The D site-binding protein (Dbp) supports the rhythmic transcription of downstream genes, in part by displaying high-amplitude cycling of its own transcripts compared to other circadian-clock genes. However, the underlying mechanism remains elusive. Here, we demonstrated that the poly(C) motif within the Dbp proximal promoter, in addition to an E-box element, provoked transcriptional activation.
KEYWORDS: circadian rhythm, Dbp, mRNA oscillation, poly(C)
ABSTRACT
The D site-binding protein (Dbp) supports the rhythmic transcription of downstream genes, in part by displaying high-amplitude cycling of its own transcripts compared to other circadian-clock genes. However, the underlying mechanism remains elusive. Here, we demonstrated that the poly(C) motif within the Dbp proximal promoter, in addition to an E-box element, provoked transcriptional activation. Furthermore, we generated a cell line with poly(C) deleted to demonstrate the endogenous effect of the poly(C) motif within the Dbp promoter. We investigated whether RNA polymerase 2 (Pol2) recruitment on the Dbp promoter was decreased in the cell line with poly(C) deleted. Next, assay for transposase-accessible chromatin (ATAC)-quantitative PCR (qPCR) showed that the poly(C) motif induced greater chromatin accessibility within the region of the Dbp promoter. Finally, we determined that the oscillation amplitude of endogenous Dbp mRNA of the cell line with poly(C) deleted was decreased, which affected the oscillation of other clock genes that are controlled by Dbp. Taken together, our results provide new insights into the function of the poly(C) motif as a novel cis-acting element of Dbp, along with its significance in the regulation of circadian rhythms.
INTRODUCTION
In studies of mood disorders and aging, disruption of the amplitude of clock gene expression has been reported (1–4). In particular, both the oscillation amplitude and the expression level of the D site-binding protein (Dbp) were decreased in fibroblasts from bipolar patients (1). Dbp has a DNA binding domain that binds specifically to the D-box (TTATG[T/C]AA) and acts as a transcription activator (5–8). In mammalian genomes, there are more than 2,000 putative D-boxes, which govern about 10% of the clock-controlled genes (9, 10). The oscillation of Dbp mRNA is regulated by the BMAL1-CLOCK complex via multiple E-boxes (11–14). Dbp mRNA oscillation is known to have a higher amplitude than other E-box-controlled clock genes (15, 16). However, little is known about the mechanism of high-amplitude Dbp mRNA oscillation among E-box-controlled clock genes.
Circadian rhythm is known to be governed by cis-acting elements, including the E-box, the D-box, and ROREs. Here, we focused on the novel cis-acting elements within the Dbp promoter, as the E-box element was the only previously reported cis-acting element in the Dbp promoter region (11–14). Thus, we investigated the unknown motif by using the MEME (multiple expectation maximum for motif elicitation) suite to find out whether it has reference to specific motifs (17, 18). First, we hypothesized that, in addition to the E-box element, a specific cis-acting element may support Dbp mRNA oscillation amplitude. Interestingly, a pair of poly(C) motifs within proximal promoter regions of Dbp were detected. Next, we deleted the poly(C) motif, using the CRISPR-Cas9 system, to demonstrate its endogenous function (19). Furthermore, we found that both the RNA polymerase 2 recruitment level and the chromatin accessibility level were decreased in the cell line with poly(C) deleted. Finally, we revealed that the mRNA oscillation of other clock genes regulated by Dbp was affected in the cell line with poly(C) deleted. Therefore, we propose that the poly(C) cis-acting element within the Dbp proximal promoter region supports the Dbp mRNA oscillation amplitude and sustains the mRNA oscillation level of Dbp-controlled clock genes.
RESULTS
Oscillation of Dbp mRNA expression is supported by its transcription at specific promoter regions.
While the high amplitude of Dbp mRNA oscillation has been reported, the mechanism behind the higher-amplitude expression of Dbp mRNA than that of other clock genes remained unclear (15, 16).
