Cytoskeleton-associated protein 4 (CKAP4) is an endoplasmic reticulum protein that is also present in the cell surface membrane, where it acts as a receptor for Dickkopf1 (DKK1). In this study, we found that CKAP4 interacts with β1 integrin and controls the recycling of α5β1 integrin independently of DKK1. In S2-CP8 cells, knockdown of CKAP4 but not DKK1 enlarged the size of cell adhesion sites and enhanced cell adhesion to fibronectin, resulting in decreased cell migration.
KEYWORDS: CKAP4, SNX17, cell adhesion, fibronectin, integrins, recycling
ABSTRACT
Cytoskeleton-associated protein 4 (CKAP4) is an endoplasmic reticulum protein that is also present in the cell surface membrane, where it acts as a receptor for Dickkopf1 (DKK1). In this study, we found that CKAP4 interacts with β1 integrin and controls the recycling of α5β1 integrin independently of DKK1. In S2-CP8 cells, knockdown of CKAP4 but not DKK1 enlarged the size of cell adhesion sites and enhanced cell adhesion to fibronectin, resulting in decreased cell migration. When CKAP4 was depleted, the levels of α5 but not β1 integrin were increased in the cell surface membrane. A similar phenotype was observed in other cells expressing low levels of DKK1. In S2-CP8 cells, α5 integrin was trafficked with β1 integrin and CKAP4 to the lysosome or recycled with β1 integrin. In CKAP4-depleted cells, the internalization of α5β1 integrin was unchanged, but its recycling was upregulated. Knockdown of sorting nexin 17 (SNX17), a mediator of integrin recycling, abrogated the increased α5 integrin levels caused by CKAP4 knockdown. CKAP4 bound to SNX17, and its knockdown enhanced the recruitment of α5β1 integrin to SNX17. These results suggest that CKAP4 suppresses the recycling of α5β1 integrin and coordinates cell adhesion sites and migration independently of DKK1.
INTRODUCTION
Cytoskeleton-associated protein 4 (CKAP4; also known as CLIMP-63 and ERGIC-63) is a nonglycosylated type II transmembrane protein that is located in the endoplasmic reticulum (ER) of all tissues (1, 2). CKAP4 plays a role in segregating ER sheets close to the nucleus. It maintains the luminal width through intermolecular binding of the luminal region of CAKP4 localized on opposing cisternal membranes (3, 4). The cytoplasmic region of CKAP4 binds to microtubules and creates a link between the ER and microtubules (5, 6). CKAP4 is a Dicer-binding protein and regulates the microRNA pathway and mRNA translation by anchoring Dicer to the ER (7). CKAP4 is also involved in gentamicin-induced apoptosis in proximal tubule cells through its binding to gentamicin in the ER lumen (8).
In addition to the functions in the ER, it has been shown that a small fraction of CKAP4 is present in the cell surface membrane of type II pneumocytes and vascular smooth muscle cells (VSMCs), where it functions as a receptor for surfactant protein A (SP-A) and tissue plasminogen activator (tPA), respectively (9, 10). In addition, it has recently been reported that CKAP4 acts as a receptor for Dickkopf1 (DKK1) (11), which can antagonize Wnt signaling (12). DKK1-CKAP4 signaling promotes cell proliferation through the phosphoinositide 3-kinase (PI3K)–AKT pathway in pancreas, lung, and esophageal cancers (11, 13–16). Because CKAP4 knockdown also inhibits cancer cell migration and invasion, CKAP4 in the cell surface membrane is thought to regulate cell migration. AKT, a potential downstream molecule of DKK1-CKAP4 signaling, is involved in cell adhesion, migration, and invasion, but its role is isoform specific and cell type dependent (17, 18). For example, overexpression of activated AKT1 and AKT2 increases invasion of renal, pancreatic, and ovarian cancer cells (19–21), whereas AKT1 suppresses the migration and invasion of breast cancer cells (22). Whether AKT is involved in the inhibition of cell migration in CKAP4-depleted cells has not yet been determined. Therefore, the molecular mechanism by which CKAP4 regulates cell migration is still unclear, and there may be a novel function for CKAP4 in the cell surface membrane other than the activation of AKT through its role as a DKK1 receptor.
Cell migration is a complex cellular behavior that involves protrusion and adhesion at the cell front and contraction and detachment at the rear (23). Cell adhesion sites, which consist of integrins and adaptor proteins, connect the cytoskeleton to the extracellular matrix through the transmission of intracellular forces to the cell exterior, ligand-dependent integrin clustering, and the formation of a macromolecular adhesion complex (24). Integrin trafficking is important for integrin-dependent cell adhesion, spreading, migration, and cancer cell invasion. Trafficking starts with integrin internalization to the endosome through micropinocytosis and clathrin-dependent and -independent endocytosis (25, 26). This internalization is an important step in the regulation of cell migration through the turnover of adhesion sites (27). Internalized integrins are primarily returned to the cell surface membrane or partially routed to the lysosome for degradation (28). In this study, we showed that CKAP4 is involved in the regulation of cell adhesion sites and migration through the recycling of α5β1 integrin independently of DKK1.
RESULTS
β1 integrin is a CKAP4-binding protein.
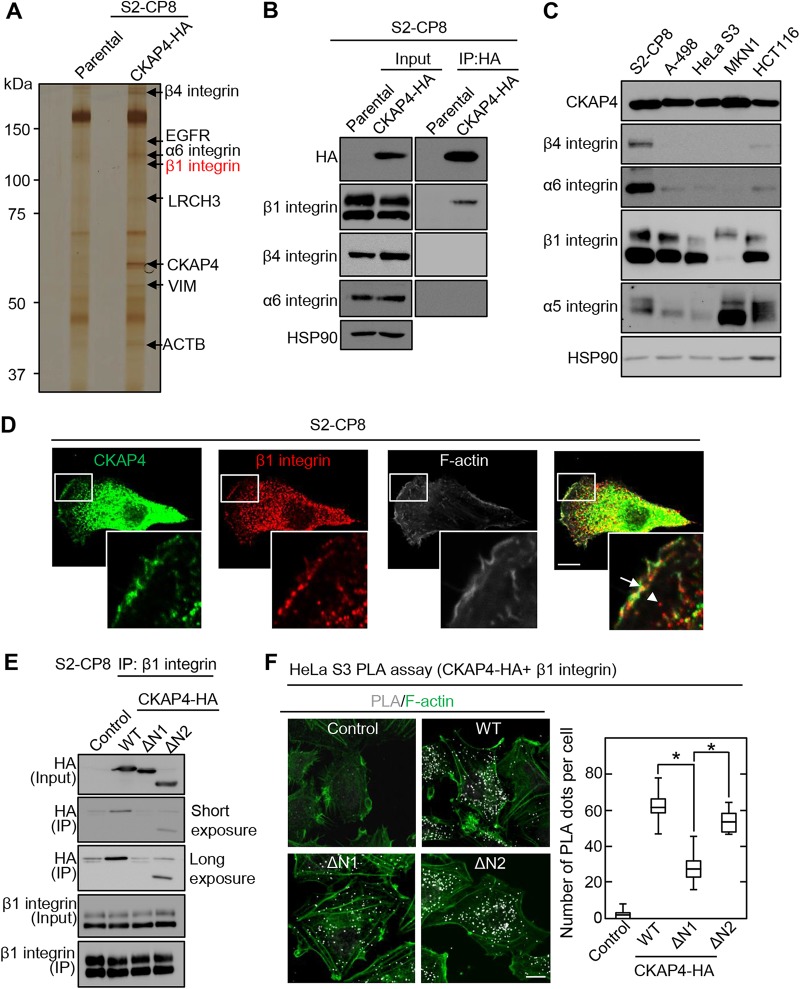
To understand the role of CKAP4 in cell migration, we investigated which proteins bind to CKAP4 in the cell surface membrane. To this end, cell surface membrane proteins in S2-CP8 cells stably expressing the CKAP4-hemagglutinin tag (S2-CP8/CKAP4-HA) were biotinylated. The CKAP4-HA-binding proteins were isolated using an anti-HA antibody and HA peptide. The isolated proteins were further precipitated using NeutrAvidin-agarose beads (Fig. 1A). The precipitated proteins were analyzed by mass spectrometry. The analysis identified integrin family members (α6, β1, and β4), epidermal growth factor receptor (EGFR), leucine-rich repeats and calponin homology domain containing 3 (LRCH3), vimentin (VIM), and actin, cytoplasmic 1 (ACTB) in the CKAP4-HA complex (Fig. 1A).
FIG 1.
β1 integrin is a CKAP4-binding protein. (A) CKAP4-interacting cell surface proteins were precipitated and detected by silver staining. The indicated bands were analyzed by mass spectrometry to identify the binding proteins. (B) Parental and CKAP4-HA-expressing S2-CP8 cells were lysed, and the lysates were immunoprecipitated using an anti-HA antibody. The immunoprecipitates were probed with the indicated antibodies. HSP90 was used as a loading control. (C) Lysates from various cancer cell lines were probed with the indicated antibodies. HSP90 was used as a loading control. (D) S2-CP8 cells were stained with anti-CKAP4 and anti-β1 integrin antibodies. An arrow and an arrowhead indicate colocalization of CKAP4 and β1 integrin and the β1 integrin-containing cytoplasmic vesicle, respectively. Scale bar, 10 μm. (E) S2-CP8 cells expressing various HA-tagged CKAP4 deletion mutants were lysed, and the lysates were immunoprecipitated with anti-β1 integrin antibody. The immunoprecipitates (IP) were probed with the indicated antibodies. (F) (Left) HeLa S3 cells expressing various HA-tagged CKAP4 deletion mutants were incubated with mouse anti-β1 integrin and rabbit anti-HA antibodies. These primary antibodies were combined with secondary PLA probes. (Right) Box and whisker plot of the number of PLA dots per cell. Scale bar, 10 μm. *, P < 0.0001.
When the interaction between CKAP4 and the integrins was examined using immunoprecipitation and Western blotting, endogenous β1 integrin, but not β4 or α6 integrin, formed a complex with CKAP4-HA in S2-CP8/CKAP4-HA cells (Fig. 1B). β1 integrin exists in an immature Golgi resident form of ∼100 kDa, which is not fully glycosylated, and its glycosylated mature form is 130 kDa. The mature β1 integrin is transferred from the Golgi apparatus to the cell surface membrane (29). CKAP4-HA primarily bound to the slowly migrating form of mature β1 integrin (Fig. 1B). Thus, glycosylation could change the three-dimensional structure of β1 integrin and the mature form of β1 integrin may expose the appropriate region to interact with CKAP4. The α6 and β4 integrins were highly expressed in S2-CP8 cells but not in other cancer cell lines, including A-498, HeLa S3, MKN1, and HCT116 cells (Fig. 1C). In contrast, the α5 and β1 integrins were detected in these cell lines. Therefore, we further analyzed the relationship between CKAP4 and β1 integrin. The relationships between CKAP4 and other candidate proteins (e.g., EGFR and LRCH) were not investigated in this study.
