Abstract
Background
This study investigated changes over time in the epidemiology of extended-spectrum β-lactamase (ESBL) producing Escherichia coli within a single equine referral hospital in the UK. Faecal samples were collected from hospitalised horses in 2008 and 2017, processed using selective media and standard susceptibility laboratory methods. A novel real-time PCR with high resolution melt analysis was used to distinguish blaCTX-M-1 and blaCTX-M-15 within CTX-M-1 group.
Results
In 2008, 457 faecal samples from 103 horses were collected, with ESBL-producing E. coli identified in 131 samples (28.7, 95% CI 24.6–33.1). In 2017, 314 faecal samples were collected from 74 horses with ESBL-producing E. coli identified in 157 samples (50.0, 95% CI 44.5–55.5). There were 135 and 187 non-duplicate ESBL-producing isolates from 2008 and 2017, respectively. In 2008, 12.6% of isolates belonged to CTX-M-1 group, all carrying blaCTX-M-1, whilst in 2017, 94.1% of isolates were CTX-M-1 group positive and of these 39.2 and 60.8% of isolates carried blaCTX-M-1 and blaCTX-M-15, respectively. In addition, the prevalence of doxycycline, gentamicin and 3rd generation cephalosporin resistance increased significantly from 2008 to 2017 while a decreased prevalence of phenotypic resistance to potentiated sulphonamides was observed.
Conclusions
The real-time PCR proved a reliable and high throughput method to distinguish between blaCTX-M-1 and blaCTX-M-15. Furthermore, its use in this study demonstrated the emergence of faecal carriage of CTX-M-15 in hospitalised horses, with an increase in prevalence of ESBL-producing E. coli as well as increased antimicrobial resistance to frequently used antimicrobials.
Keywords: ESBL-producing E. coli, Melt curve analysis, Multidrug resistance, High resolution melt, Real time PCR, CTX-M-1 group
Background
Extended spectrum β-lactamase (ESBL)-producing Escherichia coli are typically resistant to extended spectrum cephalosporins and monobactams (aztreonam), as well as non-β-lactam agents. These bacteria are of increasing concern as ESBL encoding genes are usually harboured on plasmids, which co-harbour multiple resistance genes [1] leading to multi-drug resistance (MDR, defined as resistance to 3 or more antimicrobial classes). Clinical infections caused by MDR bacteria are particularly difficult to treat and are a leading cause of morbidity and mortality in human and veterinary medicine [2, 3]. The initially identified β-lactamase enzymes such as TEM and SHV are now becoming less prevalent in ESBL-producing E. coli, while CTX-M is now the most predominant mechanism in both humans and animals [4]. Almost 170 distinct ESBL CTX-M β-lactamases have been identified mostly in Enterobacteriaceae, including in Escherichia, Klebsiella and Enterobacter species [5]. The CTX-M family includes a complex group of enzymes which have been classified into five different groups; CTX-M-1, 2, 8, 9 and 25 based on their amino acid sequences [4]. These enzymes are able to efficiently hydrolyse cefotaxime and in some cases ceftazidime (CTX-M-15, CTX-M-16, CTX-M-27) [6, 7].
The worldwide emergence of the blaCTX-M-15 gene (belonging to CTX-M-1 group) during the last two decades in humans is of concern; particularly as it is often associated with the pandemic O25/ST131 E. coli clone [8], which belongs to the highly virulent phylogenetic group B2 and often harbours the multidrug-resistant IncFII plasmids [9]. In ST648 E. coli strains in human and companion animal studies, a high proportion of blaCTX-M-15 has been identified [10]. The now widespread dissemination of CTX-M-15-producing E. coli could have two explanations. Corresponding plasmids encoding the blaCTX-M-15 gene are being transferred via horizontal transfer to multiple lineages [11], or alternatively the strains may be spreading by clonal expansion [12].
Several studies have reported blaCTX-M-1 (also belonging to group 1) to be the most common ESBL-gene in E. coli in horses [13–15], however one study also identified a low prevalence of other blaCTX-M genes [13]. CTX-M-15-producing E. coli has been identified in clinical isolates from horses in Germany [10, 16] and has been infrequently identified in E. coli from hospitalised horses in Holland [13].
