Abstract
Background
In practical production, dairy cows are frequently exposed to bacterial endotoxin (lipopolysaccharide, LPS) when they are subjected to high-concentrate diets, poor hygienic environments, as well as mastitis and metritis. Histone acetylation is an important epigenetic control of DNA transcription and a higher histone acetylation is associated with facilitated transcription. LPS might reduce histone acetylation in the mammary epithelial cells, resulting in lower transcription and mRNA expression of lactation-related genes. This study was conducted to investigate the effect of LPS on histone acetylation in bovine mammary epithelial cells and the efficacy of sodium butyrate (SB) in suppressing the endotoxin-induced adverse effect. Firstly, the bovine mammary epithelial cell line MAC-T cells were treated for 48 h with LPS at different doses of 0, 1, 10, 100, and 1000 endotoxin units (EU)/mL (1 EU = 0.1 ng), and the acetylation levels of histones H3 and H4 as well as the histone deacetylase (HDAC) activity were measured. Secondly, the MAC-T cells were treated for 48 h as follows: control, LPS (100 EU/mL), and LPS (100 EU/mL) plus SB (10 mmol/L), and the acetylation levels of histones H3 and H4 as well as milk gene mRNA expressions were determined.
Results
The results showed that HDAC activity increased linearly with increasing LPS doses (P < 0.01). The histone H3 acetylation levels were significantly reduced by LPS, while the histone H4 acetylation levels were not affected by LPS (P > 0.05). Sodium butyrate, an inhibitor of HDAC, effectively suppressed the endotoxin-induced decline of histone H3 acetylation (P < 0.05). As a result, SB significantly enhanced the mRNA expression of lactation-related genes (P < 0.05).
Conclusions
The results suggest one of the adverse effects of LPS on the lactation of bovine mammary gland epithelial cells was due to decreasing histone H3 acetylation through increasing HDAC activity, whereas the endotoxin-induced adverse effects were effectively suppressed by SB.
Electronic supplementary material
The online version of this article (10.1186/s12917-019-2007-5) contains supplementary material, which is available to authorized users.
Keywords: Endotoxin, Lipopolysaccharide, Histone acetylation, Sodium butyrate, Bovine, Mammary, Epithelial cells
Background
In practical dairy production, dairy cows are frequently fed high-concentrate diets to satisfy the high energy demand for milk production. High-concentrate diets often lead to increased yields of short-chain fatty acids (SCFAs) and lactic acid in the rumen, resulting in a decrease in rumen pH [1–3]. If rumen pH values remain below 5.6 for more than 3 h/day, subacute ruminal acidosis (SARA) occurs [4]. During high-concentrate feeding, particularly in the case of SARA, an increased amount of bacterial endotoxin (lipopolysaccharide, LPS) is released in the rumen fluid [1–3, 5]. Studies have shown that LPS in the digestive tract and blood increases significantly with increasing dietary concentrate ratios [5–8]. Our previous study also showed dairy cows fed high-concentrate diets had significantly higher LPS concentrations in the mammary arterial blood, as compared with cows fed the low-concentrate diets [9]. In practical production, dairy cows may also be exposed to LPS when they are subjected to poor hygienic environments. Furthermore, mastitis and metritis are common diseases for dairy cows, and under these conditions, LPS in the blood circulation and the mammary gland increases tremendously [3].
Once LPS enters the blood circulation, it can elicit systemic inflammatory response and local inflammatory response in the mammary gland [1]. Under the circumstances, more nutrients are used for synthesizing immune molecules, resulting in lower supply of substrate precursors for milk component synthesis and low milk production [1, 5, 10]. The entry of LPS into the mammary gland can also lead to production of an increasing amount of reactive oxygen species and NO, affecting the proliferation of mammary epithelial cells and even resulting in apoptosis of mammary epithelial cells [1, 5, 10]. LPS may also down-regulate the expression of milk genes such as fatty acid synthase (FASN) and acetyl-CoA carboxylase 1 (ACACA), resulting in lower milk yield [11, 12], probably through epigenetic manipulation.
Epigenetics is a science that studies heritable changes in gene expression that are independent of DNA sequence [13]. Major epigenetic events include histone modification, DNA methylation, and microRNA regulation. As far as histone modification is concerned, various histone modifications include acetylation, methylation, phosphorylation, and ubiquitinated succinylation, etc. [14, 15]. Histone is the basic structural protein of eukaryotic chromatin and binds most closely to DNA. Mammalian histones have five components, including H1, H2A, H2B, H3 and H4. The N-terminal of histones, especially histones H3 and H4, is very active because it extends out of the nucleosome. Therefore, histones are often chemically modified. The lysine residues in the N-terminal tail of histones are extremely active and can undergo chemical modifications such as acetylation. Acetylated histones can neutralize the positive charge of lysine residues, which reduces the binding of histones to DNA, making DNA adopt a more relaxed structure and thus facilitating transcription [16]. At present, the acetylation of histones H3 and H4 is the most frequently studied histone modification.