CKAP4 was localized to the cell surface membrane and the perinuclear ER by immunohistochemistry (Fig. 1D). β1 integrin was observed in the cell surface membrane and cytoplasmic vesicles (Fig. 1D, arrow and arrowhead) as reported previously (30). Both proteins were closely localized on the cell surface membrane and partially overlapped (Fig. 1D, arrow). When deletion mutants of CKAP4-HA (see Fig. S1 in the supplemental material) were stably expressed in S2-CP8 cells, wild-type (WT) CKAP4 and ΔN2-CKAP4, but not ΔN1-CKAP4, formed a complex with β1 integrin (Fig. 1E). The proximity ligation assay (PLA) also demonstrated that ΔN2-CKAP4 was localized with β1 integrin in HeLa S3 cells to a similar extent as WT CKAP4 (Fig. 1F and Fig. S1), suggesting that the cytoplasmic N-terminal region (amino acids [aa] 1 to 21) of CKAP4 is important for binding β1 integrin. In addition, PLA signal was also found in the cytoplasm, suggesting that CKAP4 and β1 integrin interact with each other not only in the cell surface but also in the cytoplasm.
CKAP4 regulates cell adhesion sites and migration.
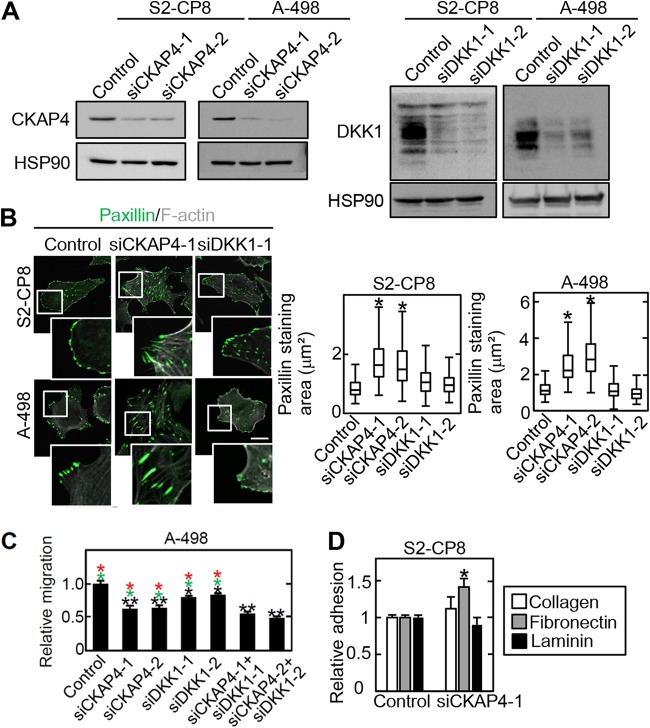
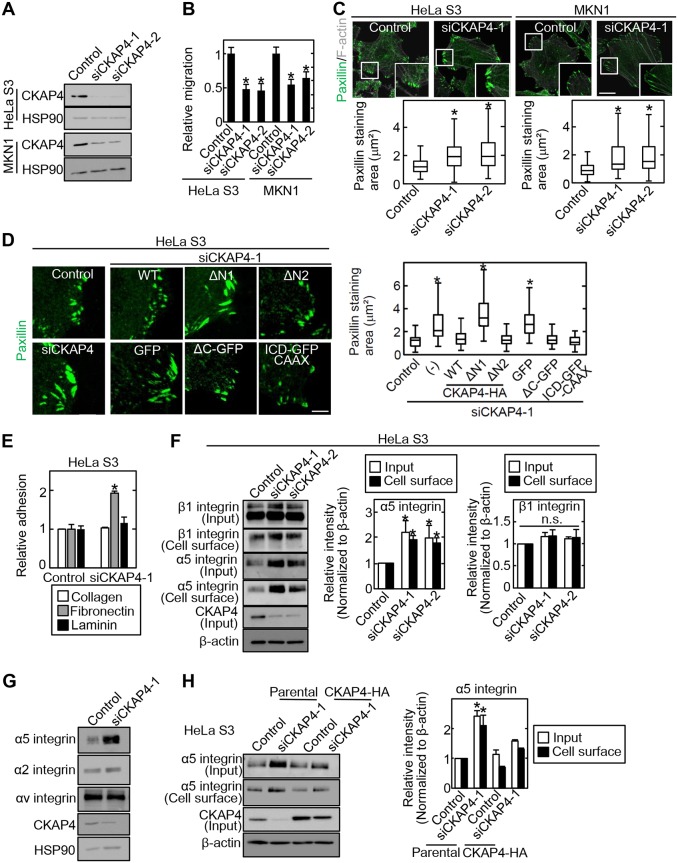
Cell surface expression of CKAP4 and total expression of CKAP4 were compared in various cancer cell lines. PANC-1, DLD-1, TMK1, MKN1, MKN45, A-498, and HeLa S3 cells expressed cell surface-localized CKAP4 to levels similar to that of S2-CP8 cells (Fig. S2). PANC-1, HCT116, Caco-2, KKLS, MKN45, and A-498 cells expressed DKK1 at higher levels than S2-CP8 cells (Fig. S2). Adhesion site turnover is important for cell migration, and there is a tight relationship between the size of cell adhesion sites and cell migration speed (24, 31); the larger the size of cell adhesion sites, the slower the migration. Therefore, the state of the cell adhesion sites was examined in CKAP4-depleted S2-CP8 and A-498 cells in this study. The size of cell adhesion sites, which was estimated by measuring the paxillin-stained areas, was increased by knockdown of CKAP4 but not DKK1 using two different small interfering RNAs (siRNAs) (Fig. 2A and B). Overexpression of CKAP4-HA decreased the size of the cell adhesion sites when CKAP4 was transiently expressed in WT S2-CP8 cells (Fig. S3). Consistent with the previous observations in S2-CP8 cells (11), knockdown of CKAP4 inhibited the migration of A-498 cells more efficiently than DKK1 knockdown (Fig. 2C). When CKAP4 and DKK1 were knocked down simultaneously in A-498 cells, cell migration was slower than when either CKAP4 or DKK1 was knocked down (Fig. 2C). These results suggest that CKAP4 and DKK1 might differentially regulate cell migration although both proteins are involved in it.
FIG 2.
Cell surface CKAP4 determines the size of cell adhesion sites. (A) S2-CP8 and A-498 cells were transfected with the indicated siRNAs. The lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (B) (Left) S2-CP8 and A-498 cells were transfected with the indicated siRNAs. The cells were fixed and stained with antipaxillin antibody and phalloidin. Scale bar, 20 μm. (Right) Box and whisker plots of the measured areas of individual paxillin-positive regions. *, P < 0.0001. (C) Migration of A-498 cells was measured using the transwell assay 4 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. Data are presented as the mean ± SD of results from three independent experiments. Black asterisks indicate a comparison between the control and the others. Red or green asterisks indicate comparisons between siCKAP4-1+siDKK1-1 or siCKAP4-2+siDKK1-2 and the others, respectively. *, P < 0.01; **, P < 0.001. (D) S2-CP8 cells transfected with CKAP4 siRNA (siCKAP4-1) were plated on collagen, fibronectin, or laminin for 5 min, and the adherent cells were stained with crystal violet. The crystal violet was eluted and measured. The results are expressed as the cell adhesion activity relative to that of cells treated with control siRNA. *, P < 0.05.
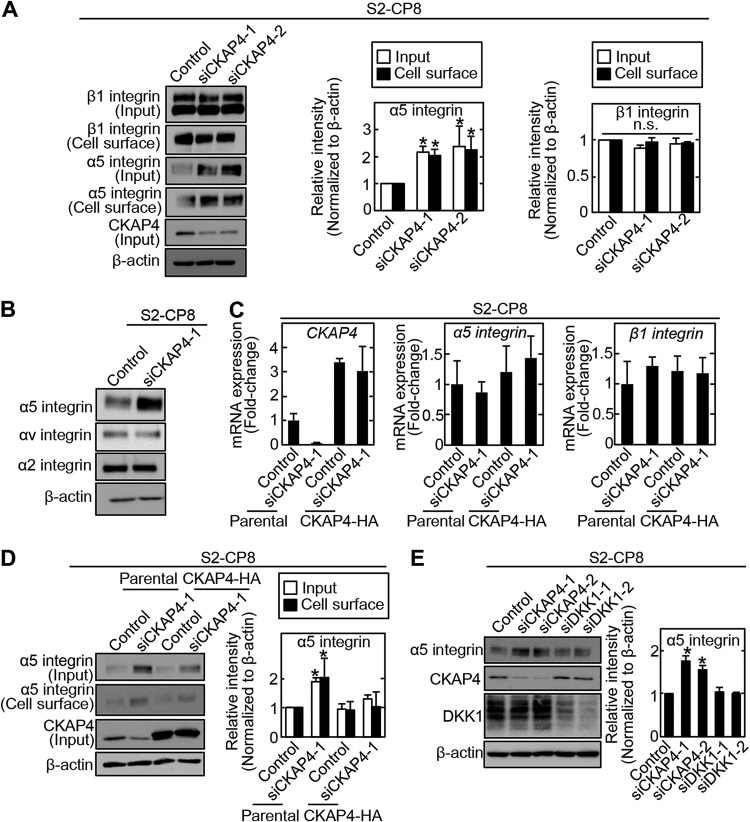
Integrin family members bind to extracellular matrix proteins (28, 32). Fibronectin-dependent adhesion was enhanced in CKAP4-depleted S2-CP8 cells, but collagen- or laminin-dependent adhesion remained unchanged (Fig. 2D). Among the different integrin family members, α5β1 integrin specifically binds to fibronectin (33). The levels of α5 integrin in the cell surface membrane and total cell lysates (input) were increased in CKAP4-depleted S2-CP8 cells, whereas the levels of β1, α2, and αv integrins were unchanged (Fig. 3A and B). The mRNA levels of α5 and β1 integrins were not affected by knockdown or overexpression of CKAP4 (Fig. 3C), indicating that the increased level of α5 integrin protein was not due to changes in transcription. The increase in α5 integrin levels in the cell surface membrane was abolished by expression of CKAP4-HA (Fig. 3D). The amount of α5 integrin was not increased by DKK1 depletion in S2-CP8 cells (Fig. 3E). Therefore, CKAP4 may regulate α5 integrin protein levels independently of DKK1. Taken together, these results suggest that CKAP4 regulates α5 integrin protein levels by binding to β1 integrin and also affects fibronectin-dependent cell adhesion and migration.
