Abstract
Purpose
Paraoxonases (PON) are calcium bound enzymes offering protection against oxidative stress by working as endogenous free-radical scavenging molecules. Oxidative stress has been implicated in pathophysiology of many diseases including cataract. Lens opacity is an age related disorder which is a principal cause of blindness in Pakistani population. Relationship of PON2 and PON3 polymorphism with genetic predisposition for incidence of cataract has not been investigated till date. Objective of the current study was to explore possible association between PON2 and PON3 polymorphism with incidence of cataract in local population.
Methods
Our study design comprised of fifty-one cataractous and fifty-nine healthy individuals. Identification of single nucleotide polymorphism (SNP) at positions (C311S and G148A) for PON2 and C133A for PON3 was conducted using restriction fragment length polymorphism (RFLP).
Results
Statistical analysis revealed significant association of PON2 G148 allele with incidence of cataract. GG allele was found to be higher in cataract patients as compared to control (p < 0.001) suggesting distribution of PON2 G148A genotype and allele frequency is linked with cataractogenesis. There was no noticeable association between PON2 C311S and PON3 C133A. Significant difference was observed in distribution of 311CS/148A combined genotype with highest frequency in control individuals (88.89%), while 311S/148G combined genotypes showed the highest frequencies among the cataract patients (71.42%).
Conclusion
Our data suggests mutation at G148A might be related with incidence of cataract in studied population.
Keywords: PON2; PON3; Polymorphism, Cataract
Introduction
Lens opacity or cataract is a leading cause of visual impairment throughout the world. WHO estimate indicated 95 million people to be effected from cataract in 2014.1 Cataract is an age-related disorder that is likely to be initiated at the age of 50 and above.2 In low and middle income countries, however, lens opacity has a higher incidence rate with an earlier onset.3 Although epidemiological studies have implicated various risk factors for age-related cataracts including smoking, exposure to UV radiation, malnutrition, use of steroids etc; involvement of genetic factors in onset of disease cannot be ruled out.4 Genetic mutations are likely to increase disposition to environmental factors thus increasing susceptibility to develop cataract. Therefore, genetic variations in relation to risk of developing age related cataract has been studied.5 Studies on association between senile cataract and genetic mutation in OGG1, EPHA2,6 FABP2, PPARG27 and MTHFR8 have been reported. Among genes regulating oxidative stress, most of the work has been conducted on glutathione S Transferases (GSTs) superfamily members, GSTM1 and GSTT1.9
Paraoxonases are calcium dependent antioxidant enzymes constituting three isoforms PON1, PON2 and PON3.10 PON1 polymorphism has been studied in different disorders including cataract,[11], [12] diabetes,13 age related macular degeneration,14 Alzheimer’s disease,15 Behcet’s disease,16 and atherosclerosis.17 Data for PON2 and PON3, however, is limited. In PON2, two polymorphic sites C311S and G148A have been studied in Alzheimer’s disease,18 ischemic stroke in type 2 diabetics,19 coronary artery disease[20], [21] and reduced bone mass in postmenopausal women.22 Most of the polymorphic sites in PON3 belong to non-coding and promoter region. There are some studies on PON3 polymorphisms in CHD,19 systemic lupus erythematosus (SLE)23 and Alzheimer’s disease.24 A handful of promoter polymorphisms have also been identified suggesting a moderate influence on circulating PON3 levels.[25], [26]
We have earlier reported PON1 polymorphism in patients suffering from cataract.11 In this communication, we studied two polymorphic sites of PON2 and one polymorphic site of PON3 gene in cataract and healthy subjects to identify whether missense mutations can be a risk factor for the development of cataract or not.
Material and methods
Sample collection
The study was conducted after approval from Institutional Review Board, National Center for Proteomics, University of Karachi. Informed written consents were obtained from all participants. We recruited fifty one age related cataract patients and fifty nine healthy individuals for this study. Blood samples were collected in K2–EDTA coated vials from cataract patients prior to cataract surgery at Department of Ophthalmology, Liaquat National Hospital, Karachi. Cataract status was ascertained through slit lamp examination. Control group comprised of subjects without any ocular disorder. Subjects were excluded from the study if they suffered from any metabolic disorder or secondary cataract due to diabetes or trauma.