Histone acetyltransferase (HAT) and histone deacetylase (HDAC) are reciprocal enzymes involved in the histone acetylation process. Under the action of HAT, the acetylation levels of histones increases, whereas HDAC deacetylates histones, resulting in a closed state of chromatin and DNA as well as gene silencing [17]. Histone deacetylase inhibitors can inhibit deacetylation by HDAC. At present, the most well-studied HDAC inhibitors include sodium butyrate (SB) [18, 19], sodium valproate [20], and trichostatin [21], and SB is of great importance for ruminant animals [22, 23].
Up to now, it remains unclear whether LPS can induce hypoacetylation of histones H3 and H4 in bovine mammary epithelial cells and in which manner LPS can induce such changes, although our previous study showed that there existed a negative relationship between LPS concentrations in the mammary arterial blood and the histone H3 acetylation level in the mammary tissue of dairy cows [9]. Therefore, we hypothesized that LPS might decrease histone acetylation through increasing HDAC activity, and SB could effectively inhibit the HDAC activity, thus antagonizing the adverse effects of LPS on histone H3 and H4 acetylation. The objective of this study was to explore the effect of LPS on the acetylation levels of histones H3 and H4 in the bovine mammary epithelial cells and the possible enzymatic mechanism. Furthermore, we also investigate the efficacy of SB in reducing LPS-induced hypoacetylation of histones H3 and H4, so as to provide an insight into possible avenues to improve lactation performance in dairy cows.
Results
Effect of LPS on acetylation of histones H3 and H4
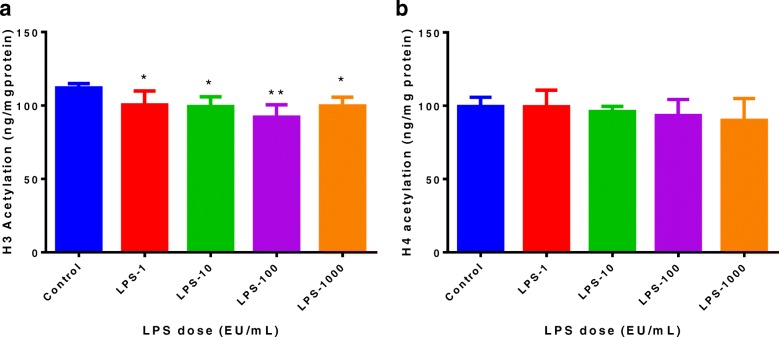
We are interested in determining the impact of LPS on the epigenetic changes in MAC-T bovine mammary epithelial cells. The results showed that the acetylation level of histone H3 decreased significantly after LPS treatment at different doses (Fig. 1a). The effect of LPS on histone H3 acetylation was not dose-dependent. LPS treatment tended to reduce the histone H4 acetylation level, but the effect was not statistically significant (P > 0.05) (Fig. 1b).
Fig. 1.
Effect of lipopolysaccharide (LPS) on histone H3 and H4 acetylation levels in MAC-T bovine mammary epithelial cells. The MAC-T bovine mammary epithelial cells were treated for 48 h with LPS at 0 (control), 1, 10, 100, 1000 endotoxin units (EU)/mL, respectively. (a) Histone H3 levels deceased significantly after LPS treatments (n = 5/treatment); (b) Histone H4 levels were not significantly (P > 0.05) affected by LPS treatments (n = 6/treatment). Data represent the mean and standard deviation and the asterisk indicates statistical difference between the indicated column and the control column (*, P < 0.05; **, P < 0.01). The raw data were shown in Additional file 1: Table S1
Effect of LPS on HDAC activity
To determine if LPS decreased histone H3 acetylation through increasing the activity of HDAC, we measured the activity of HDAC in MAC-T bovine mammary epithelial cells after LPS treatment. The activity of HDAC increased significantly (P < 0.01) after LPS treatment at different doses (Fig. 2), suggesting LPS can effectively enhance the activity of HDAC.
Fig. 2.
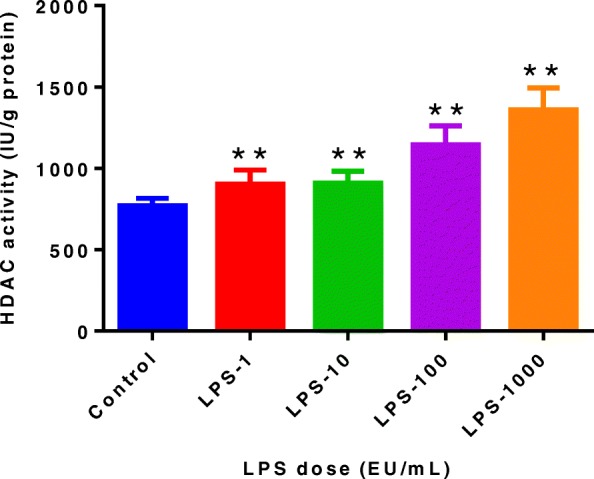
Effect of lipopolysaccharide (LPS) on histone deacetylase (HDAC) activity in MAC-T bovine mammary epithelial cells after treatment for 48 h with LPS at 0 (control), 1, 10, 100, and 1000 endotoxin units (EU)/mL, respectively. Data represent the mean and standard deviation (n = 8/treatment) and the asterisk indicates statistical difference between the indicated column and the control column (**, P < 0.01). The raw data were shown in Additional file 2: Table S2
Histone H3 acetylation levels after adding sodium butyrate
In order to determine whether SB, an HADC inhibitor, was able to counteract the adverse effect of LPS on histone H3 acetylation in MAC-T bovine mammary epithelial cells, the acetylation level of histone H3 after 48 h of treatment with LPS plus SB was determined (Fig. 3). H3 acetylation level of LPS treated cells was significantly lower than untreated control cells (P < 0.05). After adding SB into the LPS treatment, the H3 acetylation level was significantly higher (P < 0.05) than that of treatment with LPS alone or the control.