FIG 3.
CKAP4 regulates α5 integrin protein levels independently of DKK1. (A) (Left) S2-CP8 cells were transfected with the indicated CKAP4 siRNAs, and then the cell surface proteins were biotinylated and precipitated with NeutrAvidin-agarose beads. The precipitates were probed with the indicated antibodies. (Center and right) The α5 integrin and β1 integrin band intensities of input and cell surface were quantified. β-Actin was used as a loading control. *, P < 0.05. n.s., not significant. (B) S2-CP8 cells were transfected with CKAP4 siRNA (siCKAP4-1), and the lysates were probed with the indicated antibodies. β-Actin was used as a loading control. (C) Relative CKAP4 and α5- and β1-integrin mRNA levels in parental or CKAP4-HA-expressing S2-CP8 cells transfected with CKAP4 siRNA (siCKAP4-1) were measured by quantitative reverse transcription (RT)-PCR and normalized to GAPDH. Data are presented as the fold change compared to the levels in control siRNA-transfected cells. (D) (Left) S2-CP8/CKAP4-HA cells were transfected with CKAP4 siRNA (siCKAP4-1), and then the cell surface proteins were biotinylated and precipitated with NeutrAvidin-agarose beads. The precipitates were probed with the indicated antibodies. (Right) The α5 integrin band intensities of total cell lysates (input) and cell surface were quantified. β-Actin was used as a loading control. *, P < 0.05. (E) (Left) S2-CP8 cells were transfected with the indicated siRNAs, and the lysates were probed with the indicated antibodies. (Right) The α5 integrin band intensities were quantified. β-Actin was used as a loading control. *, P < 0.05.
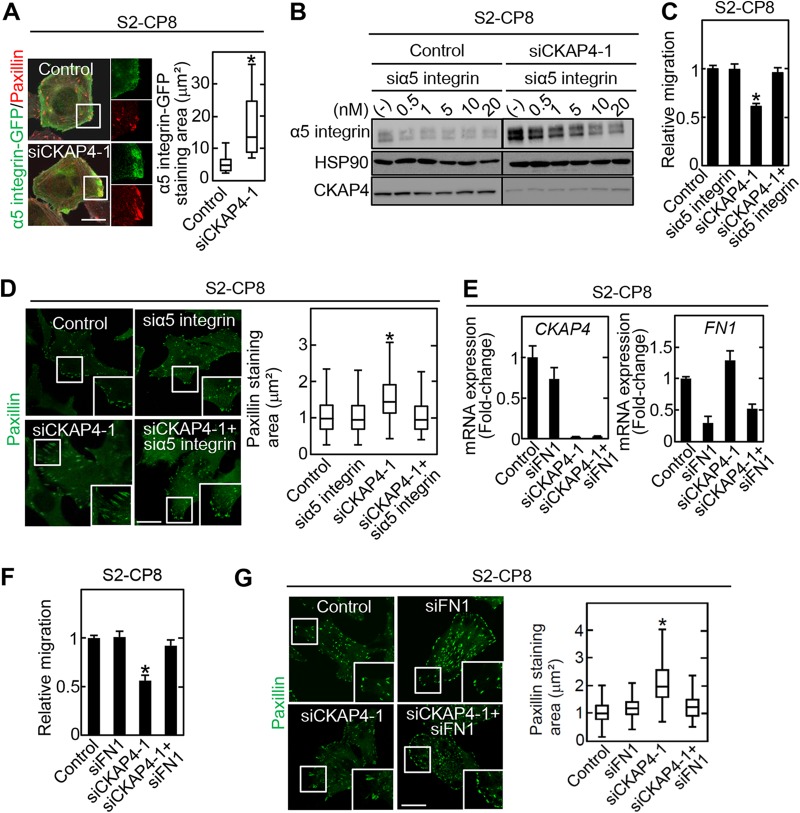
α5 integrin staining was used to examine the relationship between α5 integrin and the size of cell adhesion sites in S2-CP8 cells. The depletion of CKAP4 from S2-CP8 cells caused an increase in the α5 integrin staining areas (Fig. 4A). The α5 integrin level in the total cell lysate was increased by CKAP4 knockdown, which was suppressed by the depletion of α5 integrin (Fig. 4B). The inhibition of cell migration and enlargement of cell adhesion sites observed in the CKAP4-depleted S2-CP8 cells were rescued by α5 integrin knockdown (Fig. 4C and D). These phenotypes were also rescued by fibronectin (FN1) knockdown (Fig. 4E to G). Taken together, the phenotypes induced by CKAP4 knockdown may be at least partially caused by the increase in α5 integrin levels and its binding to fibronectin.
FIG 4.
CKAP4 is involved in cell migration through α5 integrin and fibronectin. (A) (Left) α5 integrin-GFP-expressing S2-CP8 cells were transfected with CKAP4 siRNA (siCKAP4-1) and stained with anti-GFP and antipaxillin antibodies. Scale bar, 20 μm. (Right) Box and whisker plot for the staining area of α5 integrin-GFP merged with paxillin. *, P < 0.0001. (B) S2-CP8 cells were simultaneously transfected with CKAP4 siRNA (siCKAP4-1) and the indicated concentrations of α5 integrin siRNA. The lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (C) S2-CP8 cells were simultaneously transfected with CKAP4 siRNA (siCKAP4-1) and α5 integrin siRNA. Migration of the transfected cells was measured by the transwell assay 3 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. Data are presented as the mean ± SD of results from three independent experiments. *, P < 0.05. (D) (Left) The same cells as those described for panel C were stained with antipaxillin antibody. Scale bar, 20 μm. (Right) Box and whisker plot for the measured areas of individual paxillin-positive regions. *, P < 0.0001. (E) Relative CKAP4 and FN1 mRNA expression levels in S2-CP8 cells transfected with the indicated siRNAs were measured by quantitative RT-PCR and normalized to GAPDH. Data are presented as the fold change relative to control siRNA-transfected cells. (F) S2-CP8 cells were simultaneously transfected with CKAP4 siRNA (siCKAP4-1) and fibronectin 1 siRNA (siFN1). Migration of the transfected cells was measured by the transwell assay 3 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. Data are presented as the mean ± SD of results from three independent experiments. *, P < 0.05. (G) (Left) The same cells as described for panel F were stained with antipaxillin antibody. Scale bar, 20 μm. (Right) Box and whisker plot for the measured area of individual paxillin-positive regions. *, P < 0.0001.
CKAP4 in the cell surface membrane regulates cell migration independently of DKK1.
As shown in Fig. S2, MKN1 and HeLa S3 cells express CKAP4 on the cell surface membrane but have little DKK1. To examine whether CKAP4, but not DKK1, regulates α5 integrin turnover, we examined the effects of CKAP4 knockdown on cell migration and the size of cell adhesion sites in these two cell lines. CKAP4 knockdown suppressed cell migration and enlarged the size of cell adhesion sites in both cell lines (Fig. 5A to C). The enlarged cell adhesion sites in CKAP4-depleted HeLa S3 cells was rescued by expression of WT or ΔN2 CKAP4 but not ΔN1 CKAP4 (Fig. 5D). A deletion mutant of CKAP4 extracellular domain (ΔC-GFP CKAP4 [where GFP is green fluorescent protein]), which was found at the cell surface membrane (Fig. S4), also rescued the enlarged cell adhesion sites (Fig. 5D). Therefore, the cytoplasmic N-terminal region (aa 1 to 21) of CKAP4 is important not only for the binding of β1 integrin but also for the regulation of adhesion sites. Enlargement of the cell adhesion sites was also rescued by expression of the intracellular domain (ICD) of CKAP4 fused to a cell membrane targeting domain (ICD-GFP-CAAX) (Fig. 5D and Fig. S1), suggesting that CKAP4 localized in the cell surface membrane but not ER-resident CKAP4 could regulate cell adhesion sites. Similar to S2-CP8 cells, knockdown of CKAP4 increased fibronectin-dependent cell adhesion of HeLa S3 cells (Fig. 5E). In addition, CKAP4 knockdown in HeLa S3 cells increased α5 integrin protein levels in the cell surface membrane but not that of β1, α2, and αv integrins (Fig. 5F and G). CKAP4 expression rescued the increase of α5 integrin in CKAP4-depleted HeLa S3 cells (Fig. 5H).
FIG 5.
CKAP4 regulates cell adhesion sites independently of DKK1. (A) HeLa S3 and MKN1 cells were transfected with CKAP4 siRNAs. The lysates were probed with anti-CKAP4 and anti-HSP90 antibodies. (B) HeLa S3 and MKN1 cells were transfected with CKAP4 siRNAs. Migration of the transfected cells was measured by the transwell assay 3 to 8 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. *, P < 0.0001. (C) (Top) HeLa S3 and MKN1 cells were transfected with CKAP4 siRNAs and stained with antipaxillin antibody and Alexa Fluor-phalloidin. Scale bar, 20 μm. (Bottom) Box and whisker plot for the measured areas of individual paxillin-positive regions. *, P < 0.0001. (D) (Left) CKAP4-depleted HeLa S3 cells were transfected with the indicated constructs and stained with antipaxillin antibody. Scale bar, 5 μm. (Right) Box and whisker plot for the measured areas of individual paxillin-positive regions. *, P < 0.0001. (E) HeLa S3 cells transfected with CKAP4 siRNA (siCKAP4-1) were plated on collagen, fibronectin, or laminin for 5 min, and the adherent cells were stained with crystal violet. The crystal violet was eluted and measured. The results are expressed as cell adhesion activity relative to that of cells treated with control siRNA. Data are presented as the mean ± SD of results of three independent experiments. *, P < 0.05. (F) (Left) HeLa S3 cells were transfected with CKAP4 siRNAs, and then the cell surface proteins were biotinylated and precipitated with NeutrAvidin-agarose beads. The precipitates were probed with the indicated antibodies. (Center and right) The α5 integrin and β1 integrin band intensities of total cell lysates (input) and cell surface were quantified. β-Actin was used as a loading control. *, P < 0.05; n.s., not significant. (G) HeLa S3 cells were transfected with CKAP4 siRNA (siCKAP4-1), and the lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (H) (Left) CKAP4-HA-expressing HeLa S3 cells were transfected with CKAP4 siRNA (siCKAP4-1), and then the cell surface proteins were biotinylated and precipitated with NeutrAvidin-agarose beads. The precipitates were probed with the indicated antibodies. (Right) The α5 integrin band intensities of the total cell lysates (input) and cell surface were quantified. β-Actin was used as a loading control. *, P < 0.05.
