T cells are nodal players in the adaptive immune response against pathogens and malignant cells. Alternative splicing plays a crucial role in T cell activation, which is analyzed mainly at later time points upon stimulation. Here we have discovered a 2-h time window early after stimulation where optimal splicing efficiency or, more generally, gene expression efficiency is crucial for successful T cell activation.
KEYWORDS: interleukin-2, RNA splicing, T cell activation, gene expression
ABSTRACT
T cells are nodal players in the adaptive immune response against pathogens and malignant cells. Alternative splicing plays a crucial role in T cell activation, which is analyzed mainly at later time points upon stimulation. Here we have discovered a 2-h time window early after stimulation where optimal splicing efficiency or, more generally, gene expression efficiency is crucial for successful T cell activation. Reducing the splicing efficiency at 4 to 6 h poststimulation significantly impaired murine T cell activation, which was dependent on the expression dynamics of the Egr1–Nab2–interleukin-2 (IL-2) pathway. This time window overlaps the time of peak IL-2 de novo transcription, which, we suggest, represents a permissive time window in which decreased splicing (or transcription) efficiency reduces mature IL-2 production, thereby hampering murine T cell activation. Notably, the distinct expression kinetics of the Egr1–Nab2–IL-2 pathway between mouse and human render human T cells refractory to this vulnerability. We propose that the rational temporal modulation of splicing or transcription during peak de novo expression of key effectors can be used to fine-tune stimulation-dependent biological outcomes. Our data also show that critical consideration is required when extrapolating mouse data to the human system in basic and translational research.
INTRODUCTION
T cells are crucial players in eliciting and orchestrating immune responses under a wide variety of pathophysiological conditions, ranging from pathogenic encounters to cancer immunity. The small, G0-arrested naive T cells undergo a multistep process upon activation to orchestrate an optimal response (1–3). The early phase of T cell activation is characterized by rapid gene expression changes and induction of multiple signaling pathways, resulting in cytokine production, initiation of cell growth and proliferation, cell lineage commitment, and differentiation (4). Defects in early T cell activation are often associated with immune disorders (5–8). Given the importance of the early T cell response, this process is tightly regulated in multiple layers, ranging from regulation to transcriptional and metabolic reprogramming, which, however, remain incompletely understood. Temporal gene expression analysis upon T cell activation showed the rapid de novo transcription of >2,000 genes during the first 4 h of T cell activation, which is mostly coupled to changes in translation (∼90%) (9). Temporal proteomics analysis showed a rapid reprogramming of protein phosphorylation in the early phase (in the first 2 h) followed by reprogramming of the global proteome and phosphoproteome (8 to 16 h) (10). While these rapid changes in the transcriptome and the proteome in the early activation phase of T cells are documented, a role for posttranscriptional mechanisms in mediating these changes remains largely elusive.
Splicing is a posttranscriptional process where introns are removed from pre-mRNAs to produce protein-coding mature mRNA. Additionally, by alternative splicing, a single pre-mRNA can produce multiple functional protein isoforms, thereby increasing the functionality and diversity of the proteome (11). A growing body of work shows a crucial role of splicing for optimal T cell function (12). Several studies have used a candidate approach to explore the role of specific splice isoforms in T cell activation (13–18). In addition, several groups have also explored global splicing changes upon T cell activation, but these studies focused on later phases of T cell activation (mostly at 24 h or 48 h postactivation) (19–22). Therefore, the role of splicing in early T cell activation, within hours after stimulation, is still enigmatic.
Here, we used a systematic approach to investigate the importance of splicing in the early phase of T cell activation. Using splicing inhibitors, we reversibly reduced the splicing efficiency consecutively for 2-h time windows in the first 10 h of activation in murine T cells. Our data show that T cells are vulnerable to splicing modulation at 4 to 6 h poststimulation, as hampering splicing during this time window significantly affects T cell activation at later time points. This time window overlaps with peak interleukin-2 (IL-2) production in activated mouse T cells, which we suggest becomes limiting if splicing or other steps in gene expression are temporally inhibited. Surprisingly, a human T cell line and primary human T cells were not susceptible to early splicing inhibition. We suggest that this is due to the differential expression kinetics of the Egr1–Nab2–IL-2 pathway between mouse and human, with a slower and prolonged IL-2 induction in humans. Therefore, IL-2 is not limiting for human T cells in the early activation phase and human T cells can tolerate a reduced splicing efficiency in this time window.
RESULTS
Reduced splicing efficiency in an early phase of T cell activation reduces proliferation specifically in mouse T cells.
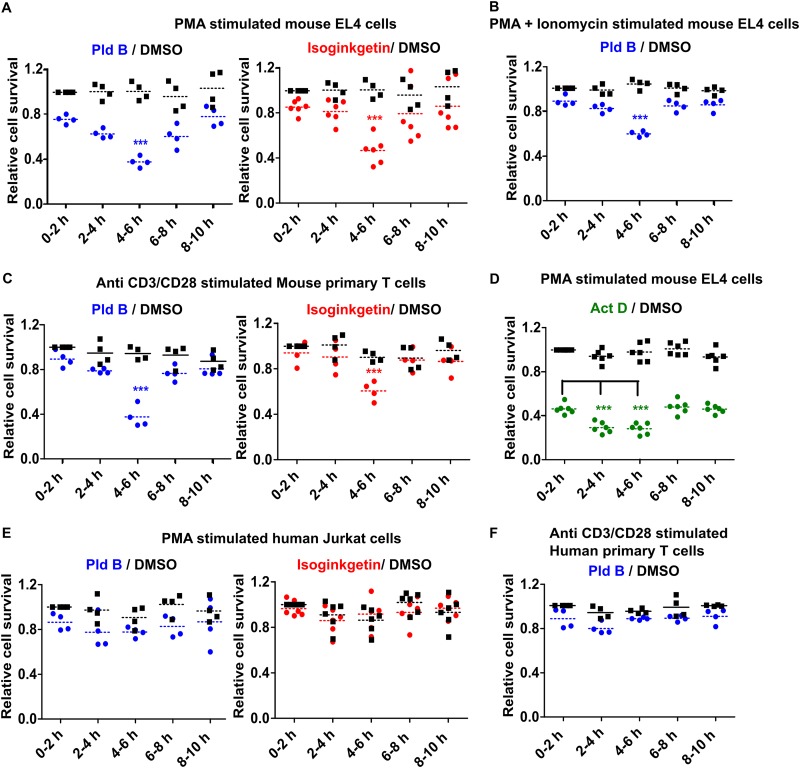
To explore the role of splicing in early T cell activation, we performed a temporal reversible splicing modulation in the EL4 mouse T cell line. We stimulated EL4 cells with phorbol myristate acetate (PMA) and reduced splicing reversibly in consecutive 2-h time windows using small-molecule splicing inhibitors until 10 h postactivation (0 to 2 h, 2 to 4 h, 4 to 6 h, 6 to 8 h, 8 to 10 h). More precisely, cells were treated with the splicing inhibitor for each 2-h time window, spun down, and resuspended, and the cells were plated in fresh medium. The cells were then allowed to grow, and at 48 h poststimulation, live cells (propidium iodide [PI] negative) were counted via flow cytometry. The data were normalized to those for the dimethyl sulfoxide (DMSO)-treated control at 0 to 2 h to give the relative live cell population. Impaired T cell activation results in defects in cell proliferation (23) and/or increased apoptosis (24), and therefore, measuring the live cell population is a simple and straightforward readout for T cell activation. We found that reducing the splicing efficiency specifically in the h 4 to 6 time window poststimulation caused an ∼50% reduction in the live cell population assessed at 48 h poststimulation, whereas this was not the case for the other time windows (Fig. 1A). As different splicing inhibitors have an intrinsic bias toward distinct kinds of splicing modulation and may have off-target effects, we used two different classes of splicing inhibitors (isoginkgetin and pladienolide B [Pld B]), and we observed the same time-dependent sensitivity for both inhibitors, further validating the results (Fig. 1A). A similar result was obtained when PMA and ionomycin were used for stimulation (Fig. 1B); for simplicity, we performed further experiments with PMA stimulation alone, unless mentioned otherwise. Next, we used mouse primary T cells and anti-CD3/anti-CD28 stimulation to investigate the dependence of T cell activation on splicing in a more physiologically relevant system. It clearly confirmed our cell line results (Fig. 1C), where we again observed fewer viable cells at 48 h poststimulation if the splicing efficiency was reduced at 4 to 6 h after stimulation.
