et al. (24) to predict that OafA-like enzymes catalyzed acyl transfer on the cytoplasmic face of the plasma membrane using acyl-CoAs as acyl donors. The rather widespread distribution of functional residues across cytoplasmic loops and transmembrane segments of these enzymes (25–26), however, makes it difficult to precisely map their catalytic site, raising the possibility that acyl transfer may perhaps occur on the periplasmic side from still unknown high-energy acyl donors. The cytoplasmic succinylation of Araf residues in mycobacteria would entail modification of Araf on the cytoplasmic arabinose donor, polyprenyl-phosphate-arabinose. In light of our failure to detect succinylated polyprenyl-phosphate-arabinose by LC/MS in total lipid fractions from M. tuberculosis (data not shown), the hypothesis of periplasmic modification of the arabinan domains of AG and LAM is preferred.
A second important question regarding the catalytic reaction catalyzed by SucT relates to the precise stage in the biosynthesis of AG and LAM at which succinylation occurs. The modification of Araf residues during elongation of the arabinan of AG/LAM could suggest a role of these substituents in governing the elongation and branching of the arabinan domain. Succinylation of AG and LAM upon completion of their synthesis, on the contrary, would rule out their involvement in biosynthesis and rather point to other functions in the physiology of the bacterium and/or its interaction with the host. The conflicting phenotypes of the M. marinum and M. smegmatis sucT mutants, where the first one displays a significantly altered LAM structure (29) and the second does not, preclude a conclusive answer to this question. It is clear from our results that succinylation is neither critical for biosynthesis of the galactan, mannan, and arabinan domains of AG and LAM nor for the capping of LAM with phosphoinositol caps in M. smegmatis. As a first step toward determining whether differences in the LAM biosynthetic pathways of slow- and fast-growing mycobacteria might have accounted for the contrasting phenotypes between the LAM structures of the M. marinum and M. smegmatis sucT mutants, we tested the ability of the sucT gene from M. tuberculosis (Rv1565c; 83% identity at the amino acid level with MMAR_2380 from M. marinum and 75% identity with MSMEG_3187 from M. smegmatis) to complement the M. smegmatis sucT mutant. The results, which are presented in Fig. S9, point to partial restoration of LAM succinylation in the complemented strain, validating Rv1565c as a LAM succinyltransferase. The reason underlying the more critical role of succinylation in elongation and branching of LAM (and perhaps AG) in slow- compared with fast-growing Mycobacterium species thus remains to be determined. Unfortunately, our attempts to develop a cell-free assay to determine whether M. smegmatis SucT acts on the arabinan domain during its elongation or upon its completion failed to detect any succinyltransferase activity associated with SucT, whatever the enzyme source (Escherichia coli lysates expressing M. smegmatis sucT, cell-free extracts prepared from M. smegmatis WT and the sucT KO) or acceptor substrates used: neoglycolipid acceptors mimicking the interior linear and branched structures of the arabinan domain of AG and LAM (Fig. S10) or intact mutant LAM devoid of succinyl substituents (data not shown).
A clear conserved effect of LAM and/or AG succinylation across slow- and fast-growing Mycobacterium species, based on reports of sucT mutants in M. avium, M. marinum, and now M. smegmatis, is on the cell surface properties of the bacilli (29–30). That these changes in surface properties are the result of alterations in the mycolylation of AG, as proposed earlier (15), was precluded by our analyses of the M. smegmatis mutant. Instead, our analyses of the cell surface composition of MsmgΔsucT point to an impact of AG and/or LAM succinylation on the production and/or export of some still unknown lipids. The control AG and LAM succinylation has on the bacilli's surface rigidity and hydrophobicity likely has significant implications for the way mycobacteria interact with their environment, including that of the infected host. A defect in biofilm formation, for instance, is expected to negatively impact the ability of mycobacteria to colonize environmental and host substrata as well as their tolerance to biocides. From a virulence standpoint, the M. avium sucT KO has been reported to be impaired in its ability to invade bronchial epithelial cells and cause lung infection in mice (32) while replicating similarly as its WT parent in THP-1 human mononuclear phagocytes (33). Whether AG/LAM succinylation contributes similarly to the adaptation of tuberculous mycobacteria to their environment and to immunopathogenesis is currently under investigation in our laboratory.
