Proliferative control in cancer cells is frequently disrupted by mutations in the retinoblastoma protein (RB) pathway. Intriguingly, RB1 mutations can arise late in tumorigenesis in cancer cells whose RB pathway is already compromised by another mutation. In this study, we present evidence for increased DNA damage and instability in cancer cells with RB pathway defects when RB1 mutations are induced.
KEYWORDS: cell cycle, chemotherapy, haploinsufficiency, metastasis, mitosis
ABSTRACT
Proliferative control in cancer cells is frequently disrupted by mutations in the retinoblastoma protein (RB) pathway. Intriguingly, RB1 mutations can arise late in tumorigenesis in cancer cells whose RB pathway is already compromised by another mutation. In this study, we present evidence for increased DNA damage and instability in cancer cells with RB pathway defects when RB1 mutations are induced. We generated isogenic RB1 mutant genotypes with CRISPR/Cas9 in a number of cell lines. Cells with even one mutant copy of RB1 have increased basal levels of DNA damage and increased mitotic errors. Elevated levels of reactive oxygen species as well as impaired homologous recombination repair underlie this DNA damage. When xenografted into immunocompromised mice, RB1 mutant cells exhibit an elevated propensity to seed new tumors in recipient lungs. This study offers evidence that late-arising RB1 mutations can facilitate genome instability and cancer progression that are beyond the preexisting proliferative control deficit.
INTRODUCTION
Loss of proliferative control is a defining feature of human cancer. Most cancer cells develop cell-intrinsic mechanisms of supplying growth-stimulatory signals as well as disrupting the response to cell cycle arrest cues (1). To this end, mutations in the retinoblastoma protein (RB) pathway are central to disrupting proliferative control in tumorigenesis (2–4). Deletion of the RB1 gene prevents cell cycle arrest in response to a broad range of signals (3). Similarly, overexpression or hyperactivation of D-type cyclins and their associated cyclin-dependent kinases (CDKs) can lead to constitutive RB phosphorylation and cell cycle entry. Finally, deletion or promoter methylation of CDKN2A that encodes the CDK inhibitor p16 serves to deregulate kinase activity, causing constitutive phosphorylation of RB. Cancer cell genomes that sustain a single mutation in this pathway are considered to have disrupted RB pathway function and are deficient for cell cycle control (3, 5, 6). Historically, this concept of RB pathway inactivation suggested that mutations in different components of the pathway are relatively equivalent and that additional mutations provide no subsequent advantage to cancer progression (2, 3, 6, 7).
A number of recent clinical observations challenge the logic of single RB pathway mutations in cancer. First, multiple studies have shown that RB1 loss is specifically predictive of a favorable response to chemotherapy (8–11), whereas p16 expression or overall proliferative rates are not (8–10). This suggests that RB pathway mutations are not necessarily equivalent. Second, a number of studies have suggested that RB1 gene loss is more prevalent in advanced cancers or mechanistically contributes to progression or dissemination (12–15), a stage where cell-autonomous proliferative control is presumably already deregulated. Collectively, these examples suggest that RB1 mutation contributes more to tumor progression than just alterations to proliferative control and that RB1 loss may confer other cancer-relevant characteristics. Remarkably, some studies even highlight that single-copy loss of RB1 may be functionally significant (12, 16–18).
Beyond RB’s role in cell cycle control through E2F transcriptional regulation, it has been reported to participate in a host of functions that contribute to genome stability (19). These include chromosome condensation through RB-dependent recruitment of condensin II and cohesin (20, 21). The RB protein also influences the repair of DNA breaks through both nonhomologous end joining (NHEJ) (22) and homologous recombination (HR) (23) and through induction of mitochondrial biogenesis that impacts cell metabolism (24–26). Some of these functions, such as repair of DNA breaks by HR, are obligatorily outside RB’s role in G1-to-S-phase regulation. In addition, other roles, such as effects on mitochondrial biogenesis and metabolism, take place in proliferating populations of cells, further suggesting that this is independent of G1/S regulation and the RB pathway. It is noteworthy that some atypical RB functions in genome stability, or late-stage cancer progression, may be sensitive to single-copy loss (16–18, 27). Thus, the existence of shallow RB1 deletions may indicate that RB’s less-well-appreciated functions in genome stability could underlie cancer-relevant characteristics that are independent of classical RB pathway function in cancer (7).
In order to test if RB1 loss is relevant to cancer cells that already possess RB pathway disruption, we induced mutations in RB1 using CRISPR/Cas9. These cells displayed spontaneous DNA damage as evidenced by γH2AX foci and elevated levels of reactive oxygen species (ROS). We also determined that RB1 mutations decreased the ability to repair DNA breaks by homologous recombination, and this is supported by elevated levels of anaphase bridges in mitosis. RB1 mutant cells were xenografted into immunocompromised mice, and this revealed similar growth kinetics in subcutaneous implantation, with RB1-null cells showing a greater propensity to colonize lungs. These experiments underscore the discovery that RB1 mutation in cells that already possess RB pathway disruption creates DNA damage and fuels cancer progression.
RESULTS
Spontaneous DNA damage in RB1-deficient cancer cells.
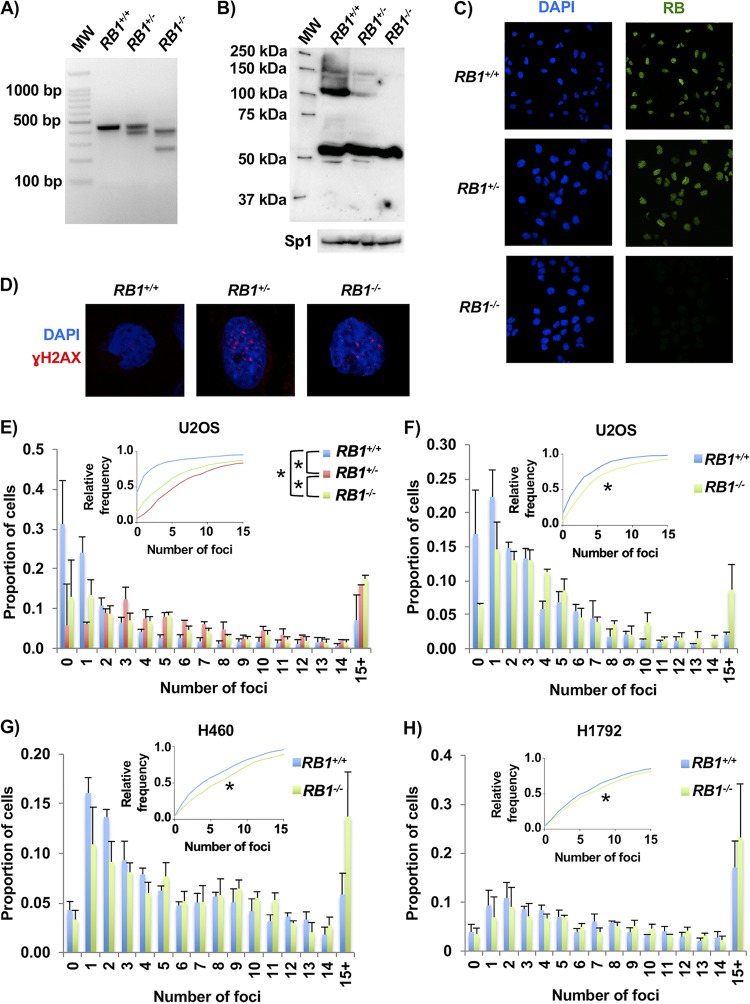
To investigate RB1 deficiency in RB pathway-disrupted cells, we utilized a number of cell lines that are reported to be defective for p16, the product of the CDKN2A gene, or that possess activation of cyclin D/CDK4. Initially, we used p16-deficient U2OS cells (28) and Cas9 with guide RNA (gRNA) pairs that target exon 22 of RB1 because loss of this exon creates null alleles in cancer (29). Cells were transfected with plasmids to deliver pairs of gRNAs and Cas9 (or the D10A mutant). Following transient drug selection, colonies were isolated, expanded, and genotyped by PCR to search for RB1 deletions (Fig. 1A). Candidates were rigorously selected by checking RB expression by Western blotting (Fig. 1B), ensuring that heterozygous clones were not mixtures of wild-type and knockout cells using fluorescent RB staining (Fig. 1C), and confirming that the most likely off-target sites were not mutated (Table 1). Using this approach, we selected four clones each for wild-type and knockout RB1 genotypes and three clones for the RB1+/− genotype, which were used in subsequent experiments.
FIG 1.
CRISPR/Cas9-induced mutations in RB1 cause DNA damage. (A) Ethidium bromide-stained agarose gel showing examples of wild-type, heterozygous, and homozygous mutant RB1 genotypes that are detected by PCR amplification of exon 22 sequences. MW, molecular weight. (B, top) Representative Western blot showing RB expression in control, heterozygous, and homozygous mutant cells. (Bottom) Sp1 loading control. (C) Immunofluorescence microscopy was used to detect RB expression (green) in cultures of control, heterozygous, or homozygous mutants. Cells were counterstained with DAPI to visualize nuclei (blue). (D) Representative confocal microscopy images of γH2AX foci (red) in control, heterozygous, and homozygous RB1 mutant cells. Cells were counterstained with DAPI to visualize nuclei (blue). (E) Counts of γH2AX foci for each of the U2OS RB1 genotypes. The average proportions of cells with discrete numbers of foci are shown as histograms, while the cumulative frequency of foci for each genotype is shown in the inset. The average distributions of foci for RB1 wild-type (4 different clones), heterozygous (3 different clones), and knockout (4 different clones) cells were compared using the Kolmogorov-Smirnov test (*, P < 0.05). (F) U2OS cells were transfected with CRISPR/Cas9 constructs targeting either a safe-harbor site in the genome or exon 2 of the RB1 gene. Three clones were selected under both control and knockout conditions, and γH2AX foci were quantified by fluorescence microscopy. The average proportions of γH2AX foci for both RB1 wild-type and knockout genotypes are shown as histograms, while the cumulative relative frequency of foci is shown in the inset. Focus distributions were again compared by a Kolmogorov-Smirnov test. (G) H460 lung cancer cells were stained for γH2AX foci and analyzed as described above for panel F. (H) H1792 non-small cell lung cancer cells were analyzed as described above for panel F. All error bars are +1 standard error of the mean (SEM). *, P < 0.05.
TABLE 1.
Characterization of the U2OS RB1 mutant clones generated by targeting exon 22a
| Clone | Cas9 form | RB1 genotypeb | Predicted proteinc | Off-target genotyped |
|
|---|---|---|---|---|---|
| ALDH1L1 | ZNF699 | ||||
| U2OS | NA | Wild type | WT | NA | NA |
| WT | |||||
| 21B2 | D10A | Wild type | WT | ND | ND |
| WT | |||||
| 21B4 | D10A | Wild type | WT | WT | WT |
| WT | |||||
| 11B5 | D10A | Wild type | WT | ND | ND |
| WT | |||||
| 4C1 | D10A | Heterozygous | WT | WT | WT |
| p.Y771fsX8 | |||||
| 21D5 | D10A | Heterozygous | WT | WT | WT |
| p.Q770HdelX10 | |||||
| 21A1 | D10A | Heterozygous | WT | WT | WT |
| p.I753delX12 | |||||
| 12C3 | D10A | Null | p.I752IfsX9 | WT | WT |
| p.E748CdelX11 | |||||
| 5A5 | WT | Null | p.N757TX31 | WT | WT |
| p.P776VdelX6 | |||||
| 6B1 | WT | Null | p.Y756YfsX14 | WT | WT |
| p.S751SfsX10 | |||||
| 5C4 | WT | Null | p.S773FfsX14 | WT | WT |
| p.Y756delX | |||||
NA, not applicable; ND, not determined; WT, wild type.
