Abstract
The retinal pigment epithelium (RPE) is a monolayer of pigmented cells between the choroid and the retina. RPE dysfunction underlies many retinal degenerative diseases, including age-related macular degeneration, the leading cause of age-related blindness. To perform its various functions in nutrient transport, phagocytosis of the outer segment, and cytokine secretion, the RPE relies on an active energy metabolism. We previously reported that human RPE cells prefer proline as a nutrient and transport proline-derived metabolites to the apical, or retinal, side. In this study, we investigated how RPE utilizes proline in vivo and why proline is a preferred substrate. By using [13C]proline labeling both ex vivo and in vivo, we found that the retina rarely uses proline directly, whereas the RPE utilizes it at a high rate, exporting proline-derived mitochondrial intermediates for use by the retina. We observed that in primary human RPE cell culture, proline is the only amino acid whose uptake increases with cellular maturity. In human RPE, proline was sufficient to stimulate de novo serine synthesis, increase reductive carboxylation, and protect against oxidative damage. Blocking proline catabolism in RPE impaired glucose metabolism and GSH production. Notably, in an acute model of RPE-induced retinal degeneration, dietary proline improved visual function. In conclusion, proline is an important nutrient that supports RPE metabolism and the metabolic demand of the retina.
Keywords: retinal metabolism, amino acid, mitochondrial metabolism, tricarboxylic acid cycle (TCA cycle) (Krebs cycle), cell metabolism, age-related macular degeneration (AMD), glucose metabolism, oxidative stress, visual function, proline, retinal pigment epithelium, retina
Introduction
The retinal pigment epithelium (RPE)3 is a monolayer of pigmented cells between the choroid and the retina. RPE dysfunction contributes to the pathogenesis of many retinal degenerative diseases, including age-related macular degeneration (AMD), the leading cause of blindness in the older population. To maintain its functions in nutrient transport, phagocytosis of outer segment, and cytokine secretion, the RPE relies on an active energy metabolism. We reported recently that the RPE has a high capacity for reductive carboxylation, a reverse tricarboxylic acid (TCA) cycle (1). Using an unbiased metabolomics screen, we found that RPE heavily consumes proline to fuel both mitochondrial oxidative phosphorylation and reductive carboxylation (2).
Proline has diverse functions in different organisms (3–5). Proline is well-known to accumulate in plants to combat various environmental stressors (6, 7). Proline is also a significant component (up to 25%) of collagen, which is the most abundant protein in extracellular matrix (ECM), such as the Bruch's membrane located underneath RPE cells (3). Proline can be catabolized through proline dehydrogenase (PRODH) to donate electrons directly to ubiquinone or into glutamate to enter the mitochondrial TCA cycle (2, 4, 8, 9). In several invertebrates, proline is the major energy source (9). In worms, proline supplementation extends their lifespans, (10), and a mutation that shifts their metabolism toward proline catabolism increases their lifespans more than 2-fold (11, 12).
Proline is not an essential amino acid. It can be produced from glutamate through pyrroline-5-carboxylate synthase (P5CS), from collagen degradation through prolidase, or from ornithine through ornithine aminotransferase (OAT). Inborn errors of genes in proline metabolism can result in retinal degeneration. A mutation in the gene encoding P5CS has been associated with retinitis pigmentosa (13). Mutations in OAT are well-documented to cause gyrate atrophy, characterized by lobular loss of the RPE/choroid and progressive retinal degeneration (14–16). OAT deficiency results in accumulation of more than 10-fold levels of ornithine in the plasma, and ornithine can inhibit P5CS in vitro (17). In RPE cell lines, supplementation of proline has been shown to rescue ornithine cytotoxicity (18, 19). A proline transporter solute carrier family 6, member 20 (SLC6A20) has been regarded as an RPE signature gene (20–22), and it is one of 22 RPE genes shared by both human and mouse RPE (20). A recent large-scale genome-wide association study reports that the locus of this proline transporter is significantly associated with AMD (22, 23). Overall, previous findings implicate proline metabolism as a potentially essential component of RPE health and function.
In this report, we investigate how proline is utilized by the RPE and retina as well as the functional roles of proline consumption. By using 13C tracing, we found that proline fuels RPE mitochondrial metabolism. The RPE also exports proline-derived intermediates to the retina ex vivo and in vivo. As RPE cells mature in vitro, they become more dependent on proline as a nutrient substrate. Proline supplementation confers resistance against oxidative damage and improves visual function.
Results
Mouse RPE/choroid but not retina utilizes proline ex vivo and exports proline-derived intermediates
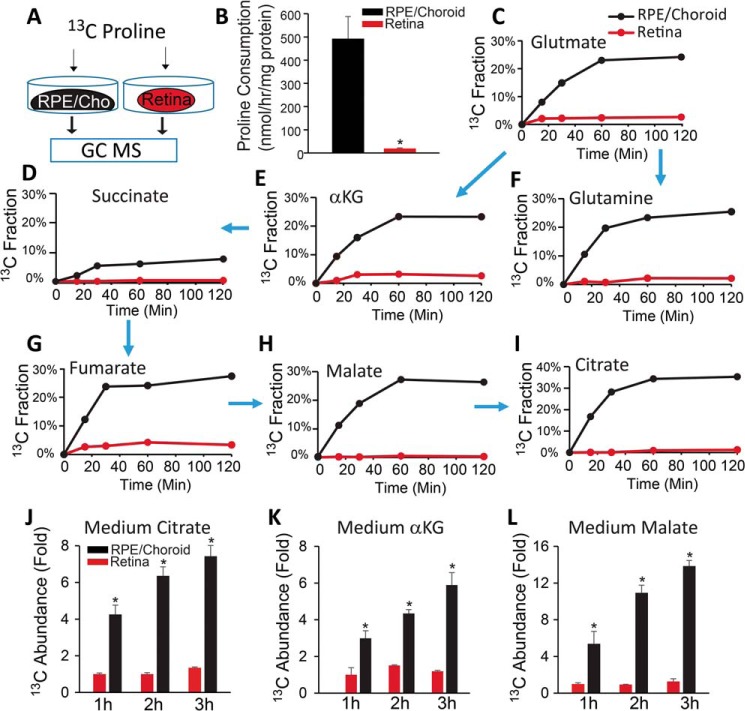
We previously reported that human fetal RPE (hRPE) in culture consumes more proline than other amino acids (2). To investigate whether native RPE utilizes proline, we isolated mouse RPE/choroid and retina and incubated them with 13C-labeled proline (Fig. 1A). We did not isolate the RPE from the choroid, as separation could disrupt RPE metabolism and influence cell viability. We found that the RPE/choroid complex consumed proline >100 times faster than the retina (Fig. 1B). This finding supports our previous reports that glucose and glutamine are major fuels for retinal mitochondrial metabolism (24). Proline can be metabolized into mitochondrial intermediates through PRODH. After incubation with [13C]proline, we found a high percentage of labeled TCA cycle intermediates (20–30%), with the exception of succinate, in total pools in the RPE/choroid. This is in contrast to only 2% of labeled intermediates in the retina (Fig. 1, C–I), confirming that proline might be a major nutrient for RPE. [13C]Proline could generate α-ketoglutarate (αKG) for either the TCA cycle to produce M4 citrate or reductive carboxylation to produce M5 citrate (Fig. S1A) (2). We found M5 citrate in the RPE/choroid but not retina, indicating that reductive carboxylation is active in native RPE cells (Fig. S1, B and C). Additionally, we found ∼2% of M3 pyruvate and ∼0.5% of M3 PEP labeled by [13C]proline, which should be generated mostly through malic enzyme rather than phosphoenolpyruvate carboxykinase (Fig. S1, A and D). To study whether RPE can release proline-derived intermediates, we measured the incubation medium and found that 13C-labeled intermediates increased in a time-dependent fashion in RPE/choroid cultures (Fig. 1, J–L).
Figure 1.
Mouse RPE/choroid utilizes [13C]proline ex vivo. A, schematic for ex vivo incubation of [13C]proline. Freshly isolated RPE/choroid or retina was incubated in 5 mm glucose and 1 mm [13C]proline for different times. B–D, RPE/choroid consumes much more proline than retina and generates mitochondrial intermediates (E–I). Metabolites were analyzed by GC MS. The 13C fraction is the percentage of labeled carbon of total isotopologues or 13C-labeled metabolite in the total pool. Arrows represent the direction in which the carbons flow in the mitochondrial Krebs cycle. J–L, proline-derived metabolites are exported into medium. The 13C abundance -fold change was calculated from the ion intensity of labeled metabolites relative to those in retina at 1 h. *, p < 0.05 versus retina. n = 4. Error bars, S.D.