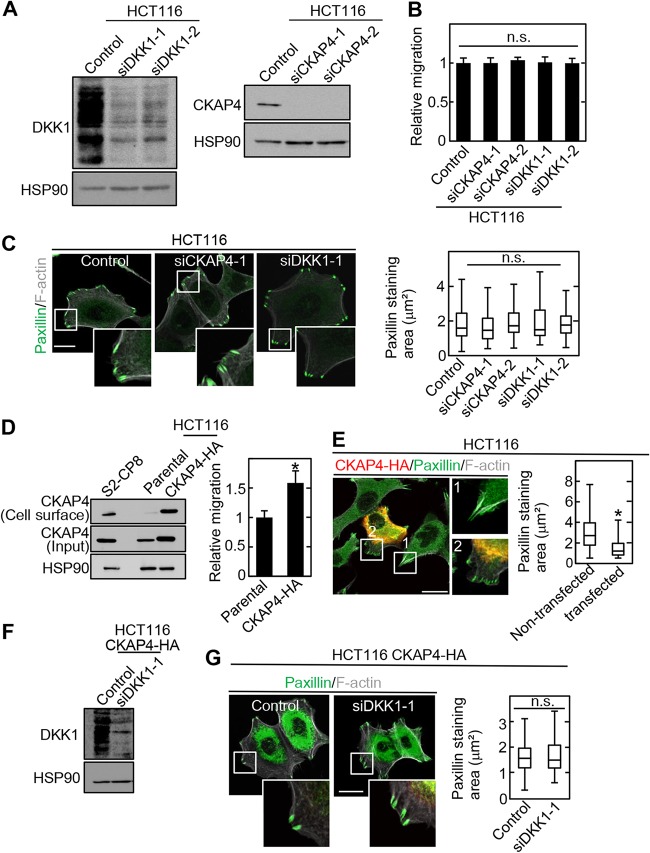
DKK1 was expressed but CKAP4 was little expressed in the cell surface membrane of HCT116 cells (Fig. S2). Knockdown of CKAP4 or DKK1 did not affect cell migration or cell adhesion sites in HCT116 cells (Fig. 6A to C). Stable overexpression of CKAP4-HA in HCT116 cells (HCT116/CKAP4-HA cells) resulted in the appearance of CKAP4 in the cell surface membrane and increased cell migration (Fig. 6D). Overexpression of CKAP4-HA decreased the size of cell adhesion sites when CKAP4 was transiently expressed in WT HCT116 cells (Fig. 6E). DKK1 knockdown in HCT116/CKAP4-HA cells did not affect cell adhesion sites (Fig. 6F and G). Taken together, these results support the idea that CKAP4 regulation of cell adhesion sites and migration is independent of DKK1.
FIG 6.
Cell surface CKAP4 regulates cell adhesion sites independently of DKK1. (A) HCT116 cells were transfected with the indicated siRNAs. The lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (B) HCT116 cells were transfected with the indicated siRNAs. Migration of the transfected cells was measured by the transwell assay 8 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. Data are presented as the mean ± SD of results for three independent experiments. n.s., not significant. (C) (Left) The same cells as those described for panel B were stained with antipaxillin antibody and Alexa Fluor-phalloidin. Scale bar, 20 μm. (Right) Box and whisker plot for the measured areas of individual paxillin-positive regions. n.s., not significant. (D) (Left) The lysates of parental and HCT116/CKAP4-HA cells were probed with the indicated antibodies. HSP90 was used as a loading control. (Right) Migration of the same cells was measured by the transwell assay 8 h after plating. Migration activity is expressed relative to that of control siRNA-transfected cells. *, P < 0.0001. (E) (Left) CKAP4-HA was transiently overexpressed in HCT116 cells, and the cells were stained with anti-HA and antipaxillin antibodies and Alexa Fluor-phalloidin. Boxed areas 1 and 2 indicate paxillin-staining areas in nontransfected and CKAP4-HA-transfected cells, respectively. The paxillin-staining area (green) in cells with (red) or without CKAP4 expression were measured. Cells expressing CKAP4-HA (red) became yellow when merged with paxillin staining (green). Scale bar, 20 μm. (Right) Box and whisker plot for the measured areas of individual paxillin-positive regions. *, P < 0.0001. (F) CKAP4-HA-expressing HCT116 cells were transfected with DKK1 siRNA (siDKK1-1), and the lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (G) (Left) CKAP4-HA-expressing HCT116 cells were transfected with DKK1 siRNA (siDKK1-1) and stained with antipaxillin antibody and Alexa Fluor-phalloidin. Scale bar, 20 μm. (Right) Box and whisker plot for areas of individual paxillin-positive regions. n.s., not significant.
CKAP4 regulates integrin recycling.
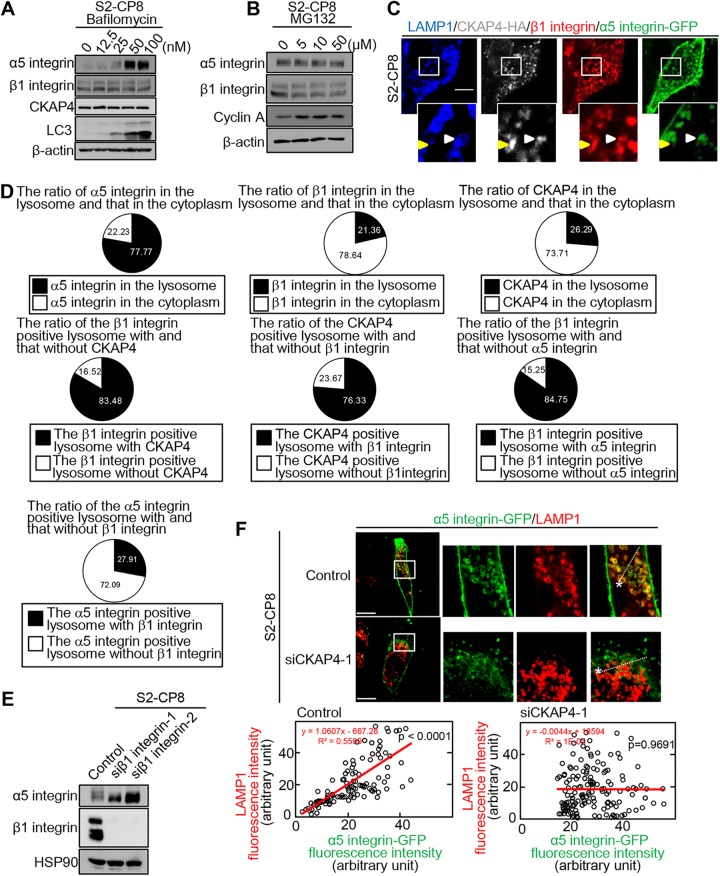
Because it has been reported that internalized α5β1 integrin is trafficked to the cell surface membrane by recycling or to the lysosome for degradation (25, 26), we examined whether CKAP4 is involved in α5β1 integrin trafficking. To this end, S2-CP8 cells were treated with the lysosome inhibitor bafilomycin or the proteasome inhibitor MG132. Consistent with the previous observations in HeLa cells (34, 35), bafilomycin increased the protein level of α5 integrin but not that of CKAP4 in S2-CP8 cells (Fig. 7A), suggesting that the increment in α5 integrin by bafilomycin is not due to a reduction in CKAP4 (Fig. 3A) and that CKAP4 is involved in the trafficking of α5 integrin to the lysosome. Although it has been reported that α5 integrin is degraded in the proteasome of renal cancer cells (36), MG132 did not affect α5 integrin protein levels in this study (Fig. 7B).
FIG 7.
CKAP4 regulates the trafficking of α5 integrin to the lysosome. (A and B) S2-CP8 cells were treated with the indicated doses of bafilomycin (A) or MG132 (B) for 12 h. The lysates were probed with the indicated antibodies. β-Actin was used as a loading control. (C) S2-CP8/CKAP4-HA cells were transfected with α5 integrin-GFP and treated with bafilomycin. The cells were stained with anti-LAMP1 (blue), anti-HA (gray), anti-β1 integrin (red), and anti-GFP (green) antibodies. The regions in the white boxes are enlarged and shown as insets. A white arrowhead indicates CKAP4 and α5 and β1 integrins colocalized with LAMP1-positive lysosomes. A yellow arrowhead indicates β1 integrin with CKAP4 in the nonlysosomal cytoplasm. Scale bar, 10 μm. (D) Colocalization of integrins or CKAP4 with lysosomes was quantified using the images obtained in panel C. (E) S2-CP8 cells were transfected with the indicated β1 integrin siRNAs, and the lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (F) S2-CP8 cells were transfected with CKAP4 siRNA (siCKAP4-1) and α5 integrin-GFP and then treated with bafilomycin. (Upper panels) Immunostaining of cells with anti-GFP and anti-LAMP1 antibodies. The regions in the white boxes are enlarged and shown as insets. (Lower panels) Scattered plot of pixel intensities along the dotted lines in the upper right panels. The regression curve was drawn to obtain a correlation coefficient. Scale bars, 10 μm.
In bafilomycin-treated cells, approximately 78% of the α5 integrin was accumulated in the LAMP1-positive lysosome (Fig. 7C and D). β1 integrin and CKAP4 were found primarily throughout the cytoplasm (Fig. 7C and D, yellow arrowhead), and approximately 21% and 26% of the β1 integrin and CKAP4, respectively, were detected in the lysosome (Fig. 7C and D, white arrowhead). In contrast, most of β1 integrin (83%) and CKAP4 (76%) were colocalized when they were present in the lysosome (Fig. 7C and D). The α5 integrin protein levels were increased in β1 integrin-depleted cells (Fig. 7E), suggesting that β1 integrin bound to CKAP4 sorts α5 integrin to the lysosome. In addition, approximately 85% of the β1 integrin in the lysosome was colocalized with α5 integrin but only 28% of lysosomal α5 integrin was colocalized with β1 integrin (Fig. 7D), suggesting that α5 integrin is sorted to the lysosome in a β1 integrin- and CKAP4-independent manner. The accumulated α5 integrin in CKAP4 knockdown cells was not detected in the lysosome (Fig. 7F). These results suggest that the fraction of β1 integrin that forms a complex with α5 integrin is trafficked with CKAP4 to the lysosome, where α5 integrin is degraded. In the same way, ICD-GFP-CAAX was also observed in the lysosome, which supports the idea that cell surface-localizing CKAP4, but not ER-resident CKAP4, is involved in the trafficking of α5β1 integrin (Fig. S5; see Discussion).