DNA extraction
DNA was isolated from whole blood within 24 h of collection as described earlier.27 Briefly, 300 μl of blood was added to 900 μl of cell lysis buffer (0.31 M sucrose, 0.01 M Tris, 0.002 M MgCl2, 0.0003 M sodium azide, pH 7.6) and centrifuged at 13,000g for one minute. The procedure was repeated twice. The pellet was broken by addition of 300 μl red cell lysis buffer (0.064 M Tris, 0.017 M Na2EDTA, 0.069 M SDS pH 8.0) followed by incubation for 30 min at 37 °C. Afterwards, 120 μl protein precipitation solution was added and centrifuged for 5 min. The supernatant was then transferred to a tube containing 300 μl iso-propanol and mixed gently for 15–20 times to visualize the DNA, followed by centrifugation. DNA thread was washed using 500 μl ethanol and air dried after centrifugation. Later, DNA was rehydrated with 50 μl of nuclease free water and stored −20 °C till further use.
Restriction fragment length polymorphism
Analysis of PON2 polymorphic site 311 was carried out as described earlier.21 Mismatch PCR assay for genotyping the polymorphic site at codon 148 in PON2 gene was performed using method of Mochizuki et al.28 In case of PON3, polymorphism at codon 133 was evaluated.19 Primer sequences, restriction enzymes and product size in each case are depicted in Table 1.
Table 1.
Primers used for PON2 and PON3 polymorphic positions C311S, G148A and C133A respectively.
| Polymorphic sites | Primer Sequence | PCR Products | Restriction Enzymes | Restricted bands | |
|---|---|---|---|---|---|
| PON 2 | C311S | 5′- ACATGCATGTACGGTGGTCT-3′ (forward) 5′-AGCAATTCATAGATTAATTGTTA-3′ (reverse) |
142 bp 120 bp* |
DdeI | C allele: 142 bp S allele: 75 bp + 67 bp |
| G148A | 5′-AGTGGAAATTTTTAAATTTGAAGCAG-3′ (forward) 5′-TTGTTTGCAAATGCTGGGGAT-3′ (reverse) |
130 bp | Fnu4HI | G allele: 130 bp A allele: 110 bp + 20 bp |
|
| PON 3 | C133A | 5′-TTACCTATCATGTAGACTGTGAGA*GT-3′ (forward) 5′-ATTCACAACATAAAGATACACAGTA-3′ (reverse) |
168 bp | HinfI | A allele: 168 bp C allele: 143 bp |
120 bp band found in all genotype because of common restriction site.
Polymerase chain reaction was performed using PCR master mix DreamTaq Green PCR Master Mix 2X (Thermo Scientific Company EU, Lithuania) containing 25 μl PCR master mix, 5 µl DNA template (>150 ng/µl), 1 µl of each forward and reverse primer (10 µM) and 16 µl of nuclease free water giving a total reaction volume of 50 μl. The cycling condition for position C311S PON2 polymorphisms was 4 min at 94 °C for initial denaturation, followed by 30 cycles of 1 min at 94 °C for denaturation, 1 min 30 s at 50 °C for annealing and 2 min at 72 °C for extension. The conditions for PON2 G148A polymorphism were essentially same as of C311S with only once exception of 40 cycles instead of 30 in denaturation step. In case of PON3 C133A, PCR condition was 5 min for 95 °C, followed by 30 cycles of 94 °C for 45 s, 53 °C for 40 s and 72 °C for 45 s with final extension for 7 min at 72 °C. Products obtained from PCR were digested with 10 U restriction enzymes of DdeI for C311S, Fun4HI for G148A and HinfI for polymorphic site of C133A respectively and visualized on 3% agarose gel electrophoresis using ethidium bromide.
Statistical analysis
Data was analyzed with SPSS version 20.0 and descriptive statistics were used. Demographic characteristics were compared by Pearson χ2 test. Gene counting method was used to calculate allele frequencies. Odds ratios were calculated with 95% confidence interval. P value < 0.05 was considered as significant.