RPE utilizes proline to fuel retinal metabolism in vivo
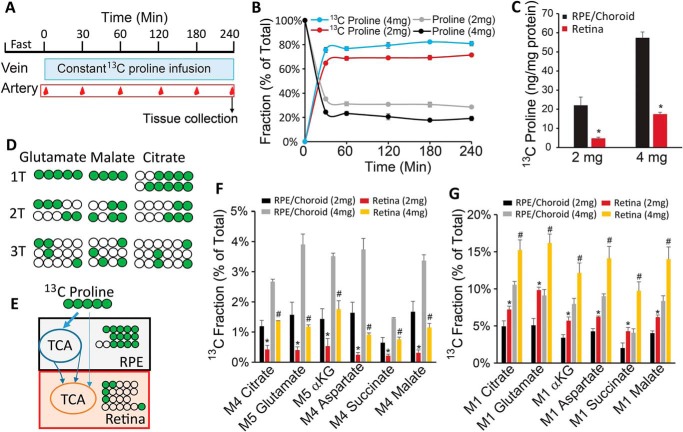
To study proline metabolism in vivo, we performed hyperinsulinemic-euglycemic clamp in conscious, unrestrained mice and infused [13C]proline through the jugular vein continuously for 4 h (25). Blood from the carotid artery was sampled at different time points to monitor [13C]proline labeling in the plasma, and the RPE/choroid and retina were collected quickly after the 4-h infusion (Fig. 2A). We infused at 2 and 4 mg/kg/min based on the concentration of proline in the plasma relative to glutamine. Plasma glutamine is slightly higher than proline, and glutamine is infused at 2 mg/kg/min in the literature (26). Both doses of proline rapidly replaced 60–80% of endogenous proline and reached a steady state within 30 min (Fig. 2B). 13C-Labeled proline is about 3–4-fold higher in the RPE/choroid than retina (Fig. 2C). To study the flow of proline-derived intermediates from RPE to retina, we analyzed the labeled patterns in the intermediates. Five-carbon–labeled 13C (M5) proline can be converted into M5 glutamate and M5 αKG to enter the TCA cycle. In the first turn, one carbon will be lost as CO2 through αKG dehydrogenase to generate M4 intermediates (Fig. 2, D and E). More carbons will be lost in the second and third turn. Interestingly, M5 glutamate, M5 αKG, and M4 mitochondrial intermediates were much higher in RPE/choroid than retina (Fig. 2F), whereas M1 intermediates were much higher in the retina than RPE/choroid (Fig. 2G). These results further support our hypothesis that RPE utilizes proline to form mitochondrial intermediates, which are then exported to fuel retinal mitochondria (Fig. 2E).
Figure 2.
Proline utilization in RPE and retina in vivo. A, schematic for [13C]proline infusion in vivo. After fasting for 6 h, [13C]proline was constantly infused through a jugular catheter in free-moving mice. Blood samples were collected through an arterial catheter. B, [13C]proline replaced unlabeled proline and reached steady state in the plasma after infusion. C, RPE/choroid had more [13C]proline than retina. D and E, schematic of 13C-labeling pattern in RPE and retina. After entering the TCA cycle, five-carbon–labeled [13C]proline was catabolized mostly into M5/M4 metabolites in the first turn (1T) of the TCA cycle, M2/M3 in 2T, and M1 in 3T. When RPE used proline initially and exported the intermediates, RPE should have more M5/M4, and retina should have more M1. F and G, M5/M4 metabolites were increased, whereas M1 metabolites were decreased in the RPE comparable with the retina. n = 4. *, p < 0.05 versus retinas with 2 mg/kg/min of [13C]proline; #, p < 0.05 versus retinas with 4 mg/kg/min [13C]proline. Error bars, S.D.
Human RPE cell switches to proline utilization during RPE maturation
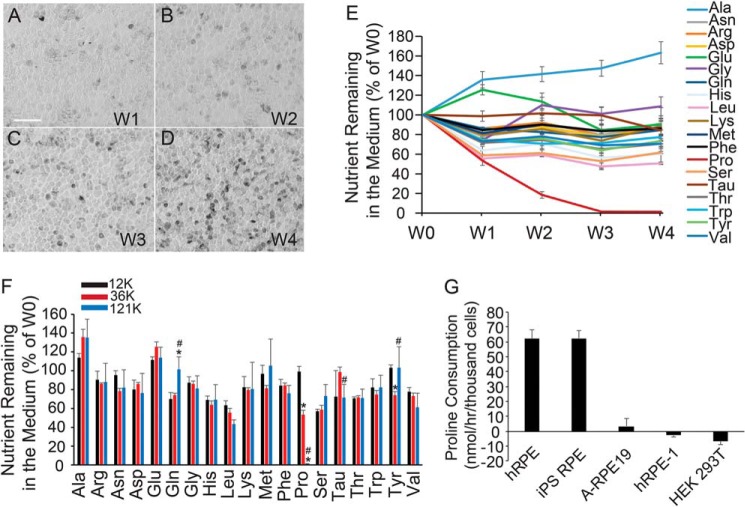
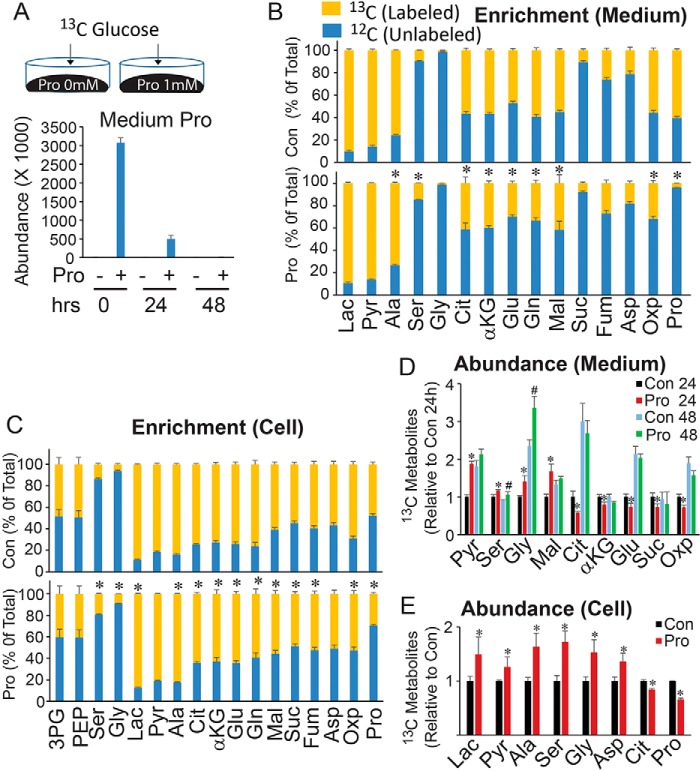
Depending on culture conditions, hRPE can take 4–6 weeks to mature, forming their characteristic cobblestone hexagonal morphology, pigmentation, and expression of typical RPE markers (Fig. 3, A–D). To study proline consumption during RPE maturation, we sampled medium 24 h after medium change in hRPE culture at weeks 1–4 and measured the metabolites by GC-MS (Fig. 3, A–E). Week 0 (W0) consisted of the control medium incubated for 24 h without hRPE cells. Proline was the only amino acid whose consumption increased with RPE maturation. Proline was almost undetectable in week 3 and 4 medium (W3 and W4; Fig. 3E), consistent with our previous report that RPE consumes a large amount of proline (2). Week 4 hRPE consumed most other amino acids at a similar rate to week 1 hRPE, with the exception of alanine, glutamine, glutamate, and taurine. To confirm this finding, we measured the nutrient consumption at the same time plated at different densities. After culture for 1 week, the hRPEs with higher initial plating densities were noted to mature more quickly than those plated at lower densities (Fig. S2, A–C). As expected, proline was the only amino acid that was consumed in a density/maturity-dependent manner, suggesting that RPE cells increase their proline consumption with cellular maturity (Fig. 3F). Consistently, both mature hRPE and induced pluripotent stem (iPS) cell–derived RPE demonstrated high proline consumption, whereas RPE cell lines (ARPE-19 and hRPE-1 cells) and human kidney epithelial cell line HEK 293T used either much less proline or none at all (Fig. 3G).
Figure 3.