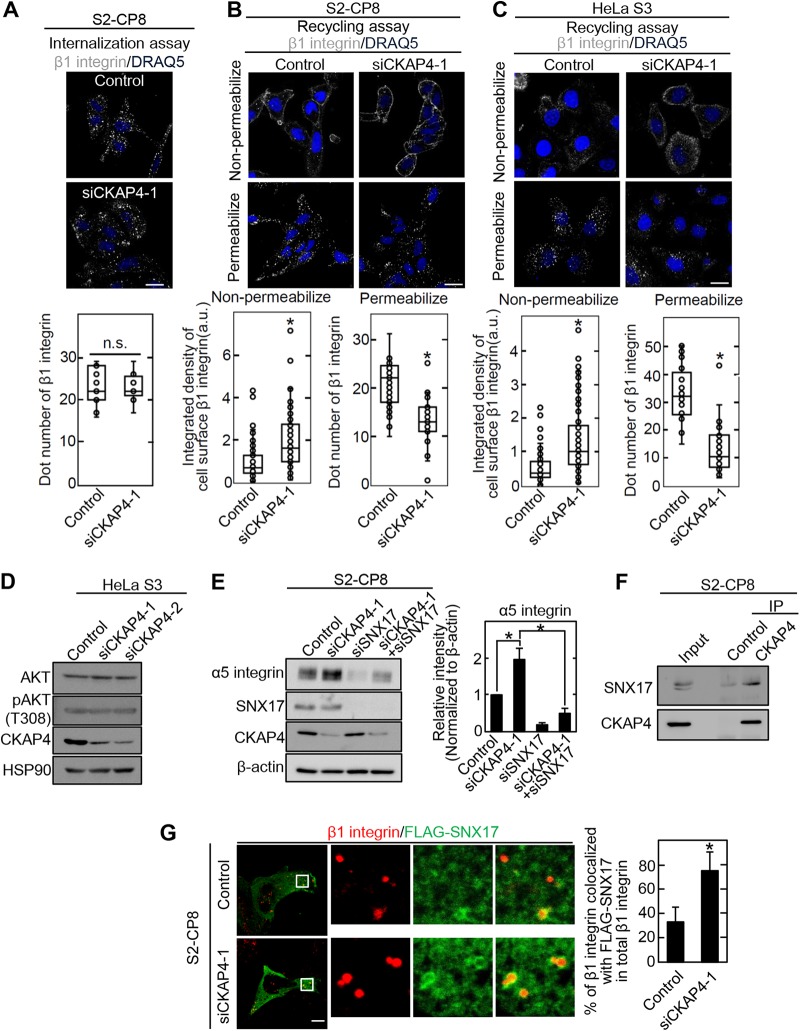
When cell surface CKAP4 was labeled with anti-CKAP4 monoclonal antibody, the CKAP4 was internalized in the cytoplasm by 30 min and recycled back to the cell surface within an additional 30 min (Fig. S6). We then examined whether CKAP4 is involved in β1 integrin recycling. After labeling cell surface β1 integrin with anti-β1 integrin (6S6) antibody, β1 integrin internalization was monitored for 30 min. The results showed that cell surface β1 integrin is internalized with the same efficiency in control and CKAP4 knockdown S2-CP8 cells (Fig. 8A). In the recycling assay for 30 min, most of the internalized β1 integrin was observed in the cytoplasm in control S2-CP8 and HeLa S3 cells, with little present in the cell surface membrane (Fig. 8B and C). In contrast, β1 integrin was clearly observed in the cell surface membrane of CKAP4 knockdown cells and decreased in the cytoplasm (Fig. 8B and C).
FIG 8.
CKAP4 inhibits α5β1 integrin recycling. (A) (Top) Integrin internalization assay for CKAP4 siRNA (siCKAP4-1)-transfected S2-CP8 cells. Cells were stained with a secondary antibody against anti-β1 integrin antibody and DRAQ5 DNA dye. Scale bar, 20 μm. (Bottom) Box and whisker plot of internalized β1 integrin levels measured using ImageJ software. n.s., not significant. (B and C) Recycling assay for CKAP4 siRNA (siCKAP4-1)-transfected S2-CP8 (B) and HeLa S3 (C) cells. Cells were stained with a secondary antibody against anti-β1 integrin antibody and DRAQ5 DNA dye. (Upper panels) Representative images of nonpermeabilized and permeabilized conditions. Scale bars, 20 μm. (Lower panels) Box and whisker plot of recycled and internalized β1 integrin levels measured using ImageJ software. Box and whisker plots of recycled (nonpermeabilized) and internalized (permeabilized) β1 integrin levels are shown as the integrated density of cell surface β1 integrin and dot number of β1 integrin, respectively. *, P < 0.0001. (D) HeLa S3 cells were transfected with CKAP4 siRNAs, and lysates were probed with the indicated antibodies. HSP90 was used as a loading control. (E) (Left) S2-CP8 cells were transfected with the indicated siRNAs. The lysates were probed with the indicated antibodies. (Right) The α5 integrin band intensities were quantified. β-Actin was used as a loading control. *, P < 0.05. (F) S2-CP8 cell lysates were immunoprecipitated with the indicated antibodies. The immunoprecipitates (IP) were probed with the indicated antibodies. (G) (Left) S2-CP8 cells were transfected with CKAP4 siRNA (siCKAP4-1) and FLAG-SNX17. The cells were stained with anti-FLAG and anti-β1 integrin antibodies. The regions in the white boxes are enlarged and shown as insets. Scale bar, 10 μm. (Right) The colocalization percentage of FLAG-SNX17 and β1 integrin in total β1 integrin was determined. *, P < 0.0001.
We also examined whether CKAP4 was involved in the known trafficking routes of α5β1 integrin. The AKT-GSK3 pathway is reported to be involved in the recycling of α5β1 integrin in NIH 3T3 fibroblasts (37). Because AKT activity was not changed in CKAP4 knockdown HeLa S3 cells (Fig. 8D), this pathway would not likely be involved in CKAP4-mediated α5β1 integrin recycling. Sorting nexin 17 (SNX17) is a β1 integrin-binding protein that prevents lysosomal degradation of α5β1 integrin (34, 38). While the α5 integrin level was increased in CKAP4 knockdown cells, it was decreased in SNX17 knockdown cells (Fig. 8E). Furthermore, knockdown of SNX17 abrogated the increased α5 integrin levels in CKAP4 knockdown cells (Fig. 8E). We found that SNX17 formed a complex with CKAP4, and the accumulation of β1 integrin into the SNX17-positive early endosomes was increased in CKAP4-depleted cells (Fig. 8F and G). These results suggest that CKAP4 suppresses the SNX17-mediated α5β1 integrin recycling pathway.
DISCUSSION
The activation of DKK1-CKAP4 signaling has been associated with the aggressiveness of various types of cancer (11, 13–16, 39, 40). Although both DKK1 and CKAP4 are involved in cell migration, the precise mechanism is unclear (11). In this study, we identified β1 integrin as a CKAP4-binding protein and found that CKAP4 regulates cell adhesion sites and cell migration through α5β1 integrin recycling independently of DKK1 (see Fig. S7 in the supplemental material).
S2-CP8 cells were used as a representative cell line to examine the novel CKAP4 functions in this study. A-498, HeLa S3, MKN1, and HCT116 cells were also used to exclude the possibility that our findings were cell line specific. S2-CP8 and A-498 cells expressed both DKK1 and cell surface CKAP4, whereas HeLa S3 and MKN1 cells expressed cell surface CKAP4 but little DKK1. HCT116 cells expressed DKK1 without cell surface CKAP4. By exploiting these features, the impact of DKK1 and cell surface CKAP4 on cell migration and cell adhesion sites was examined using gain-of-function and loss-of-function assays.
Our first finding, that CKAP4 is involved in cell migration and cell adhesion sites, was based on the observation in CKAP4-depleted S2-CP8 cells (11; this study). DKK1 knockdown also inhibited cell migration, but its effect was less than that mediated by CKAP4 knockdown. Although we believed that DKK1-CKAP4 signaling stimulated cell migration through AKT activation in the previous study (11), CKAP4 knockdown showed the same phenotypes in HeLa S3 and MKN1 cells that little express DKK1. Furthermore, overexpression of CKAP4 also reduced the size of cell adhesion sites and stimulated migration in HCT116 cells that little express cell surface CKAP4. In the rescue experiments (Fig. 5D), enlarged cell adhesion sites in CKAP4 knockdown cells were rescued by expression of a cell surface membrane-targeted form of the intracellular domain of CKAP4 (ICD-GFP-CAAX). This mutant was also localized to the lysosome (Fig. S5) and seemed to show phenotypes similar to those of WT CKAP4. Therefore, CKAP4 localized to the cell surface membrane could regulate the trafficking of α5β1 integrin and cell migration that are independent of DKK1.
The interactions between the integrins and their ligands control cell proliferation, differentiation, and migration (33). Cycles of endocytosis and recycling can regulate the availability of the integrins at the cell surface membrane. Because the degradative turnover of integrins takes time (the half-life of most integrins is approximately 12 to 24 h), the majority of internalized integrins are recycled rapidly to function as critical regulators of cell migration (26, 28, 41). Our results showed that CKAP4 binds β1 integrin and controls α5β1 integrin recycling. Among the 24 different integrin heterodimers, α5β1 integrin is one of the fibronectin receptors (42) and a component of cell adhesion sites that develop from a focal complex with focal adhesion kinase, talin, and paxillin upon membrane retraction (43). α5β1 integrin provides stable cell adhesion to the substrate (44), and the inhibition of α5β1 integrin increases persistent migration (45). Consistent with these reports, when the α5 integrin level was increased by CKAP4 knockdown, cell adhesion to fibronectin was stimulated and cell migration was reduced. These phenotypes were rescued by simultaneous knockdown of α5 integrin. Thus, the increased level of α5 integrin could be the primary cause of reduced migration in CKAP4-depleted cells. The other integrin heterodimers may be regulated independently of CKAP4, since the levels of αv and α2 integrins were not altered in CKAP4-depleted S2-CP8 and HeLa S3 cells (Fig. 3B and 5G).