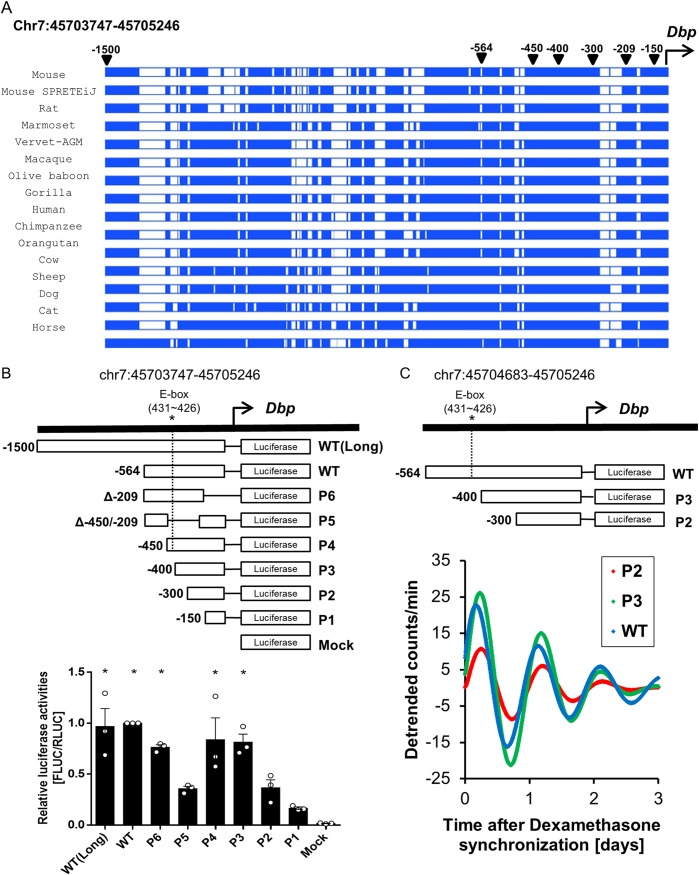
We hypothesized that the major factor that determines the high amplitude of Dbp mRNA expression is transcription efficiency via elevated transcriptional activation. To elucidate potential transcription-regulatory motifs in eutherian mammals, we aligned the proximal promoter regions (genomic regions of 1,500 bp from the transcription start site [TSS]) of the Dbp genes from multiple mammalian species using sequence data in the Ensembl database (http://www.ensembl.org). We found that certain promoter regions were highly alignable among eutherian mammals, suggesting that a possible regulatory motif could exist in the aligned promoter regions (Fig. 1A). We cloned several deletion forms of the promoter region for promoter assays with a luciferase reporter to determine the regions critical for transcriptional activation at a peak time for Dbp mRNA. Our data indicated that the P3 (−400), P4 (−450), P6 (which had lost 209 bp from the TSS [Δ−209]), wild-type (−564), and wild-type (long) (−1500) promoter regions drove significant expression of luciferase, allowing us to determine the specific regions of the proximal promoter critical for promoter activity (Fig. 1B). In particular, the P5 (which had lost the promoter region between 209 bp and 450 bp from the TSS [Δ−450/−209]) and P2 (−300) constructs had decreased promoter activity relative to that of P3 (−400), P4 (−450), P6 (Δ−209), and the wild type (−564). Next, we identified the oscillation levels of P2, P3, and the wild type using real-time luminescence (Fig. 1C). Our data showed that the region between P2 and P3 was necessary for high-amplitude oscillation of Dbp transcription. This suggests that other regions crucial for efficient promoter activity exist in the region between 300 bp and 400 bp from the TSS, apart from an E-box element. Therefore, we concluded that this specific region of the proximal promoter of Dbp could possess regions critical for Dbp transcriptional activation.
FIG 1.
Oscillation of Dbp mRNA expression is supported by its transcription at specific promoter regions. (A) Alignment of Dbp proximal promoter regions among eutherian mammals. Dbp proximal promoter regions within 0.6 kb from the TSS showed aligned genomic regions among eutherian mammals, indicating that a significant regulatory motif exists within these regions (https://www.ensembl.org/). Blue, aligned region; white, not aligned. (B) A promoter assay was conducted 24 h after dexamethasone synchronization. P3, P4, P6, wild-type (WT), and WT(Long) promoter regions showed significant promoter activities, which suggested that the region between bp −300 and −400 from the TSS is a critical promoter region. In contrast, other promoter regions had relatively little impact on promoter activity. (n = 3; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). The bars indicate means and SEM. (C) Representative detrended bioluminescence recordings from WT:LUC, P2:LUC, and P3:LUC (plasmid constructs carrying the WT, P2, and P3 promoters, respectively, expressing luciferase), transiently transfected into an NIH 3T3 cell line that was synchronized with dexamethasone, suggesting that the regions between −300 and −400 from the TSS could be crucial to the high amplitude of Dbp oscillation.
The poly(C) motif is critical for Dbp promoter activity.
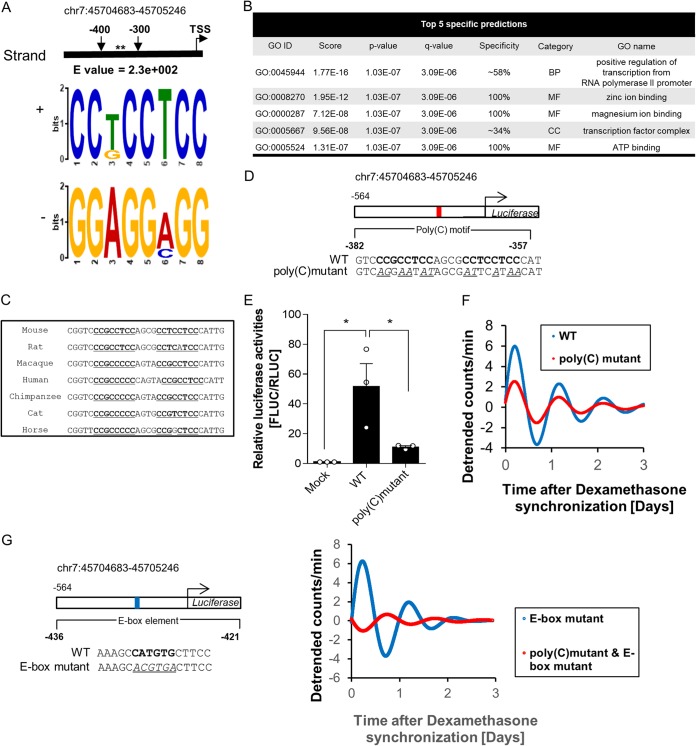
To identify transcription-regulatory motifs, we used the MEME suite, a tool for searching for a motif with recurring and fixed-length patterns within a specific genomic region. We scanned proximal promoter regions with MEME to reveal motifs between P2 (−300) and P3 (−400). The poly(C) motif (or CCT motif) (Fig. 2A) was the only motif in this region predicted to function in transcription according to the GOMo (gene ontology [GO] for motifs) tool, which showed GO terms associated with the genes whose promoters contained the poly(C) motif (Fig. 2B).
