Abstract
Hedgehog signaling pathway originally identified in the fruit fly Drosophila is an evolutionarily conserved signaling mechanism with crucial roles in embryogenesis, growth and patterning. It exerts its biological effect through a signaling mechanism that terminates at glioma-associated oncogene (GLI) transcription factors which alternate between activator and repressor forms and mediate various responses. The important components of the pathway include the hedgehog ligands (SHH), the Patched (PTCH) receptor, Smoothened (SMO), Suppressor of Fused (SuFu) and GLI transcription factors. Activating or inactivating mutations in key genes cause uncontrolled activation of the pathway in a ligand independent manner. The ligand-dependent aberrant activation of the hedgehog pathway causing overexpression of hedgehog pathway components and its target genes occurs in autocrine as well as paracrine fashion. In adults, aberrant activation of hedgehog signaling has been linked to birth defects and multiple solid cancers. In this review, we assimilate data from recent studies to understand the mechanism of functioning of the hedgehog signaling pathway, role in cancer, its association in various solid malignancies and the current strategies being used to target this pathway for cancer treatment.
Introduction
Many cell signaling processes regulate development, and advances in understanding these processes have revealed their roles in the maintenance of normal function, as well as in the development of various diseases. One such pathway is the evolutionarily conserved hedgehog (HH) signaling, which regulates cell functions that are vital for normal development and homeostasis of tissues including skin, bone, and intestine. This signaling cascade is dysregulated in a variety of diseases, and its function during embryonic development is frequently recapitulated in both disease and repair processes occurring later in life. Inactivation of this pathway during development causes birth defects whereas hyperactivation in adult life is associated with tumorigenesis. Many components of this pathway function either as a tumor suppressors or proto-oncogenes making it an important therapeutic target [1].
This signal transduction cascade was first elucidated by the developmental biologists Eric Wieschaus and Christiane Nüsslein-Volhard in the common fruit fly while studying the fly body plan [2]. During embryonic development, this pathway plays critical roles in cellular differentiation, proliferation in a tissue-specific manner to form a viable organism. It exerts its biological effects through a chain of events on the target cells that culminates in the sequestration of activator or repressor forms of GLI transcription factors in the nucleus. The main components of the HH signaling pathway include HH proteins (ligands); Sonic HH [SHH], Indian HH [IHH] and Desert HH [DHH]; Patched receptor (PTCH1, PTCH2); Smoothened receptor (SMO); Suppressor of fused homolog (SUFU); kinesin protein Kif7; protein kinase A (PKA) and GLI transcription factors. GLI transcription factors on activation migrate from the cytoplasm to the nucleus and stimulate the transcription of the target genes responsible for various physiological processes [3]. There is ample evidence to prove that hedgehog signaling is involved in various stages of carcinogenesis in vide variety of tumors and is associated with tissue invasion and metastatic potential. Several small molecule inhibitors and antibodies targeting this pathway are being developed and many more are undergoing clinical trials [4], [5], [6].
HH Signaling in Humans
Humans share many of the core components of the HH signaling pathway with Drosophila and mechanism of signal transduction is also conserved to some extent [7]. The pathway plays an essential role during embryonic development in cell proliferation, differentiation and maintaining tissue polarity. In adults, it is responsible for stem cell renewal, organ homeostasis, tissue repair and oncogenesis [4].
In humans, the HH ligands have evolved into three homologues Sonic HH, Indian HH and Desert HH contrasting a single HH protein in Drosophila. All the three proteins bind to the receptors with equal affinity but are expressed in different tissues and elicit a different biological response [8]. The most widely studied is the SHH which is expressed in a wide range of tissues including the digestive tract. The Drosophila HH and its mammalian counterparts share a similar auto processing pattern in the endoplasmic reticulum with minor differences. In mammals, the HHAT gene encodes an enzyme that acts within the secretory pathway to catalyze amino-terminal palmitoylation of Hedge domain a function performed by a Skinny HH in Drosophila [9]. In addition to receptors PTCH1, PTCH2 co-receptors Cdo, Boc and Gas1 are essential for HH pathway activation in multiple tissues. During the pathway activation ligands (SHH, IHH, DHH) bind to receptor PTCH1, PTCH2 and activates them. This relieves repression on smoothened (SMO) which is critical for activating the downstream molecules. This ultimately results in the activation of Zinc finger Glioma-associated oncogene (GLI) transcription factors [10]. The mammalian counterpart of the Drosophila Ci is GLI transcription factors which have three homologues, GLI1, GLI2 and GLI3. GLI2 and GLI3 are bifunctional transcription factors with both C-terminal activation and N- terminal repression domains and can function both as activators or repressors whereas GLI1, which lacks the N-terminal repressor domains, functions exclusively as a transcriptional activator [10]. Activator forms of GLI transcription factors on entering nucleus promote transcription of various target genes including those involved in HH pathway feedback such as GLI1, and PTCH1, proliferation-promoting genes such as Cyclin-D1, and MYC, cell cycle regulators CCND2 and CCNE1, apoptotic regulator bcl2, those involved in angiogenesis ANG1/2, epithelial-to-mesenchymal transition SNAIL and stem cell self-renewal NANOG and SOX2 [5], [11]. The ultimate outcome of the pathway depends on the balance between activator and repressor forms of GLI proteins.
The exact molecular mechanism of signal transduction cascade from SMO to the GLI proteins is not yet fully elucidated, but studies have suggested mammalian HH signaling requires the presence of primary cilia to which SMO and other downstream pathway components must translocate to accomplish the activation of GLI transcription factors [12]. In mammalian systems PTCH1, PTCH2 is present in and around primary cilia. On ligand binding, these receptors dissipate to be replaced by SMO [13]. SMO is phosphorylated by PKA and Casein Kinase 1 and translocated to the cilium [14]. The localization of SMO to the primary cilia is necessary, although not sufficient, step in its activation, in response to which the GLI transcription factors, complexed with Sufu, are transported to the tip of the Primary cilia [15]. This translocation appears to be essential for dissociation of the GLI-SuFu complex and hence for GLI activation promoting the import of their full-length forms into the nucleus where they induce transcription of target genes. The kinesin protein KIF7 coordinates HH signal transduction at the tip of cilia and prevents GLI3 cleavage into a repressor form in the presence of HH stimulation [16].
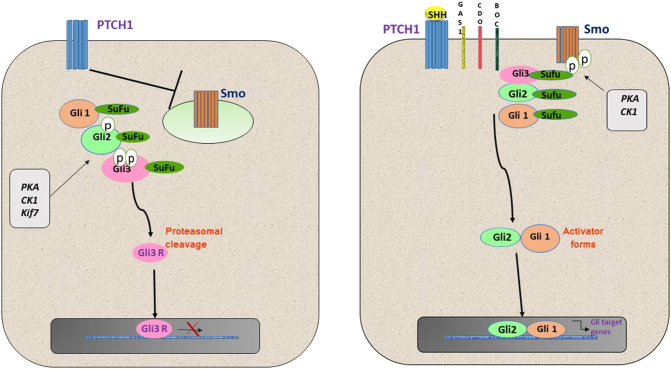
In absence of ligand binding SMO present in intracellular vesicles in the cytoplasm and its translocation to primary cilia is inhibited by Patched receptors. Under these conditions, KIF7 primarily localizes to the basal body of the primary cilium, a structure that is rich in proteasomes. GLI 2 and 3 are phosphorylated by PKA, Glycogen synthase kinase (GSK3) and Kif7. This results in converting GLI3, and GLI2, to their repressor forms via proteolytic processing which blocks transcription [17]. SuFu is a strong negative regulator of the pathway. It binds directly to GLI proteins preventing their translocation into the nucleus [18]. SuFu can also enter the nucleus where it can bind to GLI-binding sequences in the DNA and inhibit gene transcription [19]. HHIP another important transmembrane protein functions to attenuate HH signaling. It binds to HH ligand and promotes their uptake by endocytosis for lysosomal degradation [20]. The diagrammatic representation of human HH signaling pathway in OFF and ON state is shown in Figure 1. (See Table 1.)