Fig. 3.
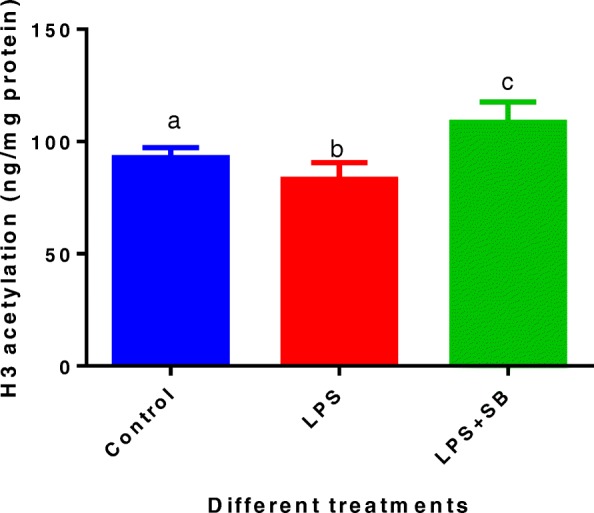
Acetylation of histone H3 in MAC-T bovine mammary epithelial cells among different treatments for 48 h: control, lipopolysaccharide (LPS, 100 EU/mL), and LPS (100 EU/mL) plus sodium butyrate (SB, 10 mmol/L). Data represent the mean and standard deviation (n = 6/treatment) and columns with different letters (a,b,c) indicate significant differences in histone H3 acetylation levels across treatments (P < 0.05). The raw data were shown in Additional file 3: Table S3
mRNA expression of lactation-related genes
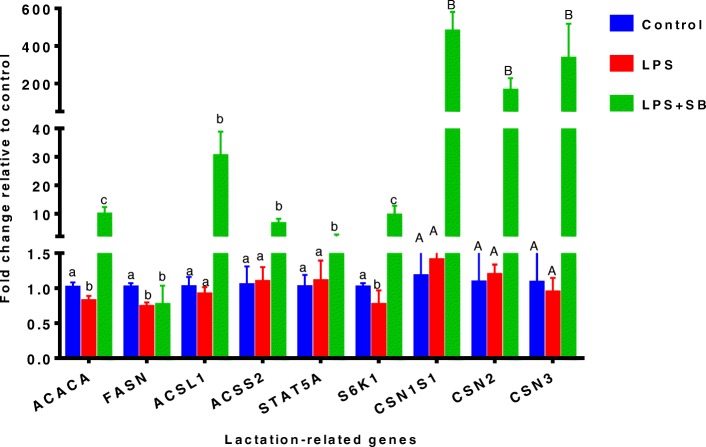
To verify the correlation between histone acetylation and mRNA expression of lactation-related genes, we next determined whether SB was able to enhance mRNA expression of lactation-related genes through improving histone H3 acetylation in MAC-T bovine mammary epithelial cells exposed to LPS (Fig. 4). The results revealed that compared with control, LPS treatment significantly reduced the mRNA expression of ACACA, FASN, and ribosomal protein S6 kinase 1 (S6K1) genes (P < 0.05) in MAC-T bovine mammary epithelial cells, whereas the mRNA expression of acyl-CoA synthetase long-chain family member 1 (ACSL1), acyl-CoA synthetase short-chain family member 2 (ACSS2), signal transducer and activator of transcription 5A (STAT5A), casein alpha s1 (CSN1S1), casein beta (CSN2), and casein kappa (CSN3) was not affected by LPS (P > 0.05). Sodium butyrate was able to significantly suppress the adverse effects of LPS on mRNA expression of lactation-related genes, resulting in a tremendous increase in the mRNA expression of all genes except that the FASN mRNA expression was only numerically increased.
Fig. 4.
mRNA expression of lactation-related genes in MAC-T bovine mammary epithelial cells under different treatments for 48 h: control, lipopolysaccharide (LPS, 100 EU/mL), and LPS (100 EU/mL) plus sodium butyrate (SB, 10 mmol/L). Data represent the mean and standard deviation (n = 3/treatment). Columns with different lowercase letters (a,b,c) indicate significantly different values among treatments (P < 0.05), and columns with different uppercase letters (A,B) indicate highly significantly different values among treatments (P < 0.01). ACACA: acetyl-CoA carboxylase 1; FASN: fatty acid synthase; ACSL1: acyl-CoA synthetase long-chain family member 1; ACSS2: acyl-CoA synthetase short-chain family member 2; STAT5A: signal transducer and activator of transcription 5A; S6K1, ribosomal protein S6 kinase 1; CSN1S1: casein alpha s1; CSN2: casein beta; CSN3: casein kappa. The raw data were shown in Additional file 4: Table S4
Discussion
In the present study, we found that histone H3 acetylation in MAC-T bovine mammary epithelial cells decreased significantly after LPS treatment for 48 h at all the doses used in the experiment, although histone H4 acetylation was not significantly affected by LPS treatment. A study by Dong et al. showed when dairy cows were fed high-concentrate diets, the histone H3 acetylation level in the mammary tissue decreased [9]. More interestingly, their study found a negative relationship between LPS concentrations in the mammary blood and the histone H3 acetylation in the mammary tissue of dairy cows. Therefore, when bovine mammary epithelial cells are exposed to LPS, the LPS-induce histone H3 hypoacetylation may affect milk gene expression due to reduced transcription. The results of the present study confirmed that LPS treatment (100 EU/mL) significantly reduced the mRNA expression of lactation-related genes such as ACACA, FASN and S6K1.
