SUMMARY
The ability of induced pluripotent stem cells (iPSCs) to differentiate into all adult cell types makes them attractive for research and regenerative medicine; however, it remains unknown when and how this capacity is established. We characterized the acquisition of developmental pluripotency in a suitable reprogramming system to show that iPSCs prior to passaging become capable of generating all tissues upon injection into preimplantation embryos. The developmental potential of nascent iPSCs is comparable to or even surpasses that of established pluripotent cells. Further functional assays and genomewide molecular analyses suggest that cells acquiring developmental pluripotency exhibit a unique combination of properties that distinguish them from canonical naive and primed pluripotency states. These include reduced clonal self-renewal potential and the elevated expression of differentiation-associated transcriptional regulators. Our observations close a gap in the understanding of induced pluripotency and provide an improved roadmap of cellular reprogramming with ramifications for the use of iPSCs.
Graphical Abstract

In Brief
Amlani et al. use a highly efficient reprogramming approach to determine when the hallmark features of self- renewal and developmental pluripotentcy are established during the derivation of induced pluripotent stem cells (iPSCs) from mouse fibroblasts. They show that nascent iPSCs appear molecularly and functionally poised for differentiation but, nevertheless, generate all adult tissues upon introduction into blastocyst-stage embryos exceptionally well.
INTRODUCTION
Pluripotent stem cells (PSCs) are endowed with the capacity to indefinitely propagate in vitro (“self-renewal”) while retaining the ability to differentiate into all somatic cell types and germ cells upon receiving environmental cues (“developmental pluripotency”). Different types of PSCs that vary in origin, molecular regulation, and functional properties have been described (Morgani et al., 2017; Weinberger et al., 2016). Among these, so-called naive PSCs most closely resemble the pluripotent inner cell mass of the mammalian embryo, can self-renew well at clonal density, and efficiently contribute to development when injected into blastocyst-stage embryos (Ying and Smith, 2017). Rodent embryonic stem cells (ESCs) and induced PSCs (iPSCs) are examples of naive PSCs. They are derived from differentiated somatic cells by enforced expression of combinations of embryonic transcription factors (TFs) such as OCT4, KLF4, SOX2 and MYC (OKSM). Human PSCs and mouse epiblast-derived stem cells (EpiSCs) more closely resemble post-implantation stages of development, self-renew poorly at clonal density, and do not integrate well into preimplantation embryos, a cellular state referred to as primed pluripotency. As the properties of primed PSCs complicate biomedical applications, efforts are being made to establish naive pluripotency in human PSCs, but no consensus on a best approach has emerged yet (Boroviak and Nichols, 2017; Weinberger et al., 2016).
The study of TF-mediated reprogramming has yielded roadmaps of transcriptional, chromatin, and cellular changes characterizing the establishment of naive pluripotency (Apostolou and Hochedlinger, 2013; Polo et al., 2012; Samavarchi-Tehrani et al., 2010; Stadtfeld et al., 2008), but it is unknown when somatic cells acquire developmental pluripotency on their trajectory to become established cell lines; a process that can take weeks to months. Consequently, the molecular features of cells at this juncture remain uncertain. The observation that chromatin features of the somatic cell of origin can persist for prolonged periods in iPSCs and influence their developmental output (Kim et al., 2010; Krijger et al., 2016; Polo et al., 2010) suggests that extensive ex vivo culture might be required before fully developmental competent cells can be attained.
Murine iPSC reprogramming is well-suited to track when developmental potency is acquired, as stringent functional assays (such as injection of cells into tetraploid [4n] blastocysts that alone cannot support embryonic development) are available (Eggan et al., 2001; Nagy et al., 1990). However, studying the acquisition of developmental pluripotency is complicated by the observation that many iPSCs never attain full developmental potency due to molecular abnormalities introduced during reprogramming (Stadtfeld et al., 2010a; Wu et al., 2014). In addition, the slow and asynchronous nature of the reprogramming process (Yamanaka, 2009) does not yield sufficient cells for meaningful functional assays at early stages of iPSC formation.
Protocols have been described that greatly facilitate reprogramming, including interference with repressive chromatinrelated factors (Rais et al., 2013), additional reprogramming factors (Di Stefano et al., 2016), and the use of chemical compounds modulating cellular signaling pathways (Bar-Nur et al., 2014; Federation et al., 2014; Vidal et al., 2014). We have taken advantage of an efficient reprogramming approach that predominantly yields fully developmentally competent PSCs to systematically assay functional properties at hallmark stages of iPSC derivation from murine fibroblasts. We find that the ability to differentiate into all essential somatic cell types after blastocyst injection is established abruptly upon reprogramming factor shutdown and does not require propagation in culture. While exhibiting reduced self-renewal potential and more rapid downregulation of core pluripotency loci in vitro, nascent iPSCs favorably compare to established naive PSCs in the 4n complementation assay. At the molecular level, the acquisition of developmental pluripotency ensues after largescale chromatin changes and coincides with the fine-tuning of chromatin accessibility and elevated expression of differentiation-associated gene loci, including regulators of primed pluripotency.