Genotypes were determined by PCR and sequencing.
Amino acid coding changes were predicted based on nucleotide sequences.
The top-scoring off-target intragenic locations determined for each gRNA, ZNF699 for gRNA X22B and ALDH1L1 for gRNA X22C, were also sequenced to probe for unwanted mutations.
To determine if RB1 mutation status affects genome stability in these engineered cell lines, DNA damage was assessed in untreated, proliferating cells by staining for γH2AX. Foci were visualized by immunofluorescence (IF) microscopy, and images were captured using confocal microscopy (Fig. 1D). The quantity of foci per nucleus was determined, and this revealed a significant increase in γH2AX in the knockout and heterozygous lines compared to those that are wild type for RB1 (Fig. 1E).
We quantified the abundance of γH2AX foci in additional RB pathway mutant cell lines to determine if increased DNA damage is a common consequence of RB loss. We used cells that were modified with control nontargeting gRNAs or that were produced by targeting exon 2 of RB1. RB deficiency in these cells was confirmed by Western blotting (data not shown). Using RB1-null U2OS cells produced in this manner, we again observed increased quantities of γH2AX foci in RB-deficient cells compared to their controls (Fig. 1F). In addition, clones of control and RB1 knockout NCI-H460 (CDKN2A-deleted) and NCI-H1792 (CDK4-amplified) lung cancer cells also displayed differences in spontaneous DNA damage (Fig. 1G and H). This analysis demonstrated an elevation in the number of γH2AX foci in each of the RB1-deleted lines.
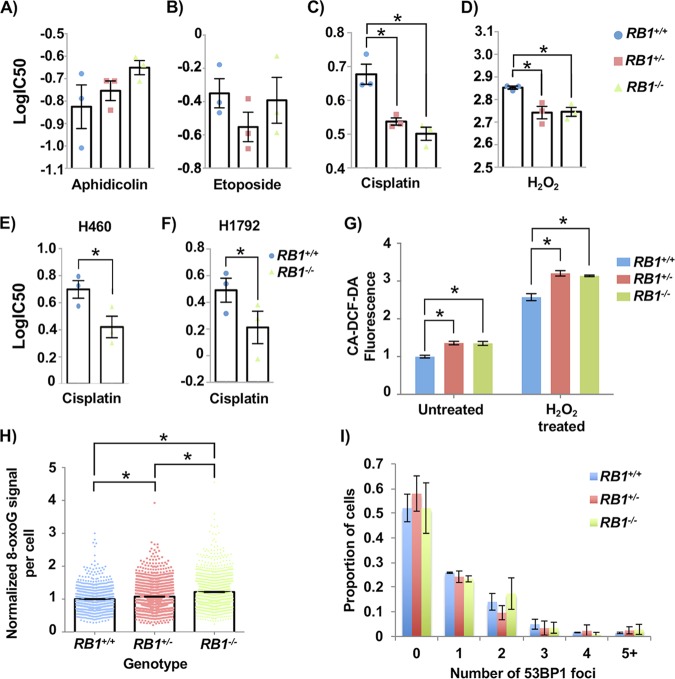
To further investigate the source of DNA damage in RB1 mutant cells, we assessed their sensitivity to a number of chemical agents to determine if specific stresses could amplify defects that cause increased DNA damage. We tested aphidicolin (APH), a DNA polymerase inhibitor that causes replication stress, and etoposide, a topoisomerase inhibitor that creates DNA double-stranded breaks. Hydrogen peroxide (H2O2) was used to induce oxidative damage, and cisplatin was used to create interstrand cross-links, among other damaging effects. Representative U2OS clones of each genotype were treated for 72 h with a range of chemical concentrations, after which alamarBlue was used to quantitate the cytotoxicity of each agent. These assays revealed that both heterozygous and homozygous RB1 mutations sensitize cells to hydrogen peroxide and cisplatin but not aphidicolin or etoposide (Fig. 2A to D). Because platinum therapeutics preferentially benefit patients with RB-deficient cancers (8, 9), we also tested control and RB1−/− H460 and H1792 cells for their sensitivity to cisplatin. This revealed increased sensitivity to cisplatin in H460 and H1792 cells upon RB1 deletion (Fig. 2E and F).
FIG 2.
Cancer cells with RB1 mutations have elevated reactive oxygen species levels and sensitivity to cisplatin. (A to D) Aphidicolin (A), etoposide (B), cisplatin (C), and hydrogen peroxide (D) were added to cultures of the indicated genotypes of U2OS cells. Viability was assessed after 72 h using alamarBlue, and dose-response curves were used to calculate IC50 values for each genotype. Both RB1 mutant genotypes have significantly lower IC50 values in response to cisplatin and hydrogen peroxide than control U2OS cells (as determined by one-way ANOVA). (E and F) H460 and H1792 cancer cells with RB1 mutations were treated with cisplatin as described above for panel C. Differences in IC50 values were determined using a paired t test. (G) To detect ROS, CA-DCF-DA was added to culture medium at the end of 72 h of mock or hydrogen peroxide treatment. Normalized fluorescence was averaged for four clones of RB1 wild-type and knockout genotypes and for three clones for the heterozygous genotype. Mean values were compared by two-way ANOVA. (H) Cells were fixed and stained for 8-oxoG and DAPI and visualized by fluorescence microscopy. The average 8-oxoG signal per nucleus was determined using ImageJ, with DAPI staining defining the nuclear area. Three clones per genotype were used, and data were normalized to the mean signal from RB1 wild-type cells in duplicate experiments. Statistical significance in staining intensity was determined by Kruskal-Wallis one-way analysis of variance and Dunn’s multiple-comparison test. (I) 53BP1 foci were quantitated for each RB1 genotype using Focinator as with γH2AX. No significant differences were observed, as determined by the Kolmogorov-Smirnov test. At least 120 cells from one U2OS clone were analyzed in duplicate experiments for each RB1 genotype. All error bars are ±1 SEM. *, P < 0.05.
Overall, these drug sensitivities suggest that oxidative damage may underlie some aspects of the DNA damage phenotype in RB1 mutant cells. We compared ROS levels in wild-type and RB1 mutant U2OS cells with and without H2O2 using an ROS indicator, 5(6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (CA-DCF-DA). For both untreated and H2O2-treated cells, there was more fluorescence of the ROS indicator in RB1 mutant cells, as RB1−/− and RB1+/− cells were equivalent (Fig. 2G). We also fixed and stained cells for 8-oxoguanine (8-oxoG), one of the most abundant lesions resulting from oxidative modification of DNA (30), and quantified the staining in 4′,6-diamidino-2-phenylindole (DAPI)-defined nuclear areas using ImageJ (31). Control U2OS values were used to normalize the 8-oxoG signal from RB1 mutants. Again, both RB1+/− and RB1−/− U2OS cells had more 8-oxoG staining than did RB1+/+ cells (Fig. 2H).
These experiments indicate that loss of RB1 in cells with preexisting RB pathway defects increases basal levels of DNA damage. Reactive oxygen species appear to be one source of this damage. A chemical agent that directly induces breaks (etoposide) did not selectively affect RB1 mutant cells; however, sensitivity to a DNA cross-linking agent (cisplatin) suggests a potential inability to repair DNA damage by homologous recombination. Both observations are consistent with a lack of 53BP1 foci, a marker of NHEJ, in RB1 mutant U2OS cells compared to controls (Fig. 2I). This suggests that the nature of DNA damage marked by γH2AX in these RB1 mutant cells is not necessarily double-stranded DNA breaks and that NHEJ is not the dominant pathway to repair damage in these cells. Overall, these experiments indicate that RB1 loss contributes to an unstable genome, regardless of the proliferative control status of the cell.
RB1 mutant cells have randomly distributed DNA damage.
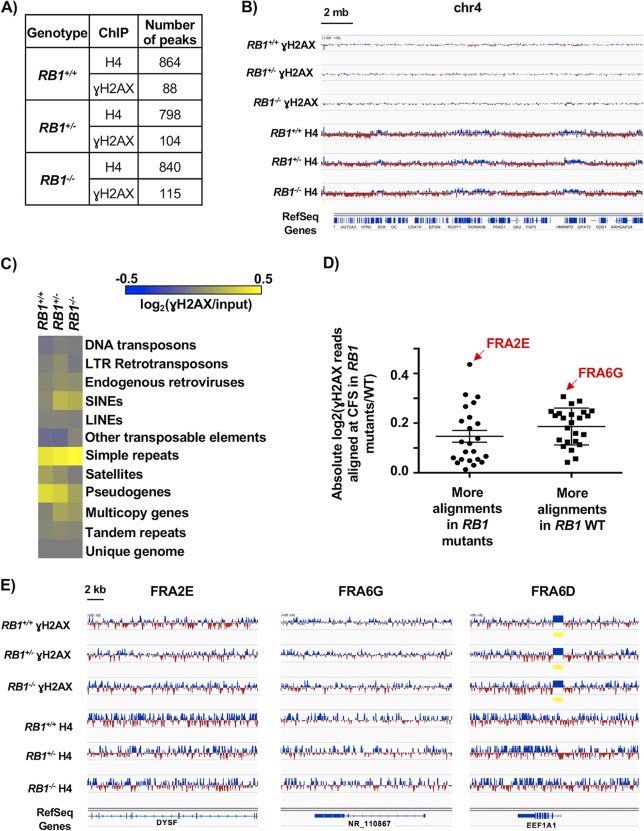
To further understand spontaneous DNA damage in RB1 mutant U2OS cells, we sought to determine if damage occurred at specific locations within the genome. We performed chromatin immunoprecipitation sequencing (ChIP-Seq) to identify DNA sequences associated with γH2AX as well as histone H4 as a control. Because spontaneous damage in untreated cell cultures is relatively rare, we pooled chromatin from 20 separate γH2AX ChIP experiments per genotype to create each sequencing library (Fig. 3A to E). We determined peak locations and numbers using model-based analysis for ChIP-Seq (MACS) (32), and the quantities of γH2AX and H4 peaks were similar between genotypes (Fig. 3A). Looking at a large region of chromosome 4 as a representative view of the genome, we did not observe consequential differences between genotypes for γH2AX or H4 peaks (Fig. 3B). Since peak finding at a genome scale failed to indicate obvious locations of DNA damage enrichment, we investigated individual genome sequence categories in search of elevated γH2AX deposition. The proportions of aligned γH2AX ChIP-Seq reads per million mapped reads versus input reads for each of the genotypes were compared within each repetitive element category and log2 transformed for display as a heat map (Fig. 3C). Some categories, such as short interspersed nuclear elements (SINEs) and multicopy genes, appear to have slight enrichment of γH2AX localization in RB1+/− and RB1−/− compared to RB1+/+ cells based on color, but a two-tailed one-sample t test with false discovery rate (FDR) multitest correction did not score these as significant. Therefore, even within repetitive sequences in the genome, there does not seem to be an enrichment of γH2AX within RB1 mutant cells compared to controls.