Experimental procedures
Bacterial strains and growth conditions
M. smegmatis mc2155 was grown under agitation at 37 °C in Middlebrook 7H9 medium supplemented with 10% albumin–dextrose–catalase (ADC) (BD Biosciences) and 0.05% Tween 80 or 0.05% tyloxapol, on Middlebrook 7H11 agar supplemented with 10% oleic acid–albumin–dextrose–catalase (OADC) (BD Biosciences), in glycerol–alanine–salts (GAS) medium with 0.05% tyloxapol, or in Sauton's medium as surface pellicles. Apramycin, kanamycin, hygromycin, and streptomycin were added to final concentrations of 25 μg ml−1, 25 μg ml−1, 50 μg ml−1, and 20 μg ml−1, respectively.
M. smegmatis knockout and complemented knockout mutants
The temperature-sensitive/sacB method was used to knock out each of the candidate succinyltransferases of M. smegmatis (34). Allelic exchange substrates consisted of an antibiotic cassette (apramycin, streptomycin, kanamycin, or hygromycin) flanked by ∼1,000 bp of upstream and downstream homologous DNA sequence flanking MSMEG_2021, MSMEG_3187, MSMEG_5041, MSMEG_5537, and MSMEG_6230. Primer sequences for these constructs are available upon request. KO mutants were confirmed by PCR using sets of primers located outside of the allelic exchange substrates. For complementation of the MSMEG_3187 KO, the entire coding sequence of MSMEG_3187 was PCR-amplified from M. smegmatis mc2155 genomic DNA and cloned into the replicative expression plasmid pMVGH1 (35), yielding pMVGH1-sucT. pMVGH1-Rv1565c, the plasmid used to complement the MSMEG_3187 KO with the Rv1565c gene from M. tuberculosis, was PCR-amplified from M. tuberculosis H37Rv genomic DNA and similarly cloned into pMVGH1.
Metabolic labeling
Radiolabeling of M. smegmatis with [1,2-14C]acetic acid (1 μCi ml−1; specific activity, 52 mCi mmol−1; PerkinElmer Life Sciences) or [U-14C]glucose (1 μCi ml−1; specific activity, 5 mCi mmol−1; American Radiolabeled Chemicals) was performed in 7H9-ADC-tyloxapol and GAS-tyloxapol, respectively, at 37 °C for 4 h with shaking.
Preparation and analysis of lipids, methylglucose lipopolysaccharides, lipoglycans, and arabinogalactan
Total lipids from M. smegmatis cells were extracted with CHCl3/CH3OH (1:2, v/v) for 2 h at 56 °C, followed by two 2-h extractions with CHCl3/CH3OH (2:1, v/v) at 56 °C. Surface-exposed lipids were extracted from whole cells in water-saturated 1-butanol as described previously (36), and the butanol-treated cells were then re-extracted twice for 2 h with CHCl3/CH3OH (2:1, v/v) at 56 °C to recover all remaining extractable lipids. Cold and radiolabeled lipids were analyzed on silica gel 60–precoated TLC plates (F254, Merck) in a variety of solvent systems to resolve lipids of various polarities. TLC plates were revealed by spraying with cupric sulfate (10% w/v in an 8% v/v phosphoric acid solution) or α-naphthol (1% w/v in ethanol) and charring. Radiolabeled products were visualized using a Sapphire Biomolecular Imager (Azure Biosystems).
Methylglucose polysaccharides were extracted from whole cells as described previously (37) and analyzed as described by De et al. (38) by ultraperformance LC on a Waters Acquity UPLC H-Class system coupled to a Bruker MaXis Plus quadrupole time-of-flight MS instrument.
Lipoglycan and mAGP complex extraction from delipidated cells followed earlier procedures (15, 39). Lipoglycans were purified by gel exclusion chromatography (40) and analyzed by SDS-PAGE on commercial NovexTM 10–20% Tricine gels stained with periodic acid–Schiff reagent. Other procedures related to the structural analysis of LAM and mAGP are detailed in the Supporting Methods.