Results
Two genetic variants in PON2 (C311S, G148A) and one in PON3 (C133A) were analyzed in blood samples from cataract patients and healthy controls using RFLP. The distribution of C311S codon for PON2 polymorphism in controls and patients is shown in Table 2. We did not observe any significant association of polymorphic marker C311S with incidence of cataract in studied groups (Table 2). The odds ratio for PON2 C vs S allele was found to be 1.448 (p = 0.179).
Table 2.
Statistical analysis of PON2 and PON3 polymorphic sites.
| Polymorphic positions | Control (F %) |
Cataract F % |
Odds ratio | 95% CI | χ2 Test | ||
|---|---|---|---|---|---|---|---|
| PON2 C311S (C- -->S) | Genotypes | CC | 19/57 (33.33%) |
15/52 (28.84%) |
1.125 | 0.502–2.519 | 0.775 |
| CS | 15/57 (26.31%) |
15/52 (28.84%) |
0.969 | 0.414–2.269 | 0.943 | ||
| SS | 23/57 (40.35%) |
22/52 (42.30%) |
1.084 | 0.505–2.326 | 0.836 | ||
| Alleles | CC | 53/114 (46.49%) |
45/104 (43.26%) |
1.448 | 0.843–2.488 | 0.179 | |
| SS | 61/114 (53.50%) |
59/104 (56.73%) |
|||||
| PON2 G148A (G- -->A) | Genotypes | GG | 18/57 (31.57%) |
29/52 (55.76%) |
0.366 | 0.167–0.800 | 0.011* |
| GA | 10/57 (17.54%) |
07/52 (13.46%) |
1.368 | 0.479–3.904 | 0.557 | ||
| AA | 29/57 (50.54%) |
16/52 (30.76%) |
2.330 | 1.063–5.110 | 0.033* | ||
| Alleles | GG | 46/114 (40.35%) |
65/104 (62.50%) |
2.464 | 1.428–4.251 | 0.001* | |
| AA | 68/114 (59.64%) |
39/104 (37.50%) |
|||||
| PON3 C133A (C- -->A) | Genotypes | CC | 20/57 (35.08%) |
14/52 (26.92%) |
1.467 | 0.647–3.329 | 0.358 |
| CA | 27/57 (47.36%) |
32/52 (61.53%) |
0.563 | 0.262–1.207 | 0.138 | ||
| AA | 10/57 (17.54%) |
06/52 (11.53%) |
1.631 | 0.548–4.855 | 0.376 | ||
| Alleles | CC | 67/114 (58.77%) |
60/104 (41.22%) |
0.957 | 0.558–1.640 | 0.872 | |
| AA | 47/114 (57.69%) |
44/104 (42.31%) |
|||||
Significance = (p < 0.05).
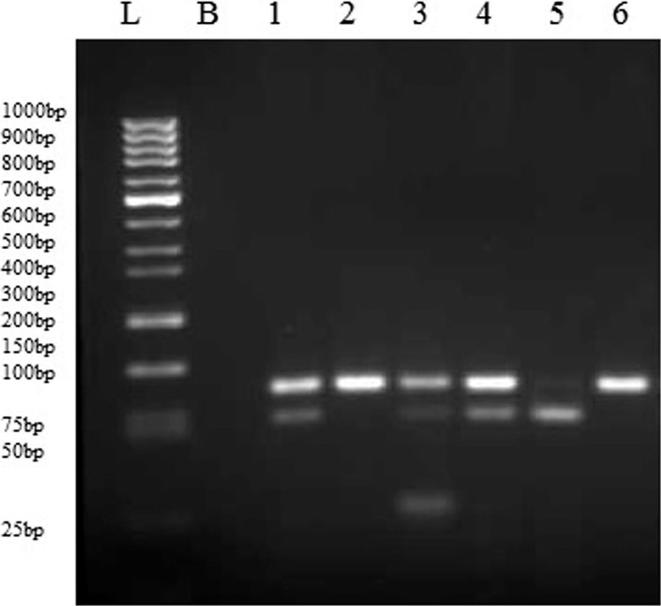
Restriction pattern of PCR amplified 130 bp product digested with Fnu4HI for genotyping at 148 position (Fig. 1). Genotype and allele frequency for PON2 G148A polymorphism is shown in Table 2. PON2 G148A polymorphism was found to be statistically significant (p < 0.05 in PON2) suggesting an association of this position with incidence of cataract. The prevalence of homozygous G allele was found to be more frequent in the cataract patients as compared to control (62.50% vs 40.35%) whereas healthy subjects had more homozygous A allele in contrast to cataract patients (59.64% vs.37.50%), as depicted in Table 2. The odds ratio for the G allele versus A allele of PON2 G148A was (OR = 2.464, p = 0.001). Allelic patterns are presented in Fig. 1.