FIG 3.
γH2AX is randomly distributed in the genomes of RB1 mutant cells. (A) Total number of MACS peaks found for H4 control and γH2AX ChIP-Seq reads for the indicated genotypes. (B) A 20-Mb region of chromosome 4, with ChIP-Seq read alignments for γH2AX and H4. Tracks were normalized by subtracting input reads. Blue indicates more reads in the ChIP versus the input, and red indicates fewer reads. (C) Determination of numbers of ChIP-Seq reads mapping to repetitive sequences as well as unique genome regions. The heat map shows the log2 ratios of the abundances of γH2AX-precipitable reads per million mapped reads versus the input for each of the respective genotypes at each element analyzed. LTR, long terminal repeat; SINEs, short interspersed nuclear elements; LINEs, long interspersed nuclear elements. (D) Aligned γH2AX ChIP-Seq read proportions within common fragile sites (CFS) were first normalized to their respective inputs, and RB1+/− and RB1−/− values were then normalized to wild-type (WT) values and log2 transformed. A two-tailed one-sample t test was performed to determine if the normalized mean read count proportions of the RB1 mutants at the various CFS are equal to the normalized read count proportions of the corresponding CFS in the wild type. CFS where the false discovery rate was less than 0.1 were grouped according to whether there were significantly more alignments in the RB1 mutants or significantly more alignments in the RB1+/+ sample. There is no significant difference between these two categories (determined by an unpaired t test). FRA2E had the most reads in RB1+/− and RB1−/− cells compared to the control, while FRA6G had the most reads in the wild type compared to the mutants. Error bars are ±1 SEM. (E) ChIP-Seq tracks for γH2AX and H4 at representative CFS. FRA2E and FRA6G from panel D are shown, while FRA6D had no change in the proportion of γH2AX reads that aligned between the genotypes. Regions of significant enrichment (MACS peaks) are denoted by yellow bars.
Previous studies suggest that γH2AX levels can be elevated at common fragile sites (CFS) of cancer cells under standard cell culture conditions because of replication stress (33). To investigate these locations, we quantified the number of γH2AX ChIP-Seq reads, scaled them to the proportions of total aligned reads, and normalized them to input levels. RB1+/− and RB1−/− cells were compared to the wild type using a two-tailed one-sample t test. This analysis revealed 23 CFS that had more γH2AX alignments than the wild type and 24 CFS that had significantly fewer in RB1 mutant cells than in controls (Fig. 3D). Figure 3E shows examples of CFS locations with the greatest increase in γH2AX in RB1 mutants (FRA2E), the greatest reduction in γH2AX in RB1 mutants (FRA6G), and unchanged γH2AX levels (FRA6D). These examples appear highly similar between genotypes. Overall, it is possible that the distribution of γH2AX within each CFS is shifting slightly between the mutants and the wild type. However, there does not appear to be more of a bias in general for γH2AX elevation at CFS in RB1 mutant cells.
Collectively, our analysis of γH2AX distribution across the genome suggests that there is no particular chromosome location or sequence category that is preferentially enriched for this mark of DNA damage. These data suggest that the increase in γH2AX foci observed in RB1+/− and RB1−/− cells is likely due to an overall increase in DNA damage and not newly arising locations, or “hot spots,” of damage. This is in contrast to primary cells with a normal RB pathway that experience RB loss and preferentially damage centromeric repeats (16). The elevated sensitivity to peroxide and cisplatin and increased ROS and 8-oxoG levels are consistent with DNA damage being randomly located in RB1 mutant U2OS cells.
Homologous recombination repair defects in RB1-deficient cancer cells.
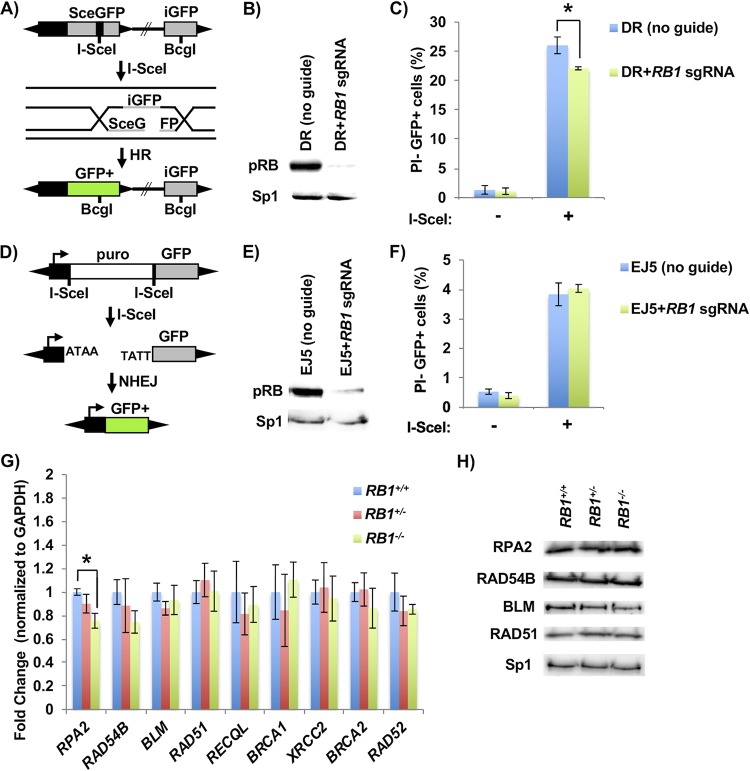
Another potential source of intrinsic DNA damage could arise from defective repair. For this reason, we investigated the efficiency of HR and NHEJ repair using fluorescent reporters (34, 35). In this assay, a promoterless, but functional, green fluorescent protein (GFP) is used to repair an adjacent break induced in a mutant, expressed, form of the GFP gene (Fig. 4A). We generated clonal U2OS lines bearing this reporter and created a population of cells deleted for RB1 with lentiviral delivery of Cas9 and an RB1-specific gRNA (Fig. 4B). Introduction of the restriction enzyme I-SceI into these cells induced breaks, and RB1-deficient cells were defective for their repair (Fig. 4C). Similarly, we generated U2OS clones that stably maintain an NHEJ reporter for repair of an induced break that links a constitutive promoter with a GFP gene. Loss of RB expression was again confirmed by Western blotting (Fig. 4E), and induction of breaks was used to test repair in an RB1-deficient background (Fig. 4F). This failed to reveal a defect in repair, suggesting that RB1 loss in U2OS cells specifically reduces HR repair.
FIG 4.
Defective homology-directed repair of DNA breaks in RB1 mutant cells. (A) Schematic of the DR-GFP homology-directed repair construct used. Cleavage of an I-SceI site integrated into an expressed, but mutant, GFP gene (SceGFP) can be repaired from a downstream internal GFP fragment (iGFP). (B) U2OS cells with clonal integration of the HR reporter construct were ablated for RB1 expression with lentiviral delivery of Cas9 and an RB1-specific sgRNA. The relative expression of RB in the population of cells was determined by Western blotting, and Sp1 serves as a loading control. (C) HR repair efficiency of RB1 mutant U2OS cells was determined by transfecting an expression vector for I-SceI endonuclease (+), or a relevant negative-control expression vector, and quantitating PI-negative (PI−) and GFP-positive (GFP+) cells by flow cytometry (n = 5). All error bars are ±1 SEM. *, P < 0.05. (D) Schematic of the EJ5 NHEJ reporter system. DNA breaks at tandem I-SceI sites release the puromycin resistance gene, allowing NHEJ repair to join a promoter to the GFP-expressing sequence. (E) After the generation of a stable U2OS clone containing the NHEJ reporter construct, RB1 was deleted as described above and confirmed by Western blotting. (F) NHEJ repair efficiency was determined by transfecting an I-SceI endonuclease expression vector (+), or the negative control, and PI-negative and GFP-positive cells were quantitated by flow cytometry (n = 3). (G) RT-qPCR was performed to assess the transcript levels of various HR factors in RB1 wild-type (4 different clones), heterozygous (3 different clones), and knockout (4 different clones) cells. Statistical differences in means were determined by one-way ANOVA. All error bars are ±1 SEM. *, P < 0.05. (H) Western blots showing expression of various HR factors in wild-type, heterozygous, and homozygous RB1 mutant clones. The Sp1 loading control is shown on the bottom.
To investigate if this reduction in HR repair is related to changes in the expression of HR factors due to RB1 loss, we examined DNA damage repair genes that are known to be regulated by E2Fs (36, 37). Of these candidates, only one gene, RPA2, was seen to have significantly lower transcript levels in RB1−/− U2OS cells (Fig. 4G). However, when protein levels of a subset of these factors, including RPA2, were investigated, no changes in expression between genotypes were evident (Fig. 4H). This suggests that the expression levels of key HR factors are not changed in these cells as a result of RB1 loss.
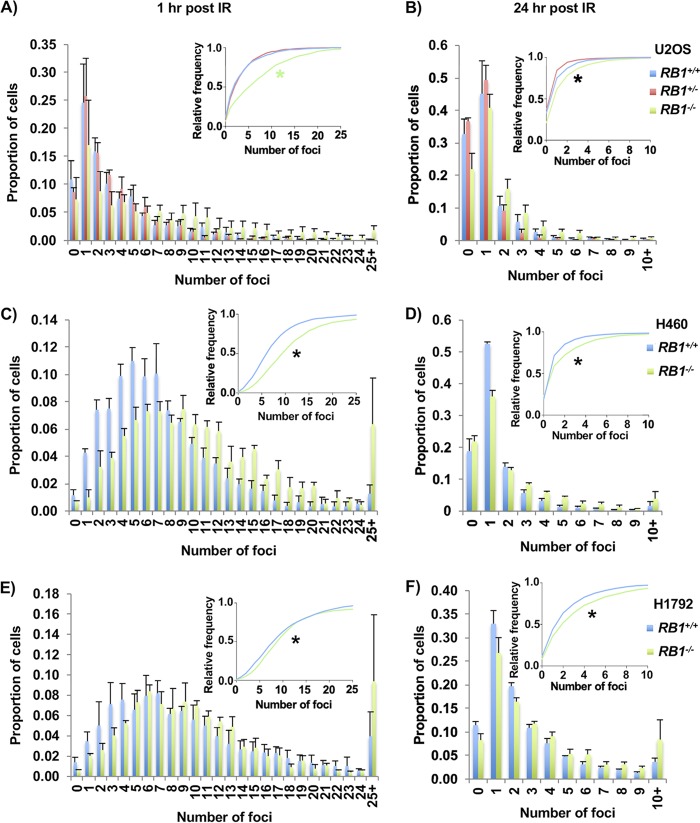
To assess how RB1 mutant U2OS cells respond to DNA breaks, each genotype was exposed to 2 Gy of gamma irradiation (γIR). Cells were fixed and stained for γH2AX and DAPI to measure the amount of DNA damage. One hour after γIR, despite pronounced γH2AX foci in all genotypes, the amount of DNA damage was significantly larger in RB1−/− clones than in heterozygous or wild-type cells (Fig. 5A). After 24 h, most of the DNA damage was repaired, and the RB1−/− cells retained more γH2AX foci than cells of the other two genotypes (Fig. 5B). This same increase in DNA damage in the RB1-null background was seen in H460 cells (Fig. 5C and D) and in H1792 cells (Fig. 5E and F), both 1 h and 24 h after exposure to 1 Gy of γIR. Overall, this indicates that cells completely lacking RB are more sensitive to γIR, likely because they are not able to repair DNA breaks as efficiently by HR repair.