Congo red binding
M. smegmatis strains were tested for Congo red binding in TS broth as described by Etienne et al. (31). Briefly, the strains were cultivated for 3 days at 37 °C with shaking in TS broth containing 100 μg/ml Congo red. Cells were collected by centrifugation and washed extensively with distilled water until the supernatant was colorless. Cells were next resuspended in 1 ml of acetone, vortexed, and gently shaken for 2 h at room temperature prior to pelleting by centrifugation. Congo red in the supernatants was quantified spectrophotometrically at 488 nm. Congo red binding was defined as the A488 nm of the acetone extracts divided by the dry weight (in milligrams) of the cell pellet.
Drug susceptibility testing
Minimum inhibitory concentration (MIC) values were determined in 7H9-ADC-tyloxapol in a total volume of 100 μl in 96-well microtiter plates. M. smegmatis cultures grown to early log phase (A600 nm ∼0.2) were diluted to a final concentration of 106 cfu/ml and incubated in the presence of serial dilutions of the drugs for 2 days at 37 °C. MICs were determined using the resazurin blue test (41).
Correlated optical fluorescence and atomic force microscopy
Preparation conditions and the technical setup for AFM experiments were conducted according to Eskandarian et al. (42). Cells of the M. smegmatis WT expressing Wag31-GFP were mixed with nonfluorescent MsmgΔsucT and deposited on a polydimethylsiloxane-coated coverslip. WT cells were distinguished from MsmgΔsucT cells by optical fluorescence microscopy. AFM measurements were made using a Dimension Icon scan head (Bruker) using ScanAsyst fluid cantilevers (Bruker) with a nominal spring constant of 0.7 n m−1 in Peak Force quantitative nanomechanical mode at a force setpoint of ∼1 nN and typical scan rates of 0.3 Hz. Indentation on the cell surface was estimated to be ∼10 nm with a range of ∼5 nm in the z axis. Height, peak force error, and Derjaguin-Muller-Toporov (DMT) modulus channels were recorded for all scanned images in the trace and retrace directions. Images were processed using Gwyddion (Department of Nanometrology, Czech Metrology Institute; http://gwyddion.net).3 ImageJ was used for extracting bacterial cell profiles from height and DMT modulus images in a tabular format. A two-sided Wilcoxon rank-sum U test was used to analyze the data, with a continuity correction and confidence level of 95% using MatLab. AFM raw data are available at https://figshare.com/s/9dac3d318fceb04653ca.3
Author contributions
Z. P., S. K. A., J. M. B., M. Joe, R. B., J. N., T. L. L., M. G., M. M., and M. Jackson conceptualization; Z. P., S. K. A., J. M. B., H. A. E., M. Joe, R. B., C.R ., J. N., T. L. L., and M. G. investigation; Z. P., S. K. A., J. M. B., H. A. E., M. Joe, R. B., C. R., V. J., J. N., T. L. L., M. G., and M. M. methodology; Z. P., J. M. B., H. A. E., C. R., T. L. L., M. G., M. M., and M. Jackson writing-original draft; Z. P., J. M. B., H. A. E., J. N., T. L. L., M. G., M. M., and M. Jackson writing-review and editing; S. K. A., J. M. B., H. A. E., M. G., M. M., and M. Jackson formal analysis; T. L. L. and M. Jackson funding acquisition; M. M. and M. Jackson supervision; M. Jackson project administration.
Supplementary Material
Acknowledgments
We thank the Integrated Screening Platform of Toulouse (PICT, IBiSA) for providing access to 600 MHz equipment and Dr. Emilie Huc-Claustre for help with the construction of the M. smegmatis MSMEG_5041 knockout mutant.
This work was supported by NIAID, National Institutes of Health Grant AI064798 (to M. J.) and the Alberta Glycomics Centre (to T. L. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S10, Tables S1–S4, and Supporting Methods.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- AG
- arabinogalactan
- LAM
- lipoarabinomannan
- AM
- d-arabino-d-mannan
- LM
- lipomannan
- mAGP
- mycolyl–AG–peptidoglycan
- OADC
- oleic acid–albumin–dextrose–catalase
- ADC
- albumin–dextrose–catalase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- AFM
- atomic force microscopy
- HMQC
- heteronuclear multiple quantum coherence
- TS
- tryptic soy.