Figure 1.
The diagrammatic representation of human hedgehog signaling pathway.
Table 1.
This table describes hedgehog inhibitors which are FDA approved or undergoing clinical trials for various cancers.
| Compound | Target molecule | Stage | Cancer | Reference |
|---|---|---|---|---|
| GDC-0449 vismodegib |
SMO | FDA approved | Basal cell carcinoma | [124] |
| GDC-0449 vismodegib |
SMO | Undergoing clinical trials | Pancreatic cancer Colorectal cancer Prostate cancer Breast cancer |
[125], [126] |
| Glasdegib | SMO | FDA approved | Acute myeloid leukemia | [127] |
| LDE225 Sonidegib |
SMO | FDA approved | Basal cell carcinoma | [128] |
| IPI-926 Saridegib |
SMO | Undergoing clinical trials | chondrosarcoma | [126] |
| Taladegib | SMO | Advanced solid tumors Esophageal cancer Colon cancer |
[129] | |
| BMS-833923 | SMO | Undergoing clinical trials | Advanced or metastatic gastric, gastroesophageal, or esophageal adenocarcinomas |
[125], [126] |
| Itraconazole | SMO | Undergoing clinical trials | Basal cell carcinoma Prostate cancer Non-small cell lung cancer |
[130] |
| Vitamin D | SMO | Undergoing clinical trials | Basal cell carcinoma Acute myeloid leukemia |
[126] |
| Arsenic trioxide | GLI | Undergoing clinical trials | Advanced Basal cell carcinoma Acute myeloid leukemia |
[131] |
Non-Canonical HH Signaling
HH signaling is complex and canonical and non-canonical signaling exists parallel to elicit various cellular responses. Evidence exists for at least two distinct mechanisms of non-canonical HH signaling [21]. Type I in which PTCH1 functions independently of SMO and Type II which functions through SMO independently of GLI [22]. Studies have proposed that the PTCH1 induces apoptosis in a non-canonical manner. In the absence of ligand binding PTCH functions as a dependence receptor inducing apoptosis. SHH functions as a survival factor and blocks the proapoptotic activity of PTCH on the binding [21]. In the absence of SHH, PTCH recruit’s adaptor protein complex that includes DRAL, the CARD containing domain proteins TUCAN or NALP1 and the apical caspase-9. PTCH triggers caspase-9 activation and enhances cell death via a caspase-9-dependent mechanism [23]. A study by Chinchilla et al. showed that none of the HH ligands is able to induce expression of GLI target genes in endothelial cells suggesting that endothelial cells do not respond to HH signaling through the canonical pathway [24]. Chang et al. showed HH binding to PTCH1 stimulate extracellular signal-regulated kinase (Erk 1/2) activation and this activation is insensitive to the small molecule SMO antagonists and occurs in a cell line that does not express SMO suggesting non-canonical signaling mechanism [25]. The PTCH1 tumor suppressor regulates cell cycle at a G2/M checkpoint by binding phosphorylated form of cyclin B1 and preventing its translocation into the nucleus [26]. SHH binding to PTCH1 induces a conformational change in PTCH11 that increases its affinity for GRK2 causing the release of cyclin B1 and allowing its nuclear translocation and consequent completion of mitosis [27]. This direct association of cyclin B1 and GRK2 with PTCH1 and the changes brought about by SHH binding suggest that this mechanism does not require SMO. Studies have also shown SMO dependent non-canonical signaling exists through the activation of small GTPases [28] although future studies designed to elucidate the exact mechanisms are needed.
HH Signaling and Cancer
Since HH signaling has been implicated in embryogenesis, cell differentiation and cell proliferation it is not difficult to understand that aberrant activation of this pathway can contribute to tumorigenesis. Over the years mounting evidence has accumulated which suggests that the HH signaling pathway is activated and plays a significant role in the initiation, progression, invasion and maintenance of cancers.
All the core components of the pathway function either as tumor suppressors or proto-oncogenes with one tightly regulated by the other. Figure 2 describes the core HH pathway components which function as proto-oncogenes and tumor suppressors. The pathway proto-oncogenes become oncogenic when up-regulated whereas tumor suppressors result in tumor growth when inactivated.
Figure 2.
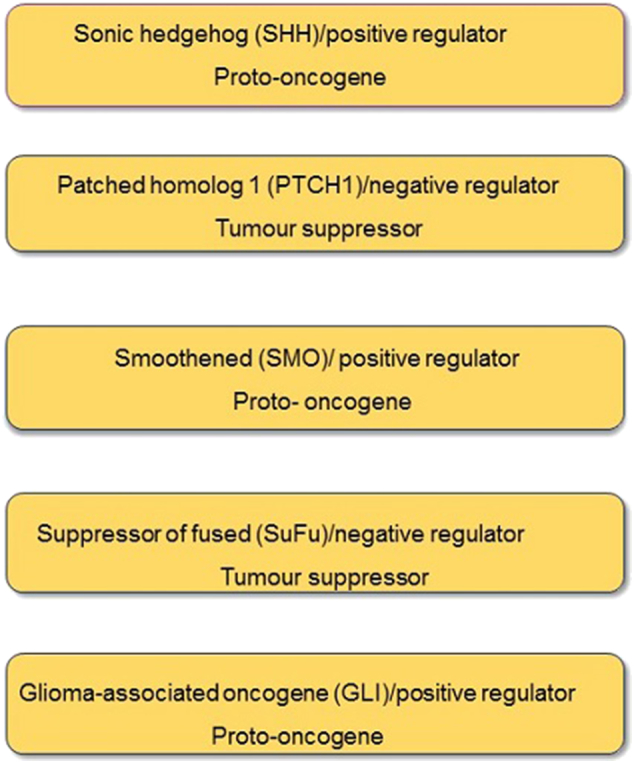
Core HH pathway components functioning as tumor suppressors or proto-oncogenes.
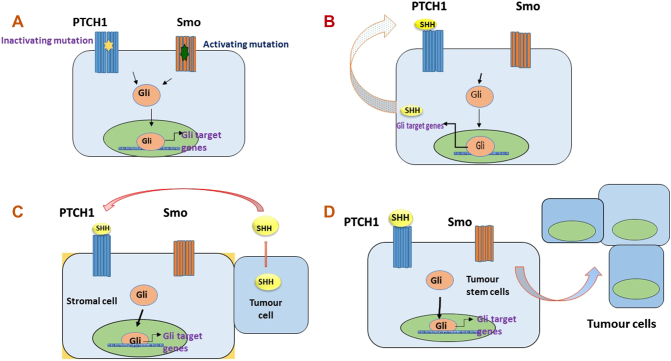
Models of HH Pathway Activation in Cancer
Four basic models have been proposed for the activation of HH signaling in cancer. Type I or ligand independent cancers are those in which loss of function mutations of inhibitory PTCH or gain of function mutations of activating SMO leads to the constitutive activation of the pathway. This results in the constitutive activation of the pathway in the presence or even in the absence of HH ligand binding. Type II is the ligand-dependent and autocrine activation in which HH ligand is overexpressed and acts on the cells producing it leading to excessive proliferation and survival of tumors. Type III is the ligand-dependent and paracrine activation in which HH ligand is overexpressed by tumor cells and acts on surrounding stromal cells which respond by producing growth factors such as IGF, etc. [29]. A reverse paracrine model has also been described in which HH ligands are secreted by stromal cells and act on neighboring tumor cells. Type IV includes aberrant activation of the pathway in cancer stem cells which may be autocrine or paracrine leading to the stem cell renewal and initiate the spread of a tumor. They are resistant to many chemotherapeutic agents and contribute to relapse [30]. Figure 3 describes the type I-IV mechanisms which aberrantly activate HH pathway in cancer.