RPE switches to utilize proline during maturation. A–D, human RPE matured after 3–4 weeks of culture. RPE cells showed typical cobblestone morphology and are pigmented under bright-field microscopy. Scale bar, 100 μm. E, nutrient consumption at different weeks of culture. For each week of culture, medium was collected for GC-MS 24 h after being changed. The y axis represents the amount of nutrient left in the medium relative to fresh medium. F, RPE cells were plated at different densities and grown for a week. Nutrients remaining in the medium 24 h after medium change were analyzed by GC MS. G, proline consumption in RPE cells and cell lines. n = 4. *, p < 0.05 versus RPE cells plated at 12,000 cells/well; #, p < 0.05 versus RPE cells plated at 36,000 cells/well. Error bars, S.D.
Inhibition of PRODH partially blocks RPE proline consumption and disrupts glucose and amino acid metabolism
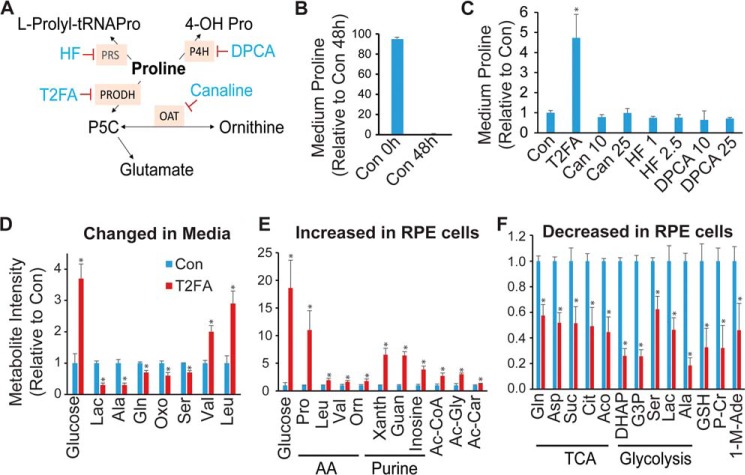
To determine the pathway responsible for high proline uptake in RPE, we used inhibitors to block several known pathways in proline catabolism, including tetrahydro-2-furoic acid (T2FA), canaline, halofuginone (HF), and 1,4-dihydrophenonthrolin-4-one-3-carboxylic acid (DPCA) (Fig. 4A). These inhibitors block PRODH, OAT, prolyl-tRNA synthetase (PRS), and prolyl-4-hydroxylase (P4H) to inhibit proline catabolism into glutamate, ornithine, an l-prolyl-tRNAPro in protein synthesis and 4-hydroxyproline (4-OH Pro) in collagen, respectively (27–30). After 48 h in RPE medium (containing 0.447 mm proline), 95% of proline was used (Fig. 4B). With the exception of T2FA, all other inhibitors could not block proline consumption, supporting our previous finding that proline is partly metabolized into glutamate for mitochondrial metabolism (Fig. 4C). To study the impact of PRODH on the consumption of other nutrients and cell metabolism, we quantified key metabolites in glucose and amino acid metabolism (Table S1) by LC MS/MS and GC MS in the medium and the cell supernatant with T2FA treatment. In addition to proline, inhibition of PRODH resulted in significant accumulation of glucose in both RPE medium and RPE cells (Fig. 4, D and E). Consistently, glucose-derived intermediates including lactate, alanine, dihydroxyacetone phosphate (DHAP), glyceraldehyde 3-phosphate (G3P), and serine were substantially decreased in medium and/or cells (Fig. 4, D and E). Purine metabolites are sensitive to the availability of glucose in RPE (31). As expected, xanthine, guanosine, and inosine dramatically increased in RPE cells (Fig. 4E). Additionally, inhibition of PRODH decreased many mitochondrial intermediates, GSH, and glutamine and increased branched-chain amino acids. These data suggest that proline metabolism may be critical for glucose metabolism and utilization of other amino acids.
Figure 4.
Inhibition of proline catabolism partially blocked proline consumption and impaired glucose metabolism. A, schematic for inhibitors and pathways in proline metabolism. B, RPE consumed most proline in medium after culture for 48 h. C, T2FA partially blocked proline consumption. Inhibitors at different concentrations were incubated for 48 h. Con was RPE medium with DMSO after 48-h culture. T2FA concentration was 5 mm, and other inhibitors were at different concentrations as labeled in micromolar. D–F, changes of metabolites in the medium or cells were measured by LC MS. n = 4. *, p < 0.05 versus Con without T2FA at 48 h. Lac, lactate; Oxo, 5-oxoproline; Xanth, xanthine; Guan, guanine; Ac, acetyl-; Ac-Car, acetyl-carnitine; Suc, succinate; Cit, citrate; Aco, aconitate; P-Cr, phosphocreatine; 1-M-Ade, 1-methyl-adenosine; HF, halofuginone; DPCA, 1,4-dihydrophenonthrolin-4-one-3-carboxylic acid; PRS, prolyl-tRNA synthetase; P4H, prolyl-4-hydroxylase; 4-OH Prom, 4-hydroxyproline; G3P, glyceraldehyde 3-phosphate. Error bars, S.D.
Proline stimulates de novo serine/glycine synthesis, glycolysis, and mitochondrial metabolism
To further examine the impact of proline on glucose metabolism, we incubated RPE cells with or without 1 mm proline in the presence of 13C glucose (Fig. 5A) and analyzed metabolites in both medium and cells (Fig. 5, B and C). ∼85% of proline in the medium was used by 24 h, and almost of all proline was consumed by 48 h (Fig. 5A). The addition of proline reduced the percentage of 13C-labeled mitochondrial TCA intermediates in the medium and cells (Fig. 5 (B and C) and Fig. S3). The labeling pattern (isotopologue) showed that M2 metabolites (derived from first turn of the TCA cycle) remained unchanged, but M4 (derived from the second turn of the TCA cycle) and M3 and M1 (derived from pyruvate carboxylase or multiple turns of the TCA cycle) intermediates were decreased (Figs. S4 and S5). These results indicate that mitochondria have a preference for utilizing proline rather than glucose as a substrate for four-carbon pools. Surprisingly, proline increased the flux of serine and glycine from [13C]glucose (Fig. 5 (B and C) and Fig. S3). Whereas the abundance (concentration or pool size) of labeled serine and glycine was significantly increased, the unlabeled serine and glycine were not (Fig. 5 (D and E) and Fig. S6), confirming that proline stimulates de novo serine biosynthesis from glucose. Additionally, proline increased [13C]lactate, [13C]pyruvate, and [13C]alanine in medium and cells (Fig. 5, C–E) and increased the overall concentration of aspartate, glutamate, and glutamine in RPE (Fig. 5E and Fig. S6). These results suggest that proline enhances glycolysis and synergizes with glucose metabolism. To further examine whether proline enhances mitochondrial energy metabolism, we measured mitochondrial O2 consumption using an extracellular flux analyzer. Proline doubled the maximum O2 consumption, comparable with glucose alone (Fig. S7). This is comparable with the combined effect of pyruvate and glutamine.
Figure 5.
Proline regulates glucose metabolism and stimulates synthesis of serine and glycine. A, schematic for [13C]glucose incubation in matured hRPE cells cultured with or without proline in DMEM. B, proline decreased the 13C fraction (13C enrichment) of mitochondrial intermediates in the medium after 24 h of culture. Top, medium without proline; bottom, medium with 1 mm proline. C, the 13C fraction of mitochondrial intermediates was decreased, but serine and glycine were increased by proline. D and E, proline regulated the levels of 13C-labeled metabolites from [13C]glucose in medium and RPE cells. Metabolites were measured by GC MS. n = 4. *, p < 0.05 versus Con without proline or Con at 24 h. #, p < 0.05 versus Con at 48 h. Lac, lactate; Pyr, pyruvate; Oxp, 5-oxoproline. Error bars, S.D.