Although conventional PCR assay can be used to determine which group a CTX-M enzyme belongs to, Sanger sequencing has, to date, been required to determine the CTX-M genotype [17], which has proved costly and time consuming, hence previous studies have often only classified genes to the group level. Whole genome or next generation sequencing, whilst also identifying genes and genetic contexts is still too expensive for routine surveillance. High resolution melt (HRM) analysis is an end-point real-time PCR detection method that differentiates amplicons based on their melt profile. Here we describe this method using novel primers to distinguish between different blaCTX-M genes within group 1 based on their different melt point. While there are studies reporting the change in antimicrobial resistance (AMR) patterns in equine clinical isolates over time, there are no studies investigating carriage of AMR genes in hospitalised horse populations over time. The aims of the present study were to investigate the trend in AMR patterns over time, to determine the prevalence of blaCTX-M-1 and blaCTX-M-15 genotypes within CTX-M-1 group and to compare changes in phenotype and genotype of ESBL-producing E. coli over time within a single equine hospital in the UK.
Results
In total 771 faecal samples were collected. In 2008 cohort, 457 faecal samples from 103 horses were collected with ESBL-producing E. coli identified in 131 samples (28.7, 95% CI 24.6–33.1) from 49 horses (47.6, 95% CI 37.7–57.6). In 2017 cohort, 314 faecal samples were collected from 72 horses with ESBL-producing E. coli identified in 157 samples (50.0, 95% CI 44.5–55.5) of samples from 47 horses (65.3, 95% CI 53.8–75.3). In the 2008 and 2017 cohorts, antimicrobials had been administered in the previous 7 days prior to sampling in 51.1% (67/131) and 67.5% (106/157) of samples respectively, where an ESBL-producer was identified. Antimicrobials had been administered in the previous 7 days in 32.5% (106/326) and 47.1% (74/157) of samples in the 2008 and 2017 cohorts respectively where no ESBL-producer was identified. Overall use of highest priority critically important antimicrobials (HPCIAs) was low and included 5.8% (6/103) horses (3 enrofloxacin and 3 ceftiofur) in the 2008 cohort and 9.7% (7/72) horses (2 enrofloxacin and 5 ceftiofur) in the 2017 cohort. Surgery was performed during hospitalisation in 53.4% (55/103) and 45.8% (33/72) of horses in the 2008 and 2017 cohorts respectively. The breakdown of type of cases in the two cohorts is shown in Table 1.
Table 1.
Clinical case type in the two cohorts. GI non-surgical cases included colic cases which were treated medically. Medical cases included general medical cases such as ophthalmology, respiratory and non-GI cases. Musculoskeletal cases included mainly lameness investigation and orthopaedic surgical cases. Soft tissue cases included mass removals, dental/sinusitis investigations and surgical airway cases. GI = Gastrointestinal
| Case type | 2008 (n = 103) | 2017 (n = 72) |
|---|---|---|
| GI non-surgical | 5 | 12 |
| GI surgical | 3 | 8 |
| Medical | 35 | 9 |
| Musculoskeletal | 29 | 27 |
| Soft tissue | 31 | 16 |
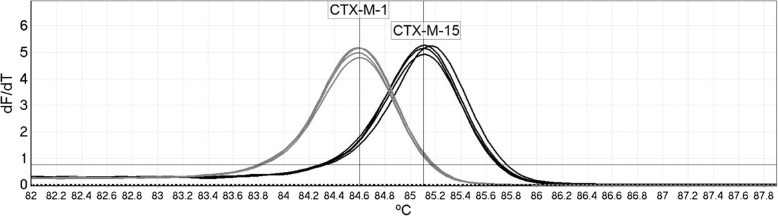
There were 135 non-duplicate ESBL isolates from 2008 cohort and 187 non-duplicate ESBL isolates from 2017 cohort. The HRM analysis demonstrated high specificity during the pilot evaluation, with no non-specific amplification generated when testing a panel of 17 non-target ESBL producers. The assay was 100% accurate at differentiating blaCTX-M-1 and blaCTX-M-15 compared with sequencing, across 44 isolates, indicating 100% agreement (Fig. 1).