Figure 3.
Type I-IV mechanisms which aberrantly activate hedgehog pathway in cancer.
Mutations in HH Signaling Pathway Genes in Cancer
Mutation-driven HH signaling genes lead to constitutive activation of the pathway promoting tumorigenesis. The Connection between HH signaling and cancer was first established when mutations of PTCH1 gene were known to cause Gorlin syndrome a rare autosomal inherited condition in which patients develop tumors on skin (basal cell carcinoma, BCC) cerebellum (medulloblastomas) and muscle (rhabdomyosarcoma) [31], [32], [33]. Gorlin syndrome is caused by loss of one functional copy of the PTCH1 gene. In tumors, both the copies of the PTCH1 gene are often found to be inactivated [34]. Dysregulated HH signaling led to increased cellular proliferation and tumor growth as observed in various mouse models. Mice with the heterozygous PTCH1 mutation have a higher incidence of developing medulloblastomas and predisposed to UV-induced BCC, comparable to patients with the Gorlin syndrome [35]. Moreover, mutations in SMO, SUFU, SHH have also been widely reported in BCC. 10% of all BCC have activating mutations in SMO and more than one-third of all medulloblastomas have increased HH signaling due to PTCH1 or SUFU mutations [36], [37], [38]. Studies have reported 30% of medulloblastomas to have aberrant HH signaling owing to mutations in PTCH1, SMO, or SuFu genes [39], [40]. PTCH1, SMO and SuFu mutations have also been observed in malignant mesothelioma [41]. Significant levels of SHH mutations have been found in Xeroderma Pigmentosum patients with a high incidence of BCC [42].
HH Ligand-Induced Cancer
Initially, HH signaling had been only implicated in cancers of the skin, skeletal muscle and brain [43] but extensive research carried out in the past decade suggests that the HH pathway is abnormally active in many solid tumors including, lung, liver, breast, prostate, stomach, colon and pancreas [44], [45], [46], [47], [48], [49], [50]. In almost all of these cancers, excessive HH signaling due to ligand (SHH) overexpression results in tumor formation and maintenance. SHH is overexpressed in prostate cancer tissues and cell lines and cell growth is blocked by cyclopamine a smoothened inhibitor both in vitro and in vivo. HH signaling is required for regeneration of prostate epithelium, and continued pathway activation transforms prostate progenitor cells and renders them tumorigenic [45]. Berman et al. have reported that wide range of gut-derived tumors, including most of those originating in the Esophagus, Stomach, Biliary Tract and Pancreas, display increased HH pathway activity, which is suppressed by cyclopamine [46]. High SHH-SMO-GLI activity is essential for tumor cell survival and maintenance in colon cancer [51]. SHH is overexpressed in metastatic breast cancer and contributes to poor prognosis and poor survival [47], [52]. Aberrant expression of SHH is also seen in non-small cell lung cancers (NSCLC) with higher expression correlating with poor survival and inhibition of SHH has marked effects on cellular invasion and cellular migration of lung cancer cells [53]. Similar results have also been observed for pancreatic cancer. SHH and GLI are overexpressed in Pancreatic cancers and can function as prognostic indicators [54]. SHH also influences tumor growth by promoting desmoplasia in PC [55]. SHH is highly expressed in Gastric cancer cell lines and promotes neoplastic transformation [56]. In hepatocarcinogenesis, SMO protein is overexpressed and correlates with increased tumor size. The blocker of oncogenic SMO reduces cell proliferation and expression of the hepatocarcinogenic oncogene, c-myc [48].
Multiple studies have suggested that tumors arise from a small population of cells known as cancer stem cells (CSCs) which have the capability of unrestrained self-renewal and differentiation along multiple lineages [57]. Dysregulation of key developmental signaling pathways such as HH, Notch, Wnt and BMP are known to transform stem cells into cancer stem cells. Studies have demonstrated that aberrant HH signaling is involved in the process of tumorigenesis, function and maintenance of CSCs [58]. HH ligand acts on CSCs secreted by adjacent tumor cells, stromal cells or the CSCs themselves. They are resistant to conventional chemotherapies, promoting survival and relapse [58]. HH signaling regulates self-renewal of CSCs in the maintenance of chronic myelogenous leukemia, several studies have also shown strong evidence of dysregulated HH signaling in CSCs of other cancers such as glioma, multiple myeloma, Gastrointestinal and Pancreatic cancers [58], [59], [60], [61], [62], [63], [64], [65].
HH Signaling in Pancreatic Cancer
Downregulation of HH signaling is critical for the embryonic development of the pancreas. Aberrant expression of HH signaling components during pancreas organogenesis results in loss of normal pancreas. Studies in Xenopus have shown that when constitutively activated SMO is injected in Xenopus embryos they fail to develop pancreas [66]. Exposure to cyclopamine, (HH pathway antagonist) that blocks SHH signaling, promotes Pancreatic expansion in embryonic chicks [67]. In adult pancreas, HH signaling is limited to β cells of the pancreas to regulate insulin secretion and is also required for the regeneration of Pancreatic tissue in disease or injury [68].
However, the darker side of HH signaling is its involvement in the development of pancreatic cancers which are highly aggressive in nature with poor response to chemotherapies. Dysregulated HH signaling has been strongly implicated in the genesis of Pancreatic cancer [69]. Some studies have suggested that HH signaling is important for maintenance but not in the initiation of Pancreatic cancer [70].
The ligand SHH is overexpressed pancreatic tumors and metastases. Overexpression of SHH has been observed at all stages of the disease. This has been confirmed on genetic mouse models of pancreatic ductal adenocarcinoma which exhibit increased SHH expression. Thayer et al have reported SHH, is overexpressed in about 70% of Pancreatic cancers [44] and can function as a prognostic biomarker in Pancreatic ductal adenocarcinomas (PDAC) [54]. Overexpression of SHH is detected in Pancreatic intraepithelial neoplasia (PanIN) and higher levels are observed throughout disease progression but are absent in normal pancreas [68]. SHH expression has crucial roles throughout Pancreatic Carcinogenesis. It enhances proliferation of pancreatic ductal cells, possibly through the transcriptional regulation of the cell cycle regulatory genes cyclin D1 and p21 and protects pancreatic ductal cells from apoptosis through the activation of phosphatidylinositol 3-kinase signaling and the stabilization of Bcl-2 and Bcl-XL. It also cooperates with activated K-Ras to promote Pancreatic tumor development and acts to provide resistance to various chemotherapeutic interventions [71]. In mouse models, simultaneous activation of Ras and HH signaling causes the widespread formation of the PanIN and enhances lethality [72].