The mechanism by which CKAP4 specifically recognizes the α5β1 integrin heterodimer among the integrin family members is currently being investigated. To target specific integrins to discrete regions of the membrane would require a heterodimer-specific mechanism. Caveolin-1 constitutively regulates endocytosis of α5β1 integrin (46), and internalized α5β1 integrin is sorted to the early endosome and subsequently to a perinuclear recycling compartment (PNRC) before returning to the cell surface membrane via a long-loop pathway that requires Rab11, the AKT-GSK3 pathway, and SNX17 (28, 34, 37, 38). The heterodimer is also routed to the lysosome for degradation by interacting with the endosomal sorting complex required for transport (ESCRT) machinery (41). Our results showed that CKAP4 is not involved in the endocytosis of α5β1 integrin but suppresses its recycling. SNX17 is localized to the early endosome and plays a role in the sorting of vesicles containing α5β1 integrin to the recycling endosome through its binding to β1 integrin (34, 38). CKAP4 formed a complex with SNX17, and β1 integrin was colocalized with SNX17 in CKAP4-depleted cells more efficiently than control S2-CP8 cells. In this context, it is intriguing to speculate that after α5β1 integrin is internalized with CKAP4, CKAP4 interferes with the binding of SNX17 to β1 integrin in the early endosome, thereby suppressing the sorting of α5β1 integrin to the recycling endosome in the long-loop pathway.
In addition, there is a short-loop pathway in which integrins, such as αvβ3, are rapidly delivered back to the plasma membrane from the early endosomes in a Rab4-dependent manner. Since our results showed that the amount of αv subunit of αvβ3 integrin is not changed in CKAP4 knockdown cells, it is unlikely that CKAP4 is involved in the short-loop pathway, but at present, the precise role of CKAP4 in this pathway is unclear.
The level of α5 integrin increased more significantly than that of β1 integrin in CKAP4-depleted cells. β1 integrin is an abundant protein among the integrin family proteins (47) and has many binding partners other than α5 integrin. Integrin heterodimers containing β1 integrin constitute the largest subgroup of the integrin family. A fraction of β1 integrin and most of α5 integrin were distributed to the lysosome in bafilomycin-treated cells (Fig. 7C and D), suggesting that a fraction of β1 integrin complexed with α5 integrin and probably CKAP4 is trafficked to and degraded in the lysosome. Thus, due to the relative abundance of β1 integrin in the integrin family (47), the impact of CKAP4 depletion on the levels of β1 and α5 integrins might be different.
Because cell surface CKAP4 is involved primarily in cancer progression through the function of the DKK1 receptor (11, 13, 14), an anti-CKAP4 antibody has been proposed as a potential treatment against cancers in which both DKK1 and cell surface membrane CKAP4 are expressed (11, 16). However, α5β1 integrin has been demonstrated to enhance invasive cancer cell migration and to represent a molecular target for cancer therapy (48–51). Therefore, it is also possible that CKAP4 has a tumor-suppressive function in a cancer context-dependent manner. Indeed, it has been reported that CKAP4 inhibits the metastasis of hepatocellular carcinoma and is associated with a good prognosis (52). In addition, cell surface CKAP4 interacts with and promotes the internalization of VE-cadherin in vascular endothelial cells (53). It is necessary to more extensively examine the relationship between the cell surface levels of CKAP4 and α5β1 integrin or VE-cadherin to understand the DKK1-independent novel CKAP4 function that regulates membrane protein trafficking.
MATERIALS AND METHODS
Materials and chemicals.
The SW480 and DLD-1 colorectal cancer cell lines and TMK1, KKLS, MKN1, and MKN45 gastric cancer cell lines were kindly provided by W. Yasui (Hiroshima University, Hiroshima, Japan). The HCT116 colorectal, AGS gastric, A-498 kidney, and HeLa S3 cervical cancer cell lines were provided by T. Kobayashi (Hiroshima University, Hiroshima, Japan), M. Hatakeyama (Tokyo University, Tokyo, Japan), T. Tanaka (Osaka City University, Osaka, Japan), and K. Matsumoto (Nagoya University, Aichi, Japan), respectively. The S2-CP8 pancreatic cancer cell line was purchased from the Cell Resource Center for Biomedical Research, Institute of Development, Aging, and Cancer, Tohoku University (Miyagi, Japan).
The PANC-1 and SUIT-2 pancreatic and Caco-2 colorectal cancer cell lines were purchased from the RIKEN Bioresource Center Cell Bank (Tsukuba, Japan). Lenti-X 293T (X293T) cells were purchased from TaKaRa Bio, Inc. (Shiga, Japan). The α5 integrin-GFP plasmid was purchased from Addgene. The antibodies and siRNAs used in this study are described in Tables S1 and S2 in the supplemental material. The following reagents were used in this study: Alexa Fluor 488-phalloidin (catalog no. A12379; Thermo Fisher Scientific), Alexa Fluor 647-phalloidin (A22287; Thermo Fisher Scientific), DRAQ5 (DR50200; BioStatus), bafilomycin A1 (B1793; Sigma-Aldrich), MG-132 (M8699; Sigma-Aldrich), and ViaFect transfection reagent (E4982; Promega).
Cell culture.
S2-CP8, SUIT-2, HCT116, A-498, HeLa S3, and X293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Caco-2 cells were grown in DMEM supplemented with 20% FBS and 1% nonessential amino acids. PANC-1, SW480, DLD-1, TMK1, KKLS, MKN1, MKN45, and AGS cells were maintained in RPMI medium supplemented with 10% FBS.
Plasmid construction.
HA-tagged CKAP4 was inserted into lentivirus vectors as described previously (11). pLVSIN-EF1α Neo/CKAP-HA (11) was used as a template to construct vectors pLVSIN-EF1α Neo/CKAP-HA (Δ2-21) and pLVSIN-EF1α Neo/CKAP-HA (Δ24-101). pLVSIN-EF1α Neo/CKAP-HA (Δ2-21) and pLVSIN-EF1α Neo/CKAP-HA (Δ24-101) were amplified with the KOD-FX mutagenesis kit (Toyobo; KFX-101). CSII-CMV-MCS-IRES2-Bsd/CKAP4-HA (Δ318-602) was constructed as described previously (11). CSII-CMV-MCS-IRES2-Bsd/CKAP4-HA was used as a template to construct CSII-CMV-MCS-IRES2-Bsd/CKAP4-HA (Δ128-317) and CKAP4-ICD (1-106)-GFP-CAAX. The CKAP4-HA (Δ128-317) PCR product was digested with EcoRI and inserted into the CSII-CMV-MCS-IRES2-Bsd vector provided by H. Miyoshi (RIKEN BioResource Center, Ibaraki, Japan). S2-CP8 cDNA was used as a template to construct pcDNA3/FLAG-SNX17. The FLAG-SNX17 PCR product was digested with EcoRI and BamHI and inserted into the pcDNA3 vector.
Identification of CKAP4-binding proteins.
To identify CKAP4-binding proteins, parental and CKAP4-HA-expressing S2-CP8 cells (5 × 106 cells in a 100-mm-diameter dish) were incubated with 0.5 mg/ml EZ-Link sulfo-NHS-biotin (catalog no. 21217; Thermo Fisher Scientific) for 30 min at 4°C. The cells were lysed in NP-40 buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 137 mM NaCl, and 1% NP-40) with protease inhibitors (10 μg/ml leupeptin, 20 μg/ml aprotinin, and 1 mM phenylmethanesulfonyl fluoride) and incubated with anti-HA antibody-conjugated magnetic beads (Wako, Japan) for 1 h at 4°C. The beads were washed three times with 1 ml of NP-40 buffer and one time with phosphate-buffered saline (PBS). To elute the proteins, the beads were incubated three times with HA peptide (4 mg/ml) in 50 μl of PBS for 30 min at 4°C. A 50% slurry of NeutrAvidin-agarose beads (29220; Thermo Fisher Scientific) (40 μl) was added to the eluted protein samples (∼150 μl), and the mixture was incubated for 2 h at 4°C. The beads were washed twice with 1 ml of NP-40 buffer and once with 1 ml of 10 mM Tris-HCl, pH 7.5. The bound complexes were dissolved in 20 μl of Laemmli sample buffer. The CKAP4-HA-interacting proteins were separated on an SDS-PAGE gel and detected by silver staining.
In-gel digestion.
In-gel digestion was performed as described previously (54). Briefly, gel pieces containing specific bands were excised from silver-stained gels and destained using a 1:1 solution of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate. The gel pieces were equilibrated in 200 mM ammonium bicarbonate for 20 min to pH 8.0. Gel pieces were soaked in acetonitrile for 5 min and then dried for 20 min in a vacuum. Prior to enzymatic digestion, the gel pieces were reduced with 10 mM dithiothreitol in 50 mM ammonium bicarbonate for 30 min at 37°C, alkylated with 55 mM iodoacetamide in 50 mM ammonium bicarbonate for 30 min, and then dehydrated using acetonitrile. The reduced and alkylated gel pieces were rehydrated in 50 mM Tris-HCl, pH 9.0, and 0.5 μg/ml trypsin. Once this solution was fully absorbed, the gel pieces were covered with enzyme-free Tris-HCl buffer and digested for 16 h at 37°C. The samples were extracted with 50% acetonitrile–5% formic acid for 20 min, and then the acetonitrile was evaporated using a SpeedVac centrifuge. The tryptic digests were desalted using C18 StageTips, concentrated using a SpeedVac centrifuge, and reconstituted in 0.1% formic acid.
LC-MS/MS analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed as described previously (54) using an UltiMate 3000 nano LC system (Thermo Fisher Scientific, Waltham, MA) coupled to a Q-Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) with a nanoelectrospray ionization source. Digested samples were injected using an autosampler and enriched on a C18 reverse-phase trap column (100-μm inside diameter by 5-mm length; Thermo Fisher Scientific) at a flow rate of 4 μl/min. The samples were separated by a 15-cm fused silica emitter packed in-house with reversed-phase resin (ReproSil-Pur C18-AQ, 3 μm; Maisch GmbH) at a flow rate of 500 nl/min with a linear gradient of 2% to 35% mobile phase B. Mobile phase B consisted of 90% acetonitrile with 0.1% formic acid. Mobile phase A consisted of 2% acetonitrile with 0.1% formic acid. The peptides were ionized using nanoelectrospray ionization in a positive ion mode.