FIG 1.
Reduced splicing efficiency in a defined time window early after stimulation hampers optimal T cell activation specifically in the murine system. (A) Temporal splicing modulation analysis in PMA-stimulated mouse EL4 T cells; (B) temporal splicing modulation analysis in PMA- and ionomycin-stimulated EL4 cells; (C) temporal splicing modulation analysis in mouse primary T cells; (D) temporal transcription inhibitor analysis of PMA-stimulated EL4 cells; (E) temporal splicing modulation analysis in PMA-stimulated human Jurkat T cells; (F) temporal splicing modulation analysis in human primary T cells. ***, P < 0.0001 (Student's unpaired t test, done using GraphPad software).
As splicing happens either co- or posttranscriptionally, we were intrigued to investigate whether there exists a similar time window that is particularly sensitive to inhibition of transcription. To address this, we used the same experimental setup with the transcription inhibitor actinomycin D (Act D) in EL4 cells and found a broader 4-h time window at between 2 and 6 h poststimulation that was especially sensitive to inhibiting transcription (Fig. 1D). This partially overlaps the time window susceptible to splicing inhibition, suggesting that interfering with gene expression in general within this defined time frame causes a defect in T cell activation.
To check whether the splicing sensitivity is general to immune activation, we performed temporal splicing modulation assays in RAW264.7 cells (a mouse macrophage cell line) stimulated with lipid A but did not observe a similar vulnerability (data not shown), indicating that it is, at least to some extent, T cell specific. We further wanted to extrapolate our finding to the human context and used temporal splicing modulation in the human Jurkat T cell line and primary human T cells. To our surprise, we did not find a similar temporal sensitivity to splicing modulation (Fig. 1E and F). This finding intrigued us, and so we explored the underlying molecular mechanism and the cause for species specificity.
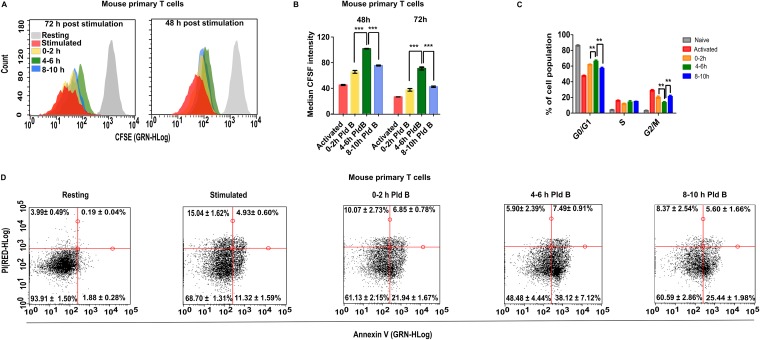
To address the molecular mechanism behind this reduced cell number, we checked cell proliferation, cell cycle, and apoptosis. To assay cell proliferation, we labeled cells with carboxyfluorescein diacetate succinimidyl ester (CFSE), which irreversibly binds to cytoplasmic proteins and which gets evenly distributed during cell division. We indeed found a significant decrease in cell proliferation after 48 and 72 h of activation (a higher CFSE intensity), if splicing was inhibited in the h 4 to 6 time window poststimulation (Fig. 2A and B). Further, the reduced cell proliferation may be due to a slightly increased percentage of cells in the G0/G1 cell cycle phase, which we observed concomitantly with a reduction in G2/M phase (Fig. 2C). Additionally, inhibiting splicing in the h 4 to 6 time window also increased apoptosis, as shown by annexin-PI staining, where we found an ∼20% increase in early apoptotic cell populations compared to the h 0 to 2 and 8 to 10 time windows (Fig. 2D). Together, these data suggest that reducing splicing efficiency in the h 4 to 6 time window interferes with T cell activation via defective cell proliferation and slightly increased apoptosis.
FIG 2.
Effect of temporal splicing inhibition on T cell proliferation, the cell cycle, and apoptosis. (A) Cell proliferation of CFSE-labeled mouse T cells at 48 h and 72 h poststimulation (the diagram is representative of those from 4 experiments); (B) quantitative analysis of cell proliferation at 48 h and 72 h poststimulation, as shown in panel A (mean ± SD; n = 4); (C) cell cycle analysis of mouse primary T cells (mean ± SD; n = 4); (D) apoptosis analysis of mouse primary T cells (mean ± SD; n = 6). GRN-HLog indicates green fluorescence intensity in log scale, and RED-HLog indicates red fluorescence intensity in log scale. ***, P < 0.0001; **, P < 0.001 (Student's unpaired t test, done using GraphPad software).
Differential expression dynamics of Egr1-Nab2 in activated murine and human T cells.