Pancreatic tumor tissues and cell lines have high expression of HH pathway components PTCH1 and GLI1 [73] and treating cell lines with cyclopamine (HH pathway antagonist) inhibit growth in Pancreatic cancers [44]. GLI mediated transcriptional activation is also required for K-Ras induced proliferation in Pan IN and PDAC formation in vivo [74]. All these studies suggest that HH signaling drives pancreatic carcinogenesis and this pathway can be an attractive target for pancreatic cancer therapy.
An important feature of pancreatic cancer is desmoplastic reaction characterized by abundant stroma and activated fibroblast cells which secrete various components of the extracellular matrix that interfere with the normal architecture of Pancreatic Tissue. Interactions between the tumor cells, stromal cells and components of the extracellular matrix leading to an extensive desmoplastic reaction with diminished vasculature which is responsible for poor response of patients to systemic therapies [75]. SHH pathway promotes stromal desmoplasia functioning in a paracrine manner. The SHH protein is overexpressed by Pancreatic tumor cells which act on surrounding non-malignant stromal cells leading to a paracrine feedback loop to the Pancreatic tumor cells and also prevents the formation of vasculature in the stroma.[70]which protects the tumor microenvironment from chemotherapy.
HH Signaling in Colorectal cancer
HH signaling has important roles in the colon embryogenesis and aberrant activation of the pathway has been linked to gastric and Pancreatic cancers [46]. However, its role in the pathogenesis of colorectal cancer is not fully understood. Some studies have shown no association between activated HH signaling and colon cancer. Chatel et al. observed HH pathway is inactive in Colorectal cancer cell lines [76] whereas Gerling et al. suggested stromal HH signaling is downregulated in colon cancer [77]. Alingar et al. proposed that HH signaling is involved in normal colonic differentiation and renewal of colonic epithelium rather than colon cancer formation [78]. However multiple studies in the last few years have produced opposite results suggesting the active participation of HH signaling pathway in colorectal tumorigenesis.
Majority of Colorectal cancers originate due to constitutive activation of Wnt pathway and many studies have suggested several crosstalk points between Wnt and HH pathways which appear to be important for colon cancer recurrence, invasion and metastasis [79]. HH signaling is activated in Colorectal cancer by a ligand-dependent mechanism with overexpression of SHH reported in many studies [80], [81]. Bian et al. have suggested SHH-GLI1 pathway is active in colon carcinogenesis and there is a positive correlation between the progression of a tumor and expression of HH pathway components [82]. SHH and activator forms of GLI3 is highly expressed in Colorectal tumor specimens and GLI 3contributes to tumorigenicity of Colorectal cancer [83]. GLI transcription factors are also activated through non-canonical routes in colon cancers. Oncogenic Kras promotes activation of GLI1 through RAF/MEK/ERK and PI3K/AKT pathways. Inactivation of PTEN and P53 also promotes GLI1 activation in colon cancer cells [84].
The expression of HH pathway components varies in colon adenoma to carcinoma progression. SMO and GLI1 expression show a gradual increase from the normal colon to colonic adenoma to colon cancer [85]. Expression of SHH, PTCH, and GLI1 is higher in Peutz-Jegher syndrome (PJP) (an intestinal condition with a very high risk of developing colon cancer) than in normal tissue and shows a gradual increase as the disease progresses from the PJP to adenoma-adenocarcinoma sequence [86]. Silencing of the PTCH1 gene by promoter methylation has also been identified as an early initiating event in colon carcinogenesis [87].
The mechanism of HH signaling in colorectal cancer is autocrine as well as paracrine. Tumor cells secrete ligand which then acts on the cells producing it to activate target genes which stimulate proliferation and survival of colon tumor cells [79]. This is supported by studies which suggest SHH, PTCH and SMO are all expressed by tumor cells [46]. Varnat et al. also proposed that active HH signaling is present in epithelial tumor cells of the colon rather than in stroma [65]. The HH ligand also functions in a paracrine fashion where the ligand produced acts on surrounding stromal cells which respond by producing growth and survival factors. [88] HH signaling is also critical for maintaining stem cell survival. Varnat et al described HH-GLI pathway is active in epithelial colon carcinoma cells, stem cells and essential for tumor growth, recurrence, metastasis, stem cell survival and expansion [65].
HH Signaling in Gastric Cancer
Hedgehog signaling is associated with gut development. SHH is expressed throughout the gut during embryonic development. In adult life, SHH is highly expressed in parietal cells of the stomach and is required to secrete gastric acid as well as gastrin [89]. SHH shapes the mucosal layer of the stomach but has to be tightly controlled during the development of the gastric glandular epithelium [90]. SHH expression appears also is critical for gastric tissue repair [91].
Gastric cancer is one of the most difficult of gastrointestinal malignancies. Aberrant activation of the SHH signaling pathway leads to the disruption of gastric cell differentiation, loss of gastric acid secretion and the neoplastic transformation [92]. Ligand-dependent hedgehog signaling has been reported in gastric cancer. Studies have reported that SHH signaling is essential for recruitment of inflammatory cells from the bone marrow to the stomach in Helicobacter pylori infection causing expansion of metaplastic cell types [93]. All gastric cancer cell lines express the ligand SHH. Both the intestinal and diffuse types of gastric cancers show dysregulated hedgehog signaling although they show different expression profiles of hedgehog pathway proteins probably due to the difference in cancer origin [94]. Dysregulated hedgehog signaling is associated with tissue invasion and increased metastatic potential in gastric cancer and blockage of hedgehog signaling reduces tumor cell proliferation [5]. Increased SHH expression is also associated with shorter survival time in gastric cancer patients, thereby providing evidence that SHH could be a beneficial biomarker or therapeutic target for gastric cancers [95]. Cyclopamine, an important smoothened inhibitor inhibits gastric cancer cell proliferation through cell cycle arrest and apoptosis. An in vivo study conducted by Zhou et al. used NOD/SCID mouse xenografts to demonstrate that cyclopamine significantly stopped tumor growth and development. Their study also specified that activated SHH signaling pathway could promote gastric cancer cell proliferation and tumor development, and blocking the pathway may provide a new opportunity in gastric cancer therapeutics [96]. Bai et al. reported that treatment of gastric cancer AGS cell line with cyclopamine, inhibited cancer cell growth, migration and invasion in a dose- and time-dependent manner. Additionally, it was revealed that several key targets of the hedgehog signaling such as GLI1 and CXCR4, were downregulated at an RNA and protein level by treatment with cyclopamine [97]. Yoo et al. reported that SHH promotes motility and invasiveness of gastric cancer cells through TGF beta-mediated activation of the ALK5-Smad 3 pathway. They also proved that signaling is responsible for metastatic potential of gastric cancer and inhibition of hedgehog signaling reduces metastasis [98]. All these studies confirm that hedgehog pathway can be targeted for the development of novel therapeutic strategies to treat gastric cancer. Lu et al. reported that high expression of GLI1 transcription factors is an indicator for highly aggressive tumor with poor prognosis in gastric cancer patients [99]. Epigenetic silencing of tumor suppressors such as PTCH1and HHIP gene due to promoter hypermethylation is also reported in gastric cancer. Treatment with demethylating agent reverses the methylation status of PTCH1 and improves its expression [100]. Hypermethylation of HHIP gene promoter in gastric cancer tissues and cells decrease its mRNA expression which may cause uncontrolled hedgehog signaling thus contributing to gastric carcinogenesis [101].
