Abstract
The transcription factor NRF2 (nuclear factor erythroid 2-related factor 2) triggers the first line of homeostatic responses against a plethora of environmental or endogenous deviations in redox metabolism, proteostasis, inflammation, etc. Therefore, pharmacological activation of NRF2 is a promising therapeutic approach for several chronic diseases that are underlined by oxidative stress and inflammation, such as neurodegenerative, cardiovascular, and metabolic diseases. A particular case is cancer, where NRF2 confers a survival advantage to constituted tumors, and therefore, NRF2 inhibition is desired. This review describes the electrophilic and nonelectrophilic NRF2 activators with clinical projection in various chronic diseases. We also analyze the status of NRF2 inhibitors, which at this time provide proof of concept for blocking NRF2 activity in cancer therapy.
1. Introduction
Nuclear factor erythroid 2-related factor 2 (NRF2) is the product of the NFE2L2 gene and belongs to the cap′n′collar transcription factor family. By sequence homology with other orthologs, the domains termed Neh1-7 have been traditionally allocated in this protein (Figure 1(a)). At the C-terminus, NRF2 contains a basic leucine-zipper (bZip) domain that participates in the formation of heterodimers with other bZip proteins, like small muscle aponeurosis fibromatosis (MAF) K, G, and F [1, 2]. These heterodimers regulate the expression of about 250 human genes that present a regulatory enhancer sequence termed Antioxidant Response Element (ARE; 5′-TGACNNNGC-3′) and participate in multiple homeostatic functions including regulation of inflammation, redox metabolism, and proteostasis [3–6].
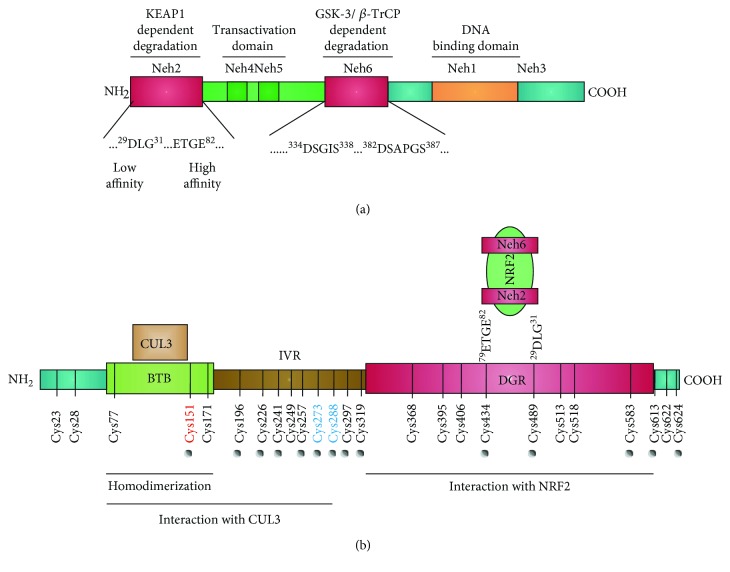
Figure 1.
Domain structures of NRF2 and KEAP1. (a) Domain structure of NRF2. NRF2 possesses six highly conserved domains called NRF2-ECH homology (Neh) domains [167]. The functional role of each Neh domain is specified. Within the Neh2 domain, the low-affinity (DLG) and high-affinity (ETGE) binding domains to KEAP1 are zoomed in. (b) Domain structure of a KEAP1 monomer showing the position of cysteine residues. The N-terminal BTB (bric-a-brac, tramtrack, broad complex) domain participates in homodimerization and binding to CUL3/RBX1. The C-terminal region, DGR (double glycine repeat) domain, contains a double glycine repeat called Kelch repeat that binds NRF2-Neh2 domain. The intervening region (IVR/LR) connects BTB and DGR domains and is particularly rich in redox-sensitive cysteine residues. Red and blue cysteine residues in KEAP1 are the most relevant for electrophile reactivity. This figure has been modified and extended from [168] to highlight the degradation domains in NRF2 and the cysteines of KEAP1.
From a clinical perspective, it is of utmost importance that NRF2 can be targeted pharmacologically in diseases underlined by oxidative stress and inflammation, such as neurodegenerative, vascular, and metabolic diseases as well as cancer [7, 8]. In models of most chronic diseases, a reinforcement of homeostasis through NRF2 activators provides a beneficial therapeutic effect. In cancer, the pharmacological regulation of NRF2 appears to be context dependent. It is generally accepted that NRF2 inhibitors not only reduce the survival and proliferative advantage of cancer cells but also sensitize tumors to chemo- and radiotherapy [9]. In this review, we describe the pharmacological activators of NRF2 that are in several stages of pharmacological development for the treatment of several chronic diseases. The most developed compounds activate NRF2 by preventing its degradation by KEAP1-dependent mechanisms. We also discuss the current state of NRF2 inhibitors which may be highly relevant for cancer therapeutics although at this time they are still in early phases of development.
2. Physiologic Regulation of NRF2
NRF2 is ubiquitously and constitutively expressed by cells, thus ensuring their prompt protective response to oxidative, inflammatory, and metabolic stresses. Under normal physiological conditions, NRF2 has a rapid turnover and presents a half-life of about 20-30 min due to its constant degradation by the ubiquitin proteasome system [10, 11]. Therefore, under nonstressed conditions, low NRF2 levels provide basal expression of its target genes.
The main control of NRF2 stability is exerted by the E3 ligase adapter Kelch-like ECH-associated protein 1 (KEAP1). KEAP1 is a homodimer protein that comprises three functional domains (Figure 1(b)): a broad complex, tramtrack, bric-a-brac (BTB) homodimerization domain, an intervening region (IVR), and a C-terminal Kelch domain with a double glycine repeat (DGR). The Kelch domain binds to the Neh2 domain of NRF2 at two amino acid sequences: DLG and ETGE. Experiments based on isothermal calorimetry have led to the conclusion that the ETGE motif exhibits about one hundred times higher affinity for KEAP1 than the DLG motif [12]. KEAP1 presents NRF2 for ubiquitination by the E3 ligase complex formed by Cullin3 and RBX1 proteins (CUL3/RBX1) [13], resulting in subsequent NRF2 degradation by the proteasome 26S [2, 14].
KEAP1 contains 27 cysteine residues in humans, converting this protein in a redox sensor for endogenous and environmental oxidative signals as well as for electrophilic reactions [15]. Under redox-challenging conditions, the cellular redox buffers comprising glutathione (GSH), thioredoxin, etc. maintain low intracellular levels of reactive oxygen species (ROS) and glutathionylated proteins. However, ROS oxidize thiols and induce glutathionylation and alkylation of macromolecules, therefore having the capacity to modify KEAP1 cysteines [16]. From a pharmacological perspective, electrophile reaction with some cysteines of KEAP1 leads to the formation of adducts that prevent the ubiquitination NRF2, resulting in its stabilization, nuclear translocation, and transcriptional induction of NRF2-target genes [7, 8].
An alternative mechanism for proteasomal degradation of NRF2 is mediated by the glycogen synthase kinase 3 (GSK-3) and the E3 ligase adapter β-TrCP. GSK-3α and β are serine/threonine protein kinases involved in several signaling pathways such as receptor tyrosine kinase, WNT, and Hedgehog that influence cell division, survival, and development [17, 18]. GSK-3α and β are maintained in an inactive state under normal conditions due to their inhibition by AKT-mediated phosphorylation at their N-terminal pseudosubstrate domain or by sequestration in protein complexes. However, in the absence of receptor signaling, active GSK-3 phosphorylates NRF2 at the Neh6 domain (DSGIS). This phosphodomain recruits β-TrCP, which recognizes pSGIpS, and the CUL1/RBX1 complex for ubiquitin-proteasome degradation [19]. β-TrCP also recognizes another motif in the Neh6 domain of NRF2 (DSAPGS) which appears to be constitutively phosphorylated in a GSK-3-independent manner [20]. Additional degradative systems are able to regulate NRF2 at posttranscriptional level, such as the inositol-requiring enzyme (IRE1)/E3 ubiquitin ligase synoviolin (HRD1) [21].
NRF2 can be regulated at the transcriptional level. The NFE2L2 gene promoter presents several regulatory sequences: (a) one xenobiotic response element (XRE; 5′-TA/TGCGTGA/C-3′) at -712 and two XRE-like sequences at +755 and +850 that are recognized by the transcription factor Aryl Hydrocarbon Receptor (AHR) [22]; (b) two ARE-like sequences at -492 (AREL1; TGACTCCGC) and -754 pb (AREL2; TGACTGTGGC), which allow NRF2 autoregulation [23]; (c) one 12-O-tetradecanoylphorbol-13-acetate-response element (TRE) (TGCGTCA) at +267 to +273 pb that is activated by the oncogenic KRAS [24], BRAF, and MYC [25] hence being critically involved in carcinogenesis; (d) one NF-κB binding site that responds to inflammatory stimuli [26]; and (e) epigenetic changes such as promoter methylation, microRNAs including miR-144 [27], miR-28 [28], miR-98-5p [29], and long noncoding RNA deregulation [30] that contribute to changes in expression of the NRF2-coding gene.
3. Pharmacologic Activators of NRF2
The so-called “NRF2 activators” should be more precisely termed “KEAP1 inhibitors” as their molecular target is in fact KEAP1 [31]. These compounds can be classified as electrophiles, protein-protein interaction (PPI) inhibitors, and multitarget drugs (Figure 2).
Figure 2.
Summary of the pharmacological strategies to modulate NRF2 activity.
3.1. Electrophilic Compounds
Most pharmacological NRF2 activators are electrophilic molecules that covalently modify cysteine residues present in the thiol-rich KEAP1 protein by oxidation or alkylation [32–34]. Many cysteines of KEAP1 are modified by different electrophiles [35–37]. Cysteines Cys-151, Cys-273, and Cys-288 [38, 39] appear to be the most susceptible to electrophile reaction [40, 41]. Other sensitive cysteines are Cys-226, Cys-434, and Cys-613. This “cysteine-code” controls KEAP1 activity when the protective response mediated by NRF2 is needed. Selected electrophilic activators of NRF2 that are in various stages of clinical development are presented in Table 1.