Fig. 1.
High resolution melt analysis results for four blaCTX-M-1 carrying isolates, four blaCTX-M-15 carrying isolates and a no template control. The calling threshold and calling bins for automatic genotyping are indicated by the horizontal and vertical lines, respectively
Conventional PCR analysis demonstrated a reduction in carriage of blaTEM and blaSHV genes of approximately 50% between the two cohorts and an increase in carriage of blaCTX-M by 37.8%. However, the blaTEM and blaSHV genes were not further confirmed in either cohort to determine if they were ESBL variants. In the 2008 cohort isolates, only 12.6% (n = 17) were CTX-M-1 group positive, all of which identified as blaCTX-M-1 using HRM analysis. In 2017 cohort isolates, 94.1% (n = 176) were CTX-M-1 group positive and of these 39.2% (n = 69) identified as blaCTX-M-1 and 60.8% (n = 107) as blaCTX-M-15 using HRM analysis. All 107 isolates identified as CTX-M-15 producers were negative for 025 and ST131 on PCR assay. There was a significant increase in carriage of qnr A (11.9%) and a small non-significant decrease in carriage of qnr B (− 1.26%) and qnr S (− 1.3%). The change in genotype between the two cohorts is shown in Table 2.
Table 2.
The prevalence and change of genotype in ESBL-producing E. coli isolated from hospitalised horses in two different cohorts from the same equine hospital sampled 10 years apart
| Genotype (322) | 2008 cohort (n = 135) | Prevalence (95% CI) | 2017 cohort (n = 187) | Prevalence (95% CI) | Change (%) | P -value |
|---|---|---|---|---|---|---|
| blaSHV (105) | 86 | 63.7 (55.3–71.3) | 19 | 10.2 (6.6–15.3) | −53.5 | < 0.001 |
| blaTEM (183) | 116 | 85.9 (79.1–90.8) | 67 | 35.8 (29.3–42.9) | −50.1 | < 0.001 |
| blaCTX-M (253) | 76 | 56.3 (47.9–64.4) | 177 | 94.6 (90.4–97.1) | + 37.8 | < 0.001 |
| blaCTX-M Gr1 (193) | 17 | 12.6 (8.0–19.2) | 176 | 94.1 (89.8–96.7) | + 81.5 | < 0.001 |
| CTX-M-1 (86) | 17 | 100 (81.6–100) | 69 | 39.2 (32.3–46.6) | −60.8 | < 0.001 † |
| CTX-M-15 (107) | 0 | 0 (0–18.4) | 107 | 60.8 (53.4–67.7) | + 60.8 | |
| blaCTX-M Gr2 (10) | 9 | 6.7 (3.6–12.2) | 1 | 0.5 (0–3.0) | −6.1 | 0.002 † |
| blaCTX-M Gr9 (50) | 50 | 37.0 (29.4–45.4) | 0 | 0.0 (0.0–2.0) | −37.0 | < 0.001 † |
| qnr A (27) | 2 | 1.5 (0.4–5.2) | 25 | 13.4 (9.2–19.0) | + 11.9 | < 0.001 † |
| qnr B (48) | 30 | 22.2 (16.0–30.0) | 18 | 9.6 (6.2–14.7) | −12.6 | 0.003 |
| qnr S (19) | 9 | 6.7 (3.6–12.2) | 10 | 5.4 (2.9–9.6) | − 1.3 | 0.8 |
P-value provided for Chi-squared test apart from where † indicates use of Fisher’s Exact Test
Furthermore, there was an increased variability in phenotypic resistance in the ESBL-producing E. coli between the two cohorts. Some of these changes were not significant (amoxicillin, enrofloxacin and MDR), while there was a significant increase in resistance to doxycycline, gentamicin and 3rd generation cephalosporins and an increase in susceptibility to trimethoprim-sulfamethoxazole. The antimicrobial resistance in non-duplicate ESBL-producing E. coli is shown in Table 3.