RESULTS AND DISCUSSION
Rapid Reprogramming Yields Developmentally Fully Competent Cells
We previously reported that combined activation of Wnt signaling (using the GSK3 antagonist CHIR99021) and inhibition of TGFb signaling (using the TGFBRI antagonist iAlk5) in the presence of ascorbic acid, a facilitator of chromatin remodeling (Monfort and Wutz, 2013), greatly increases speed and efficiency of iPSC formation (Vidal et al., 2014). We derived mouse embryonic fibroblasts (MEFs) from a “reprogrammable” transgenic mouse strain, which allows expression of OKSM in a doxycycline (dox)-dependent manner (Stadtfeld et al., 2010b). These animals also harbor a knockin of enhanced GFP into the Pou5f1 locus (Oct4-GFP), a specific marker for pluripotent cells (Lengner et al., 2007). Six days of culturing MEFs in the presence of dox and the three aforementioned compounds (3c) yielded colonies that were independent of transgenic OKSM expression (Figure 1A). By this time, each starting fibroblast had given rise to more than 100 Oct4-GFP+ cells (Figure S1A) on average, documenting the availability of large numbers of advanced intermediates for functional assays.
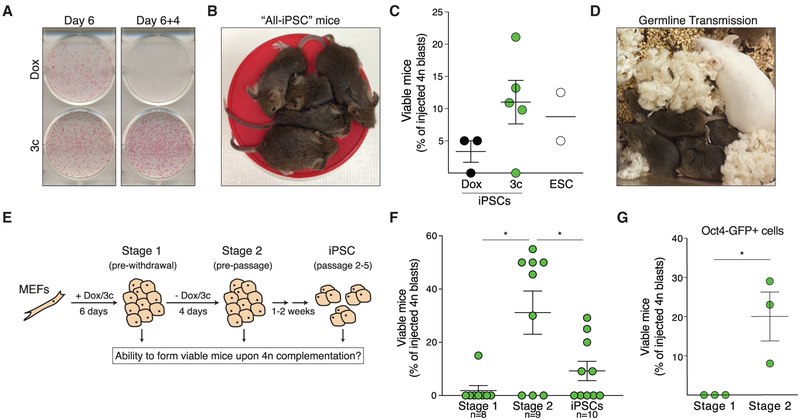
Figure 1. Acquisition of Developmental Pluripotency in a Highly Efficient Reprogramming System.
(A) Colonies after six days of reprogramming in indicated conditions (left) and after dox/3c withdrawal plus four more days of culture in mESC media (right).
(B) Agouti all-iPSC mice derived upon injection of 3c iPSCs into 4n CD1 blastocysts.
(C) Viable animals obtained upon 4n injection with indicated cell types. Circles represent the average results obtained with a clonal line.
(D) Agouti pups after breeding all-iPSC mice with CD1 females.
(E) Experimental strategy to determine when cells acquire the ability to generate all-iPSC mice.
(F) All-iPSC mice obtained with polyclonal cells at indicated reprogramming stages. A Mann-Whitney test followed by Bonferroni correction was used to measure statistical significance after accounting for technical failure as outlined in the Experimental Procedures.
(G) All-iPSC mice obtained with purified Oct4-GFP+ S1 and S2 cells. Significance is based on a Mann-Whitney test.
Significance is indicated by *p ≤ 0.05 and reflects the higher p value obtained for each comparison from two independent statistical tests.
We established five monoclonal iPSC lines in separate reprogramming experiments from the same batch of male MEFs, followed by colony picking and expansion for several weeks in ESC medium containing leukemia inhibitory factor (LIF) and 15% serum. Upon injection into 4n blastocysts, four of the five lines gave rise to “all-iPSC” mice (Figure 1B) at frequencies similar to reference ESCs (Figure 1C; Table S1). Adult all-iPSC mice generated with all four cell lines were fertile and capable of germline transmission of the original MEF genome (Figure 1D). These findings show that 3c reprogramming predominantly results in fully developmentally competent cells, a conclusion further supported by comparison to published success rates of 4n complementation (Figure S1B). While developmental assays conducted with MEFs reprogrammed with an alternative TF combination suggested an inverse relationship between reprogramming speed and iPSC quality (Buganim et al., 2014; Sebban and Buganim, 2016), our findings demonstrate that rapid induction of pluripotency does not necessarily come at a developmental cost. The epigenome-protective function of ascorbic acid during reprogramming (Stadtfeld et al., 2012) might contribute to the high quality of iPSCs generated by our approach.