FIG 5.
Defective repair of γIR-induced DNA damage in RB1 knockout cells. (A and B) U2OS cells were treated with 2 Gy γIR, fixed 1 h (A) or 24 h (B) after treatment, and stained for γH2AX. Three clones per genotype were used, and γH2AX foci were quantified. The proportions of cells with discrete numbers of foci are shown as histograms, while the cumulative frequency of foci is shown in the inset. Differences in the distributions of foci were determined using the Kolmogorov-Smirnov test. A green asterisk indicates that RB1−/− is statistically different from the other genotypes, while a black asterisk indicates that all genotypes are statistically significantly different from each other. (C and D) H460 cells were treated with 1 Gy γIR, fixed 1 h (C) or 24 h (D) after treatment, and stained for γH2AX. (E and F) H1792 cells were treated with 1 Gy γIR, fixed 1 h (E) or 24 h (F) after treatment, and stained for γH2AX. All error bars are +1 SEM. *, P < 0.05.
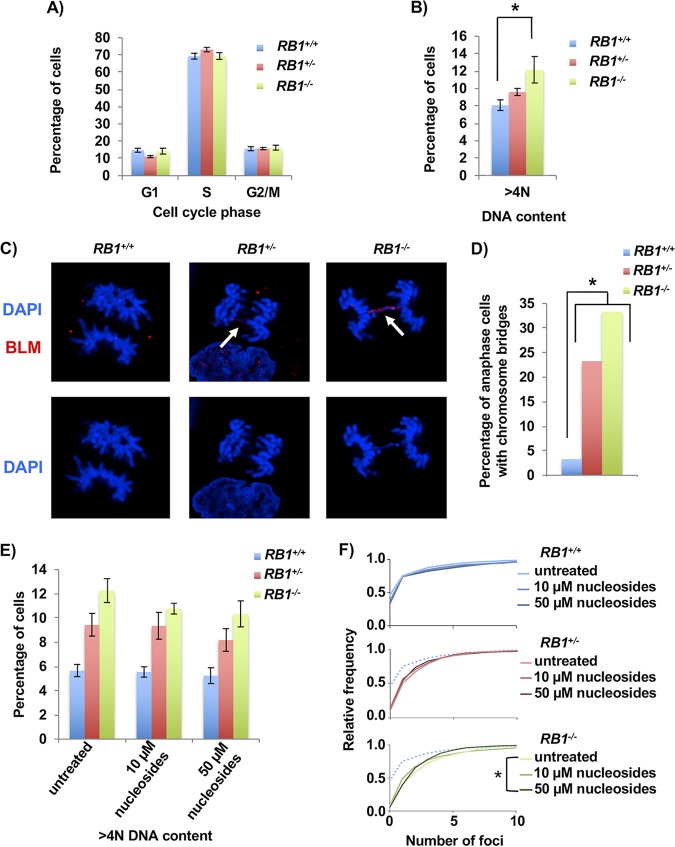
Next, we investigated the fidelity of mitosis to determine if the elevated levels of DNA damage and impaired HR repair impacted chromosome segregation and aneuploidy (38). Flow cytometry was performed on RB1-deficient cells that were labeled and stained with bromodeoxyuridine (BrdU) and propidium iodide (PI) (39). This analysis failed to show statistically different changes in cell cycle phases between the different genotypes (Fig. 6A). However, when DNA content greater than 4N was analyzed, there was a significant difference between wild-type and RB1 knockout cells, with RB1+/− cells showing an intermediate value (Fig. 6B). This suggests that mitotic errors in these cells may lead to aneuploidy.
FIG 6.
Increased mitotic errors in RB1 mutant cells. (A) BrdU and propidium iodide staining followed by flow cytometry was used to determine the cell cycle phase distribution of asynchronous cultures of U2OS cells. Four clones for the wild-type and knockout genotypes and three clones for the heterozygous genotype were analyzed. (B) Flow cytometry analysis showing the proportion of cells with >4N DNA content. Means were compared using one-way ANOVA. (C) Cells in anaphase were imaged by fluorescence microscopy using DAPI (blue) and BLM (red) in cells from each RB1 genotype. Arrows indicate anaphase bridges that are stained by both DAPI and BLM. (D) Quantitation of the numbers of anaphase cells with DAPI-stained chromosome bridges. The proportion of cells with bridges is significantly higher in the knockout and heterozygous clones than in the wild-type clone, as determined by the χ2 test. (E) Representative clones from each RB1 genotype were either left untreated or treated with 10 μM or 50 μM nucleosides for 48 h. Flow cytometry analysis of propidium iodide-stained cells shows the proportion of cells with >4N DNA content. Mean differences were compared by one-way ANOVA (n = 3). (F) Nucleoside-treated cells were fixed and stained for γH2AX after 48 h of culture. DNA damage is summarized in frequency plots. The dotted line in the RB1+/− and RB1−/− cumulative frequency plots represents the wild-type untreated cells. Statistical significance between genotypes was determined by the Kolmogorov-Smirnov test. All error bars are ±1 SEM. *, P < 0.05.
To further investigate mitotic defects and their relationship with DNA damage and replication stress, cells were stained with DAPI and antibodies to Bloom syndrome protein (BLM) to visualize chromosome bridges, and mitotic figures were imaged using confocal microscopy (Fig. 6C). We observed abundant chromosome bridges in RB1−/− and RB1+/− mutant cells (Fig. 6D). In the RB1 mutants, there were some ultrafine bridges (UFBs), which are “threadlike” DNA structures that stain only with BLM (40). In RB1+/− anaphase cells, 5% (2/43) were seen to have UFBs, and in RB1−/− cells, this was increased to 8% (3/39), whereas in wild-type cells, UFBs were not observed (0/30). However, the majority of BLM bridges were stained with DAPI, indicating that anaphase bridges were most common. Anaphase bridges are known to occur in HR-defective cells, while UFBs can be induced by replication stress (38).
Finally, to investigate the link between UFBs in the RB1 mutant cells and a possible increase in replication stress, we performed a nucleoside supplementation assay and investigated changes in DNA content and DNA damage as surrogate markers for nucleoside suppression of replication stress. Flow cytometry of cells stained with propidium iodide was performed 48 h after medium replacement, either with or without the addition of nucleosides. This analysis failed to show statistically different changes in >4N DNA content within genotypes when comparing the different nucleoside treatment groups, although there was a trend toward decreased aneuploidy after nucleoside supplementation for all genotypes (Fig. 6E). However, when we looked at DNA damage of cells grown in parallel, RB1−/− cells treated with 50 μM nucleosides had a significant reduction in γH2AX foci compared to untreated knockout cells (Fig. 6F). Nevertheless, this reduction in DNA damage after nucleoside supplementation is modest and did not restore the levels of γH2AX foci to those seen in the RB1+/+ controls.
Taken together with experiments described above, defects in mitosis are best characterized as anaphase bridges that cause aneuploidy. These types of errors are consistent with HR repair deficiency and sensitivity to a DNA cross-linking agent such as cisplatin. It appears that γH2AX foci can be suppressed slightly in RB1−/− cells with nucleosides, and RB1−/− cells exhibit some UFBs, implying that they experience modest DNA replication stress. However, the lack of sensitivity to aphidicolin and the lack of increased γH2AX deposition at repetitive genome regions argue that replication stress in these cells is quite modest. Overall, our analysis of mitotic errors is consistent with a defect in HR repair being the main source of chromosome bridges in anaphase of these RB1 mutant cells.
Increased lung metastases in RB1 mutant xenografts.
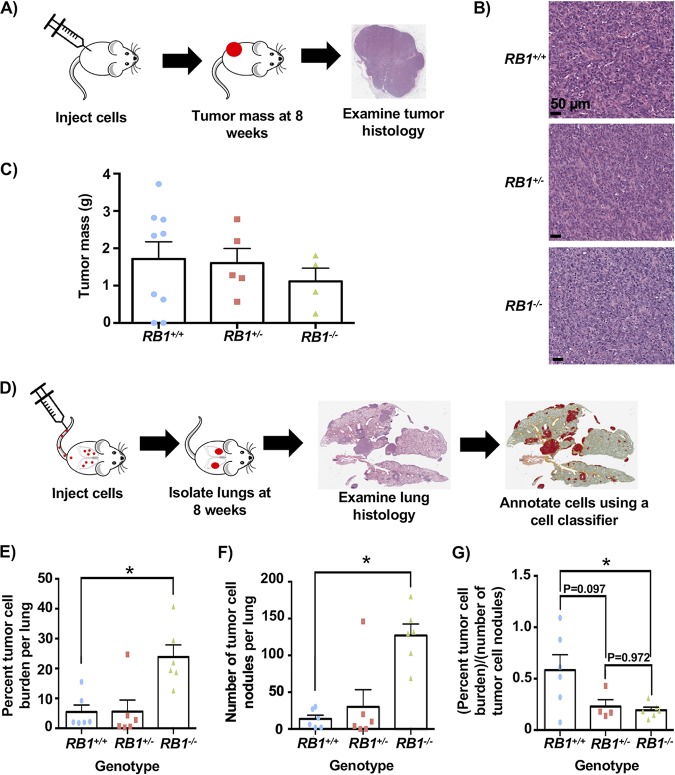
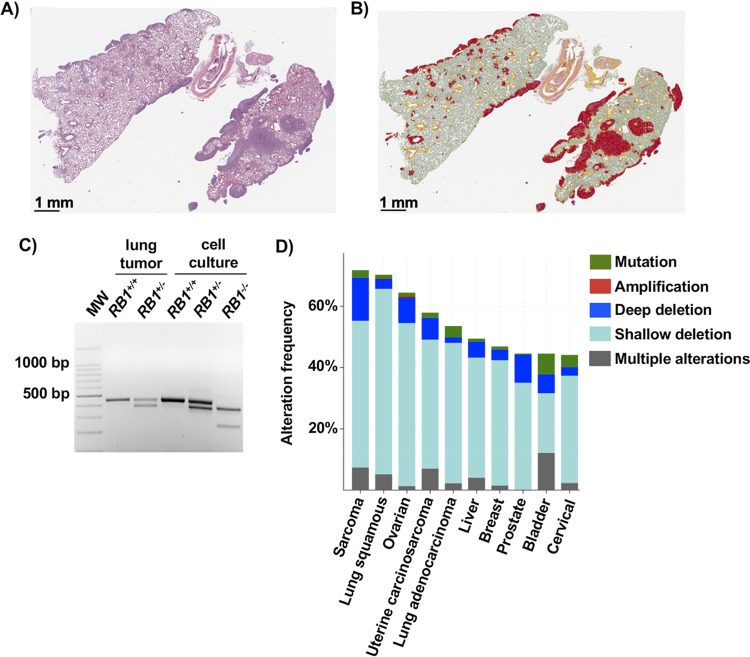
To further characterize the effects of induced RB1 mutations in cells that already possess RB pathway defects, we performed xenograft experiments to determine if new cancer-relevant properties arise upon the loss of RB. We injected cells subcutaneously into immunocompromised mice and allowed tumors to form for an 8-week period before analyzing growth by mass and histology (Fig. 7A). This analysis revealed highly cellular tumors with abundant mitotic activity and foci of necrosis. Cells appeared epithelioid, with no definite features of osteoid differentiation, and they had small areas suggesting glandular differentiation. This phenotype was consistent among all genotypes (Fig. 7B). Tumor masses were determined at the endpoint and were not statistically different between genotypes, with RB1−/− cells even trending toward a smaller size (Fig. 7C). Mice were also tail vein injected, and cell dissemination and proliferation were allowed to proceed for 8 weeks, at which time lungs were harvested, fixed, and sectioned. Hematoxylin and eosin (H&E)-stained sections were digitally analyzed to quantitate cellular infiltration of U2OS cells (Fig. 7D). This revealed a striking increase in RB1−/− U2OS cells in the lungs of these mice compared to control and RB1+/− cells, as determined by the percentage of the lung section area that is occupied by malignant cells (Fig. 7E). There were significantly more individual nodules of RB1−/− cells per lung than for the other genotypes (Fig. 7F), further suggesting that RB1 loss increased the efficiency of dissemination or establishment in the lung. Finally, the area occupied by tumor cells per nodule is lower in both RB1 mutant genotypes, indicating that control cells form rarer, larger clusters of cells, whereas RB1 mutants tend to seed more efficiently and perhaps proliferate more slowly (Fig. 7G). We note that one mouse injected with RB1+/− cells showed extensive dissemination and tumor burden that were highly reminiscent of mice injected with RB1−/− cells (Fig. 8A and B). To investigate this further, we extracted DNA from paraffin-embedded tumor material from this mouse. PCR analysis was used to genotype RB1 exon 22 in these cells (Fig. 8C). This confirmed that these cells maintained their wild-type RB1 allele, suggesting that they had not undergone loss of heterozygosity as a means to acquire an RB1−/− phenotype.