References
- 1. Raetz C. R., Reynolds C. M., Trent M. S., and Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 10.1146/annurev.biochem.76.010307.145803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swoboda J. G., Campbell J., Meredith T. C., and Walker S. (2010) Wall teichoic acid function, biosynthesis, and inhibition. ChemBioChem 11, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown S., Xia G., Luhachack L. G., Campbell J., Meredith T. C., Chen C., Winstel V., Gekeler C., Irazoqui J. E., Peschel A., and Walker S. (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914 10.1073/pnas.1209126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Needham B. D., and Trent M. S. (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11, 467–481 10.1038/nrmicro3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whitfield C., and Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 10.1146/annurev-biochem-060713-035600 [DOI] [PubMed] [Google Scholar]
- 6. Schneewind O., and Missiakas D. (2014) Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J. Bacteriol. 196, 1133–1142 10.1128/JB.01155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maldonado R. F., Sá-Correia I., and Valvano M. A. (2016) Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 40, 480–493 10.1093/femsre/fuw007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajagopal M., and Walker S. (2017) Envelope structures of Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerlach D., Guo Y., De Castro C., Kim S. H., Schlatterer K., Xu F. F., Pereira C., Seeberger P. H., Ali S., Codée J., Sirisarn W., Schulte B., Wolz C., Larsen J., Molinaro A., et al. (2018) Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 563, 705–709 10.1038/s41586-018-0730-x [DOI] [PubMed] [Google Scholar]
- 10. Angala S. K., Belardinelli J. M., Huc-Claustre E., Wheat W. H., and Jackson M. (2014) The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 49, 361–399 10.3109/10409238.2014.925420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daffé M., Crick D. C., and Jackson M. (2014) Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol. Spectr. 2, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angala S. K., Palčeková Z., Belardinelli J. M., and Jackson M. (2018) Covalent modifications of polysaccharides in mycobacteria. Nat. Chem. Biol. 14, 193–198 10.1038/nchembio.2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Škovierová H., Larrouy-Maumus G., Pham H., Belanová M., Barilone N., Dasgupta A., Mikusová K., Gicquel B., Gilleron M., Brennan P. J., Puzo G., Nigou J., and Jackson M. (2010) Biosynthetic origin of the galactosamine substituent of arabinogalactan in Mycobacterium tuberculosis. J. Biol. Chem. 285, 41348–41355 10.1074/jbc.M110.188110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wheat W. H., Dhouib R., Angala S. K., Larrouy-Maumus G., Dobos K., Nigou J., Spencer J. S., and Jackson M. (2015) The presence of a galactosamine substituent on the arabinogalactan of Mycobacterium tuberculosis abrogates full maturation of human peripheral blood monocyte-derived dendritic cells and increases secretion of IL-10. Tuberculosis 95, 476–489 10.1016/j.tube.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhamidi S., Scherman M. S., Rithner C. D., Prenni J. E., Chatterjee D., Khoo K.-H., and McNeil M. R. (2008) The identification and location of succinyl residues and the characterization of the interior arabinan region allows for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan. J. Biol. Chem. 283, 12992–13000 10.1074/jbc.M800222200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delmas C., Gilleron M., Brando T., Vercellone A., Gheorghiu M., Rivière M., and Puzo G. (1997) Comparative structural study of the mannosylated-lipoarabinomannans from Mycobacterium bovis BCG vaccine strains: characterization and localization of succinates. Glycobiology 7, 811–817 10.1093/glycob/7.6.811 [DOI] [PubMed] [Google Scholar]