Pre-passage iPSCs Complement 4n Blastocysts in a Highly Efficient Manner
We conducted 4n complementation assays with cells at three different stages of iPSC derivation that we considered likely to cover key functional changes: day six of transgenic OKSM expression (stage 1 [S1]); four days after shut-down of transgenic OKSM and washout of 3c, but before the first passage of nascent iPSCs (stage 2 [S2]); and after roughly two weeks of continuous passaging in ESC medium (early iPSC stage [iPSCs]) (Figure 1E). For each stage, we transferred more than 150 blastocysts injected with cells derived from the same MEF preparation to avoid effects caused by differences in genetic background (Table S1). As molecular maturation steps have been reported to proceed for prolonged periods of time in iPSCs (Golipour et al., 2012; Polo et al., 2010), we anticipated that the regulatory circuitry underlying pluripotency may not be stable enough in S1 and S2 cells to allow 4n complementation to succeed. Indeed, S1 cells almost invariably failed to yield all-iPSC mice (Figure 1F; Table S1). However, S2 cells complemented 4n blastocysts at efficiencies that exceeded success rates with iPSCs (Figure 1F; Table S1). No overt differences were observed between animals generated with S2 cells or iPSCs and breeding-derived “reprogrammable mice,” although we did not age animals beyond 4–8 weeks.
We confirmed the observation that developmental pluripotency is attained between S1 and S2 by using Oct4-GFP+ cells purified with a gentle microfluidics-based cell sorting technique (Figure 1G; Table S1), ruling out that the lower abundance of Oct4-GFP+ in S1 cultures (Figure S1C) had masked developmentally competent cells. This demonstrates that reactivation of Oct4 is not sufficient to endow cells with developmental pluripotency and suggests that additional molecular changes that occur between S1 and S2 are required. Of note, S1 cells, S2 cells, and iPSCs expressed similar levels of the adherence molecule E-cadherin (also known as CDH1), an important determinant for the ability to incorporate into blastocyst-stage embryos (Ohtsuka et al., 2012) (Figures S1E and S1F).
Female S2 cells also gave rise to all-iPSC mice at high frequency (Figure S1D; Table S1), suggesting that the instability of DNA methylation patterns frequently seen in female pluripotent cells (Choi et al., 2017; Ooi et al., 2010; Zvetkova et al., 2005) might not manifest at this stage. Together, these observations suggest that cells with the ability to generate all somatic tissues upon injection into tetraploid blastocysts arise abruptly and at a defined stage during reprogramming.
Reduced Self-Renewal Capacity of Nascent iPSCs
We evaluated the in vitro behavior of cells at different reprogramming stages to work toward a framework for the observed differences in developmental pluripotency. S1 cells exhibited a profound deficit to form undifferentiated colonies upon seeding at clonal density in ESC medium (Figure 2A), which we then confirmed using purified Oct4-GFP+ cells (Figure 2B). S2 cells yielded colonies 5- to 10-fold more frequently than S1 cells, but remained significantly less efficient than established iPSCs (Figure 2B). This suggests that self-renewal capacity (in contrast to the ability to complement 4n blastocysts) is gradually established during iPSC derivation and has limited predictive value for developmental potency. The observation that developmental competence peaks before self-renewal during iPSC formation is reminiscent of mammalian embryogenesis, in which the capacity of in vitro clonal growth is established later as well (Boroviak et al., 2014).
Figure 2. Self-Renewal and EB Formation of Nascent iPSCs.
(A) Colonies obtained upon seeding 500 cells from the indicated stages into mESC medium.
(B) Colony formation after single-cell sorting of Oct4-GFP+ cells. Error bars indicate SE. n = 3 complete 96-well plates per stage.
(C) EBs formed from 2,000 S2 cells and 800 established iPSCs, respectively.
(D) Flow cytometric quantification of SSEA1 expression on undifferentiated S2 cells and iPSCs and EBs, respectively. n = 3.
(E) Expression of pluripotency regulators in undifferentiated S2 cells and iPSCs (n = 2) and EBs (n = 3), measured by qPCR.
Significance is indicated by *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001, based on a one-way ANOVA with Tukey post-test (B), a t test (D), and a two-way ANOVA with Sidak post-test (E).
Both S2 cells and iPSCs aggregated into embryoid bodies (EBs) in the absence of LIF (Figure 2C), a common approach to initiate differentiation, though S2-derived EBs were slightly smaller (Figure S2A). Abundance of the pluripotency-associated surface marker SSEA1, highly expressed on undifferentiated S2 cells and iPSCs (Figure S2B), dropped in EBs derived from either cell type, indicating commencement of differentiation. However, downregulation of SSEA1 was significantly more pronounced in S2-derived EBs (Figures 2D and S2B), indicating a faster and/or more efficient exit from pluripotency. Similarly, mRNAs encoding transcriptional regulators involved in stabilization of the pluripotent state, such as POU5F1 and NANOG, were more rapidly downregulated in EBs formed by S2 cells (Figure 2E). The results of both self-renewal and in vitro differentiation assays are in agreement with the notion that the pluripotency network in S2 cells, while stabilized sufficiently to support remarkably robust in vivo developmental potency, remains less solidified than in established iPSCs and can be more easily dismantled by differentiation-inducing cues.