On the contrary, Angrisano et al. found that histone H3 acetylation at interleukin- 8 (IL-8) gene promoter increased significantly after treating human intestinal epithelial cells with LPS for 30 min and 1 h [24]. Therefore, the effect of LPS on histone acetylation may display a selective pattern, depending upon the cell type and the primary function of the cell. In the case of intestinal epithelial cells or other immune cells, exposure to LPS will lead to inflammatory processes, and synthesis of inflammatory molecules such as IL-1, IL-6, and tumor necrosis factor-a (TNF-a) takes priority. Under the circumstance, a higher histone acetylation will facilitate the synthesis of inflammatory molecules.
The results of the present study showed that with increasing LPS doses, HDAC activity keeps rising. Therefore, the histone H3 hypoacetylation after treatment of MAC-T bovine mammary epithelial cells with LPS may be due to the rise of HDAC activity induced by LPS. There are few reports about the LPS-induced increase in HDAC activity in the mammary epithelial cells or tissue of dairy cows to date. However, similar studies have been reported in the literature. Aung et al. demonstrated that the mRNA expression of HDAC-4, 6, 7, 8 and 9 increased after LPS treatment of bone marrow-derived macrophages for 24 h [25]. Xing et al. found that after treating mouse lung fibroblasts (MIC-CELL-0040) with LPS for 72 h, the gene expression of HDAC-4, HDAC-5 and HDAC-7 was significantly increased [26]. Furthermore, in their study, LPS significantly increased the expression of HDAC-4, HDAC-5 and HDAC-7 proteins, and decreased the H3 and H4 acetylation levels [26].
Milk fat and milk protein are the most important milk components. The synthesis of milk fat and milk protein involves a number of genes. Acetyl-CoA carboxylase 1 (ACACA) is a key enzyme in the first step of fatty acid synthesis, this step is not a reversible reaction, and ACACA is the rate limiting enzyme of fatty acid synthesis. Fatty acid synthase (FASN) is an essential enzyme in fatty acid synthesis, and it catalyzes the synthesis of fatty acids from acetyl CoA and malonyl CoA through prolonging the short fatty acid carbon chain to a maximum of C16:0 [27]. The synthesized fatty acids will be activated by ACSL1 and enter the pathway of triglyceride synthesis. Our results showed that after LPS treatment, mRNA expression of milk fat genes such as ACACA and FASN was reduced. The study by Liu et al. showed that after LPS treatment of dairy cow mammary epithelial cells for 24 h, the gene expression of ACACA, cluster of differentiation 36 (CD36), fatty acid binding protein (FABP), FASN, peroxisome proliferator-activated receptor gamma (PPARγ), and sterol regulatory element binding protein 1 (SREBP1) involved in milk fat synthesis was significantly reduced compared with untreated cells [28].
Casein is the principal milk protein, accounting for 80% of the total milk protein. Casein consists of CSN1S1, CSN1S2, CSN2 and CSN3. The results of the present study showed that the mRNA expression of CSN1S1, CSN2 and CSN3 was not significantly affected by LPS. However, the mRNA expression of S6K1 was significantly downregulated by LPS. Ribosomal protein S6 kinase 1 is a key protein translation regulator and is one of the most studied downstream molecules of the mammalian target of rapamycin (mTOR) pathway. The mTOR signaling pathway is not only a signal pathway for amino acid-driven protein synthesis, but also a key signal pathway for hormones such as growth hormone and insulin to regulate milk protein synthesis. The mTOR primarily regulates the translation of mRNA into protein through downstream S6K1. Valérie et al. demonstrated that the phosphorylation of mTOR increased 30 min after LPS treatment of the peripheral blood mononuclear cells, which enhanced the mTOR pathway activity [29]. In their study it was unknown whether the expression of S6K1 was increased or decreased. In the present study, our results showed that after 48 h of LPS treatment, the expression of S6K1 gene was significantly decreased, which may be one of the key factors for LPS-induced decrease of milk protein secretion. In addition, Janus kinase/Signal transducer and activator of transcription 5 (JAK/STAT5) is also an important signaling pathway involved in milk protein synthesis. During milk protein synthesis, some hormones and cytokines bind to their receptors and activate STAT5 (including STAT5A and STAT5B), and the activated STAT5 enters the nucleus to bind to target genes to regulate transcription [30]. Signal transducer and activator of transcription 5 can be used as a marker of milk protein gene transcription level [31]. However, there was no difference in STAT5A gene expression between LPS treatment and control in this present study.