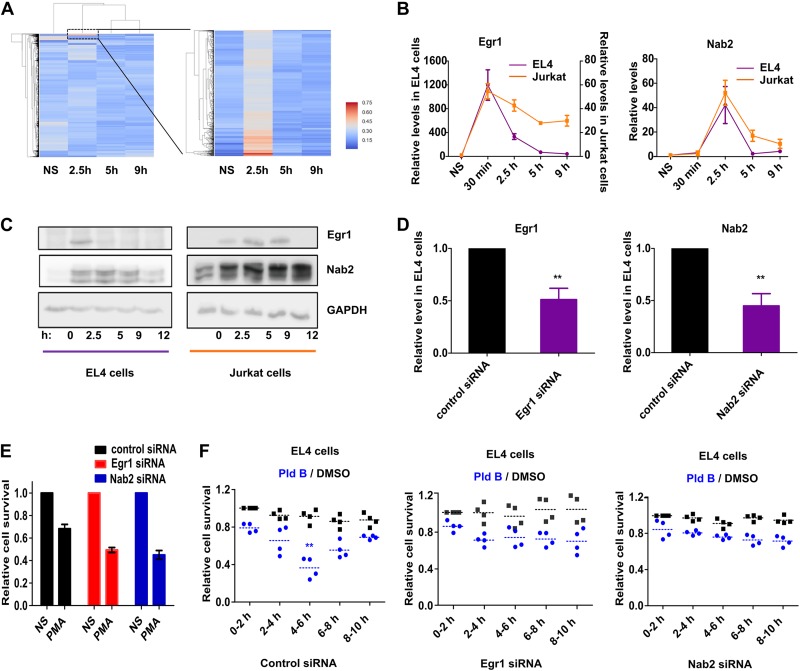
To understand the underlying molecular pathway(s), we performed global profiling of gene expression in EL4 cells upon PMA stimulation in a temporal manner. To measure transcriptional dynamics, we isolated chromatin-associated RNA from EL4 cells treated with PMA for 0, 2.5, 5, and 9 h and performed RNA sequencing. Expression analysis identified a small cluster of genes with increased de novo transcription in a small time window, 2.5 h postactivation, which was quickly switched off at later time points (Fig. 3A). Upon filtering on the read count, the strongest representatives of this group turned out to be early growth response protein 1 (Egr1) and NGFI-A-binding protein 2 (Nab2). This is of special interest, as both have well-described functions in T cell activation and are part of the same signaling cascade (25, 26). Egr1, being an immediate early transcription factor, activates the expression of the Nab2 gene (among others) (27, 28) which in turn acts as a transcriptional coactivator (29). Additionally, Nab2 inhibits Egr1 in a negative-feedback loop, thereby controlling and limiting Egr1-Nab2 signaling (30). As our sequencing data indicated that their de novo expression peak falls within the transcription-sensitive window, we wondered whether they are the upstream modulators of an effector responsible for susceptibility to splicing inhibition. Therefore, we checked the temporal expression kinetics of Egr1 and Nab2 in both stimulated EL4 and Jurkat cells, where the former responds to splicing inhibition but the latter is refractory to it. We indeed found differential expression dynamics of Egr1 and Nab2 between EL4 and Jurkat cells. Egr1 and Nab2 RNA levels peaked at 30 min and 2.5 h poststimulation, respectively, in EL4 and Jurkat cells, but in EL4 cells their expression was more transient than it was in Jurkat cells (Fig. 3B). These Egr1 and Nab2 temporal transcription dynamics in EL4 cells also correlated with previously reported findings obtained by expression analysis and ribosome profiling analysis done in mouse primary T cells (9). The differences in dynamic expression became even more evident when comparing the temporal protein levels of Egr1 and Nab2. Egr1 was detectable in EL4 cells only at 2.5 h poststimulation, while in Jurkat cells it was present until 9 h poststimulation. Nab2 similarly showed a prolonged expression in Jurkat cells compared to EL4 cells (Fig. 3C). These differential expression dynamics of Egr1 and Nab2 between murine and human T cells led us to explore their potential role in mediating the vulnerability to splicing inhibition at 4 to 6 h poststimulation.
FIG 3.
Role of Egr1 and Nab2 in mediating the sensitivity to splicing inhibition. (A) Temporal gene expression analysis in EL4 cells using RNA-Seq of chromatin-associated RNA; (B and C) temporal RNA level (mean ± SD; n = 3) (B) and protein level (C) of Egr1 and Nab2 in EL4 and Jurkat cells; (D) siRNA knockdown efficiency for Egr1 and Nab2 in EL4 cells (mean ± SD; n = 3); (E) cell viability analysis in Egr1 and Nab2 knockdown EL4 cells at 48 h poststimulation (relative to nonstimulated [NS] cells) (mean ± SD; n = 3); (F) temporal splicing modulation analysis (with Pld B) in siRNA-transfected EL4 cells. **, P < 0.001 (Student's unpaired t test, done using GraphPad software).
The Egr1-Nab2 pathway makes murine T cells susceptible to splicing inhibition.
We used small interfering RNAs (siRNAs) against Egr1 and Nab2 to investigate the potential role of Egr1 and Nab2 in mediating the effect of splicing inhibition in murine T cells. The siRNAs reduced the expression of Egr1 and Nab2 to ∼50% (Fig. 3D), and as Egr1 and Nab2 are important for T cell activation, their knockdown also reduced the live cell population poststimulation compared to that for control siRNA-treated cells (Fig. 3E). Next, we transfected EL4 cells with control siRNA or Egr1 or Nab2 siRNA and at 24 h posttransfection stimulated the cells, followed by temporal splicing modulation. As before, the data were normalized to those for the DMSO control at 0 to 2 h to give relative live cell populations. While the control siRNA-transfected cells retained their sensitivity to splicing inhibition at between 4 and 6 h, this was lost in the Egr1 and Nab2 siRNA-transfected cells (Fig. 3F), indicating that the Egr1-Nab2 pathway plays a role in mediating this effect. Since Egr1 and Nab2 are expressed and spliced before the h 4 to 6 time window susceptible to splicing modulation, we hypothesized that expression and splicing of a downstream effector of the Egr1-Nab2 pathway make the cell sensitive to splicing modulation. Based on this model, early inhibition of Egr1/Nab2 transcription and inhibition of transcription and/or splicing of a downstream effector molecule lead to inhibition of T cell activation at later time points.
Distinct IL-2 expression kinetics between mouse and human T cells.
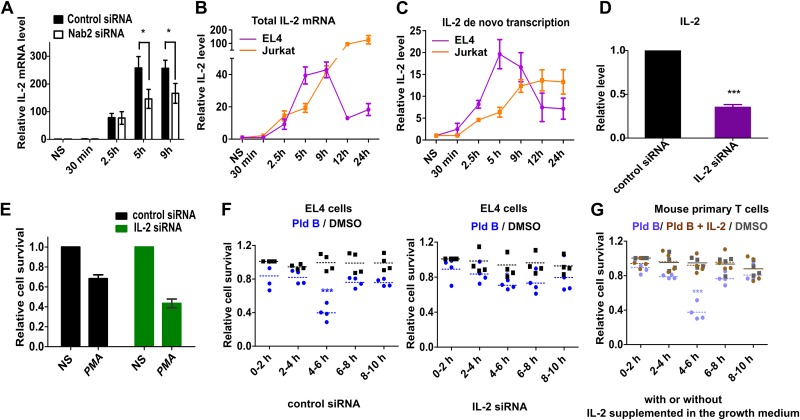
To explore for a potential downstream modulator(s), we used a literature survey and the STRING database to find potential Nab2 interactors. We choose five candidates (TRAIL, interleukin 2 [IL-2], ZFP384, ZFP362, and Erdr1) and checked their temporal expression upon stimulation of EL4 cells in the presence or absence of Nab2 (data not shown). IL-2 drew our attention, as its expression was Nab2 dependent and it peaked at between 5 h and 9 h poststimulation (Fig. 4A). We went further on with IL-2 and compared its temporal expression kinetics between EL4 and Jurkat cells. Indeed, we found a striking difference in the temporal total IL-2 level between murine and human T cell lines. In mouse EL4 cells, IL-2 mRNA peaked at 5 h, plateaued until 9 h, and strongly decreased at 12 h poststimulation, while in human Jurkat cells, the IL-2 level increased until at least 24 h poststimulation (Fig. 4B). This difference in temporal IL-2 mRNA levels suggested that it is a candidate to mediate the species-specific differences in the sensitivity to splicing inhibition.