Fig. 1.
Agarose gel electrophoresis of Restriction fragment length polymorphism for PON2. G148A PCR products show cleavage of amplified 130 bp DNA fragment. Fragment containing GG allele remained intact (lane 1). Lanes 2, 3, 4, 5 represent A/G (i.e heterozygous Alanine/Glycine). AA allele containing homozygous Alanine fragment (lane 6). L and B stand for ladder and blank, respectively.
The distribution of genotype and allele frequency for PON3 polymorphic site C133A is shown in Table 2. We did not observe any significant association at this polymorphic site. Odds ratio between C allele verses A allele was found to be 0.957 (p = 0.872). Restriction pattern of PCR product is shown in Fig. 2.
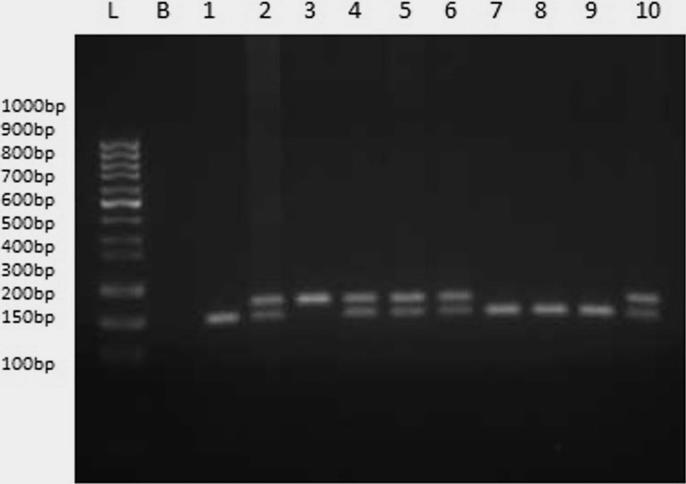
Fig. 2.
Agarose gel electrophoresis of PCR-RFLP for PON3 C133A using HinfI. Lanes 1, 7, 8, 9 represent CC allele at 2, 4, 5, 6 represent CA while 1 represent AA. L and B represents ladder and blank.
Combined genotype analysis of G148A and C311S polymorphic sites of PON2 indicated a significant association. The highest frequencies were observed in two groups: The first one is 311Cys/Ser vs 148Ala which showed higher frequency in control subjects as compared to diseased. The other genotype combination 311Ser vs 148Gly was more prevalent in cataractous group (Table 3).
Table 3.
Frequencies of combine genotype of C311S and G148A polymorphism for PON2 in control and cataract subjects.
| Combine Genotype | Control | Cataract | Odds ratio | 95% CI | χ2 Test |
|---|---|---|---|---|---|
|
N Frequency (%) |
N Frequency (%) |
||||
| 311Cys 148Gly |
09 (47.36%) |
10 (52.63%) |
0.788 | 0.292–2.122 | 0.636 |
| 311Cys 148Ala |
07 (63.63%) |
04 (36.36%) |
1.680 | 0.462–6.108 | 0.427 |
| 311Cys 148Gly/Ala |
03 (75%) |
01 (25%) |
2.833 | 0.285–28.128 | 0.354 |
| 311Cys/Ser 148 Ala |
08 (88.89%) |
01 (11.1%) |
8.327 | 1.004–69.058 | 0.022* |
| 311Ser 148Gly |
04 (28.57%) |
10 (71.42%) |
0.317 | 0.093–1.083 | 0.050* |
| 311Ser 148Ala |
14 (51.85%) |
13 (48.14%) |
0.977 | 0.409–2.333 | 0.958 |
| 311Ser 148Gly/Ala |
04 (80%) |
01 (20%) |
3.849 | 0.416–35.609 | 0.204 |
| 311Cys/Ser 148Gly/Ala |
02 928.57%) |
05 (71.42%) |
0.342 | 0.063–1.844 | 0.194 |
| 311Cys/Ser 148Gly |
05 941.67%) |
07 (58.3%) |
0.618 | 0.183–2.083 | 0.435 |
Significant value (p < 0.05).