FIG 2.
The poly(C) motif is critical for Dbp promoter activity. (A) The poly(C) motif (or CCT motif) was scanned in the region between bp −300 and −400 from the TSS using the MEME suite. The asterisks mark the position of the poly(C) motif. (B) One of the functions of the poly(C) motif predicted by GOMo was transcription. The poly(C) motif was the only motif that was predicted to function in transcription. MF, molecular function; BP, biological process; CC, cellular component. (C) Alignment of genomic regions containing a pair of poly(C) motifs showed high conservation among eutherian mammals, which suggests that a common transcription-regulatory mechanism is involved in Dbp promoter activity. Poly(C) motifs are boldface and underlined. (D) Diagram of the luciferase reporter system containing a WT promoter and a poly(C) mutant. The poly(C) motif is in boldface; mutated bases are underlined and italic. (E) Promoter activity 24 h after dexamethasone synchronization of the poly(C) mutant showed a significant decrease compared to the WT, which suggests that the poly(C) motif is crucial for promoter activity (n = 3; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). The bars indicate means and SEM. (F) Representative detrended bioluminescence recordings of WT:LUC and poly(C) mutant:LUC measured with a luciferase reporter-transfected NIH 3T3 cell line after dexamethasone synchronization, which showed a decrease in the amplitude level of the poly(C) mutant. (G) Representative detrended bioluminescence recordings of an E-box mutant:LUC and an E-box and poly(C) mutant:LUC measured with a luciferase reporter-transfected NIH 3T3 cell line after dexamethasone synchronization. The data showed a dramatic decrease in the amplitude level of the E-box and poly(C) mutant promoter.
Furthermore, high-amplitude oscillation of Dbp mRNA is often found in mammals, such as humans, mice, and rats (1, 5, 20). Therefore, we aligned the poly(C) motif sequences from several eutherian mammals, which revealed that the general transcription-regulatory mechanism could be mediated by the poly(C) motif (Fig. 2C). Next, to identify levels of promoter activity, we cloned a mutated poly(C) motif produced by random substitutions (italics) from the wild-type 5′-CCGCCTCCAGCGCCTCCTCC-3′ to mutant 5′-AGGAATATAGCGATTCATAA-3′ (Fig. 2D). We measured the activities of the wild-type and poly(C) mutant promoters and found that the Dbp promoter activity of the poly(C) mutant was significantly decreased at the peak time, which implied that the poly(C) motif played a key role in its promoter activity (Fig. 2E). Next, we triggered oscillation by synchronizing cells with dexamethasone to identify differences between wild-type and poly(C) mutant promoter activities using real-time luminescence. We found that the amplitude of luciferase expression by the poly(C) mutant was markedly decreased compared to that of the wild type (Fig. 2F). Furthermore, we confirmed that the oscillation of an E-box mutant and a poly(C) mutant promoter was more disrupted than that of the promoter of the E-box-only mutant (Fig. 2G). Also, the disruption of promoter oscillation by both the poly(C) and E-box mutations seemed very severe compared to that of the poly(C) mutation alone. This means that the poly(C) motif might not be the sole factor that drives rhythmicity, but an E-box element could also support its oscillation.
The poly(C) motif is important for RNA polymerase 2 recruitment because it induces greater chromatin accessibility.
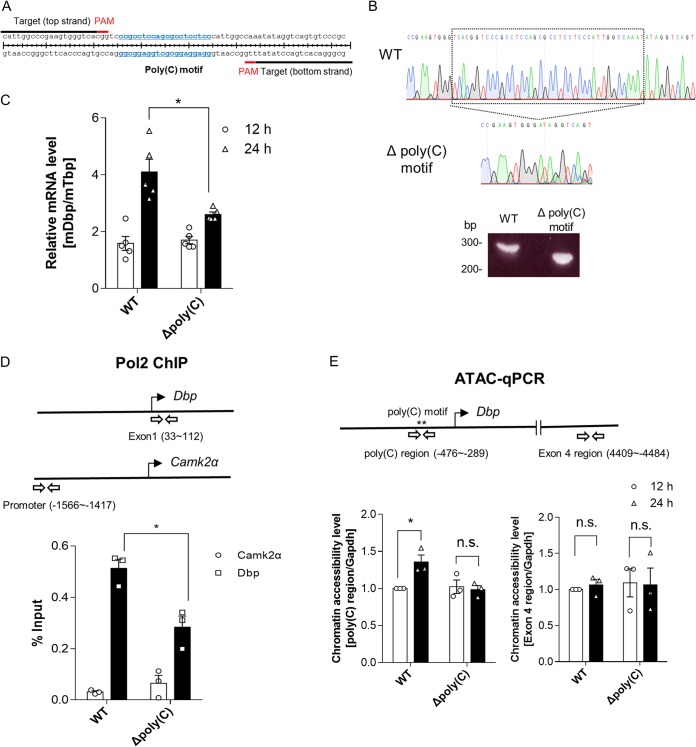
Next, using the CRISPR-Cas9 system, we designed an NIH 3T3 cell line with deletion of the poly(C) motif region within the Dbp promoter to elucidate the endogenous function of these genomic regions during Dbp mRNA oscillation (Fig. 3A). After single-cell sorting by green fluorescent protein (GFP) selection, the cell with the poly(C) motif region deletion was expanded and confirmed by genotyping and sequencing in both forward and reverse directions (Fig. 3B). We confirmed that the Dbp mRNA expression levels of cells with poly(C) deleted at 24 h after dexamethasone synchronization were decreased compared to that of wild-type cells (Fig. 3C).