FIG 4.
IL-2 mediates the sensitivity to splicing inhibition sensitivity. (A) Relative IL-2 expression in the presence and absence of Nab2 (mean ± SD; n = 3); (B) temporal IL-2 total mRNA level in EL4 and Jurkat cells (mean ± SD; n = 3); (C) IL-2 de novo transcription dynamics in EL4 and Jurkat cells (mean ± SD; n = 3); (D) siRNA knockdown efficiency for IL-2 in EL4 cells (mean ± SD; n = 3); (E) cell viability analysis in IL-2 knockdown EL4 cells at 48 h poststimulation (relative to that in nonstimulated cells) (mean ± SD; n = 3); (F) temporal splicing modulation analysis (with Pld B) in siRNA-transfected EL4 cells; (G) temporal splicing modulation analysis in mouse primary T cells in the presence of growth medium supplemented or not supplemented with IL-2. We used the data from the assay whose results are presented in Fig. 1C for direct comparison of conditions with or without IL-2 supplementation. Experiments were performed in parallel. *, P < 0.01; ***, P < 0.0001 (Student's unpaired t test, done using GraphPad software).
As the total RNA level of a transcript is affected by both the transcription rate and mRNA stability, we had a closer look at the de novo transcription dynamics. To this end, we analyzed the de novo expression kinetics of IL-2 in EL4 and Jurkat cells using reverse transcription-quantitative PCR (RT-qPCR) with chromatin-associated RNA as the template. This gave an even clearer result; while de novo IL-2 expression peaked at 5 h poststimulation and then gradually decreased in EL4 cells, in Jurkat cells the expression peaked at 9 h and then stayed constant for at least 24 h poststimulation (Fig. 4C). Thus, the time window where mouse T cells are susceptible to splicing inhibition overlaps a narrow peak of IL-2 de novo expression in mouse T cells, and therefore, splicing inhibition in that window most likely affects IL-2 expression and function.
To confirm this, for IL-2 to be the effector molecule, we used siRNA-based IL-2 knockdown, which was effective in reducing the IL-2 mRNA level down to ∼40% (Fig. 4D). Next, we did siRNA-based knockdown of IL-2 followed by temporal splicing modulation using Pld B. Reducing IL-2 levels reduced overall live cell populations poststimulation (Fig. 4E) and made stimulated EL4 cells refractory to the splicing inhibition (Fig. 4F). We further did a compensation experiment in primary mouse T cells, where the growth medium was supplemented with IL-2 in the temporal splicing modulation experiment. We did not observe an effect of splicing inhibition under the IL-2-supplemented condition, showing that exogenous IL-2 in the growth medium compensated for the reduced splicing efficiency in mouse primary T cells (Fig. 4G). Therefore, defective splicing of IL-2 appears to be the major reason for the susceptibility of murine T cells to splicing inhibition at 4 to 6 h poststimulation. IL-2 is antiapoptotic and plays an essential role in activation-induced T cell proliferation. Dysregulated IL-2 production has been reported to induce G1 cell cycle arrest and increased apoptosis (31). We found similar effects when reducing splicing efficiency in the h 4 to 6 window (Fig. 2), which further points to IL-2 as the key mediator of the temporal sensitivity to splicing modulation.
Splicing modulation reduces IL-2 expression and thereby affects T cell activation.
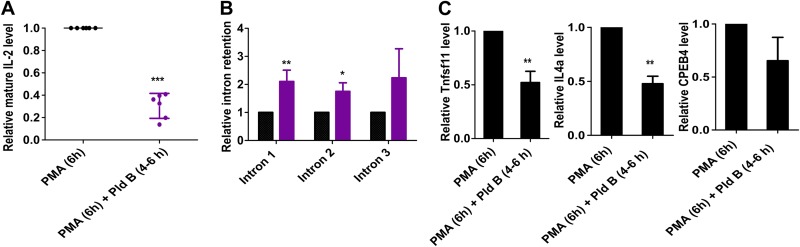
To investigate how splicing modulation affects mature IL-2 formation, we stimulated EL4 cells with PMA for 6 h in the presence and absence of Pld B in the last 2 h (i.e., 4 to 6 h postactivation). IL-2 contains 4 exons and 3 introns; while we did not find any novel splice isoforms in the presence of Pld B (data not shown), we observed an approximately 60% decrease in mature IL-2 mRNA levels when reducing splicing between 4 and 6 h poststimulation (Fig. 5A). Using an intron-exon-spanning RT-qPCR, we found that splicing inhibition by Pld B increased the relative level of all three introns, indicating that the effect is general and does not affect a specific intron (Fig. 5B). We further observed that inhibiting splicing reduced the formation of mature mRNA of multiple transcripts (Fig. 5C), showing that this effect is not specific to IL-2. However, as IL-2 is a critical activator, for example, for clonal expansion, we postulate that interfering with mature IL-2 production during its peak de novo expression is the main cause for the defect in T cell activation that we observed upon early splicing inhibition.
FIG 5.
Splicing inhibition increases intron retention and reduces mature mRNA formation. (A) Relative mature IL-2 level in EL4 cells after 6 h of PMA stimulation in the presence or absence of Pld B in the last 2 h (i.e., 4 to 6 h) after PMA stimulation (mean ± SD; n = 6); (B) relative level of IL-2 intron-retained transcripts under the same experimental conditions described for panel A (mean ± SD; n = 3); (C) relative mature Tnfsf11, IL-4a, and CPEB4 levels in EL4 cells after 6 h of PMA stimulation in the presence or absence of Pld B in the last 2 h (i.e., 4 to 6 h) after PMA stimulation (mean ± SD; n = 3). ***, P < 0.0001; **, P < 0.001; *, P < 0.01 (Student's unpaired t test, done using GraphPad software).