FIG 7.
Increased lung metastases with RB1 mutant cells in xenograft experiments. (A) Illustration of analysis of subcutaneous injections of RB1 mutant U2OS cells. Tumors were allowed to form for 8 weeks before analysis of tumor mass and histology. (B) Representative H&E-stained tissue sections from each genotype of tumor. (C) Tumor masses from subcutaneously injected cells. The means are not statistically different. (D) Schematic of tail vein injections to study dissemination to the lungs. Mice were injected with cells, and cell dissemination and proliferation were allowed to proceed for 8 weeks. Lungs were then isolated, sectioned, stained with H&E, and analyzed using QuPath. (E) The percentage of lung area occupied by cancer cells was calculated from tissue sections and averaged between mice. (F) Tumor cell nodules were counted using the assistance of QuPath and averaged among recipient mice. (G) The percent tumor cell burden was divided by the number of tumor nodules to determine the average tumor cell area per nodule. Statistical significance between genotypes was determined with a t test. All error bars are +1 SEM. *, P < 0.05.
FIG 8.
Single-copy loss of RB1 can create cancer-enabling phenotypes. (A) H&E staining of lung tissue with the highest number of RB1+/− seeding events. (B) QuPath coloring to denote tumor tissue within these lungs (red). (C) DNA was extracted from nodules of tumor cells in paraffin-embedded tissue containing these RB1+/− cells or an RB1+/+ control. PCR was performed to amplify exon 22 from recovered DNA and controls isolated from cell culture to verify the final genotype of cells in this sample. (D) The 10 most prevalent cancers were analyzed for RB1 gene alterations using TCGA and Pan-Cancer Atlas data, using cBioPortal. A deep deletion is consistent with biallelic loss of RB1, whereas a shallow deletion is suggestive of heterozygous RB1 deletion.
These xenograft experiments indicate that RB1 mutations in U2OS cells have little effect on the growth rate of primary tumors. Interestingly, RB1−/− cells are much more efficient in colonizing recipient mouse lungs, suggesting that in addition to the DNA damage and genome instability phenotypes described above, RB1 loss in RB pathway-deficient cells imparts characteristics that enable cancer progression.
DISCUSSION
To test if RB1 loss impacts cancer cells that already possess mutations disrupting the RB pathway, we used CRISPR/Cas9 to create nonfunctional RB1 alleles. Using γH2AX staining of cells, we found that even within cells deficient for one copy of RB1, there is an increase in spontaneous DNA damage. Experiments that probe drug sensitivity of RB1 mutant cells, along with ChIP-Seq analysis of γH2AX foci and analysis of DNA damage repair pathways, suggest a complex picture of cellular defects. We did not detect specific hot spots of DNA damage, but sensitivity to peroxide and cisplatin suggests that oxidative damage may cause sporadic DNA damage, as levels of reactive oxygen species are elevated in RB1-deficient cells. Mutant RB1 cells also have more abnormal mitoses characterized by anaphase bridges, reporter assays detect defects in HR repair, and nucleoside supplementation in culture medium suppresses γH2AX foci, suggesting additional endogenous sources of damage. Overall, this study reveals that RB1 mutations lead to increased DNA damage that may enhance cancer progression.
The DNA damage phenotype caused by either single-copy or homozygous mutation to RB1 is unlikely to be attributable to a single root cause, and we expect that RB1 deficiency may affect DNA damage in different cancer cells in various ways. We report that RB1 deletion compromises HR repair but not NHEJ, and this is consistent with one previous report (23) but contradicts another (22). Given that we observe more than 70% of U2OS cells incorporating BrdU in a brief pulse, they are likely biased toward HR repair pathways because of their cell cycle position. This is consistent with γH2AX foci not being accompanied by 53BP1, and the activity levels of NHEJ reporters being almost an order of magnitude lower than HR values, suggesting that U2OS cells are primed to use the HR repair pathway, and thus, phenotypes in these cells reflect this reality. We expect that the DNA damage and defective repair described in this report are relevant to cancer progression phenotypes because graded differences between RB1 wild-type, heterozygous, and homozygous genotypes are reflected in 8-oxoguanine abundance, aneuploidy, and anaphase bridges. This stepwise trend in severity of phenotype in the molecular alterations resulting from RB1 loss is similarly evident in the behavior of RB1+/+, RB1+/−, and RB1−/− mutant tail vein xenograft experiments, suggesting that this increase in metastatic burden is related to a stepwise increase in DNA damage experienced by these different genotypes.
Single-copy loss of RB1 may contribute to cancer in a number of ways. Primary RB1+/− cells from a number of sources are prone to mitotic errors (16–18), and precursor lesions to retinoblastoma are characterized by aneuploidy (41), suggesting that this is an early step in this disease. Therefore, partially defective RB1 can contribute to the early stages of cancer through a distinct set of effects. However, a number of studies have highlighted that RB1 loss is statistically enriched in advanced stages of cancer progression (12–14). In addition, analysis of the landscape of cancer alterations in The Cancer Genome Atlas (TCGA) consortia reveals that so-called “shallow deletions” of RB1 are relatively commonplace (Fig. 8D), and these events are unlikely to be explained by random, unselected events (42). From this perspective, our study offers a critical proof of concept that late-stage loss of one copy of RB1 can create cancer-enabling phenotypes in the host cell, even if it already possesses RB pathway mutations. Complete elimination of RB1 has a stronger effect on DNA damage phenotypes and dissemination to the lungs, as demonstrated in this study; however, highly abundant single-copy RB1 loss may represent a mutational compromise in which advantageous phenotypes are acquired with an economy of mutational changes (43).
A number of studies correlate the absence of RB expression with improved patient survival following treatment that includes platinum-based chemotherapy (8, 9). Loss of RB correlated with improved survival of patients with lung adenocarcinomas treated by resection and adjuvant cisplatin or carboplatin and a vinca alkaloid (8). More recently, patients with high-grade serous ovarian cancer (HGSC) who experienced exceptionally good clinical outcomes were studied (9). These patients were treated with platinum-based agents, and loss of RB was associated with long-term survival. From this perspective, U2OS cells engineered to be deficient for RB1 demonstrate that RB loss increases sensitivity to cisplatin. Given that RB1 mutant cells were not more sensitive to another agent that induces DNA breaks, etoposide, we interpret RB1 deficiency to create a unique sensitivity to cisplatin that may relate to defective HR and higher endogenous ROS levels that create a highly specific sensitivity to this class of chemotherapy.
In conclusion, although there are many cancers that have mutations in the RB pathway that spare the RB1 gene itself, further mutations to RB1 surprisingly create cancer-relevant characteristics that may influence disease progression. The frequency of shallow deletions of RB1 across many cancers suggests that disease progression may select for these characteristics.
MATERIALS AND METHODS
Cell culture.
U2OS cells and the resulting clones were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. H460 and H1792 cells and the resulting clones were grown in RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. Cells were grown at 37°C in humidified air containing 5% CO2.
Generation of RB1 deletions using CRISPR.
For creation of RB1 deletions, single guide RNAs (sgRNAs) targeting exon 22 of RB1 were selected by using the CRISPR design tool at http://crispr.mit.edu/ (44). The sgRNA sequences were as follows: 5′-CACCGTATTATAGTATTCTATAACT-3′ (X22B-top), 5′-AAACAGTTATAGAATACTATAATAC-3′ (X22B-bottom), 5′-CACCGAGGATACTTTTGACCTACCC-3′ (X22C-top), and 5′-AAACGGGTAGGTCAAAAGTATCCTC (X22C-bottom). The X22B and X22C guides were each cloned into the pX459 plasmid (with wild-type Cas9; Addgene plasmid 48139) and the pX462 plasmid (with the D10A mutant version of Cas9; Addgene plasmid 48141) Both plasmids contain a puromycin resistance cassette. Cells were seeded at a density of 105 cells per well in a 6-well dish, and the next day, a total of 1 μg per well of a 1:1 mix of X22B and X22C CRISPR plasmids (either pX459 or pX462) was transfected by use of X-tremeGENE HP DNA transfection reagent (Roche). The next day, each well was replated onto a 15-cm dish, and the day after that, cells were cultured in selection medium with 2 μg/ml of puromycin for 2 days. Following this, cells were grown in normal cell culture medium for approximately 12 days, following which single colonies were picked from the 15-cm plates using mechanical detachment with a pipette tip, placed into wells of a 48-well dish, and allowed to grow. Cells were further passaged onto larger plates and genotyped using the following primers: X22F (5′-TTACTGTTCTTCCTCAGACATTCAA-3′) and X22R (5′-GGATCAAAATAATCCCCCTCTCAT-3′). PCR products (445 bp for the wild-type band) were run in an agarose gel, and individual bands were gel purified using a Sigma GenElute gel extraction kit and sent for Sanger sequencing using the X22F and X22R primers described above. For clones with multiple PCR products, bands were purified and sent for sequencing separately to determine individual RB1 alleles in each clone. For alleles that could not easily be resolved by gel electrophoresis, PCR products were cloned into vectors using either the TOPO TA cloning kit for sequencing (Invitrogen) or the CloneJET PCR cloning kit (Thermo Scientific).