The raw data files were analyzed using Mascot Distiller v2.2 (Matrix Science, Boston, MA) to create peak lists from the recorded fragmentation spectra. Peptides and proteins were identified using Mascot v2.3 (Matrix Science) in a search against the UniProt database with a precursor mass tolerance of 10 ppm, a fragment ion mass tolerance of 0.01 Da, and strict trypsin specificity allowing for up to two missed cleavages. Carbamidomethylation of cysteine residues was set as a fixed modification, while oxidation of methionine residues was allowed as variable modifications.
Integrin internalization and recycling assays.
The integrin internalization and recycling assays were performed as described previously (34), with modifications. For the antibody-based internalization assay, cells were incubated with anti-β1 integrin antibody for 30 min at 4°C. The cells were washed with cold PBS and incubated in prewarmed culture medium for 30 min at 37°C. The cells were washed with cold PBS. The surface-bound antibody was stripped from the cells with cold PBS (pH 2.5) three times. The cells were fixed with 4% paraformaldehyde (PFA) and permeabilized in PBS containing 0.1% (wt/vol) saponin and 2 mg/ml bovine serum albumin (BSA). The cells were stained with secondary antibody and DRAQ5 DNA dye. To quantify the amount of internalized β1 integrin, the total vesicle numbers of β1 integrin in each cell were counted using ImageJ software. At least 15 cells were counted. The results are shown as box and whisker plots.
For the antibody-based recycling assay, the same labeling procedure was used. After surface-bound antibody was stripped, cells were incubated in prewarmed culture medium for an additional 30 min at 37°C to chase the internalized antibody. To detect the antibody that returned to the cell surface, cells were fixed with 4% PFA, blocked with PBS containing 2 mg/ml BSA, and incubated with secondary antibody and DRAQ5 DNA dye. Internalized β1 integrin was detected as described above. To quantify the amount of β1 integrin on the cell surface, individual images were binarized and the integrated density of five randomly selected 100-μm2 regions in the cytoplasm were measured by ImageJ software.
Knockdown of protein expression by siRNA.
Cells were transfected with a mixture of siRNAs (10 nM each unless otherwise stated) against the genes of interest using RNAiMAX (13778150; Thermo Fisher Scientific). The transfected cells were used for experiments 48 h posttransfection.
Lentivirus production and generation of stable transformants.
To generate lentiviruses, lentiviral vectors were transfected into X293T cells along with the packaging vectors, pCAG-HIV-gp and pCMV-VSV-G-RSV-Rev, using FuGENE HD transfection reagent (E2311; Promega), as described previously (55). To generate S2-CP8, HeLa S3, and HCT116 cells stably expressing CKAP4-HA, the parental cells (5 × 104 cells/well in a 12-well plate) were transduced with lentivirus-containing conditioned medium and 10 μg/ml Polybrene. The cells were centrifuged at 1,200 × g for 30 min and incubated for another 24 h.
Plasmid transfection.
To see the effect of CKAP4-HA overexpression on the size of the paxillin-stained area, cells were transiently transfected with CKAP4-HA-expressing vectors by using Viafect (Promega) according to the manufacturer’s protocol.
Detection of cell surface CKAP4.
Detection of cell surface CKAP4 was performed as described previously (11). Briefly, the cells were incubated with 0.5 mg/ml EZ-Link sulfo-NHS-biotin (sulfosuccinimidyl-6-[biotin-amido]hexanoate) for 30 min at 4°C and then lysed with 500 μl of NP-40 buffer containing protease inhibitors. Biotinylated proteins were precipitated using NeutrAvidin-agarose beads. The beads were washed twice with NP-40 buffer and once with 10 mM Tris-HCl, pH 7.5. The bound complexes were probed with anti-CKAP4 antibody.
Cell migration assay.
Cell migration was assessed using Boyden chambers (tissue culture treated, 6.5 mm in diameter, 10-μm thick, 8-μm pores; Transwell, Costar, Cambridge, MA), as described previously (56). The lower surface of the filter was coated with 10 to 40 μg/ml type I collagen for 2 h. S2-CP8, A-498, HeLa S3, or MKN1 cells (2.5 × 104) or HCT116 cells (5 × 104) were resuspended in serum-free DMEM containing 0.1% BSA and applied to the upper chamber. The same medium or culture medium was added to the lower chamber. After 3 to 8 h of incubation at 37°C, cells that migrated to the lower side of the upper chamber were fixed with PBS containing 4% PFA and stained with DRAQ5 DNA dye. At least three fields were counted for each cell line. The data are presented as the mean ± standard deviation (SD).
Immunocytochemistry.
For immunocytochemistry, the cells were fixed with PBS containing 4% PFA and then permeabilized in PBS containing 0.2% (wt/vol) Triton X-100 and 2 mg/ml BSA for 10 min. The cells were blocked in blocking buffer. Samples were incubated with antibodies diluted in PBS for 1 h or overnight, washed three times with PBS, and then stained with a secondary antibody (conjugated to Alexa Fluor 488, 546, or 633; Invitrogen) and Alexa Fluor-phalloidin (Invitrogen) diluted in PBS for 1 h. After washing, the samples were covered with PBS containing 50% glycerol.
Quantification of size of cell adhesion sites.
After cells were stained with antipaxillin antibody, areas of individual cell adhesion sites were measured using ImageJ software. At least 100 cell adhesion sites were counted for each condition. The data are presented as a box and whisker plot.
Adhesion assay.
Cells (5 × 104) were seeded in 96-well plates precoated with 40 μg/ml collagen type 1 (354236; Corning), 10 μg/ml fibronectin (F2006; Sigma-Aldrich), or 35 μg/ml laminin (354232; Corning) in PBS for 2 h. After a 5-min incubation, the cells were washed three times with PBS. Adherent cells were fixed and stained with 0.1% crystal violet in 20% methanol for 5 min at room temperature and then washed thoroughly with PBS. The crystal violet stain was eluted with 100 μl of 50% ethanol, and the absorbance at 590 nm was measured using a Synergy HTX multimode microplate reader (BioTek, USA). The results are expressed as the adhesion activity relative to that of cells treated with control siRNA. Data are presented as the mean ± SD for three independent experiments.
Immunoprecipitation.
Cells (5 × 106) were lysed in 500 μl or 1 ml of NP-40 buffer with protease inhibitors. After centrifugation, the lysates were incubated with primary antibodies and 40 μl of a 50% slurry of protein G-Sepharose beads (GE Healthcare) for 1 h at 4°C. After washing three times with NP-40 buffer, the precipitates were probed with the indicated antibodies.
PLA.
Cells grown on glass coverslips were fixed for 10 min at room temperature in PBS containing 4% PFA. The cells were permeabilized in PBS containing 0.1% (wt/vol) saponin and 2 mg/ml BSA for 10 min. The glass coverslips were blocked in blocking buffer for 30 min and incubated with primary antibodies diluted in blocking buffer for 1 h at room temperature. After washing, the coverslips were incubated with Duolink proximity ligation assay (PLA) anti-rabbit minus and PLA anti-mouse plus proximity probes (Olink Bioscience). A PLA was performed using the Duolink detection reagent kit according to the manufacturer's protocol (Olink Bioscience). PLA dots were counted using an LSM810 laser scanning microscope.
Colocalization of α5 integrin and LAMP1.
To determine the relationship between α5 integrin and LAMP1 levels, control and CKAP4 siRNA-transfected S2-CP8 cells were transfected with α5 integrin-GFP and stained with anti-GFP and anti-LAMP1 antibodies. The distributions of GFP and LAMP1 fluorescence intensities were measured along the dotted lines using LSM810. The fluorescence intensity data were graphed as scatter plots, and a regression curve was drawn to determine the correlation coefficient. The P value was calculated using simple linear regression analysis between α5 integrin-GFP and LAMP1 fluorescence intensity.
Analysis of Western blot band intensity.
The intensities of Western blot bands were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD). The band intensities of input and cell surface were normalized to HSP90 or β-actin of cell lysates. Results are shown as means ± SD of the results of three independent experiments.
Statistical analysis.
All experiments were repeated at least three times, and the results are presented as the mean ± SD. Differences between experimental groups were determined using a paired or unpaired Student t test and the Wilcoxon rank sum test (Mann-Whitney U test) (JMP version 14). P values of less than 0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Center of Medical Research and Education, Graduate School of Medicine, Osaka University, for in-gel digestion and LC-MS/MS analysis and S. Tsukita, M. Hatakeyama, Y. Matsuura, T. Tanaka, K. Matsumoto, and A. Shintani for donating cells. We also thank J. Kikuta and M. Ishii for their assistance in the observation of cell migration and H. Kimura and R. Sada for critical reading of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research (S) to A.K. (2016-2020) (no. 16H06374) and Grants-in-Aid for Scientific Research on Innovative Areas, Organelle Zone, to A.K. (2018-2019) (no. 18H04861).
Y.O. performed most of the experiments. K.F. performed some of the experiments, including the quantitative analysis of colocalization. K.F. and A.K. designed the experiments, interpreted the results, and coauthored the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00073-19.