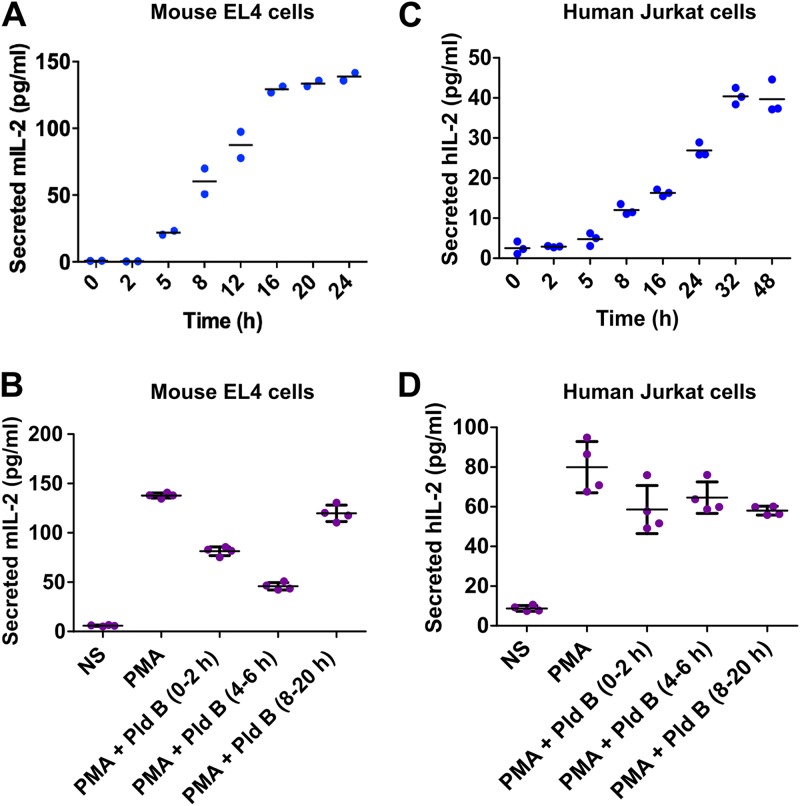
Along this line, we performed a temporal profiling of secreted IL-2 in EL4 cells using an enzyme-linked immunosorbent assay (ELISA) and found that secreted IL-2 levels plateaued at about 16 h poststimulation (Fig. 6A). Next, we stimulated EL4 cells with PMA, inhibited splicing in the h 0 to 2, 4 to 6, and 8 to 20 time windows, and checked for secreted IL-2 levels after 20 h using ELISA. We found an approximately 3-fold decrease in secreted IL-2 levels when blocking splicing in the h 4 to 6 time window. Blocking splicing in the h 0 to 2 or in the h 8 to 20 time window also reduced secreted IL-2 levels, but the effect was less strong (Fig. 6B), which is consistent with a slightly reduced cell number when reducing splicing efficiency in these time frames (e.g., see Fig. 1A). In parallel, we also performed temporal profiling of secreted IL-2 in Jurkat cells and found a different temporal kinetics, where human secreted IL-2 levels plateaued at about 32 h poststimulation (Fig. 6C). Furthermore, there was no effect on secreted IL-2 levels from Jurkat cells (32 h after stimulation) while doing the splicing inhibition at the above-mentioned time points (Fig. 6D). This again indicates that human IL-2 is refractory to early splice inhibition.
FIG 6.
Differential IL-2 production kinetics between murine and human T cells render murine cells vulnerable to splicing modulation early after activation. (A) Quantification of secreted IL-2 in EL4 cells (n = 2); (B) quantification of IL-2 levels at 20 h poststimulation with splicing inhibition by Pld B in the h 0 to 2, 4 to 6, and 8 to 20 time windows (mean ± SD; n = 4) in EL4 cells; (C) quantification of secreted IL-2 in Jurkat cells (n = 3); (D) quantification of IL-2 levels at 32 h poststimulation with splicing inhibition by Pld B in the h 0 to 2, 4 to 6, and 8 to 20 time windows (mean ± SD; n = 4) in Jurkat cells. mIL-2, mouse IL-2; hIL-2, human IL-2.
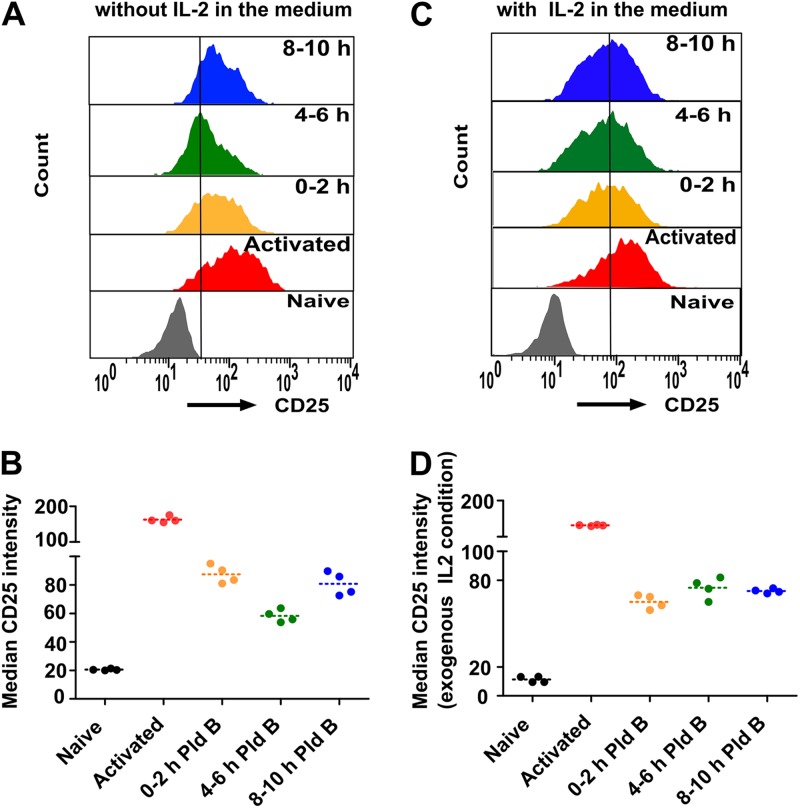
IL-2 secreted from activated T cells binds to its cell surface receptor, IL-2R, on other activated T cells, thereby boosting T cell activation. IL-2R is composed of α, β, and γ chains. IL-2Rα (also known as CD25) is the key player in mediating a high-affinity IL-2 and IL-2R interaction and thereby eliciting optimal T cell activation. The expression of CD25 strongly increases upon T cell activation in an IL-2-dependent manner (32), and hence, it is used as a standard T cell activation marker. Therefore, we checked the expression of CD25 upon splicing modulation by Pld B in mouse primary T cells as a readout for T cell activation. As before, we stimulated mouse primary T cells with anti-CD3/anti-CD28, added Pld B in the h 0 to 2, 4 to 6, and 8 to 10 time windows, and finally, used fluorescence-activated cell sorting to quantify the expression of CD25 at 48 h poststimulation. We found a sharp increase in CD25 intensity in stimulated T cells compared to naive T cells, and splicing modulation during the h 4 to 6 time window showed the strongest reduction in CD25 intensity, again indicating improper T cell activation (Fig. 7A and B) and further validating that optimal splicing efficiency is required, especially in the h 4 to 6 time window after stimulation of murine T cells. We also performed the same experiment under IL-2-supplemented conditions and found that the CD25 expression sensitivity to splicing inhibition was basically absent in the presence of exogenous IL-2 in the medium (Fig. 7C and D).
FIG 7.
Splicing modulation affects murine T cell activation in an IL-2-dependent manner. (A) Relative expression of IL-2 receptor α (CD25) in naive mouse primary T cells, 48 h poststimulation T cells, and 48 h poststimulation T cells treated with Pld B in the first 0 to 2 h, 4 to 6 h, and 8 to 10 h; (B) quantitative analysis of the experiment whose results are presented in panel A (n = 4); (C) Relative expression of IL-2 receptor α (CD25) in mouse primary T cells under IL-2-supplemented conditions as described in the legend to panel A; (D) quantitative analysis of the experiment whose results are shown in panel C (n = 4).
Taking our data together, we suggest a model in which mouse T cells are dependent on optimal IL-2 production during a narrow peak of de novo expression and that inhibiting its proper pre-mRNA processing in this time frame, e.g., through reducing splicing efficiency, has a significant impact on T cell activation.