Discussion
Identification of genetic predisposition to cataract might be beneficial for early detection and disease management. Cataract is an age-related disorder responsible for 48% of world blindness.29 Among various heterogeneous factors contributing in development of lens opacity in elderly, genetic component is reported to be critically important.30 Genetic factors are likely to increase susceptibility to environmental insult such as oxidative stress in aging population thus making them prone to develop lens opacity.31 Oxidative stress has been implicated in pathophysiology of cataract and several other eye related complications.32 PON2 and PON3 are members of Paraoxonase antioxidant family and have an established role in reducing oxidative damage through Q10.33 Both enzymes are capable to restrain LDL oxidation34 and trigger antioxidant properties along with cholesterol efflux of HDL.[35], [36] Association of PON2 and PON3 polymorphism with various disorders has sparingly been studied.[14], [15], [16], [17], [18], [19], [20], [21] Present study was conducted to explore association between senile cataract and PON2 and PON3 polymorphism.
The relationship between PON2 polymorphism and senile cataract was studied for two positions including C311S and G148A. Our results indicated an insignificant association between C311S mutation with risk of developing cataract in studied samples. Earlier investigations in coronary arterial disease for polymorphic position C311S remained inconclusive. One study stated significant association of C311 allele with the susceptibility to develop CAD,36 while other suggested 311S to be important in same disorder.21 Martinelli20 found no association at all. We also analyzed PON2 polymorphism for G148A which indicated that frequency of G148 allele was significantly higher than 148A allele in subjects suffering from cataract. Presence of G allele at 148 position has been reported in patients suffering from coronary artery disease.37 Another report showed same mutation to be the responsible for high plasma cholesterol and Apo. B.38 Newborn babies with PON2 148GA/GG genotype were also found to be at high risk of low birth weights and short birth length, when exposed to Di-n-butyl phthalate (MBP) and di-2-ethylhexyl phthalate (MEHP).39
Combine genotype analysis for PON2 polymorphism revealed the combination of G148 and 311S was more frequent in cataract subjects while heterogynous allele at 311 and 148A are more likely to be present in subjects without lens opacity. It seems likely that presence of subsequent mutations at 148 and 311 might be implicated as a risk factor for development of cataract.
Studies on PON3 mutations are limited. However, available data mainly includes investigations on promoter and intronic region polymorphism.[23], [24], [25] The most frequently studied position in coding region is C133A probably because it is located in potential binding site for transcriptional factor LF-A1T.19 The same site was also checked in current study. Our analysis did not reveal any considerable association between PON3 C133A polymorphism with senile cataract. An earlier study also reported insignificant association of this mutation with CHD.19 Still, further studies with larger dataset are needed to validate the association.
Exact physiological substrates for PON2 and PON3 have not been identified yet but initial studies have demonstrated that like PON1, PON2 and PON3 not only act as anti-atherogenic agents but can also be used as a potential target for therapy.40 PON1 and PON2 share common structural features thus it is likely that they share functional features also. However, wide expression in different tissues and multiple transcripts also suggest independent function for PON2 product38 that needs to be further explored. Some reports suggest interaction of PON2 and PON3 through coenzyme Q10 thus implicating their possible role in maintaining oxidative balance.33 PON2 can reduce levels of lipid peroxides41 and its expression is accelerated in cells during oxidative stress.42 PON3 can also inhibit LDL oxidation and its anti-atherogenic role is 100 times more effective than PON1.43 Since oxidative imbalance plays crucial role in pathophysiology of most of the diseases including cataract, the role of PON2 and PON3 in keeping oxidative homeostasis might be important. Recently, high PON2 and PON3 mRNA expression was reported in cataract probably reflecting compensatory mechanism against oxidative stress.44 Studies on polymorphism, therefore, may suggest functional outcome of genetic variation in PON3 and PON2.26
Identification of polymorphic site is gaining popularity as it may help in screening of population to predict susceptibility for the disease.45 Earlier diagnosis of population at risk through genetic testing might not only be beneficial for therapeutic intervention but may also contribute in designing strategies to delay or prevent opacities since delay of ten year in the development of opacity may reduce cataract surgeries by 50%.46 Furthermore, counseling of high risk individuals to adopt preventive measure through life style changes, healthy diet and physical activity could be additional benefit having a positive impact on patient care.