FIG 3.
The poly(C) motif is important for RNA polymerase 2 recruitment because it induces greater chromatin accessibility. (A) Schematic depiction of a pair of sgRNAs designed to delete a pair of endogenous poly(C) motif regions within the Dbp proximal promoter region. (B) A cell line with the poly(C) motif region deleted made with the CRISPR-Cas9 system was confirmed by genotyping and sequencing. The boxed region is between bp −397 and −337 from the TSS. (C) The endogenous Dbp mRNA was quantified, and the cell line with the poly(C) motif region deleted showed decreases in the mRNA expression level at 24 h after dexamethasone synchronization (n = 5; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). (D) Pol2 ChIP was conducted to measure the recruitment level of Pol2 to Dbp regions at 24 h after dexamethasone synchronization (n = 3; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). (E) ATAC-qPCR data indicated that chromatin accessibility of WT cells increased 24 h after dexamethasone synchronization; however, cells with the poly(C) motif region deleted did not show an increase in chromatin accessibility. Furthermore, the Dbp exon 4 region was not significantly changed. The Gapdh promoter region was used for normalization, which showed no oscillatory patterns of mRNA expression, in contrast to Dbp mRNA (n = 3; n.s., not significant; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). The bars indicate means and SEM.
To identify transcriptional regulation of Dbp at the peak time, we measured the level of Pol2 binding during the peak time of Dbp mRNA oscillation. The control region of Pol2 chromatin immunoprecipitation (ChIP) was the promoter region of Camk2α, which was previously reported to be encoded by a brain-specific gene (21). The data showed that the Pol2 binding level of the Dbp region was decreased in cells with poly(C) deleted (Fig. 3D).
Finally, to clarify the reason behind the reduced Pol2 binding level of Dbp in cells with poly(C) deleted, we utilized the assay for transposase-accessible chromatin (ATAC), along with real-time PCR. Interestingly, ATAC–quantitative-PCR (qPCR) data showed that chromatin accessibility levels were regulated by the poly(C) motif region depending on the circadian rhythm; however, the region of exon 4 of Dbp was not (Fig. 3E). These results imply that Dbp mRNA oscillation might be facilitated by the modulation of chromatin accessibility mediated by poly(C) motif regions.
The oscillation of other clock genes regulated by Dbp was affected by the poly(C) motif within the Dbp promoter.
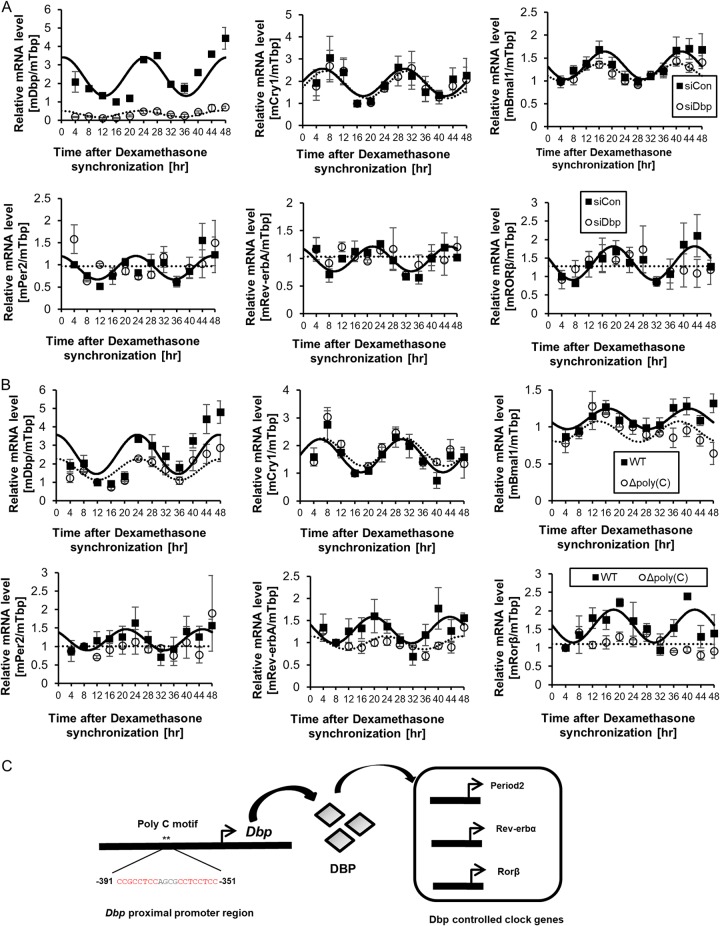
Finally, we investigated the levels of other clock genes with D-box cis-acting elements in the promoter during the knockdown of Dbp. It has been reported that Per genes, Rev-erb genes, and RAR-related orphan receptor (Ror) contain D-box elements (6, 7, 22, 23). Therefore, we triggered a circadian rhythm by dexamethasone synchronization to identify the effect of Dbp. First, we assessed endogenous Dbp mRNA oscillation during the knockdown of Dbp to demonstrate regulation by Dbp and compared it to that of Bmal1 and Cry1 mRNAs. Cells transfected with the Dbp small interfering RNA (siRNA) pool showed a decrease in Dbp mRNA oscillation amplitude compared to the control siRNA-transfected group (Fig. 4A). Furthermore, when we compared the oscillation amplitudes of Bmal1 and Cry1 during knockdown of Dbp, they showed only a marginal difference compared to the control siRNA-transfected group (Fig. 4A). Next, we found that knockdown of Dbp dramatically changed the rhythmic mRNA expression levels of the D-box-regulated genes, such as Per2, Rev-erbα, and Rorβ, compared to relatively small changes in Cry1 and Bmal1 (Fig. 4A).