Genome-wide Chromatin Remodeling Precedes the Acquisition of Developmental Pluripotency
Our results suggest that cessation of transgenic OKSM expression and withdrawal of 3c allow rapid establishment of self-renewal and success in tetraploid complementation. We conducted RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin sequencing (ATAC-seq) on MEFs and Oct4-GFP+ cells at the different reprogramming stages to identify underlying molecular events. Analyses at the genome-wide level suggested stage-specific differences, but also demonstrated an overall high degree of similarity between S1 and S2 cells and iPSCs (Figures 3A and 3B). Silencing of somatic gene expression (Figure S3A) and decommissioning of fibroblast-associated enhancer elements appeared to be largely completed by S1 (Figure S3B). Similarly, the vast majority of ESC-associated enhancer elements had acquired accessibility during the MEF to S1 transition (Figure S3C). These observations suggest that erasure of fibroblast identity and global remodeling of chromatin accessibility do not pose substantial roadblocks in our reprogramming approach, but also do not endow cells with pluripotency. Rather, the acquisition of developmental and self-renewal capacities appears associated with fine-tuning at the transcriptional or post-transcriptional level and/or accessibility changes at select regulatory regions (Figures S3D and S3E).
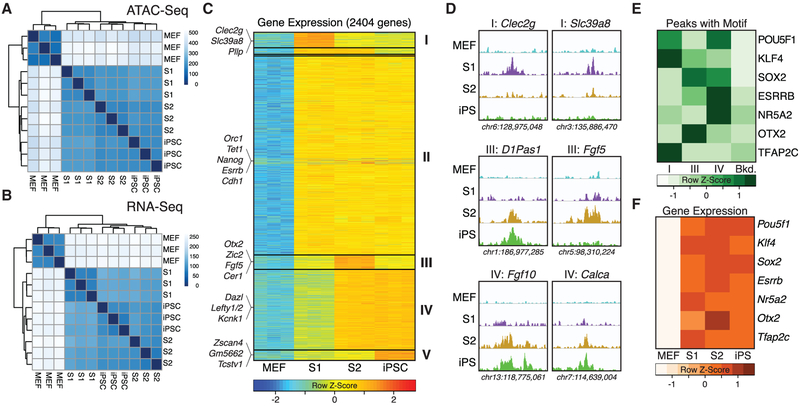
Figure 3. Molecular Characterization of Cells at Specific Reprogramming Stages.
(A and B) Unsupervised clustering of MEFs, and Oct4-GFP+ S1 cells, S2 cells and iPSCs based on ATAC-seq (A) and RNA-seq data (B).
(C) Heatmap showing expression kinetics of pluripotency-associated genes based on pairwise comparisons. Representative genes of the five major groups identified (groups I–V) are shown.
(D) ATAC-seq tracks showing examples of stage-specific chromatin accessibility at select gene loci.
(E) Row scaled prevalence of TF binding motifs enriched at ATAC-seq peaks belonging to the indicated groups and their prevalence in randomly chosen background genomic sequences matched for GC percent content.
(F) Abundance of mRNAs of TFs shown in (E) as measured by RNA-seq.
Genes upregulated at any stage of iPSC derivation were comprised of five major groups, labeled I to V based on their distinct expression kinetics (Figures 3C and S3F; Table S2). The largest (group II; equal expression in S1, S2, and iPSCs) and smallest (group V; expression highest in iPSCs) groups did not distinguish between pre-pluripotent S1 and pluripotent S2 cells. Reactivation of group II genes, which included core naive pluripotency regulators (e.g., Nanog, Sall4, Cdh1, and Esrrb) and genes associated with cell cycle progression (Polq, Chek1, Brca2) and chromatin organization (Ezh2, Tet1, Prdm14) (Figures 3C, S3F, and S3G; Table S3) is therefore insufficient to endow cells with robust self-renewal or developmental potency. Group V contained loci associated with the totipotent 2-cell stage of mammalian development (Wu et al., 2017), including members of the Zscan4 family (Figures 3C and S3F). While expression of these genes appears not to be required for the establishment of pluripotency in S2 cells, our results are consistent with the genes being beneficial upon prolonged culture.
Molecular Fine-Tuning Associated with the Acquisition of Developmental Pluripotency
Three gene groups distinguished S2 and S1 cells. Group I contained genes highest expressed in S1 cells (Figures 3C and S3F), which represent gene ontology categories associated with neither MEFs nor pluripotent cells (Figure S3G; Table S3). The majority (about 55%) of these loci contained ATAC-seq peaks exclusively seen at S1 (Figure 3D), suggesting that their expression might be controlled by regulatory elements that become transiently activated during the reprogramming process before cells acquire pluripotency. Groups III and IV became upregulated during the S1–S2 transition (Figure 3C) and, in addition to germ cell-associated factors, contained genes essential for early post-implantation development. While group IV genes, such as Lefty2, Dazl, and Dppa4, remained expressed at constant levels in iPSCs, group III transcripts peaked at S2 (Figures 3C and S3F). We observed that a substantial portion (about 40%) of group III and group IV loci gained accessible regions during the S1–S2 transition, the majority of which (but not all of) remained open in iPSCs (Figure 3D). This indicates that remodeling of gene loci encoding developmental regulators is delayed during iPSC formation compared to the bulk of regulatory elements (Figures S3C and S3E).