Proline protects RPE cells from oxidative damage
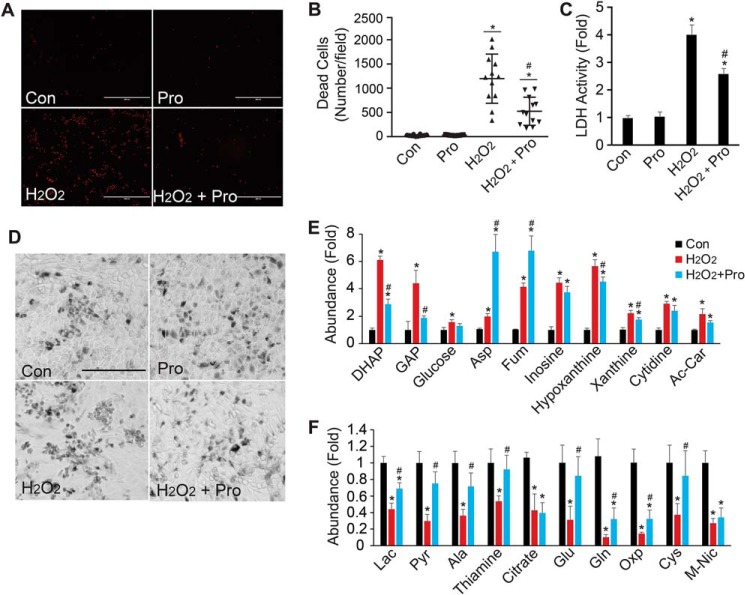
To test whether proline protects against oxidative damage in RPE, we incubated hRPE cells with proline in the presence of 1 mm hydrogen peroxide. Cell death was assessed by quantifying ethidium homodimer (EthD) staining of dead cells, lactate dehydrogenase (LDH) activity assay in the culture medium, and bright-field imaging of morphological changes. There was no difference in cell death in control hRPE with or without the addition of proline within 48 h. Hydrogen peroxide treatment increased EthD-positive cells, accumulated LDH in the medium, and decreased cell density (Fig. 6, A–D). Supplementation with proline significantly reduced the number of dead cells and decreased LDH activity compared with hydrogen peroxide treatment alone (Fig. 6, A–D). To understand the mechanism for this protection, we analyzed metabolites in the cell medium. Hydrogen peroxide increased DHAP and GAP but decreased downstream lactate and pyruvate (Fig. 6, E–F), consistent with our previous report that oxidative stress blocks glyceraldehyde-3-phosphate dehydrogenase in glycolysis (1). Proline decreased DHAP and GAP and offset the reduction of downstream metabolites, including lactate, pyruvate, alanine, glutamate, and glutamine, confirming our finding that proline is important for glucose metabolism.
Figure 6.
Proline protects oxidative damage in RPE cells. A, representative images for EthD staining. RPE cells were stained with EthD, and red fluorescent cells were dead cells. Scale bar, 400 μm. B, quantitation of dead cells stained by EthD by ImageJ. *, p < 0.05 versus Con without proline; #, p < 0.05 versus cells with H2O2. n = 12. C, proline reduced the LDH activity by H2O2 in the medium. LDH activity was -fold change relative to Con without proline. *, p < 0.05 versus Con without proline; #, p < 0.05 versus cells with H2O2. n = 4. D, proline improved cell morphology impaired by H2O2. Scale bar, 400 μm. E and F, significantly changed metabolites in RPE medium (E) and cells (F) with proline after H2O2 treatment for 24 h. n = 4. *, p < 0.05 versus Con without proline; #, p < 0.05 versus cells with H2O2. Ac-Car, acetyl-carnitine; Oxo, 5-oxoproline; M-Nic, methyl-nicotinamide. Error bars, S.D.
Proline-enriched diet improves visual function after induced oxidative damage to the RPE
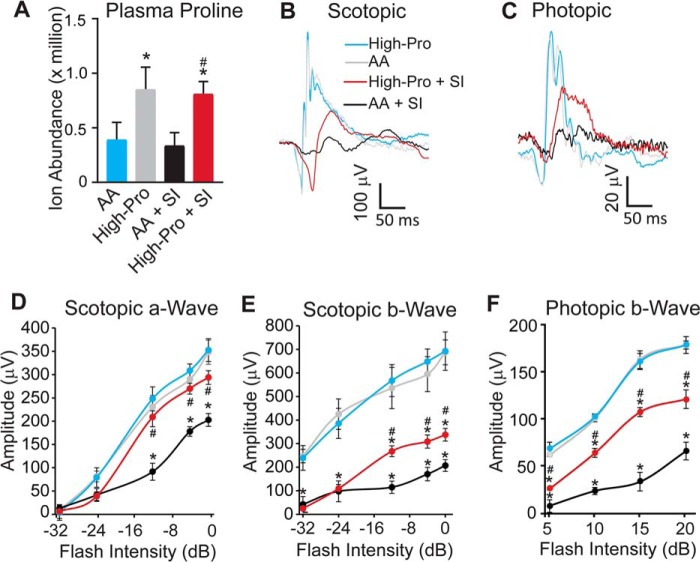
To study whether proline can reduce oxidative damage in vivo, we fed mice with a customized high proline (High-Pro) diet or regular-amino acid diet for 2 weeks. Although the total amounts of calories in the diets were similar, the High-Pro diet consisted of 2% proline (5.7 times higher proline compared with the regular-amino acid diet) (Table S2). We then injected the mice with sodium iodate (SI), which selectively damages the RPE and induces retinal degeneration (32, 33). Mice had similar body weight and food intake between the regular diet and High-Pro diet (Fig. S8). The High-Pro diet increased plasma proline ∼2-fold over the regular diet in mice (Fig. 7A). In the electroretinogram (ERG) testing, there was no difference in either the scotopic or photopic responses between the regular diet and High-Pro diet at baseline (Fig. 7, B–F). We did not quantify photopic a-wave because the response was small with more variations. As expected, SI treatment attenuated ERG responses. However, the High-Pro diet protected the decreased a-wave amplitudes and b-wave amplitudes (Fig. 7, B–F), suggesting that proline improves both rod and cone function. Furthermore, we examined glial cell activation by immunostaining with glial fibrillary acidic protein (Fig. S9). SI caused massive staining of glial fibrillary acidic protein to activate glia, but High-Pro reduced this activation (Fig. S9). To test the effect on photoreceptor viability, we stained cone photoreceptors with peanut agglutinin (PNA) in flat mount retinas. The High-Pro diet showed protection against the photoreceptor damage caused by SI treatment (Fig. 8, A and B). These results suggest that a proline-enriched diet is sufficient to improve acute retinal damage in SI-treated mice.
Figure 7.
Proline improves visual function induced by oxidative damage. A, high-proline diet doubled plasma proline in mice with or without SI. n = 6. *, p < 0.05 versus amino acid (AA) diet alone; #, p < 0.05 versus amino acid diet with SI. B–F, high-proline diet improved ERGs with SI. B, representative raw trace of scotopic response at −12 db; C, raw trace of photopic response at 10 db. n = 10. *, p < 0.05 versus amino acid diet alone; #, p < 0.05 versus amino acid diet with SI. Error bars, S.D.
Figure 8.
Proline reduces SI-induced photoreceptor cell death. A, representative images of flat mount staining with PNA. Scale bar, 10 μm. B, quantification of PNA staining with ImageJ to count stained cone photoreceptors in each field. n = 16 from four animals in each group. p < 0.05 versus amino acid diet alone; #, p < 0.05 versus amino acid diet with SI. Error bars, S.D.
Discussion
The RPE has easy access to different nutrients to support its active metabolism and to meet the energetic demand of the outer retina. However, how the RPE utilizes these nutrients and shares them with the retina is still unclear. In this study, we report that proline is an important substrate for both RPE metabolism and its metabolic communication with the retina (Fig. 9). Proline regulates glucose metabolism and can protect the RPE from oxidative damage.
Figure 9.
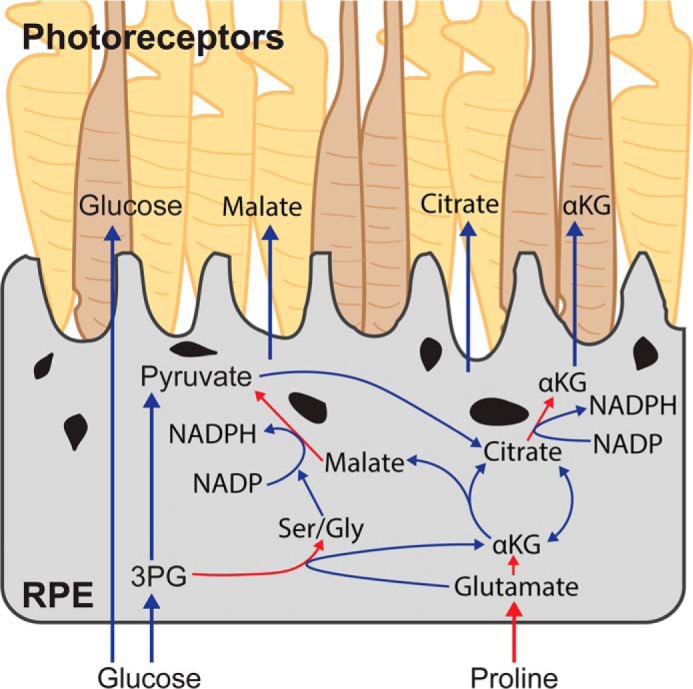
A model of proline-mediated metabolic communication between RPE and photoreceptors. RPE cells utilize proline in RPE to generate mitochondrial intermediates through the TCA cycle and NADPH. These intermediates export RPE to be used by photoreceptors. The activation of NADPH-generation pathways by proline protects RPE from oxidative damage. Additionally, the utilization of proline in RPE may spare the oxidation of glucose, which is a major nutrient in photoreceptors. Red arrows, pathways in proline utilization.