Table 3.
Prevalence of, and change in phenotypic AMR in ESBL-producing E. coli isolated from hospitalised horses in two different cohorts from the same equine hospital sampled 10 years apart (2008 and 2017) TMPS- Trimethoprim sulfamethoxazole, MDR - multidrug resistance (defined as resistance to 3 or more antimicrobial classes)
| Antimicrobial agent/phenotype | 2008 cohort (n = 135) | Prevalence (95% CI) | 2017 cohort (n = 187) | Prevalence (95% CI) | % change | Total (%) | P value |
|---|---|---|---|---|---|---|---|
| Amoxicillin (%) | 135 | 100 (97.2–100) | 187 | 100 (98.0–100) | 0 | 322 (100) | 1.0 † |
| Cefpodoxime (%) | 130 | 96.3 (91.6–98.4) | 187 | 100 (98.0–100) | + 3.7 | 317 (98.4) | 0.01† |
| Ceftiofur (%) | 123 | 91.1 (85.1–94.8) | 182 | 97.3 (93.9–98.9) | + 6.2 | 305 (94.7) | 0.03 |
| Gentamicin (%) | 118 | 87.4 (80.8–92.0) | 181 | 96.8 (93.2–98.5) | + 9.4 | 299 (92.9) | 0.0026 |
| TMPS (%) | 133 | 98.5 (94.8–99.6) | 159 | 85.0 (79.2–89.4) | −13.5 | 292 (90.7) | < 0.001 |
| Doxycycline (%) | 53 | 39.3 (31.4–47.7) | 172 | 92.0 (87.2–95.1) | + 52.7 | 225 (69.9) | < 0.001 |
| Enrofloxacin (%) | 36 | 26.7 (19.9–34.7) | 62 | 33.2 (26.8–40.2) | + 6.5 | 98 (30.4) | 0.3 |
| MDR (%) | 120 | 88.9 (82.5–93.2) | 176 | 94.1 (89.8–96.7) | + 5.2 | 296 (91.9) | 0.1 |
P-value provided for Chi-squared test apart from where † indicates use of Fisher’s Exact Test
Discussion
According to our current knowledge this study is the first to identify blaCTX-M-15 in ESBL-producing E. coli in horses at an equine hospital in the UK, as well as reporting a significant increase in prevalence of CTX-M-1 and a reduction in CTX-M-9 β-lactamase producing E. coli isolates in the same hospital over a decade. The study also demonstrated a significant increase in prevalence of overall carriage of ESBL-producing E. coli and their resistance to commonly used antimicrobials in horses, in particular, an increase in resistance to doxycycline, gentamicin and 3rd generation cephalosporins.
This is also the first study using HRM analysis to distinguish between blaCTX-M-1 and blaCTX-M-15. The assay uses a single set of primers to amplify a 213 bp region common to group 1 blaCTX-M- genes, which contains six variant bases blaCTX-M-1 and blaCTX-M-15 that are highly conserved. The presence of these six variants results in a ~ 0.5 °C difference in the melt temperature, allowing discrimination of these genes. The ability to differentiate these genes without the requirement of any post PCR processing or sequencing provides faster results and higher throughput, will be of great benefit in studies involving large sample numbers.
There was 100% agreement between the HRM analysis and the sequencing results in the 44 pilot samples, indicating complete accuracy. HRM analysis has been used previously for applications including bacterial speciation, and the detection of resistance genes [18], and has potential both as a diagnostic test and epidemiological surveillance tool. In addition to genotyping assays, HRM analysis has also been utilised to enable highly multiplexed assays, without the use of costly hydrolysis probes [18]. HRM analysis typically has a lower analytical sensitivity than probe-based PCRs, but this is not a drawback when testing high copy number samples, such as bacterial colonies. Whilst sequencing provides a greater level of resolution, HRM analysis results were sufficient to determine the gene carried by the CTX-M producers, allowing greater number of samples to be tested during the study than if relying solely on sequencing.