In our present study, SB was able to suppress the adverse effects of LPS on the mRNA expression of all milk genes (ACACA, ACSL1, ACSS2; STAT5A; S6K1; CSN1S1, CSN2 and CSN3) except that the FASN expression was only numerically increased after addition of SB into the LPS treatment. Therefore, SB, as an HDAC inhibitor, can significantly increase the lactation-related gene expression through counteracting the adverse effects of LPS on histone acetylation. Sodium butyrate appears to be likely to inhibit deacetylase activity due to its similarity to acetate. It may also be modified to butyryl-CoA, resulting in an increase in acetyl-CoA, which may alter acetylation levels through a mass action effect [32]. It is also worth mentioning that SB has proved very effective in reducing LPS-induced oxidative stress and apoptosis in cow mammary epithelial cells [33]. Sodium butyrate belongs to SCFAs, and because of its advantages in terms of high efficacy, low cost and high safety, it seems SB is a promising feed-in additive for improving lactation performance in dairy cows. More research is needed to validate and justify the use of SB for improving milk gene expression and for increasing lactation performance in cows exposed to LPS under various conditions such as high-concentrate feeding, poor hygienic environments, or/and mastitis and metritis in practical dairy production.
Conclusions
In the present study involving bovine mammary epithelial cells, we demonstrated that there existed a linear relationship between LPS doses and the HDAC activity. Moreover, the histone H3 acetylation levels were reduced by LPS, while the histone H4 acetylation levels were not significantly affected by LPS. Therefore, endotoxin was able to induce histone H3 hypoacetylation possibly through effecting an increased HDAC activity. Sodium butyrate, as an inhibitor of HDAC, effectively suppressed the endotoxin-induced decline of histone H3 acetylation, resulting in improved mRNA expression of lactation-related genes in the bovine mammary epithelial cells.
Methods
Cell culture and treatments
The bovine mammary epithelial cell line (MAC-T cells) was used in this study and the cell line establishment methods were previously described by Huynh et al. [34]. The composition of the culture medium and the cell culture method were as described in our previous study [35]. Prior to the following experiments, the MAC-T cells were checked for their viability to ensure the growth curve of the cells was normal and satisfactory (Additional file 5: Figure S1).
In the first experiment, MAC-T cells were treated (n = 6) with LPS (Escherichia coli O111:B4; Sigma, L2630, USA) at doses of 0, 1, 10, 100, 1000 endotoxin units (EU)/mL (1 EU = 0.1 ng), respectively. The treatment was repeated with another batch of MAC-T cells. The nuclear protein was extracted after 48 h treatment, and the acetylation levels of histones H3 and H4 as well as the HDAC activity were measured. The doses of LPS were set to cover a wide range of plasma LPS concentrations in cows under different conditions that vary from healthy status through SARA and diseases. Whereas LPS might not be detected in the peripheral plasma of healthy cows, it was present in the mammary plasma with a level of about 1 EU/mL in cows fed high-concentrate diets [9] and with a level of up to 870 EU/mL in severe mastitis cows (our unpublished results).
In the second experiment, the treatments (n = 6) of MAC-T cells were as follows: control, LPS (100 EU/mL), and LPS (100 EU/mL) plus SB (10 mmol/L; MedChemExpress, HY-B0350A, USA). The treatment was repeated with another batch of MAC-T cells. After 48 h treatment, the nuclear protein and RNA were extracted and the acetylation levels of histones H3 and H4 as well as milk gene mRNA expressions were determined.
Extraction of cell nuclear protein
After 48 h treatment, the cells were washed with DPBS (Solarbio, D1040–500, China) three times, digested with 0.25% trypsin/EDTA (Gibco, 25200056, USA), and centrifuged for 5 min at 3,000×g and room temperature to obtain clean cells. Histones were extracted with the cell nuclear protein extraction kit (Sangon Biotech Co., Ltd., C500009–0050, Shanghai, China). Briefly, cell membranes were lysed in a low osmotic and non-denatured system with protein phosphatases and protease inhibitors such as sodium pyrophosphate. Most of the cytoplasm protein and membrane protein were removed and the complete cell nucleus was extracted. Then the nucleus was decomposed using lysis buffer, and the cell nuclear proteins were extracted and purified. The BCA protein quantitative kit (Sangon Biotech Co., Ltd., C503021–0500, Shanghai, China) was then used to detect the extracted protein concentration in order to facilitate the subsequent accurate calculation.
Assay for acetylation levels of histones H3 and H4 and activity of HDAC
The acetylation of histones H3 and H4 was determined by the bovine histone acetylation H3/H4 ELISA kit (Jiangsu Meibiao Biological Technology Co., Ltd., MB-5290A/MB-5291A, China). The bovine HDAC ELISA kit (Jiangsu Meibiao Biological Technology Co., Ltd., MB-4783A, China) was used to determine the HDAC activity. The level of acetylated histone H3 (AH-H3) was determined by the double antibody sandwich method. The microplates are coated with the AH-H3 antibody to prepare the solid-phase antibody. Samples were added into the microplates and then combined with the horseradish peroxidase (HRP) conjugated AH-H3 antibody to form the antibody-antigen-enzyme labeled antibody complex. After several washings, the substrate tetramethylbenzidine (TMB) was added to produce color. TMB was converted to blue color under the catalysis of HRP, and finally turned yellow under acidic conditions. The color intensity was positively correlated with the AH-H3 in the samples. The optical density (OD) was measured at 450 nm by a microplate reader (Bio-Rad, xMark™, USA). The concentration of AH-H3 was calculated as per the standard curve. The principle and procedures for histone H4 acetylation and HDAC activity assays were the same as those of histone H3 acetylation assay.