Why do cells consume large amounts of free proline? A recent study examining the proteomes of bacteria, basal eukaryotes, and animals reveals that the demand for free proline increased with the emergence of animals. This may be necessary, as multicellular organisms require the production of proline-rich proteins such as collagens. The consumption of proline may also avoid the depletion of glutamate, as there is a conserved evolution of a fusion protein of glutamyl-tRNA synthetase and prolyl tRNA synthetase (34). We have found that proline increases glutamate and glutamine content in RPE. Glutamate and its products in the TCA cycle, such as αKG, can be exported to support outer retinal metabolism in vitro (2) and in vivo. The oxidation of proline may allow maximal flux through the TCA cycle in the oxidation of glucose and ensure a steady supply of these important intermediates in both the RPE and retina. RPE is active in synthesizing ECM, including different types of collagens, and exports them to maintain Bruch's membrane (35, 36). Radioactive-labeled proline was readily incorporated into collagen in feline and primate RPE cells (37, 38). The ECM remodeling occurs in both early and advanced AMD, simultaneously with RPE metabolic dysfunction (35, 39, 40). Proline metabolism provides a link between ECM remodeling and mitochondrial metabolism. It remains to be determined how cells balance the distribution of proline for collagen synthesis and catabolism in healthy and diseased RPE.
Proline is an efficient mitochondrial fuel. In fly muscles, partial oxidation of proline generates 0.52 mol of ATP/g, which is only slightly lower than that of lipids (0.65 mol ATP/g) but much higher than glucose (0.18 ATP/g) (41). Besides oxidizing proline to generate NADH, PRODH is a flavin-dependent enzyme that is capable of using FAD as a co-factor to drive ATP synthesis (42). This is consistent with our data demonstrating that proline elicits higher maximal O2 in RPE cells. Interestingly, a recent study reveals that RPE has much higher FAD levels than retina (43). In breast cancer cells, mitochondrial metabolism is shifted to proline catabolism to support their growth and form lung metastases (44). We found that hRPEs increasingly rely upon proline consumption in culture. Proline may efficiently fuel mitochondrial metabolism to meet the high metabolic demand during maturation. To control cellular growth and division, a metabolic switch from glycolysis to mitochondrial oxidative phosphorylation (OXPHOS) is a common feature during terminal differentiation (45, 46). Mitochondrial mass and OXPHOS genes are significantly increased during RPE maturation (47). Inhibition of mitochondrial OXPHOS decreases RPE maturation, resulting in RPE dedifferentiation (48). Additionally, we found that proline increases pyruvate in both medium and cells. Pyruvate has been reported to stimulate RPE differentiation and pigmentation in culture (49).
It is surprising that proline enhances serine de novo synthesis through glucose. Serine can be synthesized from the glycolytic intermediate 3-phosphoglycerate (3PG), which involves three enzymes: phosphoglycerate dehydrogenase, phosphoserine aminotransaminase (PSAT), and phosphoserine phosphatase. We did not find any difference in 3PG, indicating increased flux of the latter two enzymes. PSAT catalyzes the conversion of 3-phosphohydroxypyruvate and glutamate to 3-phosphoserine and αKG (Fig. 9). Transamination from glutamate to ketoacid is a common reaction to generate αKG and nonessential amino acids. The increase of glutamate by proline most probably stimulates the transamination reaction of PSAT to increase serine and glycine biosynthesis. Consistently, in a transcriptome database of human RPE/choroid and retina, PSAT transcripts were 30-fold higher than alanine transaminase and 2-fold higher than cytosolic aspartate transaminase (50). Interestingly, glutamine-derived glutamate contributes to PSAT activity to generate αKG, which regulates embryonic stem cell differentiation (51). We reported previously that co-culturing retina with RPE dramatically increases serine and glycine in the retina (2). A pharmacological study has also indicated that retinal glycine content comes from transport rather than de novo synthesis in retinal neurons (52). The serine biosynthesis pathway is tightly linked with the synthesis of phospholipids; the generation of glycine, cysteine, GSH, and NADPH; and the donation of a one-carbon unit (53). The retina has been found to have a higher rate of serine incorporation in phospholipids (53). Glycine and cysteine are substrates used to synthesize GSH, an important antioxidant. We found that the inhibition of proline catabolism decreases both serine and GSH in RPE. Glycine also makes up one-third of collagen, and the serine de novo pathway regulates transforming growth factor β–mediated collagen synthesis in pulmonary fibrosis (20, 54). The transcripts of phosphoglycerate dehydrogenase and PSAT are ∼6-fold higher in RPE/choroid than retina (50), which supports our finding that the RPE may be the major site of serine and glycine synthesis. Proline enhances this pathway to maintain the active metabolism of the outer retina.
How does proline protect RPE cells from oxidative damage? NADPH is needed for reduced GSH to remove reactive oxygen species and manage oxidative stress (55). Major sources of NADPH include the pentose phosphate pathway (PPP), serine-driven one-carbon metabolism, malic enzyme, and NADP-dependent isocitrate dehydrogenases (IDHs) (34, 57). In cancer cells, serine-driven NADPH is comparable with the PPP (34). IDH provides a substantial amount of NADPH in rod photoreceptors (58). The protozoan parasite, Trypanosoma brucei, adapts to its host's environment and relies on proline as a carbon source. Malic enzyme and PPP are two major pathways used by this parasite to generate NADPH through proline to combat oxidative stress (59). In human RPE, we reported that reductive carboxylation through IDH confers on RPE resistance against oxidative damage (1). Our data showed that proline stimulates the serine pathway, increases the malic enzyme pathway to generate pyruvate, and enhances reductive carboxylation in RPE (Fig. 9). Furthermore, inhibition of proline oxidation inhibits serine synthesis and reduces the level of GSH (Fig. 4). The activation of the pathways for NADPH production may contribute to the ability of proline to protect against oxidative damage.
Proline is a nonessential amino acid, but 10–115 mg/liter of proline is included in most of the widely used protocols for human RPE culture medium (60–62). Our data suggest that proline may be critical for RPE maturation and retinal function. To our knowledge, there are five known proline transporters (SLC6A20, SLC6A7, SLC3A1, SLC36A2, and SLC36A4). We analyzed a transcriptional database for maturation and differentiation of human RPE culture (63). Only SLC6A20 is significantly up-regulated in matured and differentiated RPE cells (Fig. S10). SLC6A20, a Na+- and Cl−-dependent proline transporter, is highly enriched in human RPE and mouse RPE (20–22). Further study should elucidate the importance of this transporter in proline metabolism, the synthesis of proline-rich proteins, and RPE function.
One important finding in this study is that RPE utilizes proline to generate mitochondrial intermediates for the outer retina. To maintain visual function and high turnover of outer segments, the retina has an extremely active metabolism (64). Glycolysis and mitochondrial oxidative phosphorylation are two primary pathways for energy metabolism. Like tumors, retinas prefer aerobic glycolysis, also called the Warburg effect, converting most glucose into lactate rather than into mitochondrial intermediates (24, 64). However, the retina also has active oxidative phosphorylation in its mitochondria packed in the ellipsoid (64). The proline-derived intermediates may provide substrates to retinal mitochondria for ATP synthesis and generation of neurotransmitters. Consistently, a high-proline diet improves visual function and increases photoreceptor survival in an acute model of retinal degeneration. We found that proline protects both rod and cone responses, but the scotopic a-wave is better preserved than b-wave. The a-wave reflects the hyperpolarization of the photoreceptors due to closure of sodium ion channels in the outer-segment membrane, and the b-wave originates from photoreceptor-driven bipolar cells induced by neurotransmitter glutamate and the change of potassium (65, 66). It is reported that sodium iodate could directly cause synaptic damage to diminish b-wave (67). Additionally, Müller glial cells also significantly contribute to b-wave by regulating the biosynthesis of glutamate and extracellular potassium concentration (64, 65). Our data found that proline could only partially reduce the glial activation by sodium iodate (Fig. S9). Future investigation of the protection by proline in vivo, including dose, longitude, and other models of retinal degenerations, will yield important information for potential treatment with proline supplementation.