Table 1.
Selected electrophilic activators of NRF2 under clinical development.
| Compound | Type | Mechanism of action | Disease | Clinical trial | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|
Bardoxolone-methyl (CDDO-Me)
|
Synthetic triterpenoids | Electrophilic modification of KEAP1-Cys-151 | Diabetic nephropathy | Phase II | NCT00811889 |
| IgA nephropathy CKD associated with type 1 diabetes Focal segmental glomerulosclerosis Autosomal dominant polycystic kidney |
Phase II | NCT03366337 | |||
| Chronic kidney disease Type 2 diabetes Diabetic nephropathy |
Phase III | NCT01351675 | |||
| Liver disease | Phase I/II | NCT00550849 | |||
| Hepatic impairment Healthy |
Phase I | NCT01563562 | |||
| Advanced solid tumors lymphoid malignancies | Phase I |
NCT00529438
NCT00508807 |
|||
| Alport syndrome | Phase II/III cardinal | NCT03019185 | |||
| Pulmonary hypertension | Phase III RANGER | NCT03068130 | |||
| Pulmonary arterial hypertension | Phase III | NCT02657356 | |||
| Renal insufficiency, chronic Diabetes mellitus, type 2 |
Phase II | NCT01053936 | |||
|
| |||||
RTA-408 (omaveloxolone)
|
Synthetic triterpenoids | Electrophilic modification of KEAP1-Cys-151 | Mitochondrial myopathy | Phase II | NCT02255422 |
| Friedreich's ataxia | Phase II | NCT02255435 | |||
| Inflammation and pain following ocular surgery | Phase II | NCT02065375 | |||
| Corneal endothelial cell loss Ocular pain Ocular inflammation Cataract surgery |
Phase II | NCT02128113 | |||
| Melanoma | Phase I/II | NCT02259231 | |||
| Breast cancer | Phase II | NCT02142959 | |||
|
| |||||
Dimethyl fumarate
|
Fumaric acid ester | Electrophilic modification of KEAP1-Cys-151 | Multiple sclerosis | Approved | |
| Psoriasis | Approved | ||||
| Rheumatoid arthritis | Phase II | NCT00810836 | |||
| Adult brain glioblastoma | Phase I | NCT02337426 | |||
| Cutaneous T cell lymphoma | Phase II | NCT02546440 | |||
| Obstructive sleep apnea | Phase II | NCT02438137 | |||
| Chronic lymphocytic leukemia Small lymphocytic lymphoma |
Phase I | NCT02784834 | |||
|
| |||||
ALKS-8700
|
Fumaric acid ester (MMF-derivate) | Electrophilic modification of KEAP1-Cys-151 | Multiple sclerosis | Phase III | NCT02634307 |
|
| |||||
Oltipraz
|
Organosulfur compound | Electrophilic modification of KEAP1-Cys-151 | Nonalcoholic steatohepatitis | Phase III | NCT02068339 |
| Schistosomiasis | Approved | ||||
| Lung cancer | Phase I | NCT00006457 | |||
|
| |||||
Ursodiol
|
Biliary acid | Electrophilic modification of KEAP1-Cys-151 | Cholestasis | Phase II/III | NCT00846963 |
| Diarrhea | Phase IV | NCT02748616 | |||
| Cholelithiasis | Phase III | NCT02721862 | |||
| Primary biliary cirrhosis | Phase IV | NCT01510860 | |||
| Barrett esophagus Low-grade dysplasia |
Phase II | NCT01097304 | |||
| Chronic hepatitis C | Phase III | NCT00200343 | |||
| Type 2 diabetes mellitus | Phase II | NCT02033876 | |||
|
| |||||
Sulforaphane
|
Isothiocyanate | Electrophilic modification of KEAP1-Cys-151 | Schizophrenia | Phase II/III | NCT02880462 |
| Phase II | NCT02810964 | ||||
| Phase II | NCT01716858 | ||||
| COPD | Phase II | NCT01335971 | |||
| Atopic asthmatics | Phase I | NCT01845493 | |||
| Autism spectrum disorder | Phase II | NCT01474993 | |||
| Phase II | NCT02909959 | ||||
| Phase II | NCT02677051 | ||||
| Phase II | NCT02654743 | ||||
| Phase I/II | NCT02561481 | ||||
| Healthy | Phase I | NCT01008826 | |||
| Phase I | NCT02023931 | ||||
| Melanoma | Phase I | NCT01568996 | |||
| Asthma | Phase I | NCT01845493 | |||
| Phase I/II | NCT01183923 | ||||
| Prostate cancer | Phase II | NCT01228084 | |||
| Breast cancer | Phase II | NCT00843167 | |||
| Lung cancer | Phase II | NCT03232138 | |||
| Environmental carcinogenesis | Phase II | NCT01437501 | |||
| Alcohol sensitivity | Phase II | NCT01845220 | |||
| Aging | Phase II | NCT03126539 | |||
| Rhinitis, allergic | Phase II | NCT02885025 | |||
| Helicobacter pylori infection | Phase IV | NCT03220542 | |||
| Diabetes mellitus, noninsulin-dependent | Phase II | NCT02801448 | |||
|
| |||||
Sulforadex (SFX-01)
|
Sulforaphane/alpha-cyclodextrin complex | Electrophilic modification of KEAP1-Cys-151 | Subarachnoid haemorrhage | Phase II | NCT02614742 |
| Breast neoplasm | Phase I/II | NCT02970682 | |||
| Prostate cancer | Phase I |
NCT02055716
NCT01948362 |
|||
|
| |||||
ITH12674
|
Melatonin-sulforaphane hybrid | Electrophilic modification of KEAP1-Cys-151 | Brain ischemia | Preclinical PK | No clinical trials available |
|
| |||||
Curcumin
|
Stilbene | Electrophilic modification of KEAP1-Cys-151 | Type 2 diabetes Prediabetes Insulin resistance Cardiovascular risk |
Phase IV | NCT01052025 |
| Schizophrenia Cognition Psychosis |
Phase I/II | NCT02104752 | |||
| Acute kidney injury Abdominal aortic aneurysm |
Phase II/III | NCT01225094 | |||
| Chronic kidney diseases Diabetes mellitus, type 2 Polymorphism |
Phase II/III | NCT03262363 | |||
| Alzheimer's disease | Phase I/II | NCT00164749 | |||
| Neoplasms | Phase II | NCT02944578 | |||
| Crohn's disease | Phase III | NCT02255370 | |||
| Chronic schizophrenia | Phase IV | NCT02298985 | |||
| Mild cognitive impairment | Phase II | NCT01811381 | |||
| Prostate cancer | Phase III | NCT02064673 | |||
| Major depression | Phase IV | NCT01750359 | |||
|
| |||||
Resveratrol
|
(E)-Stilbene derivate | Electrophilic modification of KEAP1-Cys-151 | Type 2 diabetes | Phase I | NCT01677611 |
| Colon cancer | Phase I | NCT00256334 | |||
| COPD | N/A | NCT02245932 | |||
| Friedreich ataxia | Phase I/II | NCT01339884 | |||
| Nonalcoholic fatty liver | Phase II/III | NCT02030977 | |||
| Nonischemic cardiomyopathy | Phase III | NCT01914081 | |||
| Endometriosis | Phase IV | NCT02475564 | |||
| Chronic renal insufficiency | Phase III | NCT02433925 | |||
| Metabolic syndrome X | Phase II | NCT02114892 | |||
| Chronic subclinical inflammation Redox status |
Phase III | NCT01492114 | |||
| Alzheimer's disease | Phase II | NCT01504854 | |||
| Phase III | NCT00743743 | ||||
| Huntington disease | Phase III | NCT02336633 | |||
|
| |||||
CXA-10
|
Nitro-fatty acid (NFA) | Electrophilic modification of KEAP1-Cys-273 and Cys-288 | Acute kidney injury | Phase I | NCT02248051 |
| Pulmonary arterial hypertension (PAH) | Phase II | NCT03449524 | |||
| Primary focal segmental glomerulosclerosis (FSGS) | Phase II | NCT03422510 | |||
One mechanism of KEAP1 inhibition is the sequestration in complexes with NRF2 that cannot be ubiquitinated. Modifications of several cysteines in KEAP1 generate a nonfunctional closed state with both Neh2 motifs (DLG and ETGE) of NRF2 interacting with the KEAP1 dimer but not leading to ubiquitination. As a result, free KEAP1 is not regenerated at a sufficient rate and newly synthesized NRF2 escapes KEAP1-mediated ubiquitination and subsequent degradation [42].
Another mechanism of KEAP1 inhibition is related to its interaction with the CUL3/RBX1 complex, required for NRF2 ubiquitination. Cys-151 located at the BTB domain influences the interaction of KEAP1 with CUL3. The crystal structure of the BTB domain bound to the pentacyclic triterpenoid 2-cyano-3,12-dioxo-oleana-1,9(11)-dien-28-oate (bardoxolone, CDDO, RTA401) indicates that adduct formation with Cys-151 most likely disrupts the interaction between KEAP1 and CUL3 [43–45]. As a result, KEAP1 is clogged in a NRF2 bound conformation, and newly formed NRF2 escapes ubiquitination. Synthetic triterpenoids have been derived from the natural compound oleanolic acid to provide them with strong Michael acceptor reactivity. This is achieved mainly through the addition of enone and ciano groups to the A ring and another enone group to the C ring [46, 47]. Bardoxolone methyl (CDDO-Me or RTA 402) reached clinical trials for the treatment of advanced chronic kidney disease (CKD) and type 2 diabetes mellitus [48]. Although phase II clinical trials demonstrated long-term increment in glomerular filtration, CDDO-Me was halted at phase III due to cardiovascular safety issues [49]. A new phase II clinical trial has recently started recruiting patients with rare chronic kidney diseases to better define the safety and efficacy profiles of CDDO-Me. Currently, CDDO-Me is also under clinical study for the Alport syndrome and pulmonary hypertension. In an effort to improve the safety profile, a second-generation difluoromethyl acetamide derivative of bardoxolone methyl, called RTA-408 (Omaveloxone), is now under clinical investigation in phase II clinical trials for Friedreich's ataxia, ocular inflammation, and pain after ocular surgery [50]. Recently, a preclinical study evaluated RTA-408 for diabetic wound recovery and pointed NRF2 upregulation as responsible for the observed improvement in regenerative capacity [51].