DISCUSSION
This study provides new insight about the importance of optimal transcriptional and splicing regulation in the early phase of murine T cell activation. We show that inhibiting splicing in a specific 2-h time window from 4 to 6 h poststimulation significantly interferes with T cell activation. We also show that there is a broader time window sensitive to the inhibition of transcription, namely, 2 to 6 h poststimulation. Inhibiting transcription showed a stronger effect on cell viability than inhibiting splicing did (Fig. 1A versus Fig. 1D), likely due to a more complete block of gene expression. While exploring the molecular mechanism, we identified IL-2 to be the prime target of this sensitivity to splicing modulation. IL-2 works in both an autocrine and a paracrine fashion and plays a multifaceted role in T cell biology (33–38). A potent T cell response requires the orchestrated production of different cytokines, including gamma interferon (IFN-γ) and IL-2. Activation-induced IL-2 production entirely depends on de novo transcription rather than the use of preformed mRNAs for early production, as is observed, e.g., for IFN-γ (39). The transient IL-2 expression in murine T cells (compared to sustained expression in human T cells) therefore renders cells vulnerable to reduced splicing (or transcription) efficiency, as they are unable to compensate for a reduction of mature IL-2 formation (Fig. 5A) during its peak de novo transcription. This transient IL-2 expression has also been shown previously in activated mouse primary cells, indicating that these IL-2 kinetics are characteristic of murine T cells and not a cell line-specific effect (39, 40). Evidence for sustained IL-2 expression in activated human CD4+ T cells has also been published, supporting our results (41). Our study highlights how different temporal expression dynamics between human and mouse in the early phase of T cell activation confers selective sensitivity to splicing modulation in murine T cells. Although the intrinsic molecular differences between the mouse and human immune systems have been explored for the last few decades, we present evidence for the distinct temporal expression of not only a single gene but also an overall pathway during early T cell activation between human and mice. It will now be interesting to explore why different expression kinetics have evolved and how different IL-2 expression dynamics impact robust immune activation. Additionally, the Egr1/Nab2 pathway is not T cell specific but plays a crucial role in different physiological and disease conditions, including cancer (42). Egr1/Nab2 is an external stimulation-dependent signaling pathway that impacts cell proliferation but that also plays a role in other physiological processes, ranging from peripheral nerve myelination (43) to angiogenesis (44). Therefore, distinct Egr1/Nab2 temporal dynamics upon activation may have additional, potentially more global effects in mediating molecular differences between human and mouse.
Taken together, the findings of our study provide another example for a molecular difference between mouse and human immune systems and highlight that careful consideration is required when extrapolating data from mouse models to the human system. Furthermore, temporarily reducing splicing efficiency during the peak expression of key mediators of this and other activation-responsive pathways also in other cellular systems may prove to represent a general concept to alter cellular responses upon activation.
MATERIALS AND METHODS
Activation of murine and human T cells.
EL4 and Jurkat cells were cultured in RPMI medium (BioWest) supplemented with 10% fetal bovine serum (FBS; Biochrom) and 1% penicillin-streptomycin (BioWest) and were activated with PMA (20 ng/ml). Mouse primary T cells were isolated from lymph nodes, and human T cells were isolated from the blood of healthy volunteers following university policies (MACSprep HLA T cell isolation kit, human). They were cultured in RPMI medium (containing 2 mM glutamine, 1 mM pyruvate, nonessential amino acids, and 50 μM 2-mercaptoethanol) supplemented with 10% FBS and 1% penicillin-streptomycin. Cells at a density of ∼2 × 105 cells/ml were activated with coated anti-CD3 antibody (1 μg/ml) and anti-CD28 antibody (1 μg/ml).
Splicing modulation assay.
Cells were activated as described above, and splicing was inhibited using either isoginkgetin (30 μM) or Pld B (20 nM) for every 2 h consecutively until 10 h poststimulation. Precisely, cells were treated with the inhibitor or the DMSO control for 2 h and then spun down and resuspended in fresh medium. At 48 h poststimulation, cells were stained with propidium iodide (PI) and were counted using a Guava easyCyte8 cytometer, and PI-negative populations were used to calculate the relative live cell population. For siRNA transfection, siRNAs (20 pmol) (Table 1) were transfected in EL4 cells using a Gene Pulser electroporation system (Bio-Rad), and at 24 h posttransfection, cells were stimulated with PMA and splicing modulation analysis was done. For the IL-2 complementation assay, cells were resuspended in RPMI containing 40 units of IL-2 after the 2-h splicing inhibitor treatment.
TABLE 1.
siRNAs
| Serial no. | Protein name | Sequence |
|---|---|---|
| 1 | Egr1 | GACUAUCUGUUUCCACAACAACAGG |
| 2 | Nab2 | AGAGAGCACCUAUCUUUCUUCCUUG |
| 3 | IL-2 | AUGUUCUGGAUUUGACUCAAAGCAA |
Cell proliferation and cell cycle and apoptosis analysis.
Cell proliferation was measured using CFSE staining, which was done as described previously (45). Mouse primary T cells were labeled with CFSE, followed by stimulation and a splicing modulation assay. At 48 and 72 h poststimulation, CFSE signals were measured using a Guava easyCyte8 cytometer, and the median CFSE intensity was used for quantification. For cell cycle analysis, mouse primary T cells were stimulated, followed by the splicing modulation assay. Cell cycle analysis was done as described previously (46) at 48 h poststimulation. Apoptosis analysis was done using a fluorescein isothiocyanate annexin V apoptosis detection kit (BD Biosciences) according to the manufacturer’s protocol.
RNA and RT-qPCR.
Total RNA isolation, reverse transcription, and quantitative PCR (qPCR) were done as described previously (47). For transcriptome sequencing (RNA-Seq) analysis and qPCR of nascent transcripts, chromatin-associated RNA was isolated as described previously (48). The primers used are listed in Table 2.
TABLE 2.
Primers for qPCR
| Serial no. | Protein name | Sequence |
Species | |
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| 1 | Egr1 | GGAGCAAAGCCAAGCAAAC | ACGGAACAACACTCTGACAC | Mouse |
| 2 | Nab2 | GAGGAGGGGTTGCTGGACC | GGCTGGAGGCAAAGTCCG | Mouse |
| 3 | IL-2 | CCTGAGCAGGATGGAGAATTACA | TCCAGAACATGCCGCAGAG | Mouse |
| 4 | TRAIL | CCTCTCGGAAAGGGCATTC | TCCTGCTCGATGACCAGCT | Mouse |
| 5 | Erdr1 | CAGTGATGTCACCCACGAAA | GGCATTTCTGTACGCAGTCA | Mouse |
| 6 | ZFP384 | CCCTGACCTCTCCAAGAAGG | GGTTTAGGAGGAGCCACAGT | Mouse |
| 7 | ZFP362 | CCCTGCTGTGAGTACATCTGA | TGGCCCTGGATTGTCTTGAT | Mouse |
| 8 | Tnfsf11 | TGTACTTTCGAGCGCAGATG | AGGCTTGTTTCATCCTCCTG | Mouse |
| 9 | IL-4a | ACACTACAGGCTGATGTTCTTCG | TGGACCGGCCTATTCATTTCC | Mouse |
| 10 | CPEB4 | GGGCTGAACGGTGGAATAAC | TGTCAGCCCTGTTCAAGCTA | Mouse |
| 11 | Egr1 | CTCTCCAGCCTGCTCGTC | AGCAGCATCATCTCCTCCAG | Human |
| 12 | Nab2 | ACCAATCCAGGGCTCTTCAG | GTGCCATGATCTCCAGAAACTCCTCCT | Human |
| 13 | IL-2 | GAATCCCAAACTCACCAGGATGCTC | TAGCACTTCCTCCAGAGGTTTGAGT | Human |
RNA-Seq analysis.