Several group III genes, such as Otx2, Fgf5, Cer1, and Zic2, are markers of the primed pluripotent state (Weinberger et al., 2016). Quantitative PCR confirmed elevated expression of primed PSC markers in S2 cells, although levels of Otx2 and Fgf5 remained lower than in EpiSCs (Figure S3H). Genomewide analysis using published RNA-seq data indicated that S2 cells are proximal to, but nevertheless distinct from, both EpiSCs and ESCs (Figure S3I). These differences might contribute to the observation that S2 cells, but not primed PSCs (Brons et al., 2007; Tesar et al., 2007), efficiently contribute to embryonic development upon blastocyst injection.
We categorized genomic regions that alter their accessibility during the S1–S2 transition into groups I (accessible in S1), III (accessible in S2), or IV (accessible in S2 and iPSCs) and then analyzed them for recurrent motifs (Table S4). We frequently found KLF4 motifs at group I peaks while OCT4 and SOX2 motifs dominated within the pluripotency-associated group IV peaks, which also showed enrichment for the naive pluripotency regulators ESRRB and NR5A2 (Figure 3E). The observation that levels of the transcripts encoding these TFs did not significantly change after the S1 stage (Figure 3F) might suggest that important aspects of the acquisition of pluripotency are driven by changes in protein abundance or in the activity of specific core reprogramming and pluripotency factors.
Expression of the Epiblast Marker Otx2 in Developmentally Competent Nascent iPSCs
We noticed that genomic regions accessible only in S2 cells were enriched for OTX2 motifs (Figure 3E), which was mirrored by the abundance of the mRNA encoding this TF (Figures 3F and S3F). OTX2 has been reported to drive exit from naive pluripotency (Acampora et al., 2013; Yang et al., 2014) and regulate neuroectoderm specification (Rhinn et al., 1998). Transient expression of differentiation-associated genes has also been seen by others studying murine (Polo et al., 2012) and human (Takahashi et al., 2014) fibroblast reprogramming. To further investigate this phenomenon, we generated an Otx2-RFP allele (Figure 4A). During reprogramming, Otx2-RFP+ cells emerged later than Oct4-GFP+ cells (Figure S4A) and were first observed as a small subset of S1 cells that expressed high levels of CD326 encoded by Epcam (Polo et al., 2012) and Oct4-GFP (Figure S4B). The abundance of Otx2-RFP+ cells increased only moderately when 3c conditions were maintained (Figure 4B). In contrast, almost all S2 cells expressed elevated Otx2 levels, but intensity of the reporter, frequency of Otx2-RFP+ cells, and Otx2 transcript levels dropped in established iPSCs (Figures 4C, 4D, and S4C–S4E). This confirms our RNA-seq kinetics and establishes reactivation of Otx2 as a late reprogramming event. We observed similar Otx2 kinetics during OKSM-mediated reprogramming in the absence of 3c compounds (Figures S4E and S4F) and during analysis of published expression data of transposon-based reprogramming (Golipour et al., 2012) (Figure S4G), suggesting our findings are not restricted to a single approach.
Figure 4. Expression of the Otx2 Locus during the Acquisition of Pluripotency.
(A) The strategy to generate the Otx2-RFP reporter.
(B) Percent of Oct4-GFP+EpCAMHIGH cells that reactivate Otx2-RFP expression at the indicated days of 3c reprogramming.
(C) Quantification of Oct4-GFP+ cells that express Otx2-RFP in S2 cells (n = 6) and iPSCs (n = 5). Significance is indicated by **p ≤ 0.01, based on a t test.
(D) Otx2-RFP expression in S2 cells and established iPSCs. iPSCs harboring only the Oct4-GFP allele were used as controls.
(E) All-iPSC mice obtained upon injection of Otx2-RFP+Oct4-GFP+ S2 cells and iPSCs, respectively. Significance is indicated by *p ≤ 0.05, based on a Mann- Whitney test.
(F) Representative image of an E14.5 all-iPSC embryo obtained with purified Otx2-RFP S2 cells. The developing brain is a known area of Otx2 expression.
We next utilized the Otx2 reporter to explore the behavior of S1 cells, S2 cells, and iPSCs when exposed to FGF2 and Activin A, which are used to convert naive PSCs into primed PSCs (Guo et al., 2009) and can also support alternative cell fates (Ohinata and Tsukiyama, 2014) (Figure S5A). The relative efficiency of colony formation (Figure S5B) was comparable to what we observed in self-renewing conditions (Figure 2B). However, while S1 cells and iPSCs seeded in presence of FGF2 and Activin A predominantly formed colonies that maintained pluripotent cell morphology (Figure S5C) and Oct4-GFP expression (Figures S5D and S5E), S2 cell-derived colonies lost their epithelial appearance and downregulated Oct4-GFP while retaining high levels of Otx2-RFP (Figures S5C–S5E). This is reminiscent to the accelerated downregulation of pluripotency-associated transcripts during EB formation (Figures 2D and 2E), suggesting transient destabilization of the pluripotent state at S2.