The most successful NRF2 activator to date is the fumaric acid ester dimethyl fumarate (DMF) (BG-12 or Tecfidera, from Biogen) that has been approved in 2013 by FDA for relapsing-remitting multiple sclerosis (MS) [52–55]. Previously, DMF was authorized for the treatment of psoriasis [56]. DMF was shown to reduce the number of peripheral T cells, CD8+ cells being more sensitive to DMF than CD4+ cells [57, 58]. DMF also reduces total B lymphocyte counts, especially memory B cells, along with a decrease in granulocyte-macrophage colony-stimulating factor, IL-6, and TNF-α production, leading to an anti-inflammatory shift in B cell responses [59, 60]. The DMF-induced activation of NRF2 in the central nervous system was described in the MS mice model of experimental allergic encephalomyelitis [61]. In this model, DMF-dependent NRF2 activation correlated with an improvement in the clinical course of MS, favored axon preservation, and increased astrocyte activation. These beneficial effects of DMF did not occur in NRF2-null mice, hence indicating that DMF was acting mainly by targeting the NRF2 pathway. DMF is mostly converted to monomethyl fumarate (MMF) by intestinal esterases, and only a small fraction is found in blood conjugated with glutathione [62]. Therefore, an oral formulation of a monomethyl fumarate (MMF) derivative, diroximel fumarate (2-(2,5-dioxo-1-pyrrolidinyl)ethyl ester; ALKS-8700; Alkermes) which exhibits improved bioavailability and efficacy, is currently under phase III trial for MS [63, 64]. However, the biological effects of these fumaric acid esters are not fully characterized and KEAP1/NRF2-independent effects are being described. For instance, it has been reported that DMF and MMF activate the nicotinic receptor hydroxycarboxylic acid receptor 2, which is expressed in immune cells and gut epithelial cells, resulting in NRF2-independent anti-inflammatory responses [65].
Oltipraz (4-methyl-5(pyrazinyl-2)-1-2-dithiole-3-thione) is a NRF2 inducer that enhances GSH biosynthesis and phase II detoxification enzymes, such as NQO1. Oltipraz is a promising chemopreventive agent [66] under phase III clinical trial for the treatment of nonalcoholic fatty liver disease.
Ursodiol (ursodeoxycholic acid) is an FDA-approved drug for the treatment of primary biliary cirrhosis. Although its cytoprotective mechanisms have not been elucidated yet, several research groups suggested that the upregulation of NRF2 by ursodiol induces detoxification and antioxidant mechanisms that play a role in its therapeutic efficacy [67, 68].
Several natural compounds have been identified as electrophilic NRF2 inducers, including sulforaphane, curcumin, resveratrol, quercetin, genistein, and more recently andrographolide [69]. For instance, sulforaphane (SFN), an isothiocyanate found in cruciferous vegetables, has been successfully used for the treatment of patients with type II diabetes mellitus [70, 71]. Due to the capacity of SFN to cross the blood-brain barrier, it protects against neurodegenerative disorders as demonstrated in murine models of disease. Regarding acute brain damage, SFN was shown to exert protective effects in hypoxic-ischemic injury in rats by reducing the infarct ratio and by upregulating NRF2 and HO-1 [72, 73]. In neurodegenerative disease models, SFN proved protective capacity against the neurotoxic Aβ 1-42 peptide in neuronal cells [74]. In vivo, SFN ameliorated cognitive impairment in an acute mouse model of Alzheimer disease (AD) [75]. In Parkinson disease (PD), SFN protected dopaminergic cells against the cytotoxic effects of 6-hydroxydopamine [76]. In the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of PD, SFN counteracted astrogliosis and microgliosis and reduced the death of dopaminergic neurons [77–79]. To improve the stability of SFN, Evgen Pharma has developed a cyclodextrin formulation, SFX-01, which is under phase II clinical trial for the treatment of subarachnoid haemorrhage. A hybrid molecule of SFN and melatonin (ITH12674) was designed to have a dual “drug-prodrug” mechanism of action for the treatment of brain ischemia [80].
Another natural compound that modifies Cys-151 in KEAP1 and has also ROS-scavenging activity is curcumin, a linear diarylheptanoid present in turmeric (Curcuma longa) [81]. It has been used for the treatment of obesity, metabolic syndrome, and prediabetes [82–84]. Furthermore, curcumin has been shown to suppress the deleterious action of carcinogens by activating NRF2 [85, 86].
9-Nitro-octadec-9-enoic acid (OA-NO2) is a nitro-fatty acid with anti-inflammatory properties. OA-NO2 reacts with several cysteine residues of KEAP1, but mainly with Cys-273 and Cys-288, and its activity seems to be independent of Cys-151 [36]. CXA-10 (10-nitro-9(E)-octadec-9-enoic acid) is an isomer of OA-NO2 which has proven efficacy the uni-nephrectomized deoxycorticosterone acetate-high salt mouse model of CKD [87] and is under several phase I clinical trials for the treatment of this disease [88] and under phase II trials for the treatment of pulmonary arterial hypertension and primary focal segmental glomerulosclerosis.
The list of electrophilic compounds able to interact with KEAP1 is continuously growing. For instance, some compounds like 15-deoxy-Δ12,14-prostaglandin J2 interact with Cys-273 and Cys-288 of the KEAP1 homodimer [40]. This prostaglandin has a cyclopentenone core that is able to modify covalently Cys-273 and induce NRF2 in models of ureteral obstruction [89], hepatic ischemia-reperfusion injury [90], and atherosclerosis [91]. However, its clinical use is still far from being demonstrated. In a recent study, the metabolite itaconate was described as a NRF2 activator that alkylates cysteines 151, 257, 288, 273, and 297 of KEAP1. A cell-permeable itaconate derivate, 4-octyl itaconate, protects against lipopolysaccharide cytotoxicity, thus providing an anti-inflammatory response. Furthermore, this compound is a more potent NRF2 activator than DMF [92]. Some other examples are tert-butylhydroquinone [93], diethyl maleate [94], TFM-735 [95], and nitric oxide [96]. However, most of these compounds have not evolved beyond proof-of-concept experiments, and a long way needs to be covered to characterize their pharmacodynamic properties, clinical safety profile, and efficacy in noncommunicable diseases.
3.2. Protein-Protein Interaction Inhibitors of the KEAP1-NRF2 System
Protein-protein interaction (PPI) inhibitors interfere with the docking of NRF2 to the Kelch propeller of KEAP1 and provide more selectivity over electrophilic compounds which may eventually form adducts with redox-sensitive cysteines other than those in KEAP1 [97]. Based on the X-ray crystal structure of KEAP1 [98], small PPI inhibitors have been designed to impede the binding of the ETGE motif to KEAP1 [99]. The ETGE motif adopts a β-hairpin structure that docks to the Kelch propeller of KEAP1 through specific hydrophobic and electrostatic interactions [98, 99]. A similar strategy is devised to prevent the interaction of the low-affinity DLG motif which is required for correct lysine ubiquitination in NRF2 [13].
The first PPI inhibitors of KEAP1 were designed from a series of truncated NRF2 peptides [100, 101]. Some selected peptides are shown in Table 2. It was found that the minimal binding sequence of NRF2 required for docking to KEAP1 is the 9-mer sequence LDEETGEFL [100–102]. A related peptide was designed to increase cell penetrance by adding the Tat sequence of the human immunodeficiency virus and the cleavage sequence of calpain (-Cal-Tat). This peptide demonstrated neuroprotection and cognition-preserving effects in a mouse model of cerebral ischemia [103]. Moreover, hybrid peptides based on both the region of interaction between KEAP1 and NRF2 (ETGE motif) and with the region of interaction between KEAP1 and p62/Sequestosome-1 (SQSTM1) exhibited superior binding activity compared to either native peptide alone [104]. Due to unfavorable drug-like properties, such as low oral bioavailability and cellular permeability of peptides, research has been lately focused on the development of small molecules. However, a cyclic peptide was used recently to improve KEAP1 binding and NRF2 accumulation in cells [105].
Table 2.
Selected peptides acting as NRF2-KEAP1 protein-protein interaction inhibitors.
| Sequence | Mechanism of action | Reference |
|---|---|---|
| LDEETGEFL-NH2 | Binding to KEAP1-Kelch domain | [100, 101] |
| DEETGE-CAL-Tat (NH2-RKKRRQRRR-PLFAERLDEETGEFLPNH2) | [103] | |
| Ac-DPETGEL-OH | [102] | |
| FITCβ-DEETGEF-OH | [102] | |
| FITC-β-LDEETGEFL-OH | [102] | |
| Ac-DEETGEF-OH | [102] | |
| Ac-DPETGEL-OH | [102] | |
| FITC-LDEETGEFL-NH2 | [100] | |
| FAM-LDEETGEFL-NH2 | [108] | |
| LQLDEETGEFLPIQGK(MR121)-OH | [107] | |
| Ac-LDEETGEFL-NH2 | [100, 101] | |
| Ac-DPETGEL-NH2 | [104] | |
| Ac-NPETGEL-OH | [104] | |
| St-DPETGEL-OH | [104] | |
| YGRKKRRQRRRLQLDEETGEFLPIQ | [162] | |
| c[GQLDPETGEFL] | [105] |
Current PPI inhibitors are tetrahydroisoquinoline [97, 106], thiopyrimidine [107], naphthalene [108], carbazone [109], and urea derivatives [110]. Recently, the naphthalene-based nonelectrophilic PPI inhibitors were modified to develop nonnaphthalene heterocyclic scaffold based on 1,4-isoquinoline that avoids the carcinogenic and mutagenic properties of naphthalenes [111]. Some patents addressing these small molecules are presented in Table 3.
Table 3.