Acknowledgments
Acknowledgement
This work was supported by Higher Education Commission (Project no: 20-2010), Islamabad, Pakistan.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Footnotes
Peer review under responsibility of Saudi Ophthalmological Society, King Saud University.
References
- 1.World Health Organization. Visual impairment and blindness; 2014. http://www.who.int/mediacentre/factsheet/fs282/en.
- 2.Liu Y.C., Wilkins M., Kim T., Malyugin B., Mehta J.S. Cataracts. Lancet. 2017;390:600–612. doi: 10.1016/S0140-6736(17)30544-5. [DOI] [PubMed] [Google Scholar]
- 3.Chua J., Koh J.Y., Tan A.G. Ancestry, socioeconomic status, and age-related cataract in Asians: the Singapore epidemiology of eye diseases study. Ophthalmology. 2015;122:2169–2178. doi: 10.1016/j.ophtha.2015.06.052. [DOI] [PubMed] [Google Scholar]
- 4.Shiels A., Hejtmancik J.F. Genetic origins of cataract. Arch Ophthalmol. 2007;125:165–173. doi: 10.1001/archopht.125.2.165. [DOI] [PubMed] [Google Scholar]
- 5.Shiels A., Bennett T.M., Hejtmancik J.F. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Zhong J., Bian Z., Fang X., Peng Y., Hu Y. Association between polymorphisms of OGG1, EPHA2 and age-related cataract risk: a meta-analysis. BMC Ophthalmol. 2016;16:168. doi: 10.1186/s12886-016-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas S., Raza S.T., Chandra A. Polymorphism of FABP2 and PPARG2 genes in risk prediction of cataract among North Indian population. Meta Gene. 2014;2:307–313. doi: 10.1016/j.mgene.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan A.G., Kifley A., Mitchell P. Associations between methylenetetrahydrofolate reductase polymorphisms, serum homocysteine levels, and incident cortical cataract. JAMA Ophthalmol. 2016;134:522–528. doi: 10.1001/jamaophthalmol.2016.0167. [DOI] [PubMed] [Google Scholar]
- 9.Liao R.-f., Ye M.-j., Liu C.-y., Ye D.-q. An updated meta-analysis: risk conferred by glutathione S-transferases (GSTM1 and GSTT1) polymorphisms to age-related cataract. J Ophthalmol. 2015;2015:1–10. doi: 10.1155/2015/103950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shunmoogam N., Naidoo P., Chilton R. Paraoxonase (PON)-1: a brief overview on genetics, structure, polymorphisms and clinical relevance. Vascular Health Risk Manage. 2018;14:137. doi: 10.2147/VHRM.S165173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baig A., Zohaib M., Rehman A.U., Zarina S. Q192R paraoxonase1 polymorphism is a risk factor for cataract in Pakistani population. Pak J Pharm Sci. 2016;29:765–771. [PubMed] [Google Scholar]
- 12.Ali O.M., Effat L.K., Amr K.S., Azeem A.A.A., Hassan S.M. Contribution of PON1 polymorphism in senile cataract among diabetic and non-diabetic Egyptian patients. J Mol Biomarkers Diagnosis. 2014;5:180. [Google Scholar]
- 13.Flekac M., Skrha J., Hilgertova J., Lacinova Z., Jarolimkova M. Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med Genet. 2008;9:30. doi: 10.1186/1471-2350-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sögüt E., Ortak H., Aydogan L., BenlI I. Association of paraoxonase 1 L55M and Q192R single-nucleotide polymorphisms with age-related macular degeneration. Retina. 2013;33:1836–1842. doi: 10.1097/IAE.0b013e318287da59. [DOI] [PubMed] [Google Scholar]
- 15.Dantoine T.F., Drouet M., Debord J., Merle L., Cogne M., Charmes J.P. Paraoxonase 1 192/55 gene polymorphisms in Alzheimer's disease. Ann N Y Acad Sci. 2002;977:239–244. doi: 10.1111/j.1749-6632.2002.tb04821.x. [DOI] [PubMed] [Google Scholar]