In conclusion, we provide evidence that proline is an important nutrient for RPE. The ability to utilize proline may promote RPE maturation, regulate glucose metabolism, increase the capacity of RPE to defend against oxidative stress, and allow for the export of critical intermediates for utilization by the outer retina.
Experimental procedures
All of the reagents, animals, and key resources are detailed in the key resources form (Table S3).
Cell culture
Human fetal RPE cells were isolated and primarily cultured as described previously (2, 68). The protocol was approved by the University of Washington Institutional Review Board. All procedures conform to the ethical principles outlined in the Declaration of Helsinki. RPE cells were plated in 12- or 24-well plates in RPE medium consisting of α-minimum Eagle's medium, nonessential amino acids, N1 supplement, 1% (v/v) FBS, taurine, hydrocortisone, triiodothryonine, and penicillin-streptomycin. The cells were changed into fresh medium 24 or 48 h before harvesting medium for metabolite analysis. For proline supplementation and the hydrogen peroxide experiment, the RPE cells were changed into clear DMEM with 5.5 mm glucose and 1% (v/v) FBS. iPS cells were differentiated to RPE using the speed differentiation protocol developed by Buchholz et al. (69). RPE cells were either manually picked or trypsinized, depending on the quality of differentiation. The RPE was then plated on Matrigel® matrix and cultured as described previously in an RPE medium containing 5% FBS and 10 μm ROCK inhibitor (Y-27632 dihydrochloride, Tocris Bioscience). After reaching confluence, the FBS concentration in the medium was decreased to 1%, and ROCK inhibitor was no longer added to the medium. hRPE and iPS RPE cells were plated at 0.25 million cells in a 24-well plate, 0.5 million cells for 12-well plates, and 0.8 million cells for 6-well plates. A-RPE 19, hRPE1, and HEK 293 T cells were plated seeded at 0.8 million cells/well in a 6-well plate and grown for a week in DMEM/F12 medium supplemented with nonessential amino acids and 5% (v/v) FBS.
Cell death staining
The cell death staining was performed as reported previously (1). RPE cells were grown for 4–5 weeks in 24-well plates and treated with 1 mm H2O2 with or without 1 mm pyruvate in DMEM. Medium was removed and replaced with 500 μl of KRB and EthD dye. Bright field and fluorescent images were taken with the Evos digital inverted microscope (AMG) and counted using Fiji or ImageJ software.
LDH activity assay
Culture medium (20 μl) was incubated with enzyme assay mix as described (1). The change of absorbance (340 nm) over time was read by a microplate reader.
Metabolite analysis
The metabolites harvested from retinal explants and human RPE were analyzed with GC MS or LC MS as we described in detail before (31, 70). For medium metabolites, 10 μl of medium or plasma was mixed with 40 μl of cold methanol to extract metabolites for analysis with GC MS or LC MS. The intracellular metabolites were extracted by using 80% cold methanol. One mouse retina or RPE/choroid was snap-frozen in liquid nitrogen and homogenized with 80% cold methanol to extract metabolites. For GC MS, the samples were derivatized by methoxyamine and N-tertbutyldimethylsilyl-N-methyltrifluoroacetamide and analyzed by the Agilent 7890B/5977B GC MS system with a DB-5MS column (30 m × 0.25 mm × 0.25-μm film). Mass spectra were collected from 80 to 600 m/z under selective ion monitoring mode. The data were analyzed by Agilent MassHunter Quantitative Analysis software, and natural abundance was corrected by ISOCOR software. LC MS used a Shimadzu LC Nexera X2 UHPLC coupled with a QTRAP 5500 LC MS/MS (AB Sciex). An ACQUITY UPLC BEH Amide analytic column (2.1 × 50 mm, 1.7 μm; Waters) was used for chromatographic separation. Each metabolite was tuned with standards for optimal transitions. The extracted MRM peaks were integrated using MultiQuant version 3.0.2 software (AB Sciex). Table S1 lists the detailed parameters for the measured metabolites.
Animals
WT male C57 B6/J mice at 6 weeks were purchased from Jackson Laboratory. The regular-amino acid diet and high-proline diet were customized at Envigo (Table S2). We monitored the food intake and animal body weight weekly to calculate food intake and change of body weight. After being fed for 2 weeks, animals received a single intraperitoneal injection of sodium iodate (35 mg/kg) or PBS. Mouse experiments were performed in accordance with National Institutes of Health guidelines, and the protocol was approved by the Institutional Animal Care and Use Committee of West Virginia University.
In vivo infusion of [13C]proline
Arterial and venous catheters were surgically implanted into the jugular veins and carotid artery of animals with a vascular access button on the back 1 week prior to infusions (25). After fasting for 6 h in the morning, mice were constantly infused with [13C]proline through the jugular vein for 4 h using the Pump 11 Elite Infuse pump and mouse infusion setup (Instech Laboratories). The mouse infusion setup included a tether and swivel system that enabled free movement of the infused mice in the cage. 10 μl of blood was collected from an arterial catheter. At the end of infusion, the mouse was euthanized by cervical dislocation. Retinas and RPE/choroid were quickly dissected with a cut and pick method (31) and snap-frozen in liquid nitrogen for metabolite analysis.
Electroretinography
Mice were dark-adapted overnight before electroretinography using the UTAS visual diagnostic system with BigShot Ganzfeld with a UBA-4200 amplifier and interface (LKC Technologies, Gaithersburg, MD) (56). Mice were anesthetized with isoflurane, and eyes were dilated with 2.5% phenylephrine (Paragon) and 1% tropicamide (Sandoz). Scotopic ERG recordings were elicited using flashes of LED white light at increasing flash intensities (−32, −24, −16, −12, −4, and 0 db) under red light. Responses were averaged at each light intensity. Values were normalized to the baseline, and each eye was evaluated separately to determine the a-wave and b-wave amplitudes.
Statistics
The significance of differences between means was determined by unpaired two-tailed t tests or analysis of variance with an appropriate post hoc test. p < 0.05 was considered to be significant using GraphPad Prism version 7.
Author contributions
J. D. conceptualization; M. Y., A. L. E., Y. W., S. Z., A. H., D. L., R. Z., J. H., M. D., C. Z., and J. D. investigation; M. Y., J. R. C., and J. D. writing; J. R. C. and J. D. funding acquisition; J. R. C. and J. D. supervision.
Supplementary Material
Acknowledgment
We thank Allison Grenell for assistance with animal electroretinography.
This work was supported by National Institutes of Health Grant EY026030 (to J. R. C. and J. D.), the BrightFocus Foundation (to J. D. and J. R. C.), the Retina Research Foundation (to J. D.), and an unrestricted grant from Research to Prevent Blindness (to J. R. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1–S10.
- RPE
- retinal pigment epithelium
- hRPE
- human fetal RPE
- αKG
- α-ketoglutarate
- 3PG
- 3-phosphoglyceric acid
- PEP
- phosphoenolpyruvate
- AMD
- age-related macular degeneration
- TCA
- tricarboxylic acid
- ECM
- extracellular matrix
- PRODH
- proline dehydrogenase
- P5CS
- pyrroline-5-carboxylate synthase
- OAT
- ornithine aminotransferase
- iPS cell
- induced pluripotent stem cell
- T2FA
- tetrahydro-2-furoic acid
- EthD
- ethidium homodimer
- LDH
- lactate dehydrogenase
- SI
- sodium iodate
- ERG
- electroretinogram
- OXPHOS
- oxidative phosphorylation
- PSAT
- phosphoserine aminotransaminase
- PPP
- pentose phosphate pathway
- IDH
- isocitrate dehydrogenase
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- Con
- RPE medium with DMSO after 48-h culture.