FIG 4.
The oscillation of other clock genes regulated by Dbp was sustained by the poly(C) motif within the Dbp promoter. (A) Dbp, Cry1, Bmal1, Per2, Rev-erbα, and Rorβ mRNA oscillation levels were quantified. The oscillation levels of Cry1 and Bmal1 mRNAs were slightly affected by Dbp knockdown; however, Per2, Rev-erbα, and Rorβ mRNA oscillation levels were disrupted by Dbp knockdown (n = 3). The error bars indicate SEM. (B) The mRNA oscillation amplitude of Dbp was decreased when its poly(C) motif was deleted. However, Cry1 and Bmal1 mRNA oscillation levels were only slightly changed. Interestingly, the oscillation levels of Per2, Rev-erbα, and Rorβ mRNAs were disrupted due to the deletion of the poly(C) motif within the Dbp promoter (n = 3). The error bars indicate SEM. (C) Our suggested model of regulation by the poly(C) motif is illustrated with Dbp, which regulates other Dbp-controlled genes. The asterisks mark the position of the poly(C) motif.
We also confirmed that the oscillation amplitude of endogenous Dbp mRNA of the cell line with poly(C) deleted was decreased compared to that of wild-type cells (Fig. 4B). Furthermore, we determined that Cry1 and Bmal1 mRNA oscillation of the cell line with poly(C) deleted was affected only slightly compared to that of wild-type cells (Fig. 4B). Next, we demonstrated that mRNA oscillation of Rev-erbα, Rorβ, and Per2 in the cell line with poly(C) deleted was disturbed (Fig. 4B). These data suggest that the poly(C) motif within the Dbp promoter supports mRNA oscillation of Dbp and other clock genes that are controlled by Dbp.
Thus, we propose a model in which the high-amplitude oscillation of Dbp mRNA, which is mediated by the poly(C) motif, affects other clock genes regulated by Dbp (Fig. 4C).
DISCUSSION
One of the main findings in this study is that high-amplitude Dbp mRNA oscillations are mediated by the poly(C) motif, which functions as a cis-acting element. Prior to this study, it was reported that the E-box element was the main regulator of rhythmic Dbp mRNA expression (11–14). Here, we uncovered an additional and novel regulating mechanism that maintains high-amplitude oscillation. We demonstrated that the poly(C) motif could indeed affect the circadian-rhythm oscillation of Dbp mRNA by acting as a cis-acting element, along with other complex regulating mechanisms.
First, we investigated the oscillation pattern of Dbp and compared it to those of other clock genes (data not shown). Next, we aligned the genomic region upstream of the TSS of Dbp among eutherian mammals to determine transcription-regulatory motifs (Fig. 1A). This suggested that certain motifs within the aligned region could function as critical regulators of circadian rhythm in eutherian mammals. Furthermore, analysis of the promoter alignment using the MEME suite revealed a poly(C) motif that was predicted to be a transcriptional activator that may interact with other activator proteins (Fig. 2). We confirmed the promoter activity and function of the poly(C) motif using luciferase reporter assays and the CRISPR-Cas9 system (Fig. 2 and 3). Thus, we found that the regulation of Dbp promoter activity was dependent on the poly(C) motif within the Dbp promoter.
Furthermore, we measured RNA polymerase 2 recruitment by the wild-type cell line and compared it to that of the cell line with the poly(C) motif deleted (Fig. 3D). We found that the poly(C) motif within the Dbp promoter enhanced RNA polymerase 2 recruitment. Next, we confirmed the chromatin accessibility levels by using ATAC, which showed that the poly(C) motif increased the level of chromatin accessibility (Fig. 3E). This may suggest that the poly(C) motif recruits unknown trans-acting factors or interacts with other chromatin regions.
Next, we measured the endogenous Dbp mRNA oscillation level of the cell line with the poly(C) motif deleted and compared it to that of the wild-type cell line (Fig. 4). The result showed a decrease in the amplitude of Dbp mRNA oscillation in the cell line with the poly(C) motif deleted (Fig. 4B). We also confirmed the mRNA oscillations of other clock genes regulated by Dbp in the cell line with the poly(C) motif deleted. In summary, we demonstrated that the poly(C) motif mediated mRNA oscillation of Dbp, which is critical for the maintenance of high-amplitude Dbp mRNA oscillation and that of other clock genes controlled by Dbp (Fig. 4C).