Selected small molecule activators of NRF2 acting as NRF2-KEAP1 protein-protein interaction inhibitors.
| Compound | Type | Ref. | Patent |
|---|---|---|---|
(SRS)-5
|
1,2,3,4-Tetrahydroisoquinoline core | [112] | WO2013/067036 |
Cpd 15
|
Benzenesulfonyl-pyrimidone | [107] | WO2016/202253 |
Cpd 16
|
1,4-Diaminonaphthalene core | [107] | WO2016/202253 |
Compound 2
|
1,4-Diaminonaphthalene core | [163] | CN105566241A |
3-(Pyridin-3-ylsulfonyl)-5-(trifluoromethyl)-2H-chromen-2-one (PSTC)
|
Sulfonyl coumarins | [164] | WO2015/092713 |
AN-465/144580
|
Other structure classes | [165] | JP2011/0167537 |
Compound 7
|
Arylcyclohexyl pyrazoles | [166] | WO2017060855 |
Several PPI inhibitors with improved selectivity over electrophiles have been identified through screening of small molecule libraries. These compounds include SRS-5, benzenesulfonyl-pyrimidone 2, N-phenyl-benzenesulfonamide, and a series of 1,4-diphenyl-1,2,3-triazole [106, 112–115]. Recently, a new protocol for identifying reversible modifiers of the NRF2/KEAP1 interaction was proposed [116]. The biochemical assays comprised time-resolved fluorescence resonance energy transfer as primary screening tool, surface plasmon resonance to evaluate the affinity of KEAP1 binders, and 1H-15N heteronuclear single-quantum coherence nuclear magnetic resonance assay to further analyze the binding mode. This protocol will help in identifying and improving the properties of reversible binders to KEAP1.
3.3. Other Mechanism of NRF2 Activation
The phosphorylation of NRF2 by GSK-3 leads to its ubiquitination by the E3 ligase β-TrCP and subsequent proteasomal degradation. An aberrant activity of GSK-3 is linked with several pathologies such as AD, cardiovascular diseases, or cancer among others [117–120]. Therefore, several clinical trials are now focused on GSK-3 inhibitors for the treatment of several pathologies [121]. For instance, the GSK-3-inhibitor Tideglusib, a thiadiazolidinone compound, was studied in phase II trials for AD in the ARGO study [122]. Another inhibitor is Enzastaurin which is intended for the treatment of solid and hematological cancers. Although Enzastaurin provided promising results at the preclinical level, treatment failed in phase II and III trials [123, 124]. GSK-3-dependent NRF2 phosphorylation was shown to be inhibited by nordihydroguaiaretic acid [125]. This compound and its derivative terameprocol are in phase I and II clinical trials for the treatment of several types of cancers, such as gliomas and leukemias (Table 4) [126].
Table 4.
Selected KEAP1-independent activators of NRF2.
| Compound | Mechanism of action | Disease | Clinical trial | ClinicalTrials.gov identifier |
|---|---|---|---|---|
Tideglusib
|
GSK-3 inhibition | Autism spectrum disorders | Phase II | NCT02586935 |
| Myotonic dystrophy 1 | Phase II | NCT02858908 | ||
| Alzheimer's disease | Phase II | NCT01350362 | ||
|
| ||||
Nordihydroguaiaretic acid (NDGA)
|
GSK-3 inhibition | Prostate cancer | Phase II | NCT00678015 |
| Phase I | NCT00313534 | |||
| Brain and central nervous system tumors | Phase I/II | NCT00404248 | ||
|
| ||||
Terameprocol (NDGA derivative)
|
GSK-3 inhibition | High-grade glioma | Phase I | NCT02575794 |
| Leukemias Acute myeloid leukemia (AML) Acute lymphocytic leukemia (ALL) |
Phase I | NCT00664677 | ||
| Refractory solid tumors Lymphoma |
Phase I | NCT00664586 | ||
|
| ||||
Enzastaurin
|
GSK-3 inhibition | Diffuse large B cell lymphoma | Phase III | NCT03263026 |
| Solid tumor Lymphoma, malignant |
Phase I | NCT01432951 | ||
|
| ||||
LS-102
|
HRD1 inhibition | — | — | No clinical trials available |
|
| ||||
Rapamycin
|
p62/SQSTM1 activation | Diabetes mellitus, type 1 | Phase III | NCT01060605 |
| Systemic lupus erythematosus (SLE) | Phase II | NCT00779194 | ||
| Autosomal dominant polycystic kidney disease | Phase II/III | NCT00920309 | ||
|
| ||||
HPP-4382
|
BACH1 inhibition | — | — | No clinical trials available |
Focusing on E3 ubiquitin ligase β-TrCP, it would be possible to develop small molecules able to disrupt the docking of NRF2 to β-TrCP, hence opening a new way regarding KEAP1-independent activators of NRF2 [127]. A novel E3 ubiquitin ligase linked to KEAP1-independent NRF2 degradation is HRD1 [21]. HRD1-dependent NRF2 degradation has been described in the context of cirrhotic liver. HRD1 is a transcriptional target of X-box-binding protein 1 (XBP1) that is upregulated upon activation of the inositol-requiring enzyme 1 (IRE1) during endoplasmic reticulum (ER) stress related to cirrhotic conditions. Inhibitors of HRD1 and IRE1 restore the NRF2 response in liver cirrhosis [21].
Several proteins contain a (E/S)TGE motif that resembles the high-affinity ETGE motif of NRF2. The motif confers to these proteins the ability to compete with NRF2 for KEAP1 binding, leading to a noncanonical mechanism of NRF2 stabilization [128]. Proteins containing the (E/S)TGE motif are dipeptidyl peptidase 3, Partner and Localizer of BRCA2, and SQSTM1/p62. SQSTM1/p62, a protein that transports specific cargos to the autophagosome, including KEAP1, sustains NRF2 stabilization and translocation to the nucleus [129–131]. Compounds which elevate SQSTM1/p62 levels, like rapamycin [132] and trehalose [133], are being therefore studied in several phase II and III trials in connection with diabetes mellitus, systemic lupus erythematosus, and autosomal dominant polycystic kidney disease.
Another way to inhibit the transcriptional activity of NRF2 is to impede its interaction with critical components in the nucleus. BTB domain and CNC homolog 1 (BACH1) is a transcriptional repressor which belongs to the cap′n′collar, b-Zip family. BACH1 competes in the nucleus with NRF2 to form heterodimers with small MAF proteins and blocks therefore the expression of ARE genes [134]. A recent study characterized the HPP-4382 compound as an inhibitor of BACH1 repression activity in vitro [135].
All these alternative mechanisms for NRF2 stabilization and activation suggest that a combinatorial pharmaceutical approach will be the best way to activate the cytoprotective responses mediated by NRF2.
4. Pharmacologic Inhibitors of NRF2
The implication of NRF2 in cancer is still controversial. Several studies described that NRF2 knockout mice are more susceptible to chemically induced carcinogenesis, pointing NRF2 as a potential tumor suppressor that limits carcinogenesis [136, 137]. On the other hand, NRF2 is overexpressed in many types of tumors, and it has been related to poor disease prognosis because it confers a survival and growth advantage to cancer cells, along with resistance to chemo- and radiotherapy [138–140]. Altogether, these results suggest a protective role of NRF2 in the first steps of cancer, but in advanced stages, NRF2 overexpression helps cancer cells to adapt to the tumorigenic demands. Cancer cells are “addicted” to NRF2 and resist treatment with chemotherapy or radiotherapy [141, 142]. Therefore, it is reasonable to assume that NRF2 inhibitors should sensitize tumor cells to anticancer therapies. In all cases, the mechanism of inhibition is either unknown or not specific, and therefore, NRF2 inhibitors are still far from being translated from bench to bedside.
4.1. Agonists of Nuclear Receptors
Ligands of the glucocorticoid receptor such as dexamethasone [143] and clobetasol propionate [144] inhibit NRF2 by blocking its transcriptional activity or preventing its nuclear translocation. All-trans-retinoic acid and bexarotene, agonists of the retinoic acid receptor-α and retinoid X receptor-α, inhibit the transcriptional activity of NRF2 [145, 146]. Retinoid X receptor-α appears to bind to the Neh7 domain of NRF2 preventing binding to the ARE enhancer [146]. The pharmacological value of this mechanism of NRF2 inhibition is limited by the multiple effects that are expected through the regulation of these nuclear receptors.
4.2. Natural Compounds
Several compounds of natural origin have been reported to inhibit NRF2. The quassinoid brusatol, extracted from Brucea javanica, inhibits the NRF2 transcriptional signature and sensitizes tumors and cancer cell lines to several chemotherapeutics [147]. However, its mechanism of action is not specific as it blocks protein translation, hence affecting other short-lived proteins as well [148–150].
The flavonoids luteolin [151] and wogonin [152] were reported to inhibit NRF2 and sensitize cells to anticancer drugs by increasing the instability to its transcript. However, later studies also indicated that these compounds may elicit NRF2 activation [153]. Therefore, their value as NRF2 inhibitor is highly controversial.
Other natural compounds such the mycotoxin ochratoxin A [154] and the coffee alkaloid trigonelline [155] prevent the nuclear translocation of NRF2. In leukemic cells, malabaricone-A, a plant-derived prooxidant, effectively inhibits NRF2 transcriptional activity as reflected by a reduction in HO-1 protein levels and leads to ROS accumulation and subsequent cell apoptosis [156]. Ascorbic acid, a well-known ROS scavenger, was found to sensitize imatinib-resistant cancer cells by decreasing the levels of the NRF2/ARE complex, hence reducing the expression of Glutamate-Cysteine Ligase Catalytic Subunit and dropping GSH levels [157]. In general, the main concern with these compounds is that their selectivity for NRF2 inhibition has not been conclusively demonstrated.