- 16.Dursun A., Cicek S., Keni F.M., Karakas-Celik S., Sezer T., Altinyazar C.H. The relation of PON1-L55M gene polymorphism and clinical manifestation of Behcet's disease. Acta Biochim Pol. 2014;61:2. [PubMed] [Google Scholar]
- 17.Imai Y., Morita H., Kurihara H. Evidence for association between paraoxonase gene polymorphisms and atherosclerotic diseases. Atherosclerosis. 2000;149:435–442. doi: 10.1016/s0021-9150(99)00340-8. [DOI] [PubMed] [Google Scholar]
- 18.Shi J., Zhang S., Tang M. Possible association between Cys311Ser polymorphism of paraoxonase 2 gene and late-onset Alzheimer's disease in Chinese. Mol Brain Res. 2004;120:201–204. doi: 10.1016/j.molbrainres.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Fan Z., Huang J. Extensive association analysis between polymorphisms of PON gene cluster with coronary heart disease in Chinese Han population. Arterioscl, Thrombosis, Vascular Biol. 2003;23:328–334. doi: 10.1161/01.atv.0000051702.38086.c1. [DOI] [PubMed] [Google Scholar]
- 20.Martinelli N., Girelli D., Olivieri O. Interaction between smoking and PON2 Ser311Cys polymorphism as a determinant of the risk of myocardial infarction. Eur J Clin Invest. 2004;34:14–20. doi: 10.1111/j.1365-2362.2004.01292.x. [DOI] [PubMed] [Google Scholar]
- 21.Sanghera D.K., Aston C.E., Saha N., Kamboh M.I. DNA polymorphisms in two paraoxonase genes (PON1 and PON2) are associated with the risk of coronary heart disease. Am J Hum Genet. 1998;62:36–44. doi: 10.1086/301669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada Y., Ando F., Niino N., Miki T., Shimokata H. Association of polymorphisms of paraoxonase 1 and 2 genes, alone or in combination, with bone mineral density in community-dwelling Japanese. J Hum Genet. 2003;48:469–475. doi: 10.1007/s10038-003-0063-x. [DOI] [PubMed] [Google Scholar]
- 23.Sanghera D.K., Manzi S., Minster R.L. Genetic variation in the paraoxonase-3 (PON3) gene is associated with serum PON1 activity. Ann Hum Genet. 2008;72(1):72–81. doi: 10.1111/j.1469-1809.2007.00400.x. [DOI] [PubMed] [Google Scholar]
- 24.Erlich P.M., Lunetta K.L., Cupples L.A. Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum Mol Genet. 2006;15:77–85. doi: 10.1093/hmg/ddi428. [DOI] [PubMed] [Google Scholar]
- 25.Aragones G., Guardiola M., Barreda M. Measurement of serum PON-3 concentration: method evaluation, reference values, and influence of genotypes in a population-based study. J Lipid Res. 2011;52:1055–1061. doi: 10.1194/jlr.D014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng C.J., Shih D.M., Hama S.Y., Villa N., Navab M., Reddy S.T. The paraoxonase gene family and atherosclerosis. Free Radic Biol Med. 2005;38:153–163. doi: 10.1016/j.freeradbiomed.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 27.Green M.R., Sambrook J. Cold Spring Harbor Laboratory Press; New York: 2012. Molecular cloning: a laboratory manual. [Google Scholar]
- 28.Mochizuki H., Scherer S.W., Xi T. PON2 gene at 7q21.3: cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene. 1998;213:149–157. doi: 10.1016/s0378-1119(98)00193-0. [DOI] [PubMed] [Google Scholar]
- 29.Dua H.S., Said D.G., Otri A.M. Are we doing too many cataract operations? Cataract surgery: a global perspective. Br J Ophthalmol. 2009;93:1–2. doi: 10.1136/bjo.2008.143685. [DOI] [PubMed] [Google Scholar]
- 30.Connell P.P., Keane P.A., O'Neill E.C. Risk factors for age-related maculopathy. J Ophthalmol. 2009;2009:39. doi: 10.1155/2009/360764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hejtmancik JF, Smaoui N. Molecular genetics of cataract. Genetics in ophthalmology. Karger Publishers; 2003;37:67–82. [DOI] [PubMed]