- 17. Weber P. L., and Gray G. R. (1979) Structural and immunochemical characterization of the acidic arabinomannan of Mycobacterium smegmatis. Carbohydr. Res. 74, 259–278 10.1016/S0008-6215(00)84781-3 [DOI] [PubMed] [Google Scholar]
- 18. Hunter S. W., Gaylord H., and Brennan P. J. (1986) Structure and antigenicity of the phosphorylated lipopolysaccharide antigens from the leprosy and tubercle bacilli. J. Biol. Chem. 261, 12345–12351 [PubMed] [Google Scholar]
- 19. Torrelles J. B., Khoo K. H., Sieling P. A., Modlin R. L., Zhang N., Marques A. M., Treumann A., Rithner C. D., Brennan P. J., and Chatterjee D. (2004) Truncated structural variants of lipoarabinomannan in Mycobacterium leprae and an ethambutol-resistant strain of Mycobacterium tuberculosis. J. Biol. Chem. 279, 41227–41239 10.1074/jbc.M405180200 [DOI] [PubMed] [Google Scholar]
- 20. Torrelles J. B., Sieling P. A., Arcos J., Knaup R., Bartling C., Rajaram M. V., Stenger S., Modlin R. L., and Schlesinger L. S. (2011) Structural differences in lipomannans from pathogenic and nonpathogenic mycobacteria that impact CD1b-restricted T cell responses. J. Biol. Chem. 286, 35438–35446 10.1074/jbc.M111.232587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torrelles J. B., Sieling P. A., Zhang N., Keen M. A., McNeil M. R., Belisle J. T., Modlin R. L., Brennan P. J., and Chatterjee D. (2012) Isolation of a distinct Mycobacterium tuberculosis mannose-capped lipoarabinomannan isoform responsible for recognition by CD1b-restricted T cells. Glycobiology 22, 1118–1127 10.1093/glycob/cws078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortalo-Magné A., Andersen A. B., and Daffé M. (1996) The outermost capsular arabinomannans and other mannoconjugates of virulent and avirulent tubercle bacilli. Microbiology 142, 927–935 10.1099/00221287-142-4-927 [DOI] [PubMed] [Google Scholar]
- 23. Guérardel Y., Maes E., Briken V., Chirat F., Leroy Y., Locht C., Strecker G., and Kremer L. (2003) Lipomannan and lipoarabinomannan from a clinical isolate of Mycobacterium kansasii: novel structural features and apoptosis-inducing properties. J. Biol. Chem. 278, 36637–36651 10.1074/jbc.M305427200 [DOI] [PubMed] [Google Scholar]
- 24. Slauch J. M., Lee A. A., Mahan M. J., and Mekalanos J. J. (1996) Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: OafA is a member of a family of integral membrane trans-acylases. J. Bacteriol. 178, 5904–5909 10.1128/jb.178.20.5904-5909.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thanweer F., Tahiliani V., Korres H., and Verma N. K. (2008) Topology and identification of critical residues of the O-acetyltransferase of serotype-converting bacteriophage, SF6, of Shigella flexneri. Biochem. Biophys. Res. Commun. 375, 581–585 10.1016/j.bbrc.2008.08.069 [DOI] [PubMed] [Google Scholar]
- 26. Thanweer F., and Verma N. K. (2012) Identification of critical residues of the serotype modifying O-acetyltransferase of Shigella flexneri. BMC Biochem. 13, 13 10.1186/1471-2091-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamaryo-Botte Y., Rainczuk A. K., Lea-Smith D. J., Brammananth R., van der Peet P. L., Meikle P., Ralton J. E., Rupasinghe T. W., Williams S. J., Coppel R. L., Crellin P. K., and McConville M. J. (2015) Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem. Biol. 10, 734–746 10.1021/cb5007689 [DOI] [PubMed] [Google Scholar]