References
- 1. Du J., Yanagida A., Knight K., Engel A. L., Vo A. H., Jankowski C., Sadilek M., Tran V. T., Manson M. A., Ramakrishnan A., Hurley J. B., and Chao J. R. (2016) Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 113, 14710–14715 10.1073/pnas.1604572113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chao J. R., Knight K., Engel A. L., Jankowski C., Wang Y., Manson M. A., Gu H., Djukovic D., Raftery D., Hurley J. B., and Du J. (2017) Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J. Biol. Chem. 292, 12895–12905 10.1074/jbc.M117.788422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phang J. M., Liu W., and Zabirnyk O. (2010) Proline metabolism and microenvironmental stress. Annu. Rev. Nutr. 30, 441–463 10.1146/annurev.nutr.012809.104638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phang J. M., Donald S. P., Pandhare J., and Liu Y. (2008) The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino acids 35, 681–690 10.1007/s00726-008-0063-4 [DOI] [PubMed] [Google Scholar]
- 5. Liang X., Zhang L., Natarajan S. K., and Becker D. F. (2013) Proline mechanisms of stress survival. Antioxid. Redox Signal. 19, 998–1011 10.1089/ars.2012.5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben Rejeb K., Abdelly C., and Savouré A. (2014) How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 80, 278–284 10.1016/j.plaphy.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L., and Becker D. F. (2015) Connecting proline metabolism and signaling pathways in plant senescence. Front. Plant Sci. 6, 552 10.3389/fpls.2015.00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hancock C. N., Liu W., Alvord W. G., and Phang J. M. (2016) Co-regulation of mitochondrial respiration by proline dehydrogenase/oxidase and succinate. Amino Acids 48, 859–872 10.1007/s00726-015-2134-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDonald A. E., Pichaud N., and Darveau C. A. (2018) “Alternative” fuels contributing to mitochondrial electron transport: importance of non-classical pathways in the diversity of animal metabolism. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 224, 185–194 10.1016/j.cbpb.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 10. Edwards C., Canfield J., Copes N., Brito A., Rehan M., Lipps D., Brunquell J., Westerheide S. D., and Bradshaw P. C. (2015) Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet. 16, 8 10.1186/s12863-015-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schroeder E. A., and Shadel G. S. (2012) Alternative mitochondrial fuel extends life span. Cell Metab. 15, 417–418 10.1016/j.cmet.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Kuhlow D., Guthke R., Platzer M., Kahn C. R., and Ristow M. (2012) Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial l-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465 10.1016/j.cmet.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolthuis D. F., van Asbeck E., Mohamed M., Gardeitchik T., Lim-Melia E. R., Wevers R. A., and Morava E. (2014) Cutis laxa, fat pads and retinopathy due to ALDH18A1 mutation and review of the literature. Eur. J. Paediatr. Neurol. 18, 511–515 10.1016/j.ejpn.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 14. O'Donnell J. J., Sandman R. P., and Martin S. R. (1978) Gyrate atrophy of the retina: inborn error of l-ornithin:2-oxoacid aminotransferase. Science 200, 200–201 10.1126/science.635581 [DOI] [PubMed] [Google Scholar]
- 15. Wang T., Lawler A. M., Steel G., Sipila I., Milam A. H., and Valle D. (1995) Mice lacking ornithine-δ-aminotransferase have paradoxical neonatal hypoornithinaemia and retinal degeneration. Nat. Genet. 11, 185–190 10.1038/ng1095-185 [DOI] [PubMed] [Google Scholar]
- 16. Wang T., Milam A. H., Steel G., and Valle D. (1996) A mouse model of gyrate atrophy of the choroid and retina: early retinal pigment epithelium damage and progressive retinal degeneration. J. Clin. Invest. 97, 2753–2762 10.1172/JCI118730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu C. A., Lin W. W., Obie C., and Valle D. (1999) Molecular enzymology of mammalian Δ1-pyrroline-5-carboxylate synthase: alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J. Biol. Chem. 274, 6754–6762 10.1074/jbc.274.10.6754 [DOI] [PubMed] [Google Scholar]
- 18. Ueda M., Masu Y., Ando A., Maeda H., Del Monte M. A., Uyama M., and Ito S. (1998) Prevention of ornithine cytotoxicity by proline in human retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 39, 820–827 [PubMed] [Google Scholar]
- 19. Ando A., Ueda M., Uyama M., Masu Y., Okumura T., and Ito S. (2000) Heterogeneity in ornithine cytotoxicity of bovine retinal pigment epithelial cells in primary culture. Exp. Eye Res. 70, 89–96 10.1006/exer.1999.0750 [DOI] [PubMed] [Google Scholar]
- 20. Bennis A., Gorgels T. G., Ten Brink J. B., van der Spek P. J., Bossers K., Heine V. M., and Bergen A. A. (2015) Comparison of mouse and human retinal pigment epithelium gene expression profiles: potential implications for age-related macular degeneration. PLoS One 10, e0141597 10.1371/journal.pone.0141597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao J. L., Yu J., Huang K., Hu J., Diemer T., Ma Z., Dvash T., Yang X. J., Travis G. H., Williams D. S., Bok D., and Fan G. (2010) Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum. Mol. Genet. 19, 4229–4238 10.1093/hmg/ddq341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strunnikova N. V., Maminishkis A., Barb J. J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A. O., Stambolian D., Abecasis G., Swaroop A., Munson P. J., and Miller S. S. (2010) Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 19, 2468–2486 10.1093/hmg/ddq129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao X. R., Huang H., and Kim H. (2019) Genome-wide association analyses identify 139 loci associated with macular thickness in the UK Biobank cohort. Hum. Mol. Genet. 28, 1162–1172 10.1093/hmg/ddy422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du J., Cleghorn W., Contreras L., Linton J. D., Chan G. C., Chertov A. O., Saheki T., Govindaraju V., Sadilek M., Satrústegui J., and Hurley J. B. (2013) Cytosolic reducing power preserves glutamate in retina. Proc. Natl. Acad. Sci. U.S.A. 110, 18501–18506 10.1073/pnas.1311193110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ayala J. E., Bracy D. P., Malabanan C., James F. D., Ansari T., Fueger P. T., McGuinness O. P., and Wasserman D. H. (2011) Hyperinsulinemic-euglycemic clamps in conscious, unrestrained mice. J. Vis. Exp., pii: 3188 10.3791/3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davidson S. M., Papagiannakopoulos T., Olenchock B. A., Heyman J. E., Keibler M. A., Luengo A., Bauer M. R., Jha A. K., O'Brien J. P., Pierce K. A., Gui D. Y., Sullivan L. B., Wasylenko T. M., Subbaraj L., Chin C. R., et al. (2016) Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 10.1016/j.cmet.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnan N., Dickman M. B., and Becker D. F. (2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 44, 671–681 10.1016/j.freeradbiomed.2007.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seiler N. (2000) Ornithine aminotransferase, a potential target for the treatment of hyperammonemias. Curr. Drug Targets 1, 119–153 10.2174/1389450003349254 [DOI] [PubMed] [Google Scholar]
- 29. Adachi R., Okada K., Skene R., Ogawa K., Miwa M., Tsuchinaga K., Ohkubo S., Henta T., and Kawamoto T. (2017) Discovery of a novel prolyl-tRNA synthetase inhibitor and elucidation of its binding mode to the ATP site in complex with l-proline. Biochem. Biophys. Res. Commun. 488, 393–399 10.1016/j.bbrc.2017.05.064 [DOI] [PubMed] [Google Scholar]
- 30. Xiong G., Deng L., Zhu J., Rychahou P. G., and Xu R. (2014) Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 14, 1 10.1186/1471-2407-14-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu S., Yam M., Wang Y., Linton J. D., Grenell A., Hurley J. B., and Du J. (2018) Impact of euthanasia, dissection and postmortem delay on metabolic profile in mouse retina and RPE/choroid. Exp. Eye Res. 174, 113–120 10.1016/j.exer.2018.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu L., Kong L., Wang J., and Ash J. D. (2018) Stimulation of AMPK prevents degeneration of photoreceptors and the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 115, 10475–10480 10.1073/pnas.1802724115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chowers G., Cohen M., Marks-Ohana D., Stika S., Eijzenberg A., Banin E., and Obolensky A. (2017) Course of sodium iodate-induced retinal degeneration in albino and pigmented mice. Invest. Ophthalmol. Vis. Sci. 58, 2239–2249 10.1167/iovs.16-21255 [DOI] [PubMed] [Google Scholar]