Finally, we wondered whether the minority population of Otx2- RFP+ cells in established iPSC cultures (Figure 4C) is functionally equivalent to S2 cells. While sorted S2 cells frequently yielded viable mice upon 4n complementation, Otx2-RFP+Oct4-GFP+ iPSCs performed significantly worse in this assay (Figures 4E and 4F). This suggests that S2 cells represent a distinct, highly developmentally potent state that is compatible with elevated expression of specific differentiation-associated transcripts.
In summary, we have described the acquisition of functional features of naive pluripotent cells during murine iPSC formation in a tractable experimental system, resulting in a refined roadmap for the establishment of pluripotency during cellular reprogramming (Figure 5). Our roadmap correlates the hallmark properties of self-renewal and developmental potency with molecular features and should provide a useful reference point for the further dissection of the molecular requirements underlying functional properties of iPSCs.
Figure 5. Kinetics of Functional, Transcriptional, and Chromatin Accessibility Changes during iPSC Formation from Murine Fibroblasts.
EXPERIMENTAL PROCEDURES
Mice
Derivation, handling, and genotyping of reprogrammable mice (JAX011001) with the Oct4-GFP allele were described previously (Stadtfeld et al., 2010b). All animal experiments were in accordance with the guidelines of the NYU School of Medicine Institutional Animal Care and Use Committee.
4n Blastocyst Complementation
Zygotes were isolated from BDF1 females as previously described (Stadtfeld et al., 2012) and cultured overnight until they reached the 2-cell stage. One hour after electro-fusion, 1-cell embryos were separated from embryos that had failed to fuse, cultured for another 2 days, then injected with cells at the indicated reprogramming stages. Monoclonal iPSCs were injected at passage 5. For micro-fluidics based sorting of Oct4-GFP+ cells prior to blastocyst injection, an On-chip Sort (On-chip Biotechnologies, Tokyo, Japan) was used. For sorting of Oct4-GFP+Otx2-RFP+ cells prior to blastocyst injection, a SH800Z equipped with a 100mm nozzle (Sony Biotechnology) was used. Viable pups were defined as those that survived for at least three days following birth.
Cell Culture
MEFs (Stadtfeld et al., 2010b) and ESCs (Czechanski et al., 2014) were derived as previously described and were heterozygous for the inducible OKSM, Rosa26-rtTA, and Oct4-GFP alleles (Stadtfeld et al., 2010b). ESCs and established iPSCs were cultured on irradiated feeder cells in KO-DMEM (Invitrogen) supplemented with L-glutamine, penicillin/streptomycin, nonessential amino acids, b-mercaptoethanol, 1,000 U/mL LIF, and 15% FBS (ESC medium). MEF cultures were established by trypsin digestion of midgestation (embryonic day [E]13.5–E15.5) embryos and maintained in DMEM supplemented with 10% FBS, L-glutamine, penicillin/streptomycin, nonessential amino acids, and b-mercaptoethanol (MEF medium). Reprogramming was carried out as previously described (Vidal et al., 2014). Briefly, 100 to 250 cells/cm2 were seeded on a layer of irradiated feeder cells in ESC medium in the presence of 1 mg/mL Dox and, if applicable, L-ascorbic acid (50 mg/mL), CHIR99021 (3 mM), and TGF-b RI Kinase Inhibitor II (250 nM) (3c). EpiSCs were a kind gift of Paul Tesar and maintained in in serum-free medium containing FGF2 (12 ng/mL) and Activin A (20 mg/mL) as previously described (Tesar et al., 2007).
In Vitro Differentiation
EB formation with the hanging drop method was conducted as previously described (Geijsen et al., 2004), followed by dissociation with 0.2% collagenase type IV and either staining with anti-SSEA1 antibody (MC-480, eBioscience) or RNA isolation. To assess the response of cells at different stages of iPSC derivation to exposure to FGF2 and Activin A, equal amounts of Oct4-GFP+ cells were seeded on feeder cells in EpiSC medium. Colonies were scored and imaged after 4 days, followed by flow cytometric analysis for Oct4-GFP and Otx2-RFP expression.
Generation of Otx2 Reporter Cells
A donor plasmid harboring the IRES-tdTomato sequence immediately downstream of the Otx2 coding region, with 800bp homology arms on each side was generated with Gibson assembly and used for transfection of reprogrammable iPSC clone #2. Clonal colonies were isolated and correct integration confirmed by PCR using primers internal and external to the donor cassette (5′ forward [F]: 5′-CTGGACAGTGCAATGTAAACT-3′; 5′ reverse [R]: 5′-AACGCACACCGGCCTTATTC-3′; 3′F: 5′-ACCAAGCTGGACATCACCTC-3′; and 3′R: 5′-CCAAATCCAGGAAGGGTTTA-3′). Reprogrammable MEFs were generated from mid-gestation embryos following 4n blastocyst injections.