- 28. Belardinelli J. M., Yazidi A., Yang L., Fabre L., Li W., Jacques B., Angala S. K., Rouiller I., Zgurskaya H. I., Sygusch J., and Jackson M. (2016) Structure-function profile of MmpL3, the essential mycolic acid transporter from Mycobacterium tuberculosis. ACS Infect. Dis. 2, 702–713 10.1021/acsinfecdis.6b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Driessen N. N., Stoop E. J., Ummels R., Gurcha S. S., Mishra A. K., Larrouy-Maumus G., Nigou J., Gilleron M., Puzo G., Maaskant J. J., Sparrius M., Besra G. S., Bitter W., Vandenbroucke-Grauls C. M., and Appelmelk B. J. (2010) Mycobacterium marinum MMAR_2380, a predicted transmembrane acyltransferase, is essential for the presence of the mannose cap on lipoarabinomannan. Microbiology 156, 3492–3502 10.1099/mic.0.037507-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamazaki Y., Danelishvili L., Wu M., Macnab M., and Bermudez L. E. (2006) Mycobacterium avium genes associated with the ability to form a biofilm. Appl. Environ. Microbiol. 72, 819–825 10.1128/AEM.72.1.819-825.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Etienne G., Villeneuve C., Billman-Jacobe H., Astarie-Dequeker C., Dupont M.-A., and Daffé M. (2002) The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148, 3089–3100 10.1099/00221287-148-10-3089 [DOI] [PubMed] [Google Scholar]
- 32. Yamazaki Y., Danelishvili L., Wu M., Hidaka E., Katsuyama T., Stang B., Petrofsky M., Bildfell R., and Bermudez L. E. (2006) The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol. 8, 806–814 10.1111/j.1462-5822.2005.00667.x [DOI] [PubMed] [Google Scholar]
- 33. Rose S. J., and Bermudez L. E. (2014) Mycobacterium avium biofilm attenuates mononuclear phagocyte function by triggering hyperstimulation and apoptosis during early infection. Infect. Immun. 82, 405–412 10.1128/IAI.00820-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelicic V., Jackson M., Reyrat J. M., Jacobs W. R. Jr., Gicquel B., and Guilhot C. (1997) Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10955–10960 10.1073/pnas.94.20.10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grzegorzewicz A. E., Pham H., Gundi V. A., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E., Korduláková J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R., and Jackson M. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morita Y. S., Velasquez R., Taig E., Waller R. F., Patterson J. H., Tull D., Williams S. J., Billman-Jacobe H., and McConville M. J. (2005) Compartmentalization of lipid biosynthesis in mycobacteria. J. Biol. Chem. 280, 21645–21652 10.1074/jbc.M414181200 [DOI] [PubMed] [Google Scholar]
- 37. Stadthagen G., Sambou T., Guerin M., Barilone N., Boudou F., Korduláková J., Charles P., Alzari P. M., Lemassu A., Daffé M., Puzo G., Gicquel B., Rivière M., and Jackson M. (2007) Genetic basis for the biosynthesis of methylglucose lipopolysaccharides in Mycobacterium tuberculosis. J. Biol. Chem. 282, 27270–27276 10.1074/jbc.M702676200 [DOI] [PubMed] [Google Scholar]
- 38. De P., McNeil M., Xia M., Boot C. M., Hesser D. C., Denef K., Rithner C., Sours T., Dobos K. M., Hoft D., and Chatterjee D. (2018) Structural determinants in a glucose-containing lipopolysaccharide from Mycobacterium tuberculosis critical for inducing a subset of protective T cells. J. Biol. Chem. 293, 9706–9717 10.1074/jbc.RA118.002582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaur D., McNeil M. R., Khoo K. H., Chatterjee D., Crick D. C., Jackson M., and Brennan P. J. (2007) New insights into the biosynthesis of mycobacterial lipomannan arising from deletion of a conserved gene. J. Biol. Chem. 282, 27133–27140 10.1074/jbc.M703389200 [DOI] [PubMed] [Google Scholar]
- 40. Berg S., Starbuck J., Torrelles J. B., Vissa V. D., Crick D. C., Chatterjee D., and Brennan P. J. (2005) Roles of the conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J. Biol. Chem. 280, 5651–5663 10.1074/jbc.M411418200 [DOI] [PubMed] [Google Scholar]
- 41. Martin A., Camacho M., Portaels F., and Palomino J.-C. (2003) Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob. Agents Chemother. 47, 3616–3619 10.1128/AAC.47.11.3616-3619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eskandarian H. A., Odermatt P. D., Ven J. X. Y., Hannebelle M. T. M., Nievergelt A. P., Dhar N., McKinney J. D., and Fantner G. E. (2017) Division site selection linked to inherited cell surface wave troughs in mycobacteria. Nat. Microbiol. 2, 17094 10.1038/nmicrobiol.2017.94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.