Further studies may be conducted to unveil trans-acting factors, such as poly(C)-binding proteins (PCBPs), including hnRNP K, that bind to the poly(C) motif within the Dbp promoter region that could potentially function as transcriptional regulators. PCBPs are also known to have a role in posttranscriptional regulation and may have a combinatorial role in Dbp expression (30). Thus, it would be interesting to further investigate the characteristics of the potential trans-acting factors, such as their rhythmic expression.
MATERIALS AND METHODS
Genome sequence analysis.
Comparative Genomics in the Ensembl database was used for genomic alignment among eutherian mammals. We extracted the mouse genomic sequence (Genome Reference Consortium mouse build 38, patch 5 [GRCm38.p5]) of chromosome 7 (chr7) 45703747 to 45705246, which is the proximal promoter region (∼1.5 kb from the transcription start site) of Dbp, to align it with those of other species for identification of common regulatory sequence motifs. We were also able to extract a poly(C) motif-containing sequence from eutherian mammals from the Ensembl database.
Cell culture and drug treatment.
NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone) supplemented with 10% fetal bovine serum (HyClone) and 1% antibiotics (WelGene) and maintained in a humidified incubator with 95% air and 5% CO2. The NIH 3T3 cells were synchronized by treatment with 100 nM dexamethasone in 12-well plates containing ∼3.0 × 105 cells per well. After 2 h, the medium was replaced with complete medium, and the cells were harvested every 4 h for each sample (24).
Plasmids.
To generate the pGL3 vector, which contains promoters of Dbp, we amplified the several forms of Dbp promoters (UCSC Genome Browser on Mouse Dec. 2011 [GRCm38/mm10] Assembly [https://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr7%3A45705088-45710203&hgsid=730668269_TXeBf0pczBFzSaLJqK28gRUluokM]) with forward and reverse primers using Pfu polymerase (Solgent, Daejeon, South Korea). The PCR products were digested with NheI and HindIII and then inserted into the pGL3 basic vector, yielding pGL3 with the short form of the Dbp promoter and pGL3 with the long form of the Dbp promoter, which was confirmed by sequencing. Serial deletion constructs (P1, P2, P3, P4, P5, and P6) and mutant constructs were cloned in the same way.
Motif search and prediction.
We searched for motifs in the proximal promoter region of Dbp (chr7 45704683 to 45705246) (UCSC Genome Browser on Mouse Dec. 2011 [GRCm38/mm10] Assembly; https://genome.ucsc.edu) using a MEME algorithm that is able to discover novel and ungapped motifs (17, 18). We selected the classic motif discovery mode and typed the genomic sequence of Dbp. For the site distribution, we selected “any number of repetitions” and allowed MEME to find three motifs. We searched critical motifs within the region by referring to the experimental promoter assay. The predicted functions were extracted with GOMo, which suggests the biological roles of designated motifs (25, 26). The score is the geometric mean score of rank sum tests for the particular GO term. For the rank sum test, the scores of genes annotated with GO terms and the scores of genes not annotated with GO terms are compared. A P value is calculated for the enrichment of the GO term. The q value is the value of the minimum false-discovery rate (FDR), which is the ratio of false positives to total positive test results, to deem significant a given GO term. The specificity refers to how specific a GO term is within the GO hierarchy. The higher the specificity of a GO term is, the more likely the GO term is to have a specific role. A tilde on a specificity value shows that the value has been rounded and that it is not exact (25, 26).
RNA extraction and quantitative PCR.
Total RNA was extracted from NIH 3T3 cells using the TRI Reagent (Molecular Research Center, BioScience Technology). We treated RQ1 RNase-free DNase (Promega) after RNA quantification to remove contaminated DNA according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed using ImProm-II (Promega) according to the manufacturer’s instructions. Endogenous mRNA levels were detected by quantitative real-time PCR using the Step One Plus real-time PCR system (Applied Biosystems) with Fast Start universal SYBR green master mix (Roche), as described previously (27). Specific primer pairs for mouse Dbp (NM_016974.3), mouse Per2 (NM_011066.3), mouse Rev-erbα (NM_145434.4), mouse Rorβ (NM_001043354.2), mouse Bmal1 (NM_007489.4), and mouse Cry1 (NM_007771.3) were used for real-time PCR (the primer sequences are shown in Table S1 in the supplemental material).
Luciferase assay.
Firefly luciferase (FLUC) and Renilla luciferase activities were determined using the Dual-Luciferase reporter assay system created by Promega (Madison, WI) according to the manufacturer’s instructions. Normalized FLUC activity was determined as the ratio of firefly luciferase activity to Renilla luciferase activity.
Real-time luminescence.
NIH 3T3 fibroblasts (5 × 105; ATCC) were plated in 35-mm dishes. The cells were transfected with 1 μg of each Dbp-luciferase construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After overnight culture, 100 nM dexamethasone was added to the cells, and the cells were incubated for 2 h. The medium was then changed to DMEM without phenol red and 1 mM luciferin (Promega). A Lumicycle device (Actimetrics) was kept in the 37°C incubator to record measured luminescence for 3 or 4 days. Lumicycle analysis software (Actimetrics) was used for Lumicycle data analysis.
Generation of cell lines with the poly(C) motif region deleted.