4.3. Other Approaches
The lack of knowledge about the fine structure of NRF2 hampers a straightforward strategy for the in silico analysis of small molecules that might dock to relevant domains of interaction with MAF proteins, ARE enhancer, etc. Therefore, a high-throughput screening was used which is helping in the identification of NRF2 inhibitors but still not providing selectivity [158]. A first-in-class compound, termed ML385, was found after the screening of a chemical library of 400,000 molecules. ML385 blocks NRF2 transcriptional activity and sensitizes KEAP1-deficient cells to carboplatin and other chemotherapeutics. ML385 interacts with the DNA-binding domain of NRF2 and most likely prevents the binding of NRF2 to AREs. However, given the similarity between AREs and other enhancers such as AP1, additional studies are needed to clearly establish if ML385 is selective for NRF2 or if it also inhibits other bZip transcription factors involved in chemoresistance.
Halofuginone, a synthetic derivate of febrifugine that is used in veterinary medicine, blocked the chemoresistance and radioresistance of cancer cells in parallel to the decrease of NRF2 protein levels [159]. It was found that halofuginone induces amino acid starvation resulting in global inhibition of protein synthesis.
Another compound, AEM1, decreased the expression of NRF2-controlled genes and sensitized KEAP1-deficient A549 lung tumor cells to various chemotherapeutic agents [160]. Although it seems that the anticancer effect of AEM1 is restricted to cell lines harboring mutations which render NRF2 constitutively active, the selectivity for NRF2 inhibition is not demonstrated yet.
In HeLa cells transfected with an ARE-driven luciferase reporter, a pyrazolyl hydroxamic acid, termed 4f, inhibited NRF2, reduced cell proliferation of myeloid cell lines, and increased apoptosis of acute myeloid leukemia cells [161]. Most likely, 4f altered the BCL2/BAX ratio and induced mitochondria-dependent apoptosis.
5. Conclusions
The NRF2/KEAP1 system represents a very promising pharmacological target to control common pathologic mechanisms of many chronic diseases characterized by low-grade oxidative stress and inflammation. A plethora of NRF2 activators, mostly of electrophilic nature, have been identified and a few are under clinical development. The pleiotropic effects of NRF2 on cell physiology together with potential off-target effects exerted by some NRF2 activators explain why drug development is moving slowly. The field of NRF2 inhibitors that may have a huge impact on cancer therapy is less advanced. Future work should be directed towards finding compounds with a good pharmacokinetic/pharmacodynamic profile for specific diseases.
Acknowledgments
This work was supported by grants SAF2016-76520-R of the Spanish Ministry of Economy and Competitiveness, B2017/BMD-3827 of the Autonomous Community of Madrid, and P_37_732/2016 REDBRAIN of the European Regional Development Fund; Competitiveness Operational Program 2014-2020. NRA and RFG are recipients of FPU and FPI contracts, respectively, of the Spanish Ministry of Economy and Competitiveness.
Abbreviations
- AD:
Alzheimer's disease
- AHR:
Aryl hydrocarbon receptor
- BACH1:
BTB domain and CNC homolog 1
- BTB:
Bric-a-brac, tramtrack, broad complex
- β-TrCP:
Beta-transducin repeat-containing E3 ubiquitin protein ligase
- CUL3:
Cullin3
- DMF:
Dimethyl fumarate
- DRG:
Double glycine repeat
- GSH:
Glutathione
- GSK-3:
Glycogen synthase kinase
- IVR:
Intervening region
- KEAP1:
Kelch-like ECH-associated protein 1
- MMF:
Monomethyl fumarate
- MS:
Multiple sclerosis
- NFE2L2:
Gene encoding NRF2
- NRF2:
Nuclear factor erythroid 2-related factor 2
- PD:
Parkinson's disease
- PPI:
Protein-protein interaction
- RBX1:
RING-box protein 1
- ROS:
Reactive oxygen species
- SFN:
Sulforaphane
- SQSTM1:
Sequestosome-1
- XRE:
Xenobiotic response element.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplementary Materials
Overview of the strategies aimed at pharmacologic regulation of NRF2. KEAP1-dependent and KEAP1-independent strategies to activate NRF2 are included in red boxes. The current strategies for inhibition of NRF2 are indicated in the grey boxes. PPI: protein-protein interaction inhibitor; DPI: DNA-protein interaction inhibitor.
References
- 1.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annual Review of Pharmacology and Toxicology. 2013;53(1):401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes J. D., Dinkova-Kostova A. T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Pajares M., Jiménez-Moreno N., García-Yagüe Á. J., et al. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pajares M., Cuadrado A., Rojo A. I. Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases. Redox Biology. 2017;11:543–553. doi: 10.1016/j.redox.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Vega M. R., Dodson M., Gross C., et al. Role of Nrf2 and autophagy in acute lung injury. Current Pharmacology Reports. 2016;2(2):91–101. doi: 10.1007/s40495-016-0053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pajares M., Jiménez-Moreno N., Dias I. H. K., et al. Redox control of protein degradation. Redox Biology. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuadrado A., Rojo A. I., Wells G., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nature Reviews Drug Discovery. 2019;18(4):295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 8.Cuadrado A., Manda G., Hassan A., et al. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacological Reviews. 2018;70(2):348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 9.Rojo de la Vega M., Chapman E., Zhang D. D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34(1):21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J. D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. Journal of Biological Chemistry. 2004;279(30):31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 11.Katoh Y., Iida K., Kang M. I., et al. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Archives of Biochemistry and Biophysics. 2005;433(2):342–350. doi: 10.1016/j.abb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Tong K. I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Molecular and Cellular Biology. 2006;26(8):2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong K. I., Padmanabhan B., Kobayashi A., et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Molecular and Cellular Biology. 2007;27(21):7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Motohashi H., Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends in Pharmacological Sciences. 2013;34(6):340–346. doi: 10.1016/j.tips.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Sihvola V., Levonen A. L. Keap1 as the redox sensor of the antioxidant response. Archives of Biochemistry and Biophysics. 2017;617:94–100. doi: 10.1016/j.abb.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Holland R., Hawkins A. E., Eggler A. L., Mesecar A. D., Fabris D., Fishbein J. C. Prospective type 1 and type 2 disulfides of Keap1 protein. Chemical Research in Toxicology. 2008;21(10):2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hur E. M., Zhou F. Q. GSK3 signalling in neural development. Nature Reviews Neuroscience. 2010;11(8):539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeflich K. P., Luo J., Rubie E. A., Tsao M. S., Jin O., Woodgett J. R. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406(6791):86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 19.Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/β-TrCP. Free Radical Biology & Medicine. 2015;88:147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J. D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32(32):3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T., Zhao F., Gao B., et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes & Development. 2014;28(7):708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao W., Hu L., Scrivens P. J., Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. Journal of Biological Chemistry. 2005;280(21):20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 23.Kwak M. K., Itoh K., Yamamoto M., Kensler T. W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Molecular and Cellular Biology. 2002;22(9):2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao S., Wang S., Moghaddam S. J., et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Research. 2014;74(24):7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeNicola G. M., Karreth F. A., Humpton T. J., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rushworth S. A., Zaitseva L., Murray M. Y., Shah N. M., Bowles K. M., MacEwan D. J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood. 2012;120(26):5188–5198. doi: 10.1182/blood-2012-04-422121. [DOI] [PubMed] [Google Scholar]
- 27.Sangokoya C., Telen M. J., Chi J. T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116(20):4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M., Yao Y., Eades G., Zhang Y., Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Research and Treatment. 2011;129(3):983–991. doi: 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X., Li X., Ma S., Guo Y., Li Y. MicroRNA-98-5p ameliorates oxygen-glucose deprivation/reoxygenation (OGD/R)-induced neuronal injury by inhibiting Bach1 and promoting Nrf2/ARE signaling. Biochemical and Biophysical Research Communications. 2018;507(1-4):114–121. doi: 10.1016/j.bbrc.2018.10.182. [DOI] [PubMed] [Google Scholar]
- 30.Fabrizio F. P., Sparaneo A., Trombetta D., Muscarella L. A. Epigenetic versus genetic deregulation of the KEAP1/NRF2 axis in solid tumors: focus on methylation and noncoding RNAs. Oxidative Medicine and Cellular Longevity. 2018;2018:21. doi: 10.1155/2018/2492063.2492063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Medicinal Research Reviews. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hur W., Gray N. S. Small molecule modulators of antioxidant response pathway. Current Opinion in Chemical Biology. 2011;15(1):162–173. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Satoh T., McKercher S. R., Lipton S. A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radical Biology & Medicine. 2013;65:645–657. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson A. J., Kerns J. K., Callahan J. F., Moody C. J. Keap calm, and carry on covalently. Journal of Medicinal Chemistry. 2013;56(19):7463–7476. doi: 10.1021/jm400224q. [DOI] [PubMed] [Google Scholar]
- 35.Wong M. H. L., Bryan H. K., Copple I. M., et al. Design and synthesis of irreversible analogues of bardoxolone methyl for the identification of pharmacologically relevant targets and interaction sites. Journal of Medicinal Chemistry. 2016;59(6):2396–2409. doi: 10.1021/acs.jmedchem.5b01292. [DOI] [PubMed] [Google Scholar]