Reads were aligned to the mm10 genome using the STAR (version 2.5.3a) program (49) with the option to count the number of reads per gene enabled. Further analysis was performed using custom Python scripts and the SciPy and Matplotlib modules. For each sample, the number of reads per gene was normalized to the total read numbers multiplied by 1 million (the number of reads per million [RPM]), and RPM values for replicates were averaged. For clustering, the expression values per gene were further normalized so that the sum of each gene row equaled 1. Clustering by expression over the time course was subsequently performed, and the most prominent cluster with an expression peak at 2.5 h was selected. For genes in this cluster, expression values were produced as the output.
Western blotting.
Protein extracts were prepared in standard lysis buffer (60 mM Tris [pH 7.5], 30 mM NaCl, 1 mM EDTA, 1% Triton X-100). SDS-PAGE and Western blotting were done using standard protocols. The antibodies used for Western blotting were as follows: anti-Egr-1 (sc-515830; Santa Cruz), anti-Nab2 (sc-23867; Santa Cruz), and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; GT239; GeneTex).
ELISA.
EL4 and Jurkat cells were stimulated with PMA, followed by the splicing modulation assay (as described above). At 20 h or 32 h poststimulation (for EL4 and Jurkat cells, respectively), cells were spun down, supernatants were collected, and ELISA was carried using a mouse or human IL-2 ELISA Max Deluxe set (BioLegend) according to the manufacturer’s protocol.
Flow cytometry.
For CD25 expression analysis, mouse primary T cells were spun down after stimulation and splicing modulation, washed 2 times with cold phosphate-buffered saline (PBS), stained with allophycocyanin (APC)-Cy7 anti-mouse CD25 antibody (BioLegend) in PBS, and analyzed using an easyCyte8 cytometer, and median APC-Cy7 intensities were used for quantification.
ACKNOWLEDGMENTS
We thank the HPC Service of ZEDAT, Freie Universität Berlin, for computing time. We also thank members of the F. Heyd laboratory for discussion and comments on the manuscript.
Funding was provided by DFG grant 278001972-TRR 186 to F.H.
D.B. performed all wet lab experiments. S.M. contributed to work with primary human T cells. A.N. performed bioinformatics analysis. B.T. performed RNA-Seq. Experimental design, data analysis, and writing of the manuscript were done by D.B. and F.H. F.H. conceived the project and supervised the work.
REFERENCES
- 1.Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, Zhang H, Huppa JB, Tsai YH, Lobry C, Xie J, Dempsey PJ, Crawford HC, Aifantis I, Davis MM, Vignali DA. 2013. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol 14:262–270. doi: 10.1038/ni.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton SE, Jameson SC. 2012. CD8 T cell quiescence revisited. Trends Immunol 33:224–230. doi: 10.1016/j.it.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J, Yamane H, Paul WE. 2010. Differentiation of effector CD4 T cell populations. Annu Rev Immunol 28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennock ND, White JT, Cross EW, Cheney EE, Tamburini BA, Kedl RM. 2013. T cell responses: naive to memory and everything in between. Adv Physiol Educ 37:273–283. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiedler W, Sykora KW, Welte K, Kolitz JE, Cunningham-Rundles C, Holloway K, Miller GA, Souza L, Mertelsmann R. 1987. T-cell activation defect in common variable immunodeficiency: restoration by phorbol myristate acetate (PMA) or allogeneic macrophages. Clin Immunol Immunopathol 44:206–218. doi: 10.1016/0090-1229(87)90066-3. [DOI] [PubMed] [Google Scholar]
- 6.Fischer MB, Hauber I, Eggenbauer H, Thon V, Vogel E, Schaffer E, Lokaj J, Litzman J, Wolf HM, Mannhalter JW. 1994. A defect in the early phase of T-cell receptor-mediated T-cell activation in patients with common variable immunodeficiency. Blood 84:4234–4241. [PubMed] [Google Scholar]
- 7.Scotese I, Gaetaniello L, Matarese G, Lecora M, Racioppi L, Pignata C. 1998. T cell activation deficiency associated with an aberrant pattern of protein tyrosine phosphorylation after CD3 perturbation in Down's syndrome. Pediatr Res 44:252–258. doi: 10.1203/00006450-199808000-00019. [DOI] [PubMed] [Google Scholar]
- 8.Shier P, Ngo K, Fung-Leung WP. 1999. Defective CD8+ T cell activation and cytolytic function in the absence of LFA-1 cannot be restored by increased TCR signaling. J Immunol 163:4826–4832. [PubMed] [Google Scholar]
- 9.Davari K, Lichti J, Gallus C, Greulich F, Uhlenhaut NH, Heinig M, Friedel CC, Glasmacher E. 2017. Rapid genome-wide recruitment of RNA polymerase II drives transcription, splicing, and translation events during T cell responses. Cell Rep 19:643–654. doi: 10.1016/j.celrep.2017.03.069. [DOI] [PubMed] [Google Scholar]
- 10.Tan H, Yang K, Li Y, Shaw TI, Wang Y, Blanco DB, Wang X, Cho JH, Wang H, Rankin S, Guy C, Peng J, Chi H. 2017. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46:488–503. doi: 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black DL. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367–370. doi: 10.1016/S0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 12.Lynch KW. 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 13.Gaudreau MC, Heyd F, Bastien R, Wilhelm B, Moroy T. 2012. Alternative splicing controlled by heterogeneous nuclear ribonucleoprotein L regulates development, proliferation, and migration of thymic pre-T cells. J Immunol 188:5377–5388. doi: 10.4049/jimmunol.1103142. [DOI] [PubMed] [Google Scholar]
- 14.Mallory MJ, Allon SJ, Qiu J, Gazzara MR, Tapescu I, Martinez NM, Fu XD, Lynch KW. 2015. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proc Natl Acad Sci U S A 112:E2139–E2148. doi: 10.1073/pnas.1423695112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel M, Wilhelmi I, Schultz AS, Preussner M, Heyd F. 2014. Activation-induced tumor necrosis factor receptor-associated factor 3 (Traf3) alternative splicing controls the noncanonical nuclear factor kappaB pathway and chemokine expression in human T cells. J Biol Chem 289:13651–13660. doi: 10.1074/jbc.M113.526269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajani DK, Walch M, Martinvalet D, Thomas MP, Lieberman J. 2012. Alterations in RNA processing during immune-mediated programmed cell death. Proc Natl Acad Sci U S A 109:8688–8693. doi: 10.1073/pnas.1201327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weg-Remers S, Ponta H, Herrlich P, König H. 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J 20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelmi I, Kanski R, Neumann A, Herdt O, Hoff F, Jacob R, Preußner M, Heyd F. 2016. Sec16 alternative splicing dynamically controls COPII transport efficiency. Nat Commun 7:12347. doi: 10.1038/ncomms12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butte MJ, Lee SJ, Jesneck J, Keir ME, Haining WN, Sharpe AH. 2012. CD28 costimulation regulates genome-wide effects on alternative splicing. PLoS One 7:e40032. doi: 10.1371/journal.pone.