We used the pSpCas9(BB)-2A-GFP (PX458) plasmid obtained from Addgene (Addgene; 48138) for CRISPR-Cas9 experiments. We designed single guide RNA (sgRNA) sequences to delete poly(C) motif-containing regions within the Dbp promoter by using the online CRISPR design tool (http://crispr.mit.edu/) (19). The sequences were as follows: target 1 Forward, 5′-CACCGCATTGGCCCGAAGTGGGTCA-3′, and Reverse, 5′-CTGACCCACTTCGGGCCAATGCAAA-3‘; target 2 Forward, 5′-CACCGGCGGGACACTGACCTATATT-3′, and Reverse, 5′-CAATATAGGTCAGTGTCCCGCCAAA-3′. Then, genomic-DNA sequencing was performed with the following primers to confirm the genomic deletion: Forward, 5′-CTAGCTAGCCGATAGCACGCGCAAAGCCA-3′; Reverse, 5′-CCCAAGCTTGGCAAGAACCAATCACGTCT-3′.
ChIP assay.
After fixation of 10 million NIH 3T3 cells with 1% formaldehyde at room temperature (RT) for 10 min, 0.125 M glycine was added for 10 min to quench the formaldehyde. Next, the fixed cells were suspended in 10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.1% SDS, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Roche). Cross-linked cells were sonicated with a sonicator (VibraCell; Sonics) with 20 cycles of sonication (30 s) and resting (30 s) at an amplitude of 8. Sonicated chromatin was analyzed by agarose gel electrophoresis to ensure that DNA fragment sizes did not exceed 500 bp. Pol2 (8WG16; Abcam) was added to 750 μl of chromatin and incubated at 4°C for 2 h with rotation; then, 30 μl of protein G magnetic beads (Thermo; catalog no. 88848) was added to the chromatin-antibody mixture and incubated at 4°C overnight with rotation. The magnetic beads were washed twice with 1 ml of ChIP wash buffer 1 (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100) for 10 min each time, twice with 1 ml of ChIP wash buffer 2 (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 0.3 M NaCl) for 10 min each time, twice with 1 ml of ChIP wash buffer 3 (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) for 10 min each time, once with 1 ml of ChIP wash buffer 4 (10 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.2% Triton X-100) for 10 min, and once with 1 ml of 1× Tris-EDTA (TE) buffer (10 mM Tris-Cl [pH 7.4], 1 mM EDTA) for 10 min. The washed beads were pelleted on a magnetic stand, and the clear supernatant was carefully removed and discarded. The beads were resuspended in 200 μl of ChIP elution buffer (50 mM Tris-Cl [pH 7.5], 10 mM EDTA, 1% SDS), and 5 μl of proteinase K (Roche; catalog no. 03 115 828 001) was added to the bead suspension. The beads and proteinase K were mixed well by pipetting and incubated at 65°C for 5 h; 200 μl of phenol-chloroform-isoamyl alcohol (25: 24:1; Affymetrix; catalog no. 75831 400 ML) was added to the bead suspension, mixed vigorously by vortexing for 30 s, and centrifuged at 13,000 rpm for 15 min at room temperature. After centrifugation, the upper aqueous phase was transferred to a fresh 1.5-ml tube; 2 μl of glycogen solution (Roche; catalog no. 10901393001), 20 μl of 3 M sodium acetate, and 500 μl of 100% ethanol were added; and the solution was incubated at −20°C overnight. DNA was pelleted by centrifuging at 13,000 rpm for 30 min at 4°C. The supernatant was carefully removed and discarded. The DNA pellet was washed with 700 μl of 70% ethanol, and DNA was pelleted by centrifuging at 13,000 rpm for 15 min at 4°C. The supernatant was carefully removed and discarded, while the DNA pellet was air dried for 15 min at room temperature. The DNA was resuspended in 50 μl of 1× TE buffer (10 mM Tris-Cl [pH 7.5], 1 mM EDTA) and used for quantification by quantitative PCR (the primer sequences are shown in Table S2 in the supplemental material).
Transient transfection and RNA interference.
The Neon transfection system (Invitrogen) was used for transient transfection as recommended by the manufacturer, and Metafectene (Bionex) was used according to the manufacturer’s instructions. We transfected cells with 2 μg of luciferase reporter plasmid and maintained the cells in 6-well plates. For Dbp knockdown, we transfected cells with 2 μl of 20 μM siRNA. We used specific siRNAs for Dbp—siGenome SMARTpool (Dharmacon) siRNAs D-046342-01 (target sequence, GAACUGAAGCCUCAACCAA), D-046342-02 (target sequence, GUCGAGAGAUGCAAGAAGA), D-046342-03 (target sequence, GUGCUGUGCUUUCACGCUA), and D-046342-04 (target sequence, GCGCAGGCUUGACAUCUAG)—to conduct the knockdown of Dbp.
ATAC-qPCR.
The assay for transpose-accessible chromatin (ATAC) was performed as described previously (28) on three wild-type NIH 3T3 cell lines and three NIH 3T3 cell lines with poly(C) deleted. Libraries were generated using the Ad1_noMX, Ad2.13, Ad2.15, Ad2.16, and Ad2.17 barcoded primers based on the study by Buenrostro et al. (29) and were amplified for 6 or 7 total cycles. The libraries were purified with SPRI beads (AccuGene; catalog no. ACN01.450) to remove the remaining primer dimers. The libraries were quantitated by using Qubit (Thermo Fisher; catalog no. Q32854) (the primer sequences are shown in Table S3 in the supplemental material).