- 36.Kansanen E., Bonacci G., Schopfer F. J., et al. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. Journal of Biological Chemistry. 2011;286(16):14019–14027. doi: 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fourquet S., Guerois R., Biard D., Toledano M. B. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. Journal of Biological Chemistry. 2010;285(11):8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levonen A. L., Landar A., Ramachandran A., et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochemical Journal. 2004;378(2):373–382. doi: 10.1042/bj20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakabayashi N., Dinkova-Kostova A. T., Holtzclaw W. D., et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto T., Suzuki T., Kobayashi A., et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Molecular and Cellular Biology. 2008;28(8):2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito R., Suzuki T., Hiramoto K., et al. Characterizations of three major cysteine sensors of Keap1 in stress response. Molecular and Cellular Biology. 2015;36(2, article MCB.00868-15) doi: 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baird L., Lleres D., Swift S., Dinkova-Kostova A. T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(38):15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleasby A., Yon J., Day P. J., et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9(6, article e98896) doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iso T., Suzuki T., Baird L., Yamamoto M. Absolute amounts and status of the Nrf2-Keap1-Cul3 complex within cells. Molecular and Cellular Biology. 2016;36(24):3100–3112. doi: 10.1128/MCB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dayalan Naidu S., Muramatsu A., Saito R., et al. C151 in KEAP1 is the main cysteine sensor for the cyanoenone class of NRF2 activators, irrespective of molecular size or shape. Scientific Reports. 2018;8(1):p. 8037. doi: 10.1038/s41598-018-26269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinkova-Kostova A. T., Liby K. T., Stephenson K. K., et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liby K. T., Sporn M. B. Synthetic oleanane triterpenoids: multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacological Reviews. 2012;64(4):972–1003. doi: 10.1124/pr.111.004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pergola P. E., Raskin P., Toto R. D., et al. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. The New England Journal of Medicine. 2011;365(4):327–336. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D. D. Bardoxolone brings Nrf2-based therapies to light. Antioxidants & Redox Signaling. 2013;19(5):517–518. doi: 10.1089/ars.2012.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch D. R., Farmer J., Hauser L., et al. Safety, pharmacodynamics, and potential benefit of omaveloxolone in Friedreich ataxia. Annals of Clinical Translational Neurology. 2019;6(1):15–26. doi: 10.1002/acn3.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rabbani P. S., Ellison T., Waqas B., et al. Targeted Nrf2 activation therapy with RTA 408 enhances regenerative capacity of diabetic wounds. Diabetes Research and Clinical Practice. 2018;139:11–23. doi: 10.1016/j.diabres.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Xu Z., Zhang F., Sun F., Gu K. F., Dong S., He D. Dimethyl fumarate for multiple sclerosis. Cochrane Database of Systematic Reviews. 2015;4, article CD011076 doi: 10.1002/14651858.CD011076.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schimrigk S., Brune N., Hellwig K., et al. Oral fumaric acid esters for the treatment of active multiple sclerosis: an open-label, baseline-controlled pilot study. European Journal of Neurology. 2006;13(6):604–610. doi: 10.1111/j.1468-1331.2006.01292.x. [DOI] [PubMed] [Google Scholar]
- 54.Gold R., Kappos L., Arnold D. L., et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. The New England Journal of Medicine. 2012;367(12):1098–1107. doi: 10.1056/NEJMoa1114287. [DOI] [PubMed] [Google Scholar]
- 55.Fox R. J., Miller D. H., Phillips J. T., et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. The New England Journal of Medicine. 2012;367(12):1087–1097. doi: 10.1056/NEJMoa1206328. [DOI] [PubMed] [Google Scholar]
- 56.Hoxtermann S., Nuchel C., Altmeyer P. Fumaric acid esters suppress peripheral CD4- and CD8-positive lymphocytes in psoriasis. Dermatology. 1998;196(2):223–230. doi: 10.1159/000017903. [DOI] [PubMed] [Google Scholar]
- 57.Mills E. A., Ogrodnik M. A., Plave A., Mao-Draayer Y. Emerging understanding of the mechanism of action for dimethyl fumarate in the treatment of multiple sclerosis. Frontiers in Neurology. 2018;9(5) doi: 10.3389/fneur.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghadiri M., Rezk A., Li R., et al. Dimethyl fumarate-induced lymphopenia in MS due to differential T-cell subset apoptosis. Neurology - Neuroimmunology Neuroinflammation. 2017;4(3, article e340) doi: 10.1212/NXI.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li R., Rezk A., Ghadiri M., et al. Dimethyl fumarate treatment mediates an anti-inflammatory shift in B cell subsets of patients with multiple sclerosis. Journal of Immunology. 2017;198(2):691–698. doi: 10.4049/jimmunol.1601649. [DOI] [PubMed] [Google Scholar]
- 60.Smith M. D., Martin K. A., Calabresi P. A., Bhargava P. Dimethyl fumarate alters B-cell memory and cytokine production in MS patients. Annals of Clinical Translational Neurology. 2017;4(5):351–355. doi: 10.1002/acn3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Linker R. A., Lee D. H., Ryan S., et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134(3):678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 62.Dibbert S., Clement B., Skak-Nielsen T., Mrowietz U., Rostami-Yazdi M. Detection of fumarate-glutathione adducts in the portal vein blood of rats: evidence for rapid dimethylfumarate metabolism. Archives of Dermatological Research. 2013;305(5):447–451. doi: 10.1007/s00403-013-1332-y. [DOI] [PubMed] [Google Scholar]
- 63.Sun H., Zhu J., Lin H., Gu K., Feng F. Recent progress in the development of small molecule Nrf2 modulators: a patent review (2012-2016) Expert Opinion on Therapeutic Patents. 2017;27(7):763–785. doi: 10.1080/13543776.2017.1325464. [DOI] [PubMed] [Google Scholar]
- 64.Zeidan T. A., Duncan S., Hencken C. P., Wynn T. A., Sanrame C. N. Prodrugs of fumarates and their use in treating various diseases. Alkermes Pharma Ireland Limited; 2014. [Google Scholar]
- 65.von Glehn F., Dias-Carneiro R. P. C., Moraes A. S., et al. Dimethyl fumarate downregulates the immune response through the HCA2/GPR109A pathway: implications for the treatment of multiple sclerosis. Multiple Sclerosis and Related Disorders. 2018;23:46–50. doi: 10.1016/j.msard.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 66.Kensler T. W., Qian G. S., Chen J. G., Groopman J. D. Translational strategies for cancer prevention in liver. Nature Reviews Cancer. 2003;3(5):321–329. doi: 10.1038/nrc1076. [DOI] [PubMed] [Google Scholar]
- 67.Okada K., Shoda J., Taguchi K., et al. Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;295(4):G735–G747. doi: 10.1152/ajpgi.90321.2008. [DOI] [PubMed] [Google Scholar]
- 68.Kawata K., Kobayashi Y., Souda K., et al. Enhanced hepatic Nrf2 activation after ursodeoxycholic acid treatment in patients with primary biliary cirrhosis. Antioxidants & Redox Signaling. 2010;13(3):259–268. doi: 10.1089/ars.2009.2903. [DOI] [PubMed] [Google Scholar]
- 69.Wong D. P. W., Ng M. Y., Leung J. Y., et al. Regulation of the NRF2 transcription factor by andrographolide and organic extracts from plant endophytes. PLoS One. 2018;13(10, article e0204853) doi: 10.1371/journal.pone.0204853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahadoran Z., Mirmiran P., Hosseinpanah F., Rajab A., Asghari G., Azizi F. Broccoli sprouts powder could improve serum triglyceride and oxidized LDL/LDL-cholesterol ratio in type 2 diabetic patients: a randomized double-blind placebo-controlled clinical trial. Diabetes Research and Clinical Practice. 2012;96(3):348–354. doi: 10.1016/j.diabres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 71.Axelsson A. S., Tubbs E., Mecham B., et al. Sulforaphane reduces hepatic glucose production and improves glucose control in patients with type 2 diabetes. Science Translational Medicine. 2017;9(394, article eaah4477) doi: 10.1126/scitranslmed.aah4477. [DOI] [PubMed] [Google Scholar]
- 72.Ping Z., Liu W., Kang Z., et al. Sulforaphane protects brains against hypoxic-ischemic injury through induction of Nrf2-dependent phase 2 enzyme. Brain Research. 2010;1343:178–185. doi: 10.1016/j.brainres.2010.04.036. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J., Kobori N., Aronowski J., Dash P. K. Sulforaphane reduces infarct volume following focal cerebral ischemia in rodents. Neuroscience Letters. 2006;393(2-3):108–112. doi: 10.1016/j.neulet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 74.Park H. M., Kim J. A., Kwak M. K. Protection against amyloid beta cytotoxicity by sulforaphane: role of the proteasome. Archives of Pharmacal Research. 2009;32(1):109–115. doi: 10.1007/s12272-009-1124-2. [DOI] [PubMed] [Google Scholar]
- 75.Kim H. V., Kim H. Y., Ehrlich H. Y., Choi S. Y., Kim D. J., Kim Y. S. Amelioration of Alzheimer’s disease by neuroprotective effect of sulforaphane in animal model. Amyloid. 2013;20(1):7–12. doi: 10.3109/13506129.2012.751367. [DOI] [PubMed] [Google Scholar]
- 76.Han J. M., Lee Y. J., Lee S. Y., et al. Protective effect of sulforaphane against dopaminergic cell death. The Journal of Pharmacology and Experimental Therapeutics. 2007;321(1):249–256. doi: 10.1124/jpet.106.110866. [DOI] [PubMed] [Google Scholar]
- 77.Jazwa A., Rojo A. I., Innamorato N. G., Hesse M., Fernández-Ruiz J., Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxidants & Redox Signaling. 2011;14(12):2347–2360. doi: 10.1089/ars.2010.3731. [DOI] [PubMed] [Google Scholar]
- 78.Tarozzi A., Angeloni C., Malaguti M., Morroni F., Hrelia S., Hrelia P. Sulforaphane as a potential protective phytochemical against neurodegenerative diseases. Oxidative Medicine and Cellular Longevity. 2013;2013:10. doi: 10.1155/2013/415078.415078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houghton C. A., Fassett R. G., Coombes J. S. Sulforaphane and other nutrigenomic Nrf2 activators: can the clinician’s expectation be matched by the reality? Oxidative Medicine and Cellular Longevity. 2016;2016:17. doi: 10.1155/2016/7857186.7857186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Egea J., Buendia I., Parada E., et al. Melatonin-sulforaphane hybrid ITH12674 induces neuroprotection in oxidative stress conditions by a ‘drug-prodrug’ mechanism of action. British Journal of Pharmacology. 2015;172(7):1807–1821. doi: 10.1111/bph.13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sreejayan, Rao M. N. A. Nitric oxide scavenging by curcuminoids. The Journal of Pharmacy and Pharmacology. 1997;49(1):105–107. doi: 10.1111/j.2042-7158.1997.tb06761.x. [DOI] [PubMed] [Google Scholar]