REFERENCES
- 1.Schweizer A, Ericsson M, Bachi T, Griffiths G, Hauri HP. 1993. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci 104:671–683. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer A, Rohrer J, Slot JW, Geuze HJ, Kornfeld S. 1995. Reassessment of the subcellular localization of p63. J Cell Sci 108:2477–2485. [DOI] [PubMed] [Google Scholar]
- 3.Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri HP. 2001. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J Cell Biol 153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. 2010. Mechanisms determining the morphology of the peripheral ER. Cell 143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klopfenstein DR, Kappeler F, Hauri HP. 1998. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J 17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vedrenne C, Hauri HP. 2006. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic 7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 7.Pepin G, Perron MP, Provost P. 2012. Regulation of human Dicer by the resident ER membrane protein CLIMP-63. Nucleic Acids Res 40:11603–11617. doi: 10.1093/nar/gks903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karasawa T, Wang Q, David LL, Steyger PS. 2010. CLIMP-63 is a gentamicin-binding protein that is involved in drug-induced cytotoxicity. Cell Death Dis 1:e102. doi: 10.1038/cddis.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razzaq TM, Bass R, Vines DJ, Werner F, Whawell SA, Ellis V. 2003. Functional regulation of tissue plasminogen activator on the surface of vascular smooth muscle cells by the type-II transmembrane protein p63 (CKAP4). J Biol Chem 278:42679–42685. doi: 10.1074/jbc.M305695200. [DOI] [PubMed] [Google Scholar]
- 10.Gupta N, Manevich Y, Kazi AS, Tao JQ, Fisher AB, Bates SR. 2006. Identification and characterization of p63 (CKAP4/ERGIC-63/CLIMP-63), a surfactant protein A binding protein, on type II pneumocytes. Am J Physiol Lung Cell Mol Physiol 291:L436–L446. doi: 10.1152/ajplung.00415.2005. [DOI] [PubMed] [Google Scholar]
- 11.Kimura H, Fumoto K, Shojima K, Nojima S, Osugi Y, Tomihara H, Eguchi H, Shintani Y, Endo H, Inoue M, Doki Y, Okumura M, Morii E, Kikuchi A. 2016. CKAP4 is a Dickkopf1 receptor and is involved in tumor progression. J Clin Invest 126:2689–2705. doi: 10.1172/JCI84658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niehrs C. 2006. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 13.Shinno N, Kimura H, Sada R, Takiguchi S, Mori M, Fumoto K, Doki Y, Kikuchi A. 2018. Activation of the Dickkopf1-CKAP4 pathway is associated with poor prognosis of esophageal cancer and anti-CKAP4 antibody may be a new therapeutic drug. Oncogene 37:3471–3484. doi: 10.1038/s41388-018-0179-2. [DOI] [PubMed] [Google Scholar]
- 14.Kajiwara C, Fumoto K, Kimura H, Nojima S, Asano K, Odagiri K, Yamasaki M, Hikita H, Takehara T, Doki Y, Morii E, Kikuchi A. 2018. p63-dependent Dickkopf3 expression promotes esophageal cancer cell proliferation via CKAP4. Cancer Res 78:6107–6120. doi: 10.1158/0008-5472.CAN-18-1749. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi A, Fumoto K, Kimura H. 2017. The Dickkopf1-cytoskeleton-associated protein 4 axis creates a novel signalling pathway and may represent a molecular target for cancer therapy. Br J Pharmacol 174:4651–4665. doi: 10.1111/bph.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura H, Yamamoto H, Harada T, Fumoto K, Osugi Y, Sada R, Maehara N, Hikita H, Mori S, Eguchi H, Ikawa M, Takehara T, Kikuchi A. 2019. CKAP4, a DKK1 receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin Cancer Res 25:1936–1947. doi: 10.1158/1078-0432.CCR-18-2124. [DOI] [PubMed] [Google Scholar]
- 17.Stambolic V, Woodgett JR. 2006. Functional distinctions of protein kinase B/Akt isoforms defined by their influence on cell migration. Trends Cell Biol 16:461–466. doi: 10.1016/j.tcb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Manning BD, Cantley LC. 2007. AKT/PKB signaling: navigating downstream. Cell 129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K, Takahashi M. 2005. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell 9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Tanno S, Tanno S, Mitsuuchi Y, Altomare DA, Xiao GH, Testa JR. 2001. AKT activation up-regulates insulin-like growth factor I receptor expression and promotes invasiveness of human pancreatic cancer cells. Cancer Res 61:589–593. [PubMed] [Google Scholar]
- 21.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. 2003. Overexpression of AKT2/protein kinase Bb leads to up-regulation of b1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res 63:196–206. [PubMed] [Google Scholar]
- 22.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. 2005. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell 20:539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. 2003. Cell migration: integrating signals from front to back. Science 302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 24.Parsons JT, Horwitz AR, Schwartz MA. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul NR, Jacquemet G, Caswell PT. 2015. Endocytic trafficking of integrins in cell migration. Curr Biol 25:R1092–1105. doi: 10.1016/j.cub.2015.09.049. [DOI] [PubMed] [Google Scholar]
- 26.Bridgewater RE, Norman JC, Caswell PT. 2012. Integrin trafficking at a glance. J Cell Sci 125:3695–3701. doi: 10.1242/jcs.095810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdembri D, Serini G. 2012. Regulation of adhesion site dynamics by integrin traffic. Curr Opin Cell Biol 24:582–591. doi: 10.1016/j.ceb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 28.De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J. 2015. Integrin traffic–the update. J Cell Sci 128:839–852. doi: 10.1242/jcs.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akiyama SK, Yamada SS, Yamada KM. 1989. Analysis of the role of glycosylation of the human fibronectin receptor. J Biol Chem 264:18011–18018. [PubMed] [Google Scholar]
- 30.Theisen U, Straube E, Straube A. 2012. Directional persistence of migrating cells requires Kif1C-mediated stabilization of trailing adhesions. Dev Cell 23:1153–1166. doi: 10.1016/j.devcel.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Wirtz D. 2013. Focal adhesion size uniquely predicts cell migration. FASEB J 27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouvard D, Pouwels J, De Franceschi N, Ivaska J. 2013. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat Rev Mol Cell Biol 14:430–442. doi: 10.1038/nrm3599. [DOI] [PubMed] [Google Scholar]
- 33.Hynes RO. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg F, Heesom KJ, Bass MD, Cullen PJ. 2012. SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J Cell Biol 197:219–230. doi: 10.1083/jcb.201111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharitidi D, Apaja PM, Manteghi S, Suzuki K, Malitskaya E, Roldan A, Gingras MC, Takagi J, Lukacs GL, Pause A. 2015. Interplay of endosomal pH and ligand occupancy in integrin a5b1 ubiquitination, endocytic sorting, and cell migration. Cell Rep 13:599–609. doi: 10.1016/j.celrep.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Hase H, Jingushi K, Ueda Y, Kitae K, Egawa H, Ohshio I, Kawakami R, Kashiwagi Y, Tsukada Y, Kobayashi T, Nakata W, Fujita K, Uemura M, Nonomura N, Tsujikawa K. 2014. LOXL2 status correlates with tumor stage and regulates integrin levels to promote tumor progression in ccRCC. Mol Cancer Res 12:1807–1817. doi: 10.1158/1541-7786.MCR-14-0233. [DOI] [PubMed] [Google Scholar]
- 37.Roberts MS, Woods AJ, Dale TC, Van Der Sluijs P, Norman JC. 2004. Protein kinase B/Akt acts via glycogen synthase kinase 3 to regulate recycling of avb3 and a5b1 integrins. Mol Cell Biol 24:1505–1515. doi: 10.1128/MCB.24.4.1505-1515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottcher RT, Stremmel C, Meves A, Meyer H, Widmaier M, Tseng HY, Fassler R. 2012. Sorting nexin 17 prevents lysosomal degradation of b1 integrins by binding to the b1-integrin tail. Nat Cell Biol 14:584–592. doi: 10.1038/ncb2501. [DOI] [PubMed] [Google Scholar]
- 39.Chen L, You C, Jin X, Zhou L, Huang L, Wang Y. 2018. Cytoskeleton-associated protein 4 is a novel serodiagnostic marker for esophageal squamous-cell carcinoma. Onco Targets Ther 11:8221–8226. doi: 10.2147/OTT.S183790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagita K, Nagashio R, Jiang SX, Kuchitsu Y, Hachimura K, Ichinoe M, Igawa S, Fukuda E, Goshima N, Satoh Y, Murakumo Y, Saegusa M, Sato Y. 2018. Cytoskeleton-associated protein 4 is a novel serodiagnostic marker for lung cancer. Am J Pathol 188:1328–1333. doi: 10.1016/j.ajpath.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Lobert VH, Brech A, Pedersen NM, Wesche J, Oppelt A, Malerod L, Stenmark H. 2010. Ubiquitination of a5b1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev Cell 19:148–159. doi: 10.1016/j.devcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Margadant C, Monsuur HN, Norman JC, Sonnenberg A. 2011. Mechanisms of integrin activation and trafficking. Curr Opin Cell Biol 23:607–614. doi: 10.1016/j.ceb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Morgan MR, Byron A, Humphries MJ, Bass MD. 2009. Giving off mixed signals–distinct functions of a5b1 and avb3 integrins in regulating cell behaviour. IUBMB Life 61:731–738. doi: 10.1002/iub.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. 2009. Clustering of a5b1 integrins determines adhesion strength whereas avb3 and talin enable mechanotransduction. Proc Natl Acad Sci U S A 106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White DP, Caswell PT, Norman JC. 2007. αvb3 and a5b1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J Cell Biol 177:515–525. doi: 10.1083/jcb.200609004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi F, Sottile J. 2008. Caveolin-1-dependent b1 integrin endocytosis is a critical regulator of fibronectin turnover. J Cell Sci 121:2360–2371. doi: 10.1242/jcs.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heino J, Ignotz RA, Hemler ME, Crouse C, Massague J. 1989. Regulation of cell adhesion receptors by transforming growth factor-b. Concomitant regulation of integrins that share a common b1 subunit. J Biol Chem 264:380–388. [PubMed] [Google Scholar]
- 48.Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B. 2011. Integrin a5b1 facilitates cancer cell invasion through enhanced contractile forces. J Cell Sci 124:369–383. doi: 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson KI, McCaffrey MW, Ozanne BW, Norman JC. 2007. Rab25 associates with a5b1 integrin to promote invasive migration in 3D microenvironments. Dev Cell 13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, Jagadeeswaran S, Montag A, Becker A, Kenny HA, Peter ME, Ramakrishnan V, Yamada SD, Lengyel E. 2008. Loss of E-cadherin promotes ovarian cancer metastasis via a5-integrin, which is a therapeutic target. Cancer Res 68:2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barkan D, Chambers AF. 2011. β1-integrin: a potential therapeutic target in the battle against cancer recurrence. Clin Cancer Res 17:7219–7223. doi: 10.1158/1078-0432.CCR-11-0642. [DOI] [PubMed] [Google Scholar]
- 52.Li SX, Liu LJ, Dong LW, Shi HG, Pan YF, Tan YX, Zhang J, Zhang B, Ding ZW, Jiang TY, Hu HP, Wang HY. 2014. CKAP4 inhibited growth and metastasis of hepatocellular carcinoma through regulating EGFR signaling. Tumour Biol 35:7999–8005. doi: 10.1007/s13277-014-2000-3. [DOI] [PubMed] [Google Scholar]
- 53.Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, Rahman A, Jin ZG, Long X, Xu Y, Miano JM. 2019. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci U S A 116:546–555. doi: 10.1073/pnas.1810729116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 55.Sato A, Kayama H, Shojima K, Matsumoto S, Koyama H, Minami Y, Nojima S, Morii E, Honda H, Takeda K, Kikuchi A. 2015. The Wnt5a-Ror2 axis promotes the signaling circuit between interleukin-12 and interferon-gamma in colitis. Sci Rep 5:10536. doi: 10.1038/srep10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, Yasui W, Kikuchi A. 2006. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res 66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.