HH Signaling in Prostate Cancer
Hedgehog signaling is activated during embryonic development of prostate and is also needed for regeneration of adult prostate epithelium, prostatic growth, branching, and proliferation [102], [103]. Several studies have pointed out the role of aberrant hedgehog signaling in prostate cancers. The hyperactivated hedgehog signaling due to overexpression of SHH is reported in prostate cancers and the mechanism of action is believed to be both autocrine as well as paracrine [104]. Tzelepi et al. demonstrated using immunohistochemistry and microarray analysis that SHH, PTCH1 and SMO were upregulated in prostate cancers and autocrine hedgehog signaling promotes the progression and pathogenesis of prostate carcinoma [105]. Paracrine Hedgehog signaling involving adjacent stromal cells is also believed to promote prostatic tumor growth [106]. Sanchez et al. reported advanced levels of SHH pathway components in the tumor as compared to normal prostatic epithelia and blockade of hedgehog signaling through treatment with smoothened inhibitor cyclopamine or GLI1 RNA interference in metastatic prostate cancer cell lines inhibited proliferation [107]. Sheng et al. proposed that hyper activated hedgehog signaling either through, through the loss of function mutations in a suppressor of fused (SuFu) or overexpression of SHH may be involved tumor progression and metastases of prostate cancer and targeted inhibition of hedgehog signaling may have substantial implications of prostate cancer therapeutics [108]. Li et al. proposed non-canonical activated hedgehog signaling independent of SMO to be involved in the pathogenesis of prostate cancers. Mediated by the binding of transcriptionally-active androgen receptors (ARs) to GLI3 [109].
High expression of all hedgehog pathway components is associated with poor prognostic parameters and SHH and SMO expressions are significantly associated with prostatic cancer recurrence [110]. The hedgehog pathway is needed for prostate epithelium development and continuous activation of the hedgehog pathway renders the prostate progenitor cells tumorigenic. The overexpression of hedgehog pathway proteins differentiates metastatic from localized prostate cancer and manipulating the pathway using inhibitors can modulate invasiveness and metastasis. Therefore targeting hedgehog pathway activity may contribute significantly in the diagnosis and treatment of prostate cancers with metastatic potential [45]. The increased hedgehog pathway activity is linked to maintenance of multidrug resistance and ATP-binding cassette (ABC) transporters play an important role in the removal of chemotherapeutic agents, including paclitaxel, from cancer cells. Upregulation of SHH enhances the resistance of PCa cell lines to paclitaxel. A higher level of SHH leads to increase in ABC transporters expression in a manner dependent on paclitaxel and silencing of SHH pathway may alter the activity of ABC proteins, thus increasing the effectiveness of conventional chemotherapy [111], [112].
HH Signaling in Breast Cancer
Breast cancer is one of the most common causes of cancer-related death among women worldwide. Although some treatment options are available the outlook of women with locally advanced or metastatic disease is poor. Few studies have indicated the involvement of hedgehog signaling in the development of normal breast. Normal mammary gland development is dependent on hedgehog pathway repression and constitutive activation of activator GLI1 or lack of functional inhibitory GLI3 have resulted in the failure of mammary bud formation in mouse models. Furthermore, overexpression of SHH in transgenic mouse embryos results in mammary bud abnormalities, including the absence of mammary buds [26]. `A possible role of aberrant hedgehog signaling activation in breast cancer formation and maintenance is proposed with mounting evidence in the last few years. Upregulated hedgehog signaling causes mammary cancer in the mouse model and many breast tumor tissues show dysregulated hedgehog signaling [113]. The expression of hedgehog pathway components SHH, PTCH1 and GLI1 show increased expression in invasive carcinomas but not in normal breast epithelium when detected by immunohistochemistry. Also, a mouse model with overexpression of GLI 1 in mammary epithelial cells was the first hedgehog pathway mouse model to develop tumors [114]. Riaz et al. demonstrated that exposure to GANT61 a potent GLI1 inhibitor significantly reduces cell viability and induces apoptosis nullifying neoplastic invasion in breast cancer cells [115]. Noman et al. established that SHH is overexpressed and is involved in mediating the aggressive phenotype of the breast cancer concluding SHH function as a novel biomarker in Breast cancers [47]. Tao et al. proved that hedgehog signaling is involved in breast ductal changes and malignant transformation and measures to inhibit hedgehog pathway activity may improve the prognosis of breast cancer patients [116]. Neelakantan et al. recognized that epithelial to mesenchymal transcription factors Twist1, Snail1 and Six1 activate paracrine hedgehog signaling by activating GLI both in canonical and non-canonical fashion resulting in breast cancer metastasis [117] and targeting the hedgehog pathway using antagonists that act downstream of SMO is effective in treating metastatic breast cancer [118]. All these studies indicate that hedgehog signaling is implicated in breast carcinogenesis and efficient inhibitors targeting this pathway may improve treatment and patient survival. More evidence that alterations in hedgehog pathway genes are implicated in breast cancer initiation comes from an epigenetic study which found PTCH1 promoter hypermethylation and loss of expression breast cancer cell lines and tissue samples. Additionally, treatment with cyclopamine, an inhibitor of the pathway, reduced cell growth and slowed the cell cycle in breast cancer cells [119]. High-resolution comparative genomic hybridization analysis performed on breast cancer cell lines and samples revealed frequent loss of the PTCH1 (9q22.1–q31) chromosomal region and concluded PTCH1 is a critical gene in establishing malignant phenotype in breast cancers [120]. Many breast tumors develop resistance to current chemotherapies available mostly due to intrinsic or acquired multi-drug resistance (MDR). Study by Mourtada et al. has made known that the hedgehog signaling activation induces chemo resistance mostly by increasing drug efflux in an ABC transporter-dependent manner and inhibition of hedgehog signaling increases the response of cancer cells to multiple structurally unrelated chemotherapies [121]. Therefore it is appropriate to consider possible effects of hedgehog signaling pathway inhibitors in combination with other drugs for breast cancer treatment.
Therapeutics
The HH signaling pathway has been studied extensively and its role in cancer well defined. The pathway has attracted a great deal of attention as a therapeutic target. The pathway can be blocked at various sites by HH inhibitors which can serve useful anticancer agents. Several small molecule inhibitors have been tried and tested. The first compound identified was cyclopamine natural alkaloid derived from corn lily Veratum californicum. It is a teratogen that functions to block Smoothened by binding to its heptahelical bundle locking it in an inactive form [122]. However, cyclopamine could not be a potent therapeutic target because of its low bioavailability short half-life and chemical instability [43]. Several synthetic, small-molecule SMO antagonists with higher potency than cyclopamine such as SANT1–SANT4, CUR-61414 GDC-0449 are now available and have been tested in preclinical models against a variety of solid tumors [43]. BCC is the first cancer to be treated with HH pathway antagonists. GDC-0449 (vismodegib) a small molecule inhibitor of SMO was the first drug to be used for the treatment of basal cell carcinomas. It is used for locally advanced and metastatic BCC and is more pharmacologically favorable than cyclopamine. LDE225 (sonidegib), discovered in 2010, is another potent selective SMO antagonist with high tissue penetration and the ability to cross the blood–brain barrier and is used for the treatment of BCC [123]. The clinical success of hedgehog inhibitors is limited to brain and skin cancers. Presently the SMO antagonists and other pathway inhibitors are also being studied for other solid tumors with activated HH pathways, such as HH-dependent medulloblastomas or ovarian cancer and metastatic pancreatic cancer, Colon cancer but have not been very successful with respect to disease-free survival. Table 1 describes the hedgehog inhibitors which are FDA approved or undergoing clinical trials for various cancers. More detailed studies of HH-dependent cancer are necessary for finding more effective drugs.