Previous studies have identified blaCTX-M-15 in five clinical samples from horses in Germany [10, 16], as well as low frequency carriage (2/123 isolates, 1.6%) in hospitalised horses in Holland [13]. Our study is the first study to report CTX-M-15 β-lactamase as the predominant enzyme in ESBL-producing E. coli carried by horses. The emergence of CTX-M-15 β-lactamase during this 10 year period in this equine hospital is in line with the global trend of the spread of this enzyme in humans [19] and more recently in veterinary studies [16, 20, 21]. CTX-M-15 producing ST131 has also been identified in a dog in Portugal [22], and in three canine clinical isolates in the UK [23] but has yet to be identified in horses, which is supported by the results from the current study. While ST131 is rare in veterinary studies [20, 22], ST648 (phylotype D) [10] and ST410 (phylotype A) [20] have more frequently been reported from clinical and environmental hospital samples. ST410 has been recently described as a new international high-risk clone [24], whilst ESBL-producing ST648 strains have also been observed globally in human patients, as well as in chicken, pigs and wild birds in Europe [25, 26]. There was also a marked difference in the carriage of CTX-M-9 group β-lactamase producing E. coli isolates between the two cohorts; it was the most common group present in the first study while in the later cohort there were no CTX-M-9 ESBL-producing E. coli positive isolates identified, which is perhaps due to the global spread if group 1 isolates (particularly blaCTX-M-15), whilst group 9 isolates (such as blaCTX-M-9 and blaCTX-M-14), commonly found in food-producing animals [4], are becoming more scarce in the equine population [13].
The increase in prevalence of faecal-carriage of ESBL-producing E. coli in horses from one hospital over a decade is perhaps not a surprising finding and hospital studies involving human patients are reporting similar results [4]. The largest increase in antimicrobial resistance between the two cohorts was to doxycycline and this is a surprising finding as tetracycline use in this equine hospital is low (data not shown). Oral antimicrobials in horses are limited to potentiated sulphonamides (authorised) and enrofloxacin (no market authorisation), but more recently oral doxycycline (no market authorisation) has become a more popular treatment as it is cost-effective and can be easily administered by the owner at home. This increased popularity of doxycycline in the equine community has perhaps led to a reduction in use of potentiated sulphonamides, which may be one reason for the change in resistance patterns in these two antimicrobials in our study. There are more than 12 tetracycline resistance genes described in E. coli and they include three main mechanisms; drug efflux pumps, ribosomal protection proteins, and drug inactivation. Some of these genes may be plasmid mediated, in particularly tetA and tetB genes which code for efflux pumps, and the increase in tetracycline resistance may be due to co-existence on the same plasmid as the ESBL genes. Further work is required to investigate such associations, including conjugation experiments, plasmid typing and sequencing. The hospital use of HPCIAs [27], including third and fourth generation cephalosporins was low in both cohorts and there was no clear reason for the increase in prevalence of ESBL-producing E. coli between the two cohorts. The use of third and fourth generation cephalosporins by referring veterinarians is unknown. However, a study in 2013 identified that third and fourth generation cephalosporins only accounted for 3% of prescriptions and that equine veterinarians in first opinion practice most commonly prescribe potentiated sulphonamides [28].
Limitations of this study include the low numbers of isolates which underwent Sanger sequencing to validate the results from the HRM analysis, but in these 44 isolates there was complete agreement. Another limitation is that the study included only two sampling periods and used slightly different sampling protocols; in 2008 cohort horses were sampled every other day while in 2017 cohort horses were sampled daily, however this is unlikely to affect the results as antimicrobial susceptibility testing was repeated on the stored isolates in 2018.
Conclusions
The present study demonstrates that HRM analysis is a reliable and low-cost method and can be used to distinguish between CTX-M-1 and CTX-M-15 β-lactamase producers in group 1 positive isolates from a conventional PCR assay without the need for sequencing of PCR amplicons. Our study also demonstrated an emergence of carriage of CTX-M-15 producing E. coli isolates in hospitalised horses in the UK. This may have public health implications as when these horses leave the hospital, they may contribute to the dissemination of ESBL-producing E. coli in the environment. It is generally accepted that carriage is a pre-requisite for infection and future studies are needed to investigate the link between faecal CTX-M-15 carriage and ESBL-producing E. coli obtained from clinical isolates in horses in the UK.