RNA isolation and cDNA synthesis
The methodology used for RNA isolation and cDNA synthesis was described in detail in our previous study [35].
Quantitative real-time PCR
Amplification and quantification of the prepared cDNA were as described in our previous study [35]. Gene-specific primer pairs were designed using Primer Premier 5.0 software for the genes including CSN1S1, CSN2, CSN3, STAT5A, ACACA, FASN, S6K1, ACSL1, and ACSS2 (Table 1). The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as the internal control gene. Calculations of relative expression levels were performed using the 2-ΔΔCT method [36] by taking into account the values of 3 replicates per treatment and at least 3 duplicates for each replicate.
Table 1.
Primer sequences used for real-time quantitative PCR
| Gene | Primer | Product size (bp) | GenBank accession No. |
|---|---|---|---|
| CSN1S1 |
F: CTTTTCAGACAATTCTACCAGCT R: AATTCACTTGACTCCTCACCAC |
171 | NM_181029.2 |
| CSN2 |
F: AGTCCAAAGTCCTGCCTGTTCC R: TGCCATATTTCCAGTCGCAGTC |
193 | XM_015471671.2 |
| CSN3 |
F: CAATACGCTGTGAGAAAGATGA R: AACTGGTTTCTGTTGGTAGTAA |
122 | NM_174294.2 |
| STAT5A |
F: CATGTCCCTCAAGAGGATCA R: TCATTGCTGCCAACACTG |
108 | NM_001012673.1 |
| ACACA |
F: GATCCAGGCCATGCTAAG R: CTGTTTCTCCAGCCACTC |
103 | XM_024979607.1 |
| FASN |
F: AGGACCTCGTGAAGGCTGTGA R: CCAAGGTCTGAAAGCGAGCTG |
85 | XM_005220997.3 |
| S6K1 |
F: GGACATGGCAGGGGTGTTT R: GGTATTTGCTCCTGTTACTTTTCG |
283 | NM_205816.1 |
| ACSL1 |
F: GCCGCATTTCACTTTTACTGC R: AGCTCTTTAGGGCAAACCCC |
136 | NM_001076085.1 |
| ACSS2 |
F: GGGCGAATGCCTCTACTGC R: GCTGGGTGATGATGGATGG |
254 | NM_001105339.1 |
| GAPDH |
F: GGGTCATCATCTCTGCACCT R: GGTCATAAGTCCCTCCACGA |
177 | NM_001034034.2 |
CSN1S1: casein alpha s1; CSN2: casein beta; CSN3: casein kappa; STAT5A: signal transducer and activator of transcription 5A; ACACA: acetyl-CoA carboxylase 1; FASN: fatty acid synthase; S6K1: ribosomal protein S6 kinase 1; ACSL1: acyl-CoA synthetase long-chain family member 1; ACSS2: acyl-CoA synthetase short-chain family member 2; GAPDH: glyceraldehyde-3-phosphate dehydrogenase
Statistical analyses
Statistical analyses were performed to compare the mean differences among treatments by using one-way ANOVA of SPSS (v.22). Mean differences for all variables were separated and compared using Duncan’s multiple comparison procedure. And data are presented as means ± standard deviation. Significance was declared at P < 0.05, and a tendency was considered if 0.05 < P < 0.10.
Additional files
Table S1. Authors’ original data for Fig. 1. (XLSX 11 kb)
Table S2. Authors’ original data for Fig. 2. (XLSX 10 kb)
Table S3. Authors’ original data for Fig. 3. (XLSX 9 kb)
Table S4. Authors’ original data for Fig. 4. (XLSX 10 kb)
Figure S1. Growth curve of the MAC-T bovine mammary epithelial cells. The growth of cells was measured by using CCK-8 (Cell Counting Kit 8, Dojindo, Japan). For each point of time, there were 12 wells in the cell culture plate (n = 12). The optical density (OD) was determined at 450 nm on a microplate reader (Bio-Rad, xMark™, USA). (PDF 6 kb)
Acknowledgements
The authors are grateful to Professors Jianxin Liu and Hongyun Liu at the Institute of Dairy Science, Zhejiang University, for kindly providing the MAC-T cell line for this study. We also thank Professor Yongju Zhao and Dr. Caode Jiang of College of Animal Science and Technology, Southwest University, for their advice in designing this study and performing the laboratory assays.