Conclusion
The hedgehog pathway is an important developmental pathway with roles in embryonic development, growth and proliferation. The hedgehog pathway begins by the binding of the ligand (SHH) to the patched transmembrane receptor (PTCH1) resulting in activation of smoothened (SMO) and transduction of signal to glioma transcription factors (GLI) which then translocate to the nucleus and induce genes responsible for growth and proliferation. Aberrant activation of the pathway in either ligand independent or dependent manner is correlated with birth defects and several cancers and inhibiting the hedgehog signaling pathway using SMO or GLI inhibitors has been found to have a substantial effect in inhibiting proliferation. Several SMO inhibitors are being successfully used in the treatment of basal cell carcinomas and are undergoing clinical trials in other cancers. In this review, we have summarized the mechanisms of hedgehog signaling, its importance in cancers such as pancreatic, colorectal prostate, breast and gastric cancers and various therapeutic options that are currently directed at this pathway. We conclude that hedgehog signaling pathway plays a significant role in the development of tumorigenesis and improving our understanding of hedgehog pathway functioning, its regulation and developing multi targeting low toxic inhibitors can help achieve better therapeutic outcomes for cancer therapy.
References
- 1.Fattahi S, Langroudi MP, Akhavan-Niaki H. Hedgehog signaling pathway: Epigenetic regulation and role in disease and cancer development. J Cell Physiol. 2018;233:5726–5735. doi: 10.1002/jcp.26506. [DOI] [PubMed] [Google Scholar]
- 2.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 3.Villavicencio EH, Walterhouse DO, Iannaccone PM (2000) The Sonic Hedgehog–Patched–Gli Pathway in Human Development and Disease. The American Journal of Human Genetics 67:1047–1054. 10.1016/S0002-9297(07)62934-6 [DOI] [PMC free article] [PubMed]
- 4.Ebrahimi A, Larijani L, Moradi A, Ebrahimi MR. Hedgehog Signalling Pathway: Carcinogenesis and Targeted Therapy. Iran J Cancer Prev. 2013;6:36–43. [PMC free article] [PubMed] [Google Scholar]
- 5.Skoda AM, Simovic D, Karin V. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn J Basic Med Sci. 2018;18:8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene. 1999;18:7844–7851. doi: 10.1038/sj.onc.1203282. [DOI] [PubMed] [Google Scholar]
- 7.Lee RTH, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 8.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 9.Katoh Y, Katoh M. Hedgehog signaling pathway and gastric cancer. Cancer Biol Ther. 2005;4:1050–1054. doi: 10.4161/cbt.4.10.2184. [DOI] [PubMed] [Google Scholar]
- 10.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 11.Gonnissen A, Isebaert S, Haustermans K. Targeting the Hedgehog signaling pathway in cancer: beyond Smoothened. Oncotarget. 2015;6:13899–13913. doi: 10.18632/oncotarget.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oro AE. The primary cilia, a “Rab-id” transit system for hedgehog signaling. Curr Opin Cell Biol. 2007;19:691–696. doi: 10.1016/j.ceb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 14.Jia J, Tong C, Wang B. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 15.Haycraft CJ, Banizs B, Aydin-Son Y. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1 doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endoh-Yamagami S, Evangelista M, Wilson D. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 17.Nozawa YI, Lin C, Chuang P-T. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Méthot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- 19.Chen M-H, Wilson CW, Li Y-J. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 21.Brennan D, Chen X, Cheng L. Noncanonical Hedgehog Signaling. Vitam Horm. 2012;88:55–72. doi: 10.1016/B978-0-12-394622-5.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins DJ, Fei DL, Riobo NA. The Hedgehog Signal Transduction Network. Sci Signal. 2012;5:6–34. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mille F, Thibert C, Fombonne J. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat Cell Biol. 2009;11:739–746. doi: 10.1038/ncb1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinchilla P, Xiao L, Kazanietz MG, Riobo NA. Hedgehog proteins activate pro-angiogenic responses in endothelial cells through non-canonical signaling pathways. Cell Cycle. 2010;9:570–579. doi: 10.4161/cc.9.3.10591. [DOI] [PubMed] [Google Scholar]
- 25.Chang H, Li Q, Moraes RC. Activation of ERK by Sonic Hedgehog Independent of Canonical Hedgehog Signalling. Int J Biochem Cell Biol. 2010;42:1462–1471. doi: 10.1016/j.biocel.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J. 2001;20:2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Yang P, Ma L. Kinase activity-independent regulation of cyclin pathway by GRK2 is essential for zebrafish early development. Proc Natl Acad Sci U S A. 2009;106:10183–10188. doi: 10.1073/pnas.0812105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polizio AH, Chinchilla P, Chen X. Sonic Hedgehog activates the GTPases Rac1 and RhoA in a Gli-independent manner via coupling of Smoothened to Gi proteins. Sci Signal. 2011;4:7–14. doi: 10.1126/scisignal.2002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 30.Cohen DJ (2012) Targeting the hedgehog pathway: role in cancer and clinical implications of its inhibition.Hematol Oncol Clin North Am 26:565–588, viii. 10.1016/j.hoc.2012.01.005 [DOI] [PubMed]
- 31.Hahn H, Wicking C, Zaphiropoulous PG. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 32.Johnson RL, Rothman AL, Xie J. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 33.Wu F, Zhang Y, Sun B. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chemical Biology. 2017;24:252–280. doi: 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazzaglia S, Tanori M, Mancuso M. Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene. 2006;25:5575–5580. doi: 10.1038/sj.onc.1209544. [DOI] [PubMed] [Google Scholar]
- 35.Aszterbaum M, Beech J, Epstein EH. Ultraviolet radiation mutagenesis of hedgehog pathway genes in basal cell carcinomas. J Investig Dermatol Symp Proc. 1999;4:41–45. doi: 10.1038/sj.jidsp.5640179. [DOI] [PubMed] [Google Scholar]
- 36.Reifenberger J, Wolter M, Weber RG. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- 37.Xie J, Murone M, Luoh SM. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 38.Taylor MD, Liu L, Raffel C. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y, Miller HL, Jensen P. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- 40.Thompson MC, Fuller C, Hogg TL. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 41.Lim CB, Prêle CM, Cheah HM. Mutational Analysis of Hedgehog Signaling Pathway Genes in Human Malignant Mesothelioma. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0066685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couvé-Privat S, Bret ML, Traiffort E. Functional Analysis of Novel Sonic Hedgehog Gene Mutations Identified in Basal Cell Carcinomas from Xeroderma Pigmentosum Patients. Cancer Res. 2004;64:3559–3565. doi: 10.1158/0008-5472.CAN-03-4040. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S, Takebe N, LoRusso P. Targeting the Hedgehog pathway in cancer. Therapeutic Advances in Medical Oncology. 2010;2:237–250. doi: 10.1177/1758834010366430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thayer SP, di Magliano MP, Heiser PW. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karhadkar SS, Bova GS, Abdallah N. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 46.Berman DM, Karhadkar SS, Maitra A. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 47.Noman AS, Uddin M, Rahman MZ. Overexpression of sonic hedgehog in the triple negative breast cancer: clinicopathological characteristics of high burden breast cancer patients from Bangladesh. Sci Rep. 2016;6:18830–18841. doi: 10.1038/srep18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sicklick JK, Li Y-X, Jayaraman A. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27:748–757. doi: 10.1093/carcin/bgi292. [DOI] [PubMed] [Google Scholar]