Methods
Faecal samples were collected from two different cohorts of hospitalised horses at a single, large referral equine hospital in North West UK which admits approximately 1500 cases annually. Animals eligible for the study were all horses hospitalised for at least one night in the hospital. Day cases were excluded as were animals receiving chemotherapy or radiotherapy and those in isolation. Samples (~ 30 g) were taken from the top of a pile of freshly passed faeces from each horse by hospital staff, placed in sterile plastic containers and transported to the laboratory on the same site as the equine hospital. In 2008 cohort (2008–2009) faecal samples were collected every other day during hospitalisation [29], while in 2017 cohort (2016–2017), recruited from the same hospital, daily faecal samples were obtained from each horse until the horse was discharged from hospital.
Isolation of resistant bacteria from faecal samples
E. coli isolation was performed according to a previously described method [29]. Briefly, 2 g of faeces were placed in a stomacher bag and 10 ml of brain heart infusion broth was added. To screen for cephalosporin-resistant E. coli (presumptive ESBL-producers), 0.5 ml of the faecal homogenate was also added to 4.5 ml buffered peptone water for aerobic incubation at 37 °C for 18–24 h. Following overnight enrichment, 5 μl of the faecal homogenate was streaked onto a selective media [Eosin Methylene Blue Agar (EMBA) or Harlequin agar)] containing 1 μg/ml cefotaxime using a 5 μl disposable sterile loop and incubated aerobically for 18–24 h at 37o C. On the selective media, if present, two colonies resembling E. coli were selected from the plate and transferred onto nutrient agar and incubated overnight at 37 °C.
Antimicrobial susceptibility and ESBL phenotypic testing
Colonies from nutrient agar plates were suspended in sterile water to make a suspension equivalent to 0.5 McFarland Turbidity Standard (0.5 MTS). Each isolate suspension was then inoculated onto a Mueller Hinton agar plates for antimicrobial susceptibility testing following the Clinical & Laboratory Standards Institute (CLSI, 2016). Double disc diffusion tests using ceftazidime (30 μg), cefotaxime (30 μg) and cefpodoxime (30 μg) ± clavulanic acid was used to confirm phenotypic ESBL-producing E. coli. Antimicrobial susceptibility testing was performed using discs of 10 μg amoxicillin (amox), 10 μg cefpodoxime (cpd), 30 μg ceftiofur (eft), 10 μg gentamicin (gent), 5 μg enrofloxacin (enf), 30 μg doxycycline (dxt), 1.25 μg trimethoprim + 23.75 μg sulfamethoxazole (TMPS). All microbiological media were from LabM, UK; antimicrobial agents were from Mast Group, UK (amox, cpd, gent, enf, dxt and tmps) or Oxoid, Basingstoke, UK (eft). After 16–18 h incubation at 37 °C, the bacterial growth inhibition zone diameter (mm) for each disc was measured. Isolates were categorised as susceptible if the diameter of the zone of inhibition was greater than the breakpoint for that drug [30], resulting in a binary outcome of susceptible or resistant. Control strain E.coli ATCC 52922 was used for susceptibility testing. MDR for E. coli was defined according to criteria which excludes intrinsic resistance; aminopenicillins and 3rd generation cephalosporins were considered separate classes for MDR calculations [31]. Long-term storage of isolates was undertaken at -80 °C using Microbank™ cryovials (Pro-Lab Diagnostics U.K, Cheshire UK) and isolates were recovered by removing a single bead from the cryovial using sterile forceps and inoculation of the bead onto nutrient agar for aerobic incubation at 37 °C for 16–18 h. There was 100% recovery of frozen isolates. All testing was performed by the same operators in 2017/2018 and the methods and interpretation criteria were the same for the two cohorts. Samples from 2008 were initially archived and retrieved in 2018.