Statistical analysis.
Graphs were plotted as means and standard errors of the means (SEM). Two-way analysis of variance (ANOVA) was used to conduct grouped analyses with multiple groups by Tukey's multiple-comparison test analyzed with PRISM software (GraphPad Software). We also used CircWave software (https://www.euclock.org/results/item/circ-wave.html), which could fit a sine wave to confirm the rhythmicity to be determined.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Bio and Medical Technology Development Program (2017M3C7A1023478) and a grant (2019R1A2C2009440) from the National Research Foundation (NRF), funded by the Korean government (MSIT); the Cooperative Research Program for Agriculture Science and Technology Development (project no. PJ01324801) of the Rural Development Administration; and BK21 Plus, funded by the Ministry of Education, Republic of Korea (10Z20130012243).
P.K.K., H.-M.K., S.W.K., and B.K. conducted and analyzed the experiments; H.Y. and H.-O.K. analyzed the lumicycle data; T.-Y.R. and K.-T.K. designed the experiments; P.K.K. and K.-T.K. wrote the paper.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00101-19.
REFERENCES
- 1.Yang S, Van Dongen HPA, Wang K, Berrettini W, Bućan M. 2009. Assessment of circadian function in fibroblasts of patients with bipolar disorder. Mol Psychiatry 14:143–155. doi: 10.1038/mp.2008.10. [DOI] [PubMed] [Google Scholar]
- 2.Luo WY, Chen WF, Yue ZF, Chen DC, Sowcik M, Sehgal A, Zheng XZ. 2012. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell 11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gloston GF, Yoo SH, Chen ZJ. 2017. Clock-enhancing small molecules and potential applications in chronic diseases and aging. Front Neurol 8:100. doi: 10.3389/fneur.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY, Logan RW, Ma T, Lewis DA, Tseng GC, Sibille E, McClung CA. 2016. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc Natl Acad Sci U S A 113:206–211. doi: 10.1073/pnas.1508249112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P, Schibler U. 1992. The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci Suppl 16:123–127. [DOI] [PubMed] [Google Scholar]
- 6.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 7.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. 2001. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev 15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falvey E, Marcacci L, Schibler U. 1996. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem 377:797–809. [PubMed] [Google Scholar]
- 9.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. 2008. Analysis and synthesis of high-amplitude cis-elements in the mammalian circadian clock. Proc Natl Acad Sci U S A 105:14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozek K, Relogio A, Kielbasa SM, Heine M, Dame C, A K, Herzel H. 2009. Regulation of clock-controlled genes in mammals. PLoS One 4:e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripperger JA, Shearman LP, Reppert SM, Schibler U. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev 14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 12.Ripperger JA, Schibler U. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 13.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. 2012. Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell 48:277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA. 2010. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev 24:1317–1328. doi: 10.1101/gad.578810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuarin J, Schibler U. 1990. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 63:1257–1266. doi: 10.1016/0092-8674(90)90421-A. [DOI] [PubMed] [Google Scholar]
- 16.Mueller CR, Maire P, Schibler U. 1990. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell 61:279–291. doi: 10.1016/0092-8674(90)90808-R. [DOI] [PubMed] [Google Scholar]
- 17.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 19.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyohara YB, Nishii K, Ukai-Tadenuma M, Ueda HR, Uchiyama Y, Yagita K. 2008. Detection of a circadian enhancer in the mDbp promoter using prokaryotic transposon vector-based strategy. Nucleic Acids Res 36:e23. doi: 10.1093/nar/gkn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett MK, Erondu NE, Kennedy MB. 1983. Purification and characterization of a calmodulin-dependent protein kinase that is highly concentrated in brain. J Biol Chem 258:12735–12744. [PubMed] [Google Scholar]
- 22.Ohno T, Onishi Y, Ishida N. 2006. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res 35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamajuku D, Shibata Y, Kitazawa M, Katakura T, Urata H, Kojima T, Nakata O, Hashimoto S. 2010. Identification of functional clock-controlled elements involved in differential timing of Per1 and Per2 transcription. Nucleic Acids Res 38:7964–7973. doi: 10.1093/nar/gkq678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT. 2012. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol 32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boden M, Bailey TL. 2008. Associating transcription factor-binding site motifs with target GO terms and target genes. Nucleic Acids Res 36:4108–4117. doi: 10.1093/nar/gkn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buske FA, Boden M, Bauer DC, Bailey TL. 2010. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics 26:860–866. doi: 10.1093/bioinformatics/btq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KH, Kim SH, Kim HJ, Kim W, Lee HR, Jung Y, Choi JH, Hong KY, Jang SK, Kim KT. 2014. AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res 42:3590–3606. doi: 10.1093/nar/gkt1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, Cho SW, Mumbach MR, Carter AC, Kasowski M, Orloff LA, Risca VI, Kundaje A, Khavari PA, Montine TJ, Greenleaf WJ, Chang HY. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi HS, Hwang CK, Song KY, Law P-Y, Wei L-N, Loh HH. 2009. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem Biophys Res Commun 380:431–436. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.