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigoryev YA, Kurian SM, Nakorchevskiy AA, Burke JP, Campbell D, Head SR, Deng J, Kantor AB, Yates JR III, Salomon DR. 2009. Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes. PLoS One 4:e7906. doi: 10.1371/journal.pone.0007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. 2007. Global analysis of alternative splicing during T-cell activation. RNA 13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, Zhu W, Ajith S, Blencowe BJ, Lynch KW. 2012. Alternative splicing networks regulated by signaling in human T cells. RNA 18:1029–1040. doi: 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson-White S, McDuffie M. 2003. Defective T-cell activation is associated with augmented transforming growth factor beta sensitivity in mice with mutations in the Sno gene. Mol Cell Biol 23:5446–5459. doi: 10.1128/mcb.23.15.5446-5459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duke RC, Cohen JJ. 1986. IL-2 addiction: withdrawal of growth factor activates a suicide program in dependent T cells. Lymphokine Res 5:289–299. [PubMed] [Google Scholar]
- 25.Collins S, Lutz MA, Zarek PE, Anders RA, Kersh GJ, Powell JD. 2008. Opposing regulation of T cell function by Egr-1/NAB2 and Egr-2/Egr-3. Eur J Immunol 38:528–536. doi: 10.1002/eji.200737157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins S, Wolfraim LA, Drake CG, Horton MR, Powell JD. 2006. Cutting edge: TCR-induced NAB2 enhances T cell function by coactivating IL-2 transcription. J Immunol 177:8301–8305. doi: 10.4049/jimmunol.177.12.8301. [DOI] [PubMed] [Google Scholar]
- 27.Kumbrink J, Gerlinger M, Johnson JP. 2005. Egr-1 induces the expression of its corepressor Nab2 by activation of the Nab2 promoter thereby establishing a negative feedback loop. J Biol Chem 280:42785–42793. doi: 10.1074/jbc.M511079200. [DOI] [PubMed] [Google Scholar]
- 28.Kumbrink J, Kirsch KH, Johnson JP. 2010. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J Cell Biochem 111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya S, Wei J, Melichian DS, Milbrandt J, Takehara K, Varga J. 2009. The transcriptional cofactor Nab2 is induced by TGF-beta and suppresses fibroblast activation: physiological roles and impaired expression in scleroderma. PLoS One 4:e7620. doi: 10.1371/journal.pone.0007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. 1996. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol 16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iacobelli M, Rohwer F, Shanahan P, Quiroz JA, McGuire KL. 1999. IL-2-mediated cell cycle progression and inhibition of apoptosis does not require NF-kappa B or activating protein-1 activation in primary human T cells. J Immunol 162:3308–3315. [PubMed] [Google Scholar]
- 32.Sereti I, Gea-Banacloche J, Kan MY, Hallahan CW, Lane HC. 2000. Interleukin 2 leads to dose-dependent expression of the alpha chain of the IL-2 receptor on CD25-negative T lymphocytes in the absence of exogenous antigenic stimulation. Clin Immunol 97:266–276. doi: 10.1006/clim.2000.4929. [DOI] [PubMed] [Google Scholar]
- 33.Burchill MA, Yang J, Vang KB, Farrar MA. 2007. Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol Lett 114:1–8. doi: 10.1016/j.imlet.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao W, Lin JX, Leonard WJ. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malek TR. 2008. The biology of interleukin-2. Annu Rev Immunol 26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 36.Malek TR, Castro I. 2010. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paetkau V. 1985. Molecular biology of interleukin 2. Can J Biochem Cell Biol 63:691–699. doi: 10.1139/o85-086. [DOI] [PubMed] [Google Scholar]
- 38.Ross SH, Cantrell DA. 2018. Signaling and function of interleukin-2 in T lymphocytes. Annu Rev Immunol 36:411–433. doi: 10.1146/annurev-immunol-042617-053352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salerno F, Paolini NA, Stark R, von Lindern M, Wolkers MC. 2017. Distinct PKC-mediated posttranscriptional events set cytokine production kinetics in CD8(+) T cells. Proc Natl Acad Sci U S A 114:9677–9682. doi: 10.1073/pnas.1704227114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. 2004. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J Immunol 172:6136–6143. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- 41.Peter D, Jin SL, Conti M, Hatzelmann A, Zitt C. 2007. Differential expression and function of phosphodiesterase 4 (PDE4) subtypes in human primary CD4+ T cells: predominant role of PDE4D. J Immunol 178:4820–4831. doi: 10.4049/jimmunol.178.8.4820. [DOI] [PubMed] [Google Scholar]
- 42.Shimoyamada H, Yazawa T, Sato H, Okudela K, Ishii J, Sakaeda M, Kashiwagi K, Suzuki T, Mitsui H, Woo T, Tajiri M, Ohmori T, Ogura T, Masuda M, Oshiro H, Kitamura H. 2010. Early growth response-1 induces and enhances vascular endothelial growth factor-A expression in lung cancer cells. Am J Pathol 177:70–83. doi: 10.2353/ajpath.2010.091164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams KW, Kletsov S, Lamm RJ, Elman JS, Mullenbrock S, Cooper GM. 2017. Role for Egr1 in the transcriptional program associated with neuronal differentiation of PC12 cells. PLoS One 12:e0170076. doi: 10.1371/journal.pone.0170076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucerna M, Mechtcheriakova D, Kadl A, Schabbauer G, Schafer R, Gruber F, Koshelnick Y, Muller HD, Issbrucker K, Clauss M, Binder BR, Hofer E. 2003. NAB2, a corepressor of EGR-1, inhibits vascular endothelial growth factor-mediated gene induction and angiogenic responses of endothelial cells. J Biol Chem 278:11433–11440. doi: 10.1074/jbc.M204937200. [DOI] [PubMed] [Google Scholar]
- 45.Herdt O, Neumann A, Timmermann B, Heyd F. 2017. The cancer-associated U2AF35 470A>G (Q157R) mutation creates an in-frame alternative 5′ splice site that impacts splicing regulation in Q157R patients. RNA 23:1796–1806. doi: 10.1261/rna.061432.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen Y, Vignali P, Wang R. 2017. Rapid profiling cell cycle by flow cytometry using concurrent staining of DNA and mitotic markers. Bio Protoc 7:e2517. doi: 10.21769/BioProtoc.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preußner M, Goldammer G, Neumann A, Haltenhof T, Rautenstrauch P, Müller-McNicoll M, Heyd F. 2017. Body temperature cycles control rhythmic alternative splicing in mammals. Mol Cell 67:433–446.e4. doi: 10.1016/j.molcel.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Conrad T, Orom UA. 2017. Cellular fractionation and isolation of chromatin-associated RNA. Methods Mol Biol 1468:1–9. doi: 10.1007/978-1-4939-4035-6_1. [DOI] [PubMed] [Google Scholar]
- 49.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]