- 82.Qiu P., Man S., Li J., et al. Overdose intake of curcumin initiates the unbalanced state of bodies. Journal of Agricultural and Food Chemistry. 2016;64(13):2765–2771. doi: 10.1021/acs.jafc.6b00053. [DOI] [PubMed] [Google Scholar]
- 83.Na L. X., Li Y., Pan H. Z., et al. Curcuminoids exert glucose-lowering effect in type 2 diabetes by decreasing serum free fatty acids: a double-blind, placebo-controlled trial. Molecular Nutrition & Food Research. 2013;57(9):1569–1577. doi: 10.1002/mnfr.201200131. [DOI] [PubMed] [Google Scholar]
- 84.Chuengsamarn S., Rattanamongkolgul S., Phonrat B., Tungtrongchitr R., Jirawatnotai S. Reduction of atherogenic risk in patients with type 2 diabetes by curcuminoid extract: a randomized controlled trial. The Journal of Nutritional Biochemistry. 2014;25(2):144–150. doi: 10.1016/j.jnutbio.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 85.Hatcher H., Planalp R., Cho J., Torti F. M., Torti S. V. Curcumin: from ancient medicine to current clinical trials. Cellular and Molecular Life Sciences. 2008;65(11):1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balogun E., Hoque M., GONG P., et al. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. The Biochemical Journal. 2003;371(3):887–895. doi: 10.1042/bj20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arbeeny C. M., Ling H., Smith M. M., et al. CXA-10, a nitrated fatty acid, is renoprotective in deoxycorticosterone acetate-salt nephropathy. The Journal of Pharmacology and Experimental Therapeutics. 2019;369(3):503–510. doi: 10.1124/jpet.118.254755. [DOI] [PubMed] [Google Scholar]
- 88.Batthyany C. I., Lopez G. V. Nitroalkene tocopherols and analogs thereof for use in the treatment and prevention of inflammation related conditions. Complexa Inc.; 2015. [Google Scholar]
- 89.Nilsson L., Palm F., Nørregaard R. 15-Deoxy-Δ12,14-prostaglandin J2 exerts antioxidant effects while exacerbating inflammation in mice subjected to ureteral obstruction. Mediators of Inflammation. 2017;2017:10. doi: 10.1155/2017/3924912.3924912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen K., Li J. J., Li S. N., et al. 15-Deoxy-Δ12,14-prostaglandin J2 alleviates hepatic ischemia-reperfusion injury in mice via inducing antioxidant response and inhibiting apoptosis and autophagy. Acta Pharmacologica Sinica. 2017;38(5):672–687. doi: 10.1038/aps.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu J., Guo S., Xue X., et al. Identification of a novel series of anti-inflammatory and anti-oxidative phospholipid oxidation products containing the cyclopentenone moiety in vitro and in vivo: implication in atherosclerosis. Journal of Biological Chemistry. 2017;292(13):5378–5391. doi: 10.1074/jbc.M116.751909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mills E. L., Ryan D. G., Prag H. A., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556(7699):113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X. J., Hayes J. D., Higgins L. G., Wolf C. R., Dinkova-Kostova A. T. Activation of the NRF2 signaling pathway by copper-mediated redox cycling of para- and ortho-hydroquinones. Chemistry & Biology. 2010;17(1):75–85. doi: 10.1016/j.chembiol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 94.Lee J. M., Moehlenkamp J. D., Hanson J. M., Johnson J. A. Nrf2-dependent activation of the antioxidant responsive element by tert-butylhydroquinone is independent of oxidative stress in IMR-32 human neuroblastoma cells. Biochemical and Biophysical Research Communications. 2001;280(1):286–292. doi: 10.1006/bbrc.2000.4106. [DOI] [PubMed] [Google Scholar]
- 95.Higashi C., Kawaji A., Tsuda N., et al. The novel Nrf2 inducer TFM-735 ameliorates experimental autoimmune encephalomyelitis in mice. European Journal of Pharmacology. 2017;802:76–84. doi: 10.1016/j.ejphar.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki T., Yamamoto M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. Journal of Biological Chemistry. 2017;292(41):16817–16824. doi: 10.1074/jbc.R117.800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richardson B. G., Jain A. D., Speltz T. E., Moore T. W. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorganic & Medicinal Chemistry Letters. 2015;25(11):2261–2268. doi: 10.1016/j.bmcl.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Padmanabhan B., Tong K. I., Ohta T., et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Molecular Cell. 2006;21(5):689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 99.Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO Journal. 2006;25(15):3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Inoyama D., Chen Y., Huang X., Beamer L. J., Kong A. N. T., Hu L. Optimization of fluorescently labeled Nrf2 peptide probes and the development of a fluorescence polarization assay for the discovery of inhibitors of Keap1-Nrf2 interaction. Journal of Biomolecular Screening. 2012;17(4):435–447. doi: 10.1177/1087057111430124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y., Inoyama D., Kong A. N. T., Beamer L. J., Hu L. Kinetic analyses of Keap1-Nrf2 interaction and determination of the minimal Nrf2 peptide sequence required for Keap1 binding using surface plasmon resonance. Chemical Biology & Drug Design. 2011;78(6):1014–1021. doi: 10.1111/j.1747-0285.2011.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hancock R., Bertrand H. C., Tsujita T., et al. Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction. Free Radical Biology & Medicine. 2012;52(2):444–451. doi: 10.1016/j.freeradbiomed.2011.10.486. [DOI] [PubMed] [Google Scholar]
- 103.Tu J., Zhang X., Zhu Y., et al. Cell-permeable peptide targeting the Nrf2-Keap1 interaction: a potential novel therapy for global cerebral ischemia. The Journal of Neuroscience. 2015;35(44):14727–14739. doi: 10.1523/JNEUROSCI.1304-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hancock R., Schaap M., Pfister H., Wells G. Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction with improved binding and cellular activity. Organic & Biomolecular Chemistry. 2013;11(21):3553–3557. doi: 10.1039/c3ob40249e. [DOI] [PubMed] [Google Scholar]
- 105.Lu M. C., Jiao Q., Liu T., et al. Discovery of a head-to-tail cyclic peptide as the Keap1-Nrf2 protein-protein interaction inhibitor with high cell potency. European Journal of Medicinal Chemistry. 2018;143:1578–1589. doi: 10.1016/j.ejmech.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 106.Jnoff E., Albrecht C., Barker J. J., et al. Binding mode and structure-activity relationships around direct inhibitors of the Nrf2-Keap1 complex. ChemMedChem. 2014;9(4):699–705. doi: 10.1002/cmdc.201300525. [DOI] [PubMed] [Google Scholar]
- 107.Marcotte D., Zeng W., Hus J. C., et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorganic & Medicinal Chemistry. 2013;21(14):4011–4019. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 108.Jiang Z.-Y., Lu M. C., Xu L.−. L., et al. Discovery of potent Keap1-Nrf2 protein-protein interaction inhibitor based on molecular binding determinants analysis. Journal of Medicinal Chemistry. 2014;57(6):2736–2745. doi: 10.1021/jm5000529. [DOI] [PubMed] [Google Scholar]
- 109.Ranjan N., Fulcrand G., King A., et al. Selective inhibition of bacterial topoisomerase I by alkynyl-bisbenzimidazoles. MedChemComm. 2014;5(6):816–825. doi: 10.1039/C4MD00140K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sato M., Aoki T., Inoue H., et al. TORAY INDUSTRIES Inc.; 2013. Keap1 protein binding compound, cristal of complex between the same and Keap1 protein, and method for producing the same. [Google Scholar]
- 111.Richardson B. G., Jain A. D., Potteti H. R., et al. Replacement of a naphthalene scaffold in Kelch-like ECH-associated protein 1 (KEAP1)/nuclear factor (erythroid-derived 2)-like 2 (NRF2) inhibitors. Journal of Medicinal Chemistry. 2018;61(17):8029–8047. doi: 10.1021/acs.jmedchem.8b01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu L., Magesh S., Chen L., et al. Discovery of a small-molecule inhibitor and cellular probe of Keap1-Nrf2 protein-protein interaction. Bioorganic & Medicinal Chemistry Letters. 2013;23(10):3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wen X., Thorne G., Hu L., Joy M. S., Aleksunes L. M. Activation of NRF2 signaling in HEK293 cells by a first-in-class direct KEAP1-NRF2 inhibitor. Journal of Biochemical and Molecular Toxicology. 2015;29(6):261–266. doi: 10.1002/jbt.21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bertrand H. C., Schaap M., Baird L., et al. Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1-Nrf2 protein-protein interaction. Journal of Medicinal Chemistry. 2015;58(18):7186–7194. doi: 10.1021/acs.jmedchem.5b00602. [DOI] [PubMed] [Google Scholar]
- 115.Nasiri H. R., Linge S., Ullmann D. Thermodynamic profiling of inhibitors of Nrf2:Keap1 interactions. Bioorganic & Medicinal Chemistry Letters. 2016;26(2):526–529. doi: 10.1016/j.bmcl.2015.11.082. [DOI] [PubMed] [Google Scholar]
- 116.Bresciani A., Missineo A., Gallo M., et al. Nuclear factor (erythroid-derived 2)-like 2 (NRF2) drug discovery: biochemical toolbox to develop NRF2 activators by reversible binding of Kelch-like ECH-associated protein 1 (KEAP1) Archives of Biochemistry and Biophysics. 2017;631:31–41. doi: 10.1016/j.abb.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 117.Hooper C., Killick R., Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. Journal of Neurochemistry. 2008;104(6):1433–1439. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silva T., Reis J., Teixeira J., Borges F. Alzheimer’s disease, enzyme targets and drug discovery struggles: from natural products to drug prototypes. Ageing Research Reviews. 2014;15:116–145. doi: 10.1016/j.arr.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 119.Luo J. Glycogen synthase kinase 3β (GSK3β) in tumorigenesis and cancer chemotherapy. Cancer Letters. 2009;273(2):194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lal H., Ahmad F., Woodgett J., Force T. The GSK-3 family as therapeutic target for myocardial diseases. Circulation Research. 2015;116(1):138–149. doi: 10.1161/CIRCRESAHA.116.303613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saraswati A. P., Ali Hussaini S. M., Krishna N. H., Babu B. N., Kamal A. Glycogen synthase kinase-3 and its inhibitors: potential target for various therapeutic conditions. European Journal of Medicinal Chemistry. 2018;144:843–858. doi: 10.1016/j.ejmech.2017.11.103. [DOI] [PubMed] [Google Scholar]