- 32.Vinson J.A. Oxidative stress in cataracts. Pathophysiology. 2006;13:151–162. doi: 10.1016/j.pathophys.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Witte I., Foerstermann U., Devarajan A., Reddy S.T., Horke S. Protectors or traitors: the roles of PON2 and PON3 in atherosclerosis and cancer. J Lipids. 2012;2012:342806. doi: 10.1155/2012/342806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng C.J., Hama S.Y., Bourquard N., Navab M., Reddy S.T. Adenovirus mediated expression of human paraoxonase 2 protects against the development of atherosclerosis in apolipoprotein E-deficient mice. Mol Genet Metab. 2006;89:368–373. doi: 10.1016/j.ymgme.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Ng C.J., Wadleigh D.J., Gangopadhyay A. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J Biol Chem. 2001;276:44444–44449. doi: 10.1074/jbc.M105660200. [DOI] [PubMed] [Google Scholar]
- 36.Ng C.J., Bourquard N., Hama S.Y. Adenovirus-mediated expression of human paraoxonase 3 protects against the progression of atherosclerosis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:1368–1374. doi: 10.1161/ATVBAHA.106.134189. [DOI] [PubMed] [Google Scholar]
- 37.Chi D., Ling W., Ma J. Relationship between paraoxonase 1 55 Met/Leu, paraoxonase 2 148 Ala/Gly genetic polymorphisms and coronary artery disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23(3):289–293. [PubMed] [Google Scholar]
- 38.Shin B.S., Oh S.Y., Kim Y.S., Kim K.W. The paraoxonase gene polymorphism in stroke patients and lipid profile. Acta Neurol Scand. 2008;117:237–243. doi: 10.1111/j.1600-0404.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 39.Xie C., Jin R., Zhao Y. Paraoxonase 2 gene polymorphisms and prenatal phthalates' exposure in Chinese newborns. Environ Res. 2015;140:354–359. doi: 10.1016/j.envres.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 40.She Z.-G., Chen H.-Z., Yan Y., Li H., Liu D.-P. The human paraoxonase gene cluster as a target in the treatment of atherosclerosis. Antioxidants Redox Signal. 2012;16:597–632. doi: 10.1089/ars.2010.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenblat M., Draganov D., Watson C.E., Bisgaier C.L., La Du B.N. Mouse macrophage paraoxonase 2 activity is increased whereas cellular paraoxonase 3 activity is decreased under oxidative stress. Art Thromb Vasc Biol. 2003;23:468–474. doi: 10.1161/01.ATV.0000059385.95664.4D. [DOI] [PubMed] [Google Scholar]
- 42.Aviram M., Rosenblat M. Paraoxonases 1, 2, and 3, oxidative stress, and macrophage foam cell formation during atherosclerosisdevelopment. Free Radic Biol Med. 2004;37:1304–1316. doi: 10.1016/j.freeradbiomed.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 43.Draganov D.I., Stetson P.L., Watson C.E., Billecke S.S., La Du B.N. Rabbit serum paraoxonase 3 (PON3) is a high density lipoprotein – associated lactonase and protects low density lipoprotein against oxidation. J Biol Chem. 2000;275:33435–33442. doi: 10.1074/jbc.M004543200. [DOI] [PubMed] [Google Scholar]
- 44.Bharathidevi S.R., Babu K.A., Jain N. Ocular distribution of antioxidant enzyme paraoxonase & its alteration in cataractous lens & diabetic retina. Indian J Med Res. 2017;145:513–520. doi: 10.4103/ijmr.IJMR_1284_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X., Jiao X., Ma Z., Hejtmancik J.F. Polymorphism rs7278468 is associated with Age- related cataract through decreasing transcriptional activity of the CRYAA promoter. Sci Rep. 2016;6:23206. doi: 10.1038/srep23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brian G., Taylor H. Cataract blindness – challenges for the 21st century. Bull World Health Organ. 2001;79:249–256. [PMC free article] [PubMed] [Google Scholar]