RNA-Seq
Oct4-GFP+ cells were isolated to >95% purity using flow cytometry and total RNA prepared with the miRNeasy Mini Kit (QIAGEN). Polyclonal iPSC cultures used for RNA-seq were at passage 5. Libraries were synthesized using ScriptSeq Complete Gold Kit, Low Input using 500ng of RNA depleted of ribosomal RNA (Ribo-Zero, Epicenter). Libraries were run on three lanes of an Illumina HiSeq 2500 flow cell (v4 chemistry) as paired end, 50 nucleotides per read. Reads were aligned to mm10 using TopHat 2.0.9 with Bowtie 2.2.6. Samtools was used to select paired, unique reads. Reads aligning to mitochondrial DNA were removed. Remaining reads were quantified using HTSeq and analyzed using the R package DESeq2. An FPKM cut off of 1 (estimated with DESeq2′s FPKM function and the longest annotated isoform) was used when analyzing differentially expressed genes (fold change > 2; adjusted p value < 0.05). Enriched gene ontology terms were identified using GOrilla (Eden et al., 2009).
ATAC-Seq
Libraries were synthesized from 50,000 input cells as previously described and analyzed in a similar manner (Buenrostro et al., 2013). Specifically, reads aligned to mm10 (Bowtie 2.2.6, -X 2000) were filtered using Picard Tools and Samtools (-F 1804) with mitochondrial DNA alignments excluded. Peaks were called using PeaKDEck and quantified using HTSeq. Differential peaks were identified (DESeq2: fold change > 1.5, adjusted p value < 0.1). Percent of target sequences bearing selected motifs were measured using HOMER 4.9 (Heinz et al., 2010). Polyclonal iPSC cultures used for ATAC-seq were at passage 5.
qPCR
cDNA samples were prepared with the Transcriptor First Strand cDNA synthesis Kit (Roche) and run on a LightCycler 480 (Roche). Specific mRNA levels were determined using gene-specific primer pairs: Pou5f1 (5′-ACCTGGCTTCAGACTTCGC-3′, 5′-TGAGCCTGGTCCGATTCCA-3′), Esrrb (5′-CA AGAGAACCATTCAAGGCAACA-3′, 5′-CATCCCCACTTTGAGGCATTT-3′), Nanog (5′-TTGCTTACAAGGGTCTGCTACT-3′, 5′-ACTGGTAGAAGAATCAG GGCT-3′), Otx2 (5′-GAATCCAGGGTGCAGGTATGG-3′, 5′-CTGAACTCACTT CCCGAGCTG-3′), Zic2 (5′-GGCAAACCCTTTAAGGCCAAA-3′, 5′-AGGTTTCT CCCCTGTATGAGTTCT-3′), Fgf5 (5′-TGTGTCTCAGGGGATTGTAGG-3′, 5′-AG CTGTTTTCTTGGAATCTCTCC-3′), and Gapdh (5′-AGGTCGGTGTGAACGGA TTTG-3′, 5′-TGTAGACCATGTAGTTGAGGTCA-3′).
Statistical Analysis
Experimental data were analyzed for statistical significance using GraphPad Prism 7.0c, with either a t test, a one-way ANOVA (parametric data), or a Mann-Whitney test (injection data). Raw data are presented for analysis of 4n injections conducted at different stages of iPSC derivation (Figure 1F and Table S1). The generation of chimeric mice in our hands has a 20% failure rate independent of the developmental capacity of injected cells, calculated from injection of ESCs into diploid blastocysts which failed to yield viable mice (S.Y.K.). Therefore, 20% of injections at each stage are expected to not yield pups for technical reasons and must be considered missing values for statistical testing. To assess significance, two separate sets of statistical tests were carried out on either the data with imputed values (calculated based on the median of remaining values within each stage) or on complete case analysis (missing values from each stage were excluded from statistical testing). Mann-Whitney tests, followed by Bonferroni correction to account for multiple testing resulting from stage-wise comparisons, were performed. The higher p value obtained from both independent statistical tests is shown for each stage-wise comparison in Figure 1F.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the ATAC-seq and RNA-seq data reported in this paper are GEO: GSE106331 and GSE106332, respectively. All sequencing datasets can be referenced from GEO SuperSeries: GSE106334.
Supplementary Material
Highlights
Rapid reprogramming yields developmentally highly competent iPSCs
Developmental pluripotency is established before the first passage of nascent iPSCs
Nascent iPSCs transit through a unique cellular state poised toward differentiation
ACKNOWLEDGMENTS
We would like to thank members of the Stadtfeld lab for helpful discussion and suggestions on this project. We are grateful to Olivier Elemento, Ashley Doane, Aris Tsirigos, Igor Dolgalev, and Yixiao Gong for helpful suggestions on bioinformatics analysis and Xiaochun Li and Zhengming Chen for consultations on biostatistics. We thank Jin Akagi for performing On-chip-based cell sorting. M.S. was supported by a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research, a Basil O’Connor Starter Scholar Research Award (5-FY14–114) from the March of Dimes Foundation, the New York State Department of Health (C092546 and C028130), a Junior Faculty Scholar Award from the American Society of Hematology, and the National Institutes of Health (1R21HD079883–01 and 1R01GM111852–01). The Laura and Isaac Perlmutter Cancer Center support grant (NIH/NCI P30CA016087) partially funds the NYULMC Core Cytometry Facility and the GTC.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, five figures, and four tables and can be found within this article online at https://doi.org/10.1016/j.celrep.2017.12.098.