- 122.Lovestone S., Boada M., Dubois B., et al. A phase II trial of tideglusib in Alzheimer’s disease. Journal of Alzheimer's Disease. 2015;45(1):75–88. doi: 10.3233/JAD-141959. [DOI] [PubMed] [Google Scholar]
- 123.Bourhill T., Narendran A., Johnston R. N. Enzastaurin: a lesson in drug development. Critical Reviews in Oncology/Hematology. 2017;112:72–79. doi: 10.1016/j.critrevonc.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 124.Lombardi G., Pambuku A., Bellu L., et al. Effectiveness of antiangiogenic drugs in glioblastoma patients: a systematic review and meta-analysis of randomized clinical trials. Critical Reviews in Oncology/Hematology. 2017;111:94–102. doi: 10.1016/j.critrevonc.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 125.Rojo A. I., Medina-Campos O. N., Rada P., et al. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radical Biology & Medicine. 2012;52(2):473–487. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 126.Palomo V., Martinez A. Glycogen synthase kinase 3 (GSK-3) inhibitors: a patent update (2014-2015) Expert Opinion on Therapeutic Patents. 2017;27(6):657–666. doi: 10.1080/13543776.2017.1259412. [DOI] [PubMed] [Google Scholar]
- 127.Rada P., Rojo A. I., Evrard-Todeschi N., et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/β-TrCP axis. Molecular and Cellular Biology. 2012;32(17):3486–3499. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hast B. E., Goldfarb D., Mulvaney K. M., et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Research. 2013;73(7):2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jain A., Lamark T., Sjøttem E., et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. Journal of Biological Chemistry. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lau A., Wang X. J., Zhao F., et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Molecular and Cellular Biology. 2010;30(13):3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Komatsu M., Kurokawa H., Waguri S., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 132.Sarkar S., Rubinsztein D. C. Small molecule enhancers of autophagy for neurodegenerative diseases. Molecular BioSystems. 2008;4(9):895–901. doi: 10.1039/b804606a. [DOI] [PubMed] [Google Scholar]
- 133.Mizunoe Y., Kobayashi M., Sudo Y., et al. Trehalose protects against oxidative stress by regulating the Keap1-Nrf2 and autophagy pathways. Redox Biology. 2018;15:115–124. doi: 10.1016/j.redox.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dhakshinamoorthy S., Jain A. K., Bloom D. A., Jaiswal A. K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. Journal of Biological Chemistry. 2005;280(17):16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 135.Attucks O. C., Jasmer K. J., Hannink M., et al. Induction of heme oxygenase I (HMOX1) by HPP-4382: a novel modulator of Bach1 activity. PLoS One. 2014;9(7, article e101044) doi: 10.1371/journal.pone.0101044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramos-Gomez M., Dolan P. M., Itoh K., Yamamoto M., Kensler T. W. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24(3):461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 137.Xu C., Huang M. T., Shen G., et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Research. 2006;66(16):8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 138.Solis L. M., Behrens C., Dong W., et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clinical Cancer Research. 2010;16(14):3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shibata T., Ohta T., Tong K. I., et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ohta T., Iijima K., Miyamoto M., et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Research. 2008;68(5):1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 141.Milkovic L., Zarkovic N., Saso L. Controversy about pharmacological modulation of Nrf2 for cancer therapy. Redox Biology. 2017;12:727–732. doi: 10.1016/j.redox.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kitamura H., Motohashi H. NRF2 addiction in cancer cells. Cancer Science. 2018;109(4):900–911. doi: 10.1111/cas.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ki S. H., Cho I. J., Choi D. W., Kim S. G. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPβ TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Molecular and Cellular Biology. 2005;25(10):4150–4165. doi: 10.1128/MCB.25.10.4150-4165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Choi E. J., Jung B. J., Lee S. H., et al. A clinical drug library screen identifies clobetasol propionate as an NRF2 inhibitor with potential therapeutic efficacy in KEAP1 mutant lung cancer. Oncogene. 2017;36(37):5285–5295. doi: 10.1038/onc.2017.153. [DOI] [PubMed] [Google Scholar]
- 145.Wang X. J., Hayes J. D., Henderson C. J., Wolf C. R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang H., Liu K., Geng M., et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Research. 2013;73(10):3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 147.Ren D., Villeneuve N. F., Jiang T., et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(4):1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harder B., Tian W., la Clair J. J., et al. Brusatol overcomes chemoresistance through inhibition of protein translation. Molecular Carcinogenesis. 2017;56(5):1493–1500. doi: 10.1002/mc.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vartanian S., Ma T. P., Lee J., et al. Application of mass spectrometry profiling to establish brusatol as an inhibitor of global protein synthesis. Molecular & Cellular Proteomics. 2016;15(4):1220–1231. doi: 10.1074/mcp.M115.055509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhu J., Wang H., Chen F., et al. An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radical Biology & Medicine. 2016;99:544–556. doi: 10.1016/j.freeradbiomed.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 151.Tang X., Wang H., Fan L., et al. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radical Biology & Medicine. 2011;50(11):1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 152.Zhong Y., Zhang F., Sun Z., et al. Drug resistance associates with activation of Nrf2 in MCF-7/DOX cells, and wogonin reverses it by down-regulating Nrf2-mediated cellular defense response. Molecular Carcinogenesis. 2013;52(10):824–834. doi: 10.1002/mc.21921. [DOI] [PubMed] [Google Scholar]
- 153.Pandurangan A. K., Ananda Sadagopan S. K., Dharmalingam P., Ganapasam S. Luteolin, a bioflavonoid inhibits Azoxymethane-induced colorectal cancer through activation of Nrf2 signaling. Toxicology Mechanisms and Methods. 2014;24(1):13–20. doi: 10.3109/15376516.2013.843111. [DOI] [PubMed] [Google Scholar]
- 154.Limonciel A., Jennings P. A review of the evidence that ochratoxin A is an Nrf2 inhibitor: implications for nephrotoxicity and renal carcinogenicity. Toxins. 2014;6(1):371–379. doi: 10.3390/toxins6010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arlt A., Sebens S., Krebs S., et al. Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene. 2013;32(40):4825–4835. doi: 10.1038/onc.2012.493. [DOI] [PubMed] [Google Scholar]
- 156.Manna A., de Sarkar S., de S., Bauri A. K., Chattopadhyay S., Chatterjee M. The variable chemotherapeutic response of Malabaricone-A in leukemic and solid tumor cell lines depends on the degree of redox imbalance. Phytomedicine. 2015;22(7-8):713–723. doi: 10.1016/j.phymed.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 157.Tarumoto T., Nagai T., Ohmine K., et al. Ascorbic acid restores sensitivity to imatinib via suppression of Nrf2-dependent gene expression in the imatinib-resistant cell line. Experimental Hematology. 2004;32(4):375–381. doi: 10.1016/j.exphem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 158.Singh A., Venkannagari S., Oh K. H., et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chemical Biology. 2016;11(11):3214–3225. doi: 10.1021/acschembio.6b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tsuchida K., Tsujita T., Hayashi M., et al. Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radical Biology & Medicine. 2017;103:236–247. doi: 10.1016/j.freeradbiomed.2016.12.041. [DOI] [PubMed] [Google Scholar]
- 160.Bollong M. J., Yun H., Sherwood L., Woods A. K., Lairson L. L., Schultz P. G. A small molecule inhibits deregulated NRF2 transcriptional activity in cancer. ACS Chemical Biology. 2015;10(10):2193–2198. doi: 10.1021/acschembio.5b00448. [DOI] [PubMed] [Google Scholar]
- 161.Zhang J., Su L., Ye Q., et al. Discovery of a novel Nrf2 inhibitor that induces apoptosis of human acute myeloid leukemia cells. Oncotarget. 2017;8(5):7625–7636. doi: 10.18632/oncotarget.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Steel R., Cowan J., Payerne E., O'Connell M. A., Searcey M. Anti-inflammatory effect of a cell-penetrating peptide targeting the Nrf2/Keap1 interaction. ACS Medicinal Chemistry Letters. 2012;3(5):407–410. doi: 10.1021/ml300041g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jiang Z. Y., Xu L.−. L., Lu M. C., et al. Structure-activity and structure-property relationship and exploratory in vivo evaluation of the nanomolar Keap1-Nrf2 protein-protein interaction inhibitor. Journal of Medicinal Chemistry. 2015;58(16):6410–6421. doi: 10.1021/acs.jmedchem.5b00185. [DOI] [PubMed] [Google Scholar]
- 164.Yonchuk J. G., Foley J. P., Bolognese B. J., et al. Characterization of the potent, selective Nrf2 activator, 3-(pyridin-3-ylsulfonyl)-5-(trifluoromethyl)-2H-chromen-2-one, in cellular and in vivo models of pulmonary oxidative stress. The Journal of Pharmacology and Experimental Therapeutics. 2017;363(1):114–125. doi: 10.1124/jpet.117.241794. [DOI] [PubMed] [Google Scholar]
- 165.Sun H.-P., Jiang Z.-Y., Zhang M.-Y., et al. Novel protein–protein interaction inhibitor of Nrf2–Keap1 discovered by structure-based virtual screening. MedChemComm. 2014;5(1):93–98. doi: 10.1039/C3MD00240C. [DOI] [Google Scholar]
- 166.Davies T. G., Wixted W. E., Coyle J. E., et al. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. Journal of Medicinal Chemistry. 2016;59(8):3991–4006. doi: 10.1021/acs.jmedchem.6b00228. [DOI] [PubMed] [Google Scholar]
- 167.Itoh K., Wakabayashi N., Katoh Y., et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & Development. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes to Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the strategies aimed at pharmacologic regulation of NRF2. KEAP1-dependent and KEAP1-independent strategies to activate NRF2 are included in red boxes. The current strategies for inhibition of NRF2 are indicated in the grey boxes. PPI: protein-protein interaction inhibitor; DPI: DNA-protein interaction inhibitor.