The top-scoring off-target intragenic locations determined for each gRNA using the CRISPR Design tool were also sequenced to probe for mutations. gRNA X22B had a potential off-target site in ZNF699 (X22B_OT_ZNF699_F [5′-GTGCCCTAAAACACTGAGGGA-3′] and X22B_OT_ZNF699_R [5′-TTTATGATCAACAAGGACCAGAGC-3′]), while X22C has a potential off-target site in ALDH1L1 (X22C_OT_ALDH1L1_F [5′-GCCACGCTATGCTTGTGATG-3′] and X22C_OT_ALDH1L1_R [5′-CACCCCAGAGAAGGGAACAC-3′]). PCR products were gel purified as described above and sent for Sanger sequencing using their respective primers.
Nuclear extracts were prepared from U2OS CRISPR clones of interest, and Western blotting was carried out using previously described protocols (45). Antibodies raised against RB (clones G3-245 [BD Pharmingen] and C-15 [Santa Cruz]) and Sp1 (clone H-225; Santa Cruz) were used for Western blotting. Samples were Western blotted using standard techniques.
To generate additional RB1 knockout and control cell lines, sgRNAs targeting either exon 2 of RB1 (5′-GGAGAAAGTTTCATCTG-3′) or a gene desert region of the genome (5′-TGAGCCTATATTAATTGG-3′) were utilized. The sgRNAs were cloned into the lentiCRISPR v2 vector (Addgene plasmid 52961), which also encodes Cas9. To generate lentivirus, HEK293T cells were transfected with the sgRNA vector and a 1:1:1 mixture of lentiviral packaging constructs (Addgene plasmids 12251, 12253, and 8454) using polyethylenimine transfection reagent. Twenty-four hours after transfection, the HEK293T medium was replaced, and recipient cells (U2OS, NCI-H1792, and NCI-H460) were seeded for infection. The following day, medium on the recipient cells was replaced with lentiviral medium, and Polybrene was added at a final concentration of 8 μg/ml. A second infection was performed the next day. Infected cells were then selected with 2 μg/ml puromycin for 3 days. To generate isogenic clones, populations of knockout (or control) cells were sorted using a fluorescence-activated cell sorter (FACS) as single cells in 96-well plates (BD FACSAria II) and allowed to grow for approximately 2 weeks. Colonies were then expanded and screened for loss of RB by an immunoassay using the Simple Western system according to the manufacturer’s instructions. Successful knockout clones were also genotyped to confirm clonogenic origin. Genomic DNA was extracted using the PureLink genomic DNA minikit (Invitrogen), and the region surrounding the cut site was amplified by PCR using the following primers: X2F (5′-TCACAGAAGTGTTTTGCTGCTT-3′) and X2R (5′-TTTGGTGGGAGGCATTTATGGA-3′). PCR products were purified using the DNA Clean and Concentrator kit (Zymo Research) and sent for Sanger sequencing.
Fluorescence microscopy.
Cells grown either on glass coverslips or in glass-bottom plates were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 10 min and then permeabilized with PBS–0.3% Triton X-100 for 10 min at room temperature. The fixed cells were blocked in blocking buffer (PBS–0.3% Triton X-100 with either 5% donkey or goat serum depending on the species in which the secondary antibodies were raised) for at least 1 h at room temperature. Cells were then incubated with primary antibody in blocking buffer at room temperature for 1 h or at 4°C overnight. Antibodies raised against RB (clone G3-245; BD Pharmingen), γH2AX (clone JBW301; EMD Millipore), 53BP1 (H-300; Santa Cruz), and BLM (C-18; Santa Cruz) were used for IF. After 3 washes with PBS–0.3% Triton X-100, cells were incubated with secondary antibody diluted in blocking buffer for 1 h at room temperature. Cells were washed twice with PBS–0.3% Triton X-100, incubated with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS–0.3% Triton X-100 for 5 min, washed twice more with PBS–0.3% Triton X-100, and then washed once with PBS before mounting with Slowfade gold antifade mountant (catalog number S36936; Thermo Fisher Scientific).
For 8-oxoG staining, cells were fixed and blocked as described above, washed with PBS–0.3% Triton X-100, and incubated in an RNase solution (0.2 mg/ml RNase A, 10 mM Tris-HCl [pH 7.5], 15 mM NaCl, and 0.1% Triton X-100 in 1× PBS) for 1 h at room temperature. Cells were washed with PBS–0.3% Triton X-100 and then incubated in 2 M HCl for 10 min at room temperature, followed by a rinse with 50 mM Tris-HCl (pH 8.0). Cells were washed with PBS–0.3% Triton X-100, primary antibody incubation was then performed using anti-DNA/RNA damage antibody raised against 8-oxoG (clone 15A3, catalog number ab62623; Abcam), and all subsequent steps were completed as described above.
For confocal microscopy of RB, γH2AX, and BLM, cells were examined on an Olympus FluoView FV1000 confocal microscope system. For confocal microscopy of 53BP1, a Nikon A1R confocal microscope was used. For nonconfocal microscopy, images were acquired using a Zeiss Axioskop 40 microscope and a Spot flex camera. Foci were quantified using Focinator (46), while overall staining intensity in cells was quantified by using ImageJ (31).
Gamma irradiation of cells.
Cells subjected to γIR were plated at 100,000 cells per well in 6-well dishes with glass coverslips on the bottom. The next day, cells were exposed to a cobalt 60 source until a dose of either 2 Gy or 1 Gy was received. Cells were placed back in the cell culture incubator until the appropriate time point after treatment to fix cells for IF.
NHEJ and HR repair assays.
For the HR repair assay, pDRGFP was used, which was a gift from Maria Jasin (Addgene plasmid 26475 [http://n2t.net/addgene:26475]; Research Resource identifier [RRID], Addgene_26475), and for the NHEJ assay, pimEJ5GFP was used, which was a gift from Jeremy Stark (Addgene plasmid 44026 [http://n2t.net/addgene:44026]; RRID, Addgene_44026). pDRGFP was linearized using EcoRV, and pimEJ5GFP was linearized using XhoI. These linearized fragments were then individually used for transfection using Lipofectamine 3000 transfection reagent (Invitrogen) into U2OS cells. The next day, each well was replated onto a 10-cm dish, and a day later, cells were cultured in selection medium with 2 μg/ml of puromycin for 3 days. To isolate single-cell colonies, limiting dilutions were then used to seed cells into 96-well plates. After approximately 3 weeks, wells with growth from single-cell isolates were transferred to single wells of 12-well plates and after a few days were treated with puromycin again to ensure that the selected clones still contained either the NHEJ or HR constructs.
To determine the reporter efficiency in the isolated clones, 2 sets of transfections were performed per clone, again using Lipofectamine 3000 transfection reagent (Invitrogen). For the first set of transfections, each clone was transfected with a plasmid expressing the I-SceI endonuclease, pCBASceI, which was a gift from Maria Jasin (Addgene plasmid 26477 [http://n2t.net/addgene:26477]; RRID, Addgene_26477), and a blasticidin marker, pMSCV-Blasticidin, which was a gift from David Mu (Addgene plasmid 75085 [http://n2t.net/addgene:75085]; RRID, Addgene_75085). The second set of transfections was performed with an empty backbone plasmid, pCAG-FALSE, which was a gift from Wilson Wong (Addgene plasmid 89689 [http://n2t.net/addgene:89689]; RRID, Addgene_89689), and pMSCV-Blasticidin. For both of these transfections, the blasticidin resistance plasmid was used in a 1:3 ratio with the complementary plasmid. The next day, each well was replated onto a 10-cm dish, and the day after that, cells were cultured in selection medium with 10 μg/ml of blasticidin for 1 week. GFP-positive cells were then quantified by flow cytometric (FACS) analysis. To prepare cells for FACS analysis, they were washed with PBS, trypsinized, resuspended in culture medium, and washed twice with PBS. Cell pellets were then resuspended in 0.5 ml of flow cytometry staining buffer with propidium iodide (0.05% sodium azide and 0.5% bovine serum albumin [BSA] in 1× PBS with 0.01 mg/ml propidium iodide). For each reporter construct, the clone with the highest ratio of the GFP signal when transfected with pCBASceI to the GFP signal when transfected with pCAG-FALSE was selected for future studies.
To introduce CRISPR constructs into selected clones for each repair reporter, lentivirus particles were generated in HEK293T cells. Lentivirus was created for both lentiCRISPR v2 with no guide RNA inserted and lentiCRISPR v2 with the X22B sgRNA sequences for RB1 (described above) inserted. lentiCRISPR v2 was a gift from Feng Zhang (Addgene plasmid 52961 [http://n2t.net/addgene:52961]; RRID, Addgene_52961). The X22B RB1 guide sequences were inserted into the lentiCRISPR v2 plasmid as previously described (47, 48). Culture media containing lentiviral particles were transferred to appropriate U2OS HR and NHEJ reporter clones for 48 h, followed by selection with 4 μg/ml puromycin for at least 5 days.
The populations of U2OS HR and NHEJ reporter clones that were infected with lentiCRISPR v2 plasmids, either with or without the RB1 guide, were then transfected with both pCBASceI and pMSCV-Blasticidin or pCAG-FALSE and pMSCV-Blasticidin, selected as described above, and analyzed by FACS analysis to determine repair efficiency. These transfections were performed in the same transduced population of cells, but for experimental replicates, the cells were seeded at different times and then subsequently transfected with plasmids. Cells grown in parallel to the transfected cells were used to prepare nuclear extracts for Western blotting.
Expression of HR factors.