- 34. Eswarappa S. M., Potdar A. A., Sahoo S., Sankar S., and Fox P. L. (2018) Metabolic origin of the fused aminoacyl tRNA synthetase, glutamyl-prolyl tRNA synthetase. J. Biol. Chem. 293, 19148–19156 10.1074/jbc.RA118.004276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nita M., Strzałka-Mrozik B., Grzybowski A., Mazurek U., and Romaniuk W. (2014) Age-related macular degeneration and changes in the extracellular matrix. Med. Sci. Monit. 20, 1003–1016 10.12659/MSM.889887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kigasawa K., Ishikawa H., Obazawa H., Minamoto T., Nagai Y., and Tanaka Y. (1998) Collagen production by cultured human retinal pigment epithelial cells. Tokai J. Exp. Clin. Med. 23, 147–151 [PubMed] [Google Scholar]
- 37. Li W., Stramm L. E., Aguirre G. D., and Rockey J. H. (1984) Extracellular matrix production by cat retinal pigment epithelium in vitro: characterization of type IV collagen synthesis. Exp. Eye Res. 38, 291–304 10.1016/0014-4835(84)90167-2 [DOI] [PubMed] [Google Scholar]
- 38. Hirata A., and Feeney-Burns L. (1992) Autoradiographic studies of aged primate macular retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 33, 2079–2090 [PubMed] [Google Scholar]
- 39. Hillenkamp J., Hussain A. A., Jackson T. L., Cunningham J. R., and Marshall J. (2004) The influence of path length and matrix components on ageing characteristics of transport between the choroid and the outer retina. Invest. Ophthalmol. Vis. Sci. 45, 1493–1498 10.1167/iovs.03-0765 [DOI] [PubMed] [Google Scholar]
- 40. Fisher C. R., and Ferrington D. A. (2018) Perspective on AMD pathobiology: a bioenergetic crisis in the RPE. Invest. Ophthalmol. Vis. Sci. 59, AMD41–AMD47 10.1167/iovs.18-24289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bursell E., Billing K. J., Hargrove J. W., McCabe C. T., and Slack E. (1973) The supply of substrates to the flight muscle of tsetse flies. Trans. R. Soc. Trop. Med. Hyg. 67, 296 10.1016/0035-9203(73)90222-8 [DOI] [PubMed] [Google Scholar]
- 42. Liu L. K., Becker D. F., and Tanner J. J. (2017) Structure, function, and mechanism of proline utilization A (PutA). Arch. Biochem. Biophys. 632, 142–157 10.1016/j.abb.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sinha T., Makia M., Du J., Naash M. I., and Al-Ubaidi M. R. (2018) Flavin homeostasis in the mouse retina during aging and degeneration. J. Nutr. Biochem. 62, 123–133 10.1016/j.jnutbio.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elia I., Broekaert D., Christen S., Boon R., Radaelli E., Orth M. F., Verfaillie C., Grünewald T. G. P., and Fendt S. M. (2017) Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 8, 15267 10.1038/ncomms15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agathocleous M., and Harris W. A. (2013) Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 23, 484–492 10.1016/j.tcb.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 46. Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Hamm M., Gage F. H., and Hunter T. (2016) Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife 5, e13374 10.7554/eLife.13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iacovelli J., Rowe G. C., Khadka A., Diaz-Aguilar D., Spencer C., Arany Z., and Saint-Geniez M. (2016) PGC-1α induces human RPE oxidative metabolism and antioxidant capacity. Invest. Ophthalmol. Vis. Sci. 57, 1038–1051 10.1167/iovs.15-17758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adijanto J., and Philp N. J. (2014) Cultured primary human fetal retinal pigment epithelium (hfRPE) as a model for evaluating RPE metabolism. Exp. Eye Res. 126, 77–84 10.1016/j.exer.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahmado A., Carr A. J., Vugler A. A., Semo M., Gias C., Lawrence J. M., Chen L. L., Chen F. K., Turowski P., da Cruz L., and Coffey P. J. (2011) Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Invest. Ophthalmol. Vis. Sci. 52, 7148–7159 10.1167/iovs.10-6374 [DOI] [PubMed] [Google Scholar]
- 50. Whitmore S. S., Wagner A. H., DeLuca A. P., Drack A. V., Stone E. M., Tucker B. A., Zeng S., Braun T. A., Mullins R. F., and Scheetz T. E. (2014) Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res. 129, 93–106 10.1016/j.exer.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hwang I. Y., Kwak S., Lee S., Kim H., Lee S. E., Kim J. H., Kim Y. A., Jeon Y. K., Chung D. H., Jin X., Park S., Jang H., Cho E. J., and Youn H. D. (2016) Psat1-dependent fluctuations in α-ketoglutarate affect the timing of ESC differentiation. Cell Metabol. 24, 494–501 10.1016/j.cmet.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 52. Gu H., Du J., Carnevale Neto F., Carroll P. A., Turner S. J., Chiorean E. G., Eisenman R. N., and Raftery D. (2015) Metabolomics method to comprehensively analyze amino acids in different domains. Analyst 140, 2726–2734 10.1039/C4AN02386B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hirabayashi Y., and Furuya S. (2008) Roles of l-serine and sphingolipid synthesis in brain development and neuronal survival. Prog. Lipid Res. 47, 188–203 10.1016/j.plipres.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 54. Marmorstein L. Y., McLaughlin P. J., Peachey N. S., Sasaki T., and Marmorstein A. D. (2007) Formation and progression of sub-retinal pigment epithelium deposits in Efemp1 mutation knock-in mice: a model for the early pathogenic course of macular degeneration. Hum. Mol. Genet. 16, 2423–2432 10.1093/hmg/ddm199 [DOI] [PubMed] [Google Scholar]
- 55. Fan J., Ye J., Kamphorst J. J., Shlomi T., Thompson C. B., and Rabinowitz J. D. (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 10.1038/nature13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Y., Grenell A., Zhong F., Yam M., Hauer A., Gregor E., Zhu S., Lohner D., Zhu J., and Du J. (2018) Metabolic signature of the aging eye in mice. Neurobiol. Aging 71, 223–233 10.1016/j.neurobiolaging.2018.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ribas de Pouplana L. (2018) Genetic code and metabolism: the perpetual waltz. J. Biol. Chem. 293, 19157–19158 10.1074/jbc.H118.006600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Newman A. M., Gallo N. B., Hancox L. S., Miller N. J., Radeke C. M., Maloney M. A., Cooper J. B., Hageman G. S., Anderson D. H., Johnson L. V., and Radeke M. J. (2012) Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 4, 16 10.1186/gm315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Allmann S., Morand P., Ebikeme C., Gales L., Biran M., Hubert J., Brennand A., Mazet M., Franconi J. M., Michels P. A., Portais J. C., Boshart M., and Bringaud F. (2013) Cytosolic NADPH homeostasis in glucose-starved procyclic Trypanosoma brucei relies on malic enzyme and the pentose phosphate pathway fed by gluconeogenic flux. J. Biol. Chem. 288, 18494–18505 10.1074/jbc.M113.462978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fronk A. H., and Vargis E. (2016) Methods for culturing retinal pigment epithelial cells: a review of current protocols and future recommendations. J. Tissue Eng. 7, 2041731416650838 10.1177/2041731416650838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maminishkis A., Chen S., Jalickee S., Banzon T., Shi G., Wang F. E., Ehalt T., Hammer J. A., and Miller S. S. (2006) Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest. Ophthalmol. Vis. Sci. 47, 3612–3624 10.1167/iovs.05-1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hu J., and Bok D. (2001) A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol. Vis. 7, 14–19 [PubMed] [Google Scholar]
- 63. Radeke M. J., Radeke C. M., Shih Y. H., Hu J., Bok D., Johnson L. V., and Coffey P. J. (2015) Restoration of mesenchymal retinal pigmented epithelial cells by TGFβ pathway inhibitors: implications for age-related macular degeneration. Genome Med. 7, 58 10.1186/s13073-015-0183-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hurley J. B., Lindsay K. J., and Du J. (2015) Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 93, 1079–1092 10.1002/jnr.23583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bui B. V., Hu R. G., Acosta M. L., Donaldson P., Vingrys A. J., and Kalloniatis M. (2009) Glutamate metabolic pathways and retinal function. J. Neurochem. 111, 589–599 10.1111/j.1471-4159.2009.06354.x [DOI] [PubMed] [Google Scholar]
- 66. Reichenbach A., Henke A., Eberhardt W., Reichelt W., and Dettmer D. (1992) K+ ion regulation in retina. Can. J. Physiol. Pharmacol. 70, S239–S247 10.1139/y92-267 [DOI] [PubMed] [Google Scholar]
- 67. Wang J., Iacovelli J., Spencer C., and Saint-Geniez M. (2014) Direct effect of sodium iodate on neurosensory retina. Invest. Ophthalmol. Vis. Sci. 55, 1941–1953 10.1167/iovs.13-13075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sonoda S., Spee C., Barron E., Ryan S. J., Kannan R., and Hinton D. R. (2009) A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat. Protoc. 4, 662–673 10.1038/nprot.2009.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Buchholz D. E., Pennington B. O., Croze R. H., Hinman C. R., Coffey P. J., and Clegg D. O. (2013) Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl. Med. 2, 384–393 10.5966/sctm.2012-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Du J., Linton J. D., and Hurley J. B. (2015) Probing metabolism in the intact retina using stable isotope tracers. Methods Enzymol. 561, 149–170 10.1016/bs.mie.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.