Abbreviations
- ACACA
Acetyl-CoA carboxylase 1
- ACSL1
Acyl-CoA synthetase long-chain family member 1
- ACSS2
Acyl-CoA synthetase short-chain family member 2
- AH-H3
Acetylated histone H3
- CD36
Cluster of differentiation 36
- CSN1S1
Casein alpha s1
- CSN2
Casein beta
- CSN3
Casein kappa
- EU
Endotoxin unit
- FABP
Fatty acid binding protein
- FASN
Fatty acid synthase
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- HRP
Horseradish peroxidase
- IL
Interleukin
- LPS
Lipopolysaccharide
- mTOR
Mammalian target of rapamycin
- PPARγ
Peroxisome proliferator-activated receptor gamma
- S6K1
Ribosomal protein S6 kinase 1
- SARA
Subacute ruminal acidosis
- SB
Sodium butyrate
- SCFA
short-chain fatty acid
- SREBP
Sterol regulatory element binding protein 1
- STAT
Signal transducer and activator of transcription
- TMB
Tetramethylbenzidine
- TNF
tumor necrosis factor
Authors’ contributions
JC, YW, YS, XD and GD conceived the study and designed the experiment. JC, YW, YS, ZW, ZZ and YX conducted the experiments and performed the laboratory analyses. JC, YW and GD wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant number: 31672448) and Fundamental Research Funds for the Central Universities (Grant number: XDJK2018D008). The whole study including experiment design, cell culture, sample detection, data analysis, and writing of this manuscript was supported by Grant 31672448. Part of the sample detection was supported by Grant XDJK2018D008.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
The study was approved by Southwest University Laboratory Animal Welfare and Ethics Committee. This study did not involve the use of animals. The bovine mammary epithelial cell line (MAC-T cells) used in this experiment was provided by Professors Jianxin Liu and Hongyun Liu at the Institute of Dairy Science, Zhejiang University, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jingbo Chen and Yongjiang Wu contributed equally to this work.
Contributor Information
Jingbo Chen, c10417511@email.swu.edu.cn.
Yongjiang Wu, wuyongjiang@email.swu.edu.cn.
Yawang Sun, Email: syaw507@swu.edu.cn.
Xianwen Dong, chenghuafen@email.swu.edu.cn.
Zili Wang, Email: wzl9698@swu.edu.cn.
Zhu Zhang, Email: b20020901102@swu.edu.cn.
Yanli Xiao, xyl616856826@email.swu.edu.cn.
Guozhong Dong, Email: gzdong@swu.edu.cn.
References
- 1.Dong GZ, Liu SM, Wu YX, Lei CL, Zhou J, Zhang S. Diet-induced bacterial immunogens in the gastrointestinal tract of dairy cows: impacts on immunity and metabolism. Acta Vet Scand. 2011;53:48. doi: 10.1186/1751-0147-53-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plaizier JC, Khafipour E, Li S, Gozho GN, Krause DO. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim Feed Sci Technol. 2012;172:9–21. doi: 10.1016/j.anifeedsci.2011.12.004. [DOI] [Google Scholar]
- 3.Eckel EF, Ametaj BN. Invited review: role of bacterial endotoxins in the etiopathogenesis of periparturient diseases of transition dairy cows. J Dairy Sci. 2016;99:5967–5990. doi: 10.3168/jds.2015-10727. [DOI] [PubMed] [Google Scholar]
- 4.Plaizier JC, Krause DO, Gozho GN, McBride BW. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. 2009;176:21–31. doi: 10.1016/j.tvjl.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Dong GZ, Ao CJ, Zhang S, Qiu M, Wang X, et al. Feeding a high-concentrate corn straw diet increased the release of endotoxin in the rumen and pro-inflammatory cytokines in the mammary gland of dairy cows. BMC Vet Res. 2014;10:172. doi: 10.1186/s12917-014-0172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khafipour E, Krause DO, Plaizier JC. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J Dairy Sci. 2009;92:1060–1070. doi: 10.3168/jds.2008-1389. [DOI] [PubMed] [Google Scholar]
- 7.Emmanuel DGV, Dunn SM, Ametaj BN. Feeding high proportions of barley grain stimulates an inflammatory response in dairy cows. J Dairy Sci. 2008;91:606–614. doi: 10.3168/jds.2007-0256. [DOI] [PubMed] [Google Scholar]
- 8.Gozho GN, Krause DO, Plaizier JC. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J Dairy Sci. 2007;90:856–866. doi: 10.3168/jds.S0022-0302(07)71569-2. [DOI] [PubMed] [Google Scholar]
- 9.Dong GZ, Qiu M, Ao C, Zhou J, Erdene K, Wang X, et al. Feeding a high-concentrate corn straw diet induced epigenetic alterations in the mammary tissue of dairy cows. PLoS One. 2014;9:e107659. doi: 10.1371/journal.pone.0107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ning LT, Dong GZ, Ao C, Zhang DG, Erdene K, Zhang FQ, et al. Effects of continuous low dose infusion of lipopolysaccharide on inflammatory responses, milk production and milk quality in dairy cows. J Anim Physiol Anim Nutr. 2018;102:e262–e269. doi: 10.1111/jpn.12737. [DOI] [PubMed] [Google Scholar]
- 11.Weerapan K, Min-Sun K, Memon RA, Shigenaga JK, Moser AH, Feingold KR, et al. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J Lipid Res. 2004;45:1169–1196. doi: 10.1194/jlr.R300019-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;12:1997–2006. doi: 10.1194/jlr.R200015-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 14.Tony K. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Tchurikov NA. Molecular mechanisms of epigenetics. Biochemistry. 2005;70:406–423. doi: 10.1007/s10541-005-0131-2. [DOI] [PubMed] [Google Scholar]