- 49.Watkins DN, Berman DM, Burkholder SG. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 50.Kubo M, Nakamura M, Tasaki A. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 51.Wu C, Zhu X, Liu W. Hedgehog signaling pathway in colorectal cancer: function, mechanism, and therapy. Onco Targets Ther. 2017;10:3249–3259. doi: 10.2147/OTT.S139639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noman AS, Uddin M, Chowdhury AA. Serum sonic hedgehog (SHH) and interleukin-(IL-6) as dual prognostic biomarkers in progressive metastatic breast cancer. Sci Rep. 2017;7:1796–1808. doi: 10.1038/s41598-017-01268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang WG, Ye L, Ruge F. Expression of Sonic Hedgehog (SHH) in Human Lung Cancer and the Impact of YangZheng XiaoJi on SHH-mediated Biological Function of Lung Cancer Cells and Tumor Growth. Anticancer Res. 2015;35:1321–1331. [PubMed] [Google Scholar]
- 54.Maréchal R, Bachet J-B, Calomme A. Sonic Hedgehog and Gli1 Expression Predict Outcome in Resected Pancreatic Adenocarcinoma. Clin Cancer Res. 2015;21:1215–1224. doi: 10.1158/1078-0432.CCR-14-0667. [DOI] [PubMed] [Google Scholar]
- 55.Bailey JM, Swanson BJ, Hamada T. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akyala AI, Peppelenbosch MP. Gastric cancer and Hedgehog signaling pathway: emerging new paradigms. Genes Cancer. 2018;9:1–10. doi: 10.18632/genesandcancer.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 58.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers (Basel) 2015;7:1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehtesham M, Sarangi A, Valadez JG. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 60.Bar EE, Chaudhry A, Lin A. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peacock CD, Wang Q, Gesell GS. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dierks C, Beigi R, Guo G-R. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 63.Huang F-T, Zhuan-Sun Y-X, Zhuang Y-Y. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int J Oncol. 2012;41:1707–1714. doi: 10.3892/ijo.2012.1597. [DOI] [PubMed] [Google Scholar]
- 64.Yoon C, Park DJ, Schmidt B. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin Cancer Res. 2014;20:3974–3988. doi: 10.1158/1078-0432.CCR-14-0011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Varnat F, Duquet A, Malerba M. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Rosenthal A, de Sauvage FJ, Shivdasani RA. Downregulation of Hedgehog signaling is required for organogenesis of the small intestine in Xenopus. Dev Biol. 2001;229:188–202. doi: 10.1006/dbio.2000.9953. [DOI] [PubMed] [Google Scholar]
- 67.Kim SK, Melton DA. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc Natl Acad Sci U S A. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu D, Schlotman KE, Xie J. Deciphering the role of hedgehog signaling in pancreatic cancer. J Biomed Res. 2016;30:353–360. doi: 10.7555/JBR.30.20150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lau J, Kawahira H, Hebrok M. Hedgehog signaling in pancreas development and disease. Cell Mol Life Sci. 2006;63:642–652. doi: 10.1007/s00018-005-5357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog Signaling in Development and Cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morton JP, Mongeau ME, Klimstra DS. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci U S A. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasca di Magliano M, Sekine S, Ermilov A. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 74.Rajurkar M, De Jesus-Monge WE, Driscoll DR. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc Natl Acad Sci U S A. 2012;109:E1038–E1047. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neesse A, Michl P, Frese KK. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 76.Chatel G, Ganeff C, Boussif N. Hedgehog signaling pathway is inactive in colorectal cancer cell lines. Int J Cancer. 2007;121:2622–2627. doi: 10.1002/ijc.22998. [DOI] [PubMed] [Google Scholar]
- 77.Gerling M, Büller NVJA, Kirn LM. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat Commun. 2016;7:12321–12356. doi: 10.1038/ncomms12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alinger B, Kiesslich T, Datz C. Hedgehog signaling is involved in differentiation of normal colonic tissue rather than in tumor proliferation. Virchows Arch. 2009;454:369–379. doi: 10.1007/s00428-009-0753-7. [DOI] [PubMed] [Google Scholar]
- 79.Song L, Li Z-Y, Liu W-P, Zhao M-R. Crosstalk between Wnt/β-catenin and Hedgehog/Gli signaling pathways in colon cancer and implications for therapy. Cancer Biol Ther. 2015;16:1–7. doi: 10.4161/15384047.2014.972215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshikawa K, Shimada M, Miyamoto H. Sonic hedgehog relates to colorectal carcinogenesis. J Gastroenterol. 2009;44:1113–1117. doi: 10.1007/s00535-009-0110-2. [DOI] [PubMed] [Google Scholar]
- 81.Douard R, Moutereau S, Pernet P. Sonic Hedgehog-dependent proliferation in a series of patients with colorectal cancer. Surgery. 2006;139:665–670. doi: 10.1016/j.surg.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Bian Y-H, Huang S-H, Yang L. Sonic hedgehog-Gli1 pathway in colorectal adenocarcinomas. World J Gastroenterol. 2007;13:1659–1665. doi: 10.3748/wjg.v13.i11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwasaki H, Nakano K, Shinkai K, et al Hedgehog Gli3 activator signal augments tumorigenicity of colorectal cancer via upregulation of adherence-related genes. Cancer Science 104:328–336. 10.1111/cas.12073 [DOI] [PMC free article] [PubMed]
- 84.Varnat F, Siegl-Cachedenier I, Malerba M. Loss of WNT-TCF addiction and enhancement of HH-GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol Med. 2010;2:440–457. doi: 10.1002/emmm.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang X, Zhang S-S, Wei G-J. Dysregulation of hedgehog signaling pathway related components in the evolution of colonic carcinogenesis. Int J Clin Exp Med. 2015;8:21379–21385. [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X, Su J, Li R. Aberrant expression of Sonic hedgehog signaling in Peutz-Jeghers syndrome. Hum Pathol. 2016;50:153–161. doi: 10.1016/j.humpath.2015.09.044. [DOI] [PubMed] [Google Scholar]
- 87.Peng L, Hu J, Li S. Aberrant methylation of the PTCH1 gene promoter region in aberrant crypt foci. Int J Cancer. 2013;132:E18–E25. doi: 10.1002/ijc.27812. [DOI] [PubMed] [Google Scholar]
- 88.Theunissen J-W, de Sauvage FJ. Paracrine Hedgehog signaling in cancer. Cancer Res. 2009;69:6007–6010. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- 89.van den Brink GR, Hardwick JC, Tytgat GN. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology. 2001;121:317–328. doi: 10.1053/gast.2001.26261. [DOI] [PubMed] [Google Scholar]