Total RNA from cells was isolated using TRIzol reagent according to a standard protocol (Invitrogen). First-strand cDNA synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad). Isolated cDNA was used in reverse transcription-quantitative PCRs (qRT-PCRs) with iQ SYBR green supermix (Bio-Rad), using the following primers: RPA2_F (5′-CGAAAGCTATGGCAGCTCCT-3′), RPA2_R (5′-GGCTCGGGCTCTTGATTTCT-3′), RAD54B_ F (5′-GCGAGGGGATAGCTGGTTAC-3′), RAD54B_R (5′-AGTCGTGACCGGCGAAAAT-3′), BLM_F (5′-GAGTCTGCGTGCGAGGATTA-3′), BLM_R (5′-AGTGTTCTGGCTGAGTGACG-3′), RAD51_F (5′-AGCTGGGAACTGCAACTCAT-3′), RAD51_R (5′-CCACACTGCTCTAACCGTGA-3′), RECQL_F (5′-AGAGAAAGCCTATGAAGCAAGGA-3′), RECQL_R (5′-GGCTTCTGCCGAACCTCATA-3′), BRCA1_F (5′-CTGAAGACTGCTCAGGGCTATC-3′), BRCA1_R (5′-AGGGTAGCTGTTAGAAGGCTGG-3′), XRCC2_F (5′-GCGATGTGTAGTGCCTTCCA-3′), XRCC2_R (5′-TTCAAGAATATCACCATGCACAGG-3′), BRCA2_F (5′-AAGCACTCCAGATGGCACAAT-3′), BRCA2_R (5′-GGGTACACAGGTAATCGGCT-3′), RAD52_F (5′-ATGCTTTGGACAGTGCCAGT-3′), RAD52_R (5′-ACATTCTGCTGCGTGATGGA-3′), GAPDH_F (5′-ATGACCACAGTCCATGCCAT-3′), and GAPDH_R (5′-TTGAAGTCAGAGGAGACCAC-3′). The resulting target quantification cycle (Cq) values were normalized to the value for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and then expressed as fold changes relative to the global wild-type mean.
Nuclear extracts were prepared from clones for Western blots, and the following antibodies were used: RPA32 (catalog number A300-244A; Bethyl), RAD54B (catalog number ab83311; Abcam), BLM (C-18; Santa Cruz), and RAD51 (catalog number ab63801; Abcam) antibodies.
Determination of IC50s.
For 50% inhibitory concentration (IC50) assays, U2OS cells were seeded at a density of 1,200 cells per well, and H460 and H1792 cells were seeded at a density of 1,500 cells per well, in 96-well dishes. Twenty-four hours after plating cells, medium was replaced with medium containing the drugs of interest at the appropriate concentrations. Technical triplicates were analyzed for each biological replicate. Serial dilutions of stock solutions of aphidicolin (APH), hydrogen peroxide (H2O2), etoposide, hydroxyurea (HU), and cisplatin were created so that a constant amount of drug was added to the medium for each drug concentration used. After 72 h, alamarBlue was added to an amount equal to 10% of the volume in the well (i.e., 10 μl per well with 100 μl of medium and drug). After 4 h of incubation, cytotoxicity was measured using a Synergy H4 hybrid reader (BioTek, USA), using excitation/emission wavelengths of 560 nm/590 nm. Values were corrected using a blank of medium and alamarBlue only. The amount of fluorescence of alamarBlue for each well of drug-treated cells was then normalized to the fluorescence value obtained for the untreated cells of the same technical replicate. These normalized fluorescence values relative to untreated cells were then analyzed using Prism. The drug concentrations were log transformed, and the data were subsequently fit to a curve using nonlinear regression [log(inhibitor) versus response (three parameters)]. IC50 values were obtained from the best-fit values, and IC50 values from three biological replicates were compared using ordinary one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test or paired t test.
ChIP sequencing.
ChIP was conducted according to protocols adapted from the ones described previously by Cecchini et al. (49). Briefly, cross-linked chromatin was sonicated so that most chromatin was ≤400 bp. Sheared chromatin was then normalized between experimental groups and precleared with protein G Dynabeads and IgG. Precleared chromatin was then incubated with protein G Dynabeads and ChIP antibodies to immunoprecipitate proteins. Antibodies raised against γH2AX (clone JBW301; EMD Millipore) and H4 (clone 62-141-13; EMD Millipore) were used for ChIP. Cross-links were reversed at 65°C, and samples were treated with RNase and proteinase K. DNA was isolated for library preparation, and 20 replicates per genotype for γH2AX ChIP-Seq were pooled to achieve the DNA yield required for library preparation (NEBNext Ultra II DNA library prep kit). ChIP libraries were sequenced using an Illumina NextSeq system (high-output 75-cycle kit).
The resulting FASTQ reads were aligned to human genome build hg19 using Bowtie2 version 2.3.0 (50). The following command was used: bowtie2 -t -p 4 -D 15 -R 2 -L 32 -i S,1,0.75 -x hg19 -U <reads>.fastq -S <output>.sam. Peaks were identified using MACS2 version macs2 2.1.1.20160309, according to parameters stated below and with the options to detect broad peak distributions for histone marks (32). For H4 ChIP-Seq, the corresponding inputs were used as the control, and for γH2AX ChIP-Seq, the first input replicate was used as the control. The following command was used: macs2 callpeak -t <ChIP>.bam -c <input>.bam -n <output> –outdir ./macs2/-g hs –broad –broad-cutoff 0.1.
To find the abundance of ChIP-Seq reads in common fragile sites (CFS), the cytogenetically determined locations of CFS, as determined previously by Lukusa and Fryns, were converted to human genomic coordinates (hg19) using the UCSC Genome Browser (51, 52). Bedtools coverage was then used to find the number of alignments for each ChIP-Seq sample within the individual CFS (53). The abundances of reads that mapped to CFS were then converted to proportions by dividing by the total number of mapped reads. The proportion data were further normalized against the input control, and ratios were then made by comparing the mutant proportions to the wild-type proportions. A two-tailed one-sample t test was performed to test if the normalized mean read count proportions for the RB1+/− and the RB1−/− ChIP-Seq assays at each of the CFS are equal to the normalized read count proportions of the corresponding CFS from the wild-type control. A multitest correction was applied to the calculated P values (using the “fdr” method from the “p.adjust” function in R). Statistical analysis of sequence data was performed using R (version 3.4.2), and the plotting function used was lattice (v0.20-35).
For repeat analysis, another set of alignments was performed. For this analysis, reads were mapped using Bowtie version 1.2.1.1 with high stringency to the hg19 genome (54). The following command was used: bowtie -S –best -m 1 –chunkmbs 500 -p 4 -t –un <not_aligned_unique> –max <multiple_reads_unique> hg19 <reads>.fastq <output>.sam. All remaining reads were mapped to repeat-containing indices using previously reported methods (55), with indices also being derived from the Repbase and Tandem Repeats Databases (56, 57). For these remaining repeat alignments, the –m 1 parameter of the Bowtie mapping was changed to –k 1. Finally, all remaining reads were remapped to hg19 at low stringency to exhaustively match sequence tags to the mouse genome. The abundances of sequence tags that mapped to nonunique regions of the genome were compared by using log2 ratios of γH2AX-precipitable tags per million mapped reads in the mutant versus the wild type and converted into heat maps using matrix2png (58). To test for the significance of enrichment of reads mapped to various repeat categories, the same analysis to test for significance within CFS was used (see above).
Flow cytometry.
Cells were plated on 6-cm plates at a density of 100,000 cells per plate. Approximately 24 h later, cells were pulsed with BrdU for a duration of 30 min. Cell cycle analysis was then carried out as previously described (39).
Nucleoside supplementation.
For nucleoside complementation, cells were seeded at a density of 50,000 cells per 6-cm plate for flow cytometry and at 25,000 cells per 6-well plate with glass coverslips for immunofluorescence. Approximately 24 h after seeding of cells, medium was replaced, either with or without the addition of nucleosides. To prepare nucleosides, uridine (Sigma) and cytidine (Sigma) were dissolved in autoclaved Milli-Q water to make 10 mM stocks, while adenosine (Sigma) and guanosine (Sigma) were dissolved to make 2 mM stocks. The suspensions were briefly boiled, filter sterilized, and added to complete medium at a final concentration of either 50 μM or 10 μM. Forty-eight hours after nucleoside addition, cells were either fixed for γH2AX IF using nonconfocal microscopy and Focinator (as above) or fixed for flow cytometry. For analysis of DNA content by flow cytometry, propidium iodide-stained DNA content was analyzed.
Measurement of reactive oxygen species.
Cells were plated in 96-well plates at a density of 1,200 cells per well in DMEM without phenol red (catalog number 31053-028; Thermo Fisher Scientific). H2O2 was added 24 h after seeding of cells. Seventy-two hours later, 5(6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; CA-DCF-DA) (catalog number C400; Thermo Fisher Scientific) at a stock concentration of 20 mM in dimethyl sulfoxide (DMSO) was diluted in DMEM without phenol red to a concentration of 40 μM. This master mix of medium and CA-DCF-DA was added directly to the wells already containing medium and the drug of interest, to obtain a final concentration of 20 μM CA-DCF-DA. The cells were then placed in the incubator for 45 min, and readings were obtained using a Synergy H4 hybrid reader (BioTek, USA), using excitation/emission wavelengths of 492 nm/525 nm. Technical triplicates were analyzed for each biological replicate, and the average background readings (cells treated with the highest concentration of the drug of interest for 72 h and DMSO in place of CA-DCF-DA) from each cell line were subtracted from the average of each treatment reading for analysis of fluorescence.
Mouse xenografts.
U2OS clones were grown in cell culture to approximately 80% confluence. Cells were washed with PBS, trypsinized, centrifuged, and washed 3 times with Hanks’ balanced salt solution (HBSS) (1×). Cells were then resuspended in HBSS at a concentration of 5 × 106 cells/ml so that 200 μl contained the 1 × 106 cells required for each injection.
For subcutaneous injections, mice were approximately 8 weeks old, and for the tail vein injections, mice were approximately 13 weeks old when injected. All mice were given at least 3 days to acclimatize. All mice were female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (stock number 005557; The Jackson Laboratory) and were housed and handled as approved by the Canadian Council on Animal Care, under an approved protocol (2016-068).
For subcutaneous injections, 1 × 106 RB1+/+ cells were injected into the left flank of all mice, and 1 × 106 RB1+/− or RB1−/− cells were injected into the right flank. The mice used for the subcutaneous injections were euthanized approximately 8 weeks after injection. Necropsies were performed, and tumor mass was determined. Tumors were then fixed in formalin for 48 h and processed for histological assessment.
For tail vein injections, 1 × 106 cells were injected into the lateral tail vein. Mice were euthanized 8 weeks after tail vein injection. All animals were subjected to a thorough necropsy, and all lungs as well as any abnormal tissues or organs were fixed in formalin for 48 h and processed for histological assessment.
All tissues of interest from both studies were embedded, sectioned, and stained with hematoxylin and eosin according to standard methods. Slides were imaged using an Aperio ScanScope slide scanner (Leica Biosystems).
For quantitative pathology of lungs from tail vein-injected mice, images were analyzed using QuPath (59). Briefly, annotations were drawn around each individual lung. Within these annotated lungs, cells were detected using the cell detection command. The features within these cells were then smoothed by using the add smoothed features command (using 25 μm as the radius). Within the lungs, regions containing different cell types were annotated, and these annotations were used to train a cell classifier. All 67 possible cell features were then used to build the random-trees classifier, using default parameters. A script was then made to determine the total cell area of all cell types called by the classifier within each lung, and the percentages of tumor cell area were calculated from these values. Tumor cell nodules were manually counted using the cell types determined by the classifier; anything thought to have derived from a single-cell seeding event was considered a tumor cell nodule.
To determine the RB1 genotype of seeded U2OS cells of interest, embedded mouse lung tissue was deparaffinized and lysed, formalin cross-links were reversed, and DNA was isolated according to the manufacturer’s instructions (QIAamp DNA FFPE tissue kit; Qiagen). DNA was genotyped as described above by PCR using genotyping primers (X22F and X22R).