DECLARATION OF INTERESTS
M.S. and B.A. have filed for a patent based partially on results presented in this manuscript. The other authors declare no competing interests.
REFERENCES
- Acampora D, Di Giovannantonio LG, and Simeone A (2013). Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development 140, 43–55. [DOI] [PubMed] [Google Scholar]
- Apostolou E, and Hochedlinger K (2013). Chromatin dynamics during cellular reprogramming. Nature 502, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Brumbaugh J, Verheul C, Apostolou E, Pruteanu-Malinici I, Walsh RM, Ramaswamy S, and Hochedlinger K (2014). Small molecules facilitate rapid and synchronous iPSC generation. Nat. Methods 11, 1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, and Nichols J (2017). Primate embryogenesis predicts the hallmarks of human naı¨ve pluripotency. Development 144, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, and Nichols J (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, and Vallier L (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. (2014). The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell 15, 295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Clement K, Huebner AJ, Webster J, Rose CM, Brumbaugh J, Walsh RM, Lee S, Savol A, Etchegaray JP, et al. (2017). DUSP9 Modulates DNA Hypomethylation in Female Mouse Pluripotent Stem Cells. Cell Stem Cell 20, 706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechanski A, Byers C, Greenstein I, Schrode N, Donahue LR, Hadjan tonakis AK, and Reinholdt LG (2014). Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc 9, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Collombet S, Jakobsen JS, Wierer M, Sardina JL, Lackner A, Stadhouders R, Segura-Morales C, Francesconi M, Limone F, et al. (2016). C/EBPa creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat. Cell Biol. 18, 371–381. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM 3rd, Yanagimachi R, and Jaenisch R (2001). Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA 98, 6209–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federation AJ, Bradner JE, and Meissner A (2014). The use of small molecules in somatic-cell reprogramming. Trends Cell Biol. 24, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, and Daley GQ (2004). Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427, 148–154. [DOI] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, and Wrana JL (2012). A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, and Smith A (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineagedetermining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PH, Di Stefano B, de Wit E, Limone F, van Oevelen C, de Laat W, and Graf T (2016). Cell-of-Origin-Specific 3D Genome Structure Acquired during Somatic Cell Reprogramming. Cell Stem Cell 18, 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, and Jaenisch R (2007). Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1, 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, and Wutz A (2013). Breathing-in epigenetic change with vitamin C. EMBO Rep. 14, 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S, Nichols J, and Hadjantonakis AK (2017). The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev. Biol 17, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, Markkula M, and Rossant J (1990). Embryonic stem cells alone are able to support fetal development in the mouse. Development 110, 815–821. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, and Tsukiyama T (2014). Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS ONE 9, e107308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka S, Nishikawa-Torikai S, and Niwa H (2012). E-cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS ONE 7, e45220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, and Bestor TH (2010). Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol 28, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. (2013). Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502, 65–70. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dierich A, Shawlot W, Behringer RR, Le Meur M, and Ang SL (1998). Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development 125, 845–856. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, and Wrana JL (2010). Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77. [DOI] [PubMed] [Google Scholar]
- Sebban S, and Buganim Y (2016). Nuclear Reprogramming by Defined Factors: Quantity Versus Quality. Trends Cell Biol. 26, 65–75. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, and Hochedlinger K (2008). Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, and Hochedlinger K (2010a). Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, and Hochedlinger K (2010b). A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods 7, 53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, et al. (2012). Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet 44, 398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Sasaki A, Yamamoto M, Nakamura M, Sutou K, Osafune K, and Yamanaka S (2014). Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun 5, 3678. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, and McKay RD (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. [DOI] [PubMed] [Google Scholar]
- Vidal SE, Amlani B, Chen T, Tsirigos A, and Stadtfeld M (2014). Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Reports 3, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, and Hanna JH (2016). Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol 17, 155–169. [DOI] [PubMed] [Google Scholar]
- Wu T, Liu Y, Wen D, Tseng Z, Tahmasian M, Zhong M, Rafii S, Stadtfeld M, Hochedlinger K, and Xiao A (2014). Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell 15, 281–294. [DOI] [PubMed] [Google Scholar]
- Wu G, Lei L, and Schöler HR (2017). Totipotency in the mouse. J. Mol. Med. (Berl.) 95, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S (2009). Elite and stochastic models for induced pluripotent stem cell generation. Nature 460, 49–52. [DOI] [PubMed] [Google Scholar]
- Yang SH, Kalkan T, Morissroe C, Marks H, Stunnenberg H, Smith A, and Sharrocks AD (2014). Otx2 and Oct4 drive early enhancer activation during embryonic stem cell transition from naive pluripotency. Cell Rep. 7, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, and Smith A (2017). The Art of Capturing Pluripotency: Creating the Right Culture. Stem Cell Reports 8, 1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, and Brockdorff N (2005). Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet 37, 1274–1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the ATAC-seq and RNA-seq data reported in this paper are GEO: GSE106331 and GSE106332, respectively. All sequencing datasets can be referenced from GEO SuperSeries: GSE106334.