- 90.van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- 91.Engevik AC, Feng R, Yang L, Zavros Y. The acid-secreting parietal cell as an endocrine source of Sonic Hedgehog during gastric repair. Endocrinology. 2013;154:4627–4639. doi: 10.1210/en.2013-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Konstantinou D, Bertaux-Skeirik N, Zavros Y. Hedgehog Signaling in the Stomach. Curr Opin Pharmacol. 2016;31:76–82. doi: 10.1016/j.coph.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Merchant JL, Ding L. Hedgehog Signaling Links Chronic Inflammation to Gastric Cancer Precursor Lesions. Cell Mol Gastroenterol Hepatol. 2017;3:201–210. doi: 10.1016/j.jcmgh.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akyala AI, Peppelenbosch MP. Gastric cancer and Hedgehog signaling pathway: emerging new paradigms. Genes Cancer. 2018;9:1–10. doi: 10.18632/genesandcancer.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ertao Z, Jianhui C, Chuangqi C. Autocrine Sonic hedgehog signaling promotes gastric cancer proliferation through induction of phospholipase Cγ1 and the ERK1/2 pathway. J Exp Clin Cancer Res. 2016;35:63. doi: 10.1186/s13046-016-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wan J, Zhou J, Zhao H. Sonic Hedgehog Pathway Contributes to Gastric Cancer Cell Growth and Proliferation. Biores Open Access. 2014;3:53–59. doi: 10.1089/biores.2014.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bai R, Zhao H, Zhang X, Du S. Characterization of sonic hedgehog inhibition in gastric carcinoma cells. Oncol Lett. 2014;7:1381–1384. doi: 10.3892/ol.2014.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoo YA, Kang MH, Kim JS, Oh SC. Sonic hedgehog signaling promotes motility and invasiveness of gastric cancer cells through TGF-beta-mediated activation of the ALK5-Smad 3 pathway. Carcinogenesis. 2008;29:480–490. doi: 10.1093/carcin/bgm281. [DOI] [PubMed] [Google Scholar]
- 99.Lu L, Wu M, Zhao F. Prognostic and clinicopathological value of Gli-1 expression in gastric cancer: A meta-analysis. Oncotarget. 2016;7:69087–69096. doi: 10.18632/oncotarget.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zuo Y, Song Y, Zhang M. Role of PTCH1 gene methylation in gastric carcinogenesis. Oncol Lett. 2014;8:679–682. doi: 10.3892/ol.2014.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song Y, Zuo Y. Occurrence of HHIP gene CpG island methylation in gastric cancer. Oncol Lett. 2014;8:2340–2344. doi: 10.3892/ol.2014.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Freestone SH, Marker P, Grace OC. Sonic hedgehog regulates prostatic growth and epithelial differentiation. Dev Biol. 2003;264:352–362. doi: 10.1016/j.ydbio.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 103.Podlasek CA, Barnett DH, Clemens JQ. Prostate development requires Sonic hedgehog expressed by the urogenital sinus epithelium. Dev Biol. 1999;209:28–39. doi: 10.1006/dbio.1999.9229. [DOI] [PubMed] [Google Scholar]
- 104.Gonnissen A, Isebaert S, Haustermans K. Hedgehog Signaling in Prostate Cancer and Its Therapeutic Implication. Int J Mol Sci. 2013;14:13979–14007. doi: 10.3390/ijms140713979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tzelepi V, Karlou M, Wen S. Expression of hedgehog pathway components in prostate carcinoma microenvironment: shifting the balance towards autocrine signalling. Histopathology. 2011;58:1037–1047. doi: 10.1111/j.1365-2559.2011.03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shaw A, Gipp J, Bushman W. The Sonic Hedgehog Pathway Stimulates Prostate Tumor Growth by Paracrine Signaling and Recaptures Embryonic Gene Expression in Tumor Myofibroblasts. Oncogene. 2009;28:4480–4490. doi: 10.1038/onc.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanchez P, Hernández AM, Stecca B. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheng T, Li C, Zhang X. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li N, Truong S, Nouri M. Non-canonical activation of hedgehog in prostate cancer cells mediated by the interaction of transcriptionally active androgen receptor proteins with Gli3. Oncogene. 2018;37:2313. doi: 10.1038/s41388-017-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim T-J, Lee JY, Hwang T-K. Hedgehog signaling protein expression and its association with prognostic parameters in prostate cancer: a retrospective study from the view point of new 2010 anatomic stage/prognostic groups. J Surg Oncol. 2011;104:472–479. doi: 10.1002/jso.21988. [DOI] [PubMed] [Google Scholar]
- 111.Statkiewicz M, Maryan N, Lipiec A. The role of the SHH gene in prostate cancer cell resistance to paclitaxel. Prostate. 2014;74:1142–1152. doi: 10.1002/pros.22830. [DOI] [PubMed] [Google Scholar]
- 112.Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288:156–166. doi: 10.1016/j.jns.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 113.Kasper M, Jaks V, Fiaschi M, Toftgård R. Hedgehog signalling in breast cancer. Carcinogenesis. 2009;30:903–911. doi: 10.1093/carcin/bgp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fiaschi M, Rozell B, Bergström A. Targeted expression of GLI1 in the mammary gland disrupts pregnancy-induced maturation and causes lactation failure. J Biol Chem. 2007;282:36090–36101. doi: 10.1074/jbc.M704280200. [DOI] [PubMed] [Google Scholar]
- 115.Riaz SK, Ke Y, Wang F. Influence of SHH/GLI1 axis on EMT mediated migration and invasion of breast cancer cells. Sci Rep. 2019;9:6620. doi: 10.1038/s41598-019-43093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tao Y, Mao J, Zhang Q, Li L. Overexpression of Hedgehog signaling molecules and its involvement in triple-negative breast cancer. Oncol Lett. 2011;2:995–1001. doi: 10.3892/ol.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Neelakantan D, Zhou H, Oliphant MUJ. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8 doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Benvenuto M, Masuelli L, De Smaele E. In vitro and in vivo inhibition of breast cancer cell growth by targeting the Hedgehog/GLI pathway with SMO (GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget. 2016;7:9250–9270. doi: 10.18632/oncotarget.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolf I, Bose S, Desmond JC. Unmasking of epigenetically silenced genes reveals DNA promoter methylation and reduced expression of PTCH in breast cancer. Breast Cancer Res Treat. 2007;105:139–155. doi: 10.1007/s10549-006-9440-4. [DOI] [PubMed] [Google Scholar]
- 120.Naylor TL, Greshock J, Wang Y. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sims-Mourtada J, Izzo JG, Ajani J, Chao KSC. Sonic Hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 2007;26:5674–5679. doi: 10.1038/sj.onc.1210356. [DOI] [PubMed] [Google Scholar]
- 122.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rimkus TK, Carpenter RL, Qasem S. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 2016;8:22–45. doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Axelson M, Liu K, Jiang X. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res. 2013;19:2289–2293. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- 125.Hui M, Cazet A, Nair R. The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast Cancer Res. 2013;15:203. doi: 10.1186/bcr3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xie H, Paradise BD, Ma WW, Fernandez-Zapico ME. Recent Advances in the Clinical Targeting of Hedgehog/GLI Signaling in Cancer. Cell. 2019;8 doi: 10.3390/cells8050394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Norsworthy KJ, By K, Subramaniam S, et al (2019) FDA Approval Summary: Glasdegib for newly-diagnosed acute myeloid leukemia. Clin Cancer Res clincanres.0365.2019. 10.1158/1078-0432.CCR-19-0365 [DOI] [PubMed]
- 128.Casey D, Demko S, Shord S. FDA Approval Summary: Sonidegib for Locally Advanced Basal Cell Carcinoma. Clin Cancer Res. 2017;23:2377–2381. doi: 10.1158/1078-0432.CCR-16-2051. [DOI] [PubMed] [Google Scholar]
- 129.M.D. Anderson Cancer Center Taladegib, Paclitaxel, Carboplatin, and Radiation Therapy in Treating Patients with Localized Esophageal or Gastroesophageal Junction Cancer - Full Text View - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02530437. Accessed 1 Jul 2019
- 130.Pounds R, Leonard S, Dawson C, Kehoe S. Repurposing itraconazole for the treatment of cancer. Oncol Lett. 2017;14:2587–2597. doi: 10.3892/ol.2017.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clinical Trials.gov. https://clinicaltrials.gov/