- 16.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;1:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 17.Cao DS, Wang ZG, Zhang CL, Oh J, Xing W, Li S, et al. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Elsasser TH, Li RW. Epigenetic regulation in bovine cells: nutrient-induced modulation of gene expression and cellular functions. In: Rechi LJ, editor. Animal genetics. New York: Nova Science Publishers, Inc.; 2009. pp. 153–173. [Google Scholar]
- 19.Li CJ, Li RW, Elsasser TH. MicroRNA (miRNA) expression is regulated by butyrate-induced epigenetic modulation of gene expression in bovine cells. Genet Epigenet. 2010;3:23–32. doi: 10.4137/GEG.S6144. [DOI] [Google Scholar]
- 20.Kumar P, Mohan V, Sinha R, Chagtoo M, Godbole MM. Histone deacetylase inhibition reduces hypothyroidism-induced neurodevelopmental defects in rats. J Endocrinol. 2015;227:83–92. doi: 10.1530/JOE-15-0168. [DOI] [PubMed] [Google Scholar]
- 21.Doherty R, O’Farrelly C, Meade KG. Epigenetic regulation of the innate immune response to LPS in bovine peripheral blood mononuclear cells (PBMC) Vet Immunol Immunopathol. 2013;154:102–110. doi: 10.1016/j.vetimm.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Wu S, Li RW, Li W, Li CJ. Transcriptome characterization by RNA-seq unravels the mechanisms of butyrate-induced epigenomic regulation in bovine cells. PLoS One. 2012;7:e36940. doi: 10.1371/journal.pone.0036940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Li C, Huang W, Li W, Li RW. Alternative splicing regulated by butyrate in bovine epithelial cells. PLoS One. 2012;7:e39182. doi: 10.1371/journal.pone.0039182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angrisano T, Pero R, Peluso S, Keller S, Sacchetti S, Bruni CB, et al. LPS-induced IL-8 activation in human intestinal epithelial cells is accompanied by specific histone H3 acetylation and methylation changes. BMC Microbiol. 2010;10:172. doi: 10.1186/1471-2180-10-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aung HT, Schroder K, Himes SR, Brion K, van Zuylen W, Trieu A, et al. LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J. 2006;20:1315–1327. doi: 10.1096/fj.05-5360com. [DOI] [PubMed] [Google Scholar]
- 26.Xing SP, Nie F, Xu QY, Deng YX, Li W, Yang ZW, et al. HDAC is essential for epigenetic regulation of Thy-1 gene expression during LPS/TLR4-mediated proliferation of lung fibroblasts. Lab Investig. 2015;95:1105–1116. doi: 10.1038/labinvest.2015.97. [DOI] [PubMed] [Google Scholar]
- 27.Wakil SJ, Stoops JK, Joshi VC. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;1:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 28.Liu LX, Lin Y, Liu LL, Bian YJ, Zhang L, Gao XJ, et al. 14-3-3γ regulates lipopolysaccharide-induced inflammatory responses and lactation in dairy cow mammary epithelial cells by inhibiting NF-κB and MAPKs and up-regulating mTOR signaling. Int J Mol Sci. 2015;16:16622–16641. doi: 10.3390/ijms160716622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valérie S, Saman A, Garcia IA, Knoll ML, Joseph C, Bulger EM, et al. Role of the mTOR pathway in LPS-activated monocytes: influence of hypertonic saline. J Surg Res. 2011;171:769–776. doi: 10.1016/j.jss.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- 31.Selvaggi M, Dario C, Normanno G, Celano GV, Dario M. Genetic polymorphism of STAT5A protein: relationships with production traits and milk composition in Italian Brown cattle. J Dairy Res. 2009;76:441–445. doi: 10.1017/S0022029909990070. [DOI] [PubMed] [Google Scholar]
- 32.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–121. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Wang HH, Nie XT, Jiang WR, Zhang YS. Sodium butyrate ameliorates lipopolysaccharide-induced cow mammary epithelial cells from oxidative stress damage and apoptosis. J Cell Biochem. 2019;120:2370–2381. doi: 10.1002/jcb.27565. [DOI] [PubMed] [Google Scholar]
- 34.Huynh HT, Robitaille G, Turner JD. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp Cell Res. 1991;197:191–199. doi: 10.1016/0014-4827(91)90422-Q. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Wu Y, Sun Y, Dong X, Wang Z, Zhang Z, Xiao Y, Dong G. Bacterial lipopolysaccharide induced alterations of genome-wide DNA methylation and promoter methylation of lactation-related genes in bovine mammary epithelial cells. Toxins. 2019;11:298. doi: 10.3390/toxins11050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2−△△CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Authors’ original data for Fig. 1. (XLSX 11 kb)
Table S2. Authors’ original data for Fig. 2. (XLSX 10 kb)
Table S3. Authors’ original data for Fig. 3. (XLSX 9 kb)
Table S4. Authors’ original data for Fig. 4. (XLSX 10 kb)
Figure S1. Growth curve of the MAC-T bovine mammary epithelial cells. The growth of cells was measured by using CCK-8 (Cell Counting Kit 8, Dojindo, Japan). For each point of time, there were 12 wells in the cell culture plate (n = 12). The optical density (OD) was determined at 450 nm on a microplate reader (Bio-Rad, xMark™, USA). (PDF 6 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.