Data extraction from cBioPortal.
Only TCGA studies used for the Pan-Cancer Atlas with 150 samples or more on cBioPortal were selected to query (60, 61). Data from cBioPortal were obtained in January 2019. Mutation and copy number alteration (CNA) data were analyzed, with the gene set user-defined list being entered as “RB1: AMP HOMDEL HETLOSS mut.”
Data availability.
Processed reads have been deposited in the GEO database (accession number GSE125379).
ACKNOWLEDGMENTS
We thank colleagues for discussions and encouragement over the course of this study.
A.E.M. was a recipient of fellowship support from the NSERC and OGS. This work was supported by funding from the Cancer Research Society and the CIHR (MOP 324579) as well as the NIH (CA228413). F.A.D. is the Wolfe senior fellow in tumor suppressor genes at Western University.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ, McCormick F. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103–112. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 3.Knudsen ES, Knudsen KE. 2008. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer 8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhart DL, Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson NJ. 2016. RB1: a prototype tumor suppressor and an enigma. Genes Dev 30:1492–1502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ. 1996. Cancer cell cycles. Science 274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 7.Dick FA, Goodrich DW, Sage J, Dyson NJ. 2018. Non-canonical functions of the RB protein in cancer. Nat Rev Cancer 18:442–451. doi: 10.1038/s41568-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecchini MJ, Ishak CA, Passos DT, Warner A, Palma DA, Howlett CJ, Driman DK, Dick FA. 2015. Loss of the retinoblastoma tumor suppressor correlates with improved outcome in patients with lung adenocarcinoma treated with surgery and chemotherapy. Hum Pathol 46:1922–1934. doi: 10.1016/j.humpath.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Garsed DW, Alsop K, Fereday S, Emmanuel C, Kennedy CJ, Etemadmoghadam D, Gao B, Gebski V, Gares V, Christie EL, Wouters MCA, Milne K, George J, Patch AM, Li J, Arnau GM, Semple T, Gadipally SR, Chiew YE, Hendley J, Mikeska T, Zapparoli GV, Amarasinghe K, Grimmond SM, Pearson JV, Waddell N, Hung J, Stewart CJR, Sharma R, Allan PE, Rambau PF, McNally O, Mileshkin L, Hamilton A, Ananda S, Grossi M, Cohen PA, Leung YC, Rome RM, Beale P, Blomfield P, Friedlander M, Brand A, Dobrovic A, Kobel M, Harnett P, Nelson BH, Bowtell DDL, deFazio A, Traficante N, for the Australian Ovarian Cancer Study Group. 2018. Homologous recombination DNA repair pathway disruption and retinoblastoma protein loss are associated with exceptional survival in high-grade serous ovarian cancer. Clin Cancer Res 24:569–580. doi: 10.1158/1078-0432.CCR-17-1621. [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Huang CC, Otterson GA, Leon ME, Tang Y, Shilo K, Villalona MA. 2012. Altered p16(INK4) and RB1 expressions are associated with poor prognosis in patients with nonsmall cell lung cancer. J Oncol 2012:957437. doi: 10.1155/2012/957437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludovini V, Gregorc V, Pistola L, Mihaylova Z, Floriani I, Darwish S, Stracci F, Tofanetti FR, Ferraldeschi M, Di Carlo L, Ragusa M, Daddi G, Tonato M. 2004. Vascular endothelial growth factor, p53, Rb, Bcl-2 expression and response to chemotherapy in advanced non-small cell lung cancer. Lung Cancer 46:77–85. doi: 10.1016/j.lungcan.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 12.McNair C, Xu K, Mandigo AC, Benelli M, Leiby B, Rodrigues D, Lindberg J, Gronberg H, Crespo M, De Laere B, Dirix L, Visakorpi T, Li F, Feng FY, de Bono J, Demichelis F, Rubin MA, Brown M, Knudsen KE. 2018. Differential impact of RB status on E2F1 reprogramming in human cancer. J Clin Invest 128:341–358. doi: 10.1172/JCI93566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson DR, Wu Y-M, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, Schuetze S, Alva A, Siddiqui J, Chugh R, Worden F, Zalupski MM, Innis J, Mody RJ, Tomlins SA, Lucas D, Baker LH, Ramnath N, Schott AF, Hayes DF, Vijai J, Offit K, Stoffel EM, Roberts JS, Smith DC, Kunju LP, Talpaz M, Cieślik M, Chinnaiyan AM. 2017. Integrative clinical genomics of metastatic cancer. Nature 548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, Van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis F. 2016. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thangavel C, Boopathi E, Liu Y, Haber A, Ertel A, Bhardwaj A, Addya S, Williams N, Ciment SJ, Cotzia P, Dean JL, Snook A, McNair C, Price M, Hernandez JR, Zhao SG, Birbe R, McCarthy JB, Turley EA, Pienta KJ, Feng FY, Dicker AP, Knudsen KE, Den RB. 2017. RB loss promotes prostate cancer metastasis. Cancer Res 77:982–995. doi: 10.1158/0008-5472.CAN-16-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, Cecchini MJ, Martens AL, Percy V, Welch I, Boutros PC, Brown GW, Dick FA. 2014. Haploinsufficiency of an RB-E2F1-condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov 4:840–853. doi: 10.1158/2159-8290.CD-14-0215. [DOI] [PubMed] [Google Scholar]
- 17.Zheng L, Flesken-Nikitin A, Chen PL, Lee WH. 2002. Deficiency of retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res 62:2498–2502. [PubMed] [Google Scholar]
- 18.Gonzalez-Vasconcellos I, Anastasov N, Sanli-Bonazzi B, Klymenko O, Atkinson MJ, Rosemann M. 2013. Rb1 haploinsufficiency promotes telomere attrition and radiation-induced genomic instability. Cancer Res 73:4247–4255. doi: 10.1158/0008-5472.CAN-12-3117. [DOI] [PubMed] [Google Scholar]
- 19.Velez-Cruz R, Johnson DG. 2017. The retinoblastoma (RB) tumor suppressor: pushing back against genome instability on multiple fronts. Int J Mol Sci 18:E1776. doi: 10.3390/ijms18081776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning AL, Yazinski SA, Nicolay B, Bryll A, Zou L, Dyson NJ. 2014. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell 53:993–1004. doi: 10.1016/j.molcel.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longworth MS, Herr A, Ji JY, Dyson NJ. 2008. RBF1 promotes chromatin condensation through a conserved interaction with the condensin II protein dCAP-D3. Genes Dev 22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook R, Zoumpoulidou G, Luczynski MT, Rieger S, Moquet J, Spanswick VJ, Hartley JA, Rothkamm K, Huang PH, Mittnacht S. 2015. Direct involvement of retinoblastoma family proteins in DNA repair by non-homologous end-joining. Cell Rep 10:2006–2018. doi: 10.1016/j.celrep.2015.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velez-Cruz R, Manickavinayaham S, Biswas AK, Clary RW, Premkumar T, Cole F, Johnson DG. 2016. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev 30:2500–2512. doi: 10.1101/gad.288282.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolay BN, Danielian PS, Kottakis F, Lapek JD Jr, Sanidas I, Miles WO, Dehnad M, Tschop K, Gierut JJ, Manning AL, Morris R, Haigis K, Bardeesy N, Lees JA, Haas W, Dyson NJ. 2015. Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev 29:1875–1889. doi: 10.1101/gad.264127.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, Ju Y, Chung PE, Pellecchia G, Fell VL, Bae S, Muthuswamy L, Datti A, Egan SE, Jiang Z, Leone G, Bader GD, Schimmer A, Zacksenhaus E. 2016. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest 126:3739–3757. doi: 10.1172/JCI81568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benevolenskaya EV, Frolov MV. 2015. Emerging links between E2F control and mitochondrial function. Cancer Res 75:619–623. doi: 10.1158/0008-5472.CAN-14-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. 2010. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horowitz JM, Park SH, Bogenmann E, Cheng JC, Yandell DW, Kaye FJ, Minna JD, Dryja TP, Weinberg RA. 1990. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci U S A 87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furtado C, Kunrath-Lima M, Rajao MA, Mendes IC, de Moura MB, Campos PC, Macedo AM, Franco GR, Pena SD, Teixeira SM, Van Houten B, Machado CR. 2012. Functional characterization of 8-oxoguanine DNA glycosylase of Trypanosoma cruzi. PLoS One 7:e42484. doi: 10.1371/journal.pone.0042484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP. 2011. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennardo N, Cheng A, Huang N, Stark JM. 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. 2007. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelot C, Magdalou I, Lopez BS. 2015. Replication stress in mammalian cells and its consequences for mitosis. Genes (Basel) 6:267–298. doi: 10.3390/genes6020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cecchini MJ, Amiri M, Dick FA. 2012. Analysis of cell cycle position in mammalian cells. J Vis Exp 59:e3491. doi: 10.3791/3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan KL, Hickson ID. 2011. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol 22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Dimaras H, Khetan V, Halliday W, Orlic M, Prigoda NL, Piovesan B, Marrano P, Corson TW, Eagle RC Jr, Squire JA, Gallie BL. 2008. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet 17:1363–1372. doi: 10.1093/hmg/ddn024. [DOI] [PubMed] [Google Scholar]
- 42.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, et al. . 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cecchini MJ, Dick FA. 2011. The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem J 434:297–308. doi: 10.1042/BJ20101210. [DOI] [PubMed] [Google Scholar]
- 46.Oeck S, Malewicz NM, Hurst S, Rudner J, Jendrossek V. 2015. The Focinator—a new open-source tool for high-throughput foci evaluation of DNA damage. Radiat Oncol 10:163. doi: 10.1186/s13014-015-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cecchini MJ, Thwaites M, Talluri S, Macdonald JI, Passos DT, Chong JL, Cantalupo P, Stafford P, Saenz-Robles MT, Francis SM, Pipas JM, Leone G, Welch I, Dick FA. 2014. A retinoblastoma allele that is mutated at its common E2F interaction site inhibits cell proliferation in gene-targeted mice. Mol Cell Biol 34:2029–2045. doi: 10.1128/MCB.01589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukusa T, Fryns JP. 2008. Human chromosome fragility. Biochim Biophys Acta 1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, Fischer CM, Gibson D, Gonzalez JN, Guruvadoo L, Haeussler M, Heitner S, Hinrichs AS, Karolchik D, Lee BT, Lee CM, Nejad P, Raney BJ, Rosenbloom KR, Speir ML, Villarreal C, Vivian J, Zweig AS, Haussler D, Kuhn RM, Kent WJ. 2017. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45:D626–D634. doi: 10.1093/nar/gkw1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Day DS, Luquette LJ, Park PJ, Kharchenko PV. 2010. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gelfand Y, Rodriguez A, Benson G. 2007. TRDB—the Tandem Repeats Database. Nucleic Acids Res 35:D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pavlidis P, Noble WS. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 59.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. 2017. QuPath: open source software for digital pathology image analysis. Sci Rep 7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Processed reads have been deposited in the GEO database (accession number GSE125379).