Genotypic analysis
Cell lysates were prepared by adding two to three colonies of pure 24-h cultures to 0.5 mL sterile water and heating at 100 °C for 20 min. All isolates phenotypically consistent with E. coli were confirmed using uidA gene primers in a standard PCR assay [29, 32]. All isolates confirmed as EBSL-producers using double disc diffusion test were tested by conventional PCR for blaCTX-M genes using universal blaCTX-M primers as previously described [33]. To determine the CTX-M group, all CTX-M positive isolates were tested using primers specific to blaCTX-M groups 1, 2 [34] and 9 [33]. All isolates demonstrating an ESBL-phenotype were also tested for blaTEM, blaSHV and blaOXA encoding beta-lactamase genes [35] and plasmid-mediated qnrA, B and S genes conferring quinolone resistance using further multiplex PCR assays [36].
HRM analysis
All isolates identified as carrying CTX-M-1 group ESBL genes were categorised as blaCTX-M-1 or blaCTX-M-15 producers using a novel HRM analysis based real time PCR assay. Primers were designed from CTX-M-1 group sequences aligned in MEGA, using ClustalX. A 213 bp region was selected with the maximum variation between blaCTX-M-1 or blaCTX-M-15 gene sequences, to allow for the greatest Tm shift during melt analysis. Primers were designed using Primer3 (http://primer3.ut.ee/), and amplicon Tm was estimated using the nearest neighbour method in OligoCalc (http://biotools.nubic.northwestern.edu/OligoCalc.html). E. coli 13353 was used as a control strain in the assay validation (CTX-M-15). The primer details are shown in Table 4.
Table 4.
Forward and reverse sequence, amplicon size and melt temperature for the high-resolution melt curve analysis distinguishing between blaCTX-M-1 or blaCTX-M-15 within CTX-M group 1
| Primer | Sequence 5 > 3 | Amplicon size | Melt Tm |
|---|---|---|---|
| CTXMG1 F | TGTTGTTAGGAAGTGTGCCG | 213 bp | 84.75 C (CTX-M-1) |
| CTXMG1 R | CGCTTTCACTTTTCTTCAGCAC | 85.25 (CTX-M-15) |
Reactions were carried out in the Rotor-Gene Q 6000 (Qiagen, Germany) using Type-It HRM mix (Qiagen, Germany), and 400 nM of both forward and reverse primers. Cycling conditions were 95 °C for 5 min, followed by 30 cycles of 95 °C for 10 s, 58 °C for 30 s and 72 °C for 10 s. Reactions were monitored in the FAM channel, and end point high resolution melt analysis was carried out using the HRM channel at 0.1 °C increments from 72 °C to 95 °C. The calling bins for blaCTX-M-1 and blaCTX-M-15 gene were set at 84.75 °C (+/− 0.1 °C) and 85.25 °C (+/− 0.1 °C), respectively, by the predicted melt temperatures. The specificity of the primers was confirmed by testing against a bank of 17 non-target β-lactamase producers, including CTX-M-9 (n = 5), CTX-M-2 (n = 1), CTX-M-8 (n = 1) group, TEM (n = 5) and SHV (n = 5) positive isolates. The accuracy of the melt analysis method was validated by Sanger sequencing (Source Bioscience, UK), where the blaCTX-M gene was confirmed in 44 CTX-M-1 group carrying isolates (9 blaCTX-M-1, 35 blaCTX-M-15) and then testing these isolates using HRM analysis.
The assay was then used to test all CTX-M-1 group producers identified in the study. Isolates carrying blaCTX-M-15 were tested further by conventional PCR assay to determine whether they belonged to serogroup O25 [37] and for markers for sequence type (ST)131 [38], representing the pandemic E. coli clone associated with CTX-M-15 in human clinical infections [8].
Data analysis
The 95% confidence intervals around proportions were calculated using Wilson’ Score interval [39]. Differences in proportions between the two cohorts were compared using a Chi-squared test. Where a comparison had less than five in any category a Fishers exact test was used. All analysis was performed using EpiTools [39]. A P value of < 0.05 was regarded as significant.
Acknowledgements
The authors wish to thank the staff and clients of all the University of Liverpool Equine Hospital and Ruth Ryvar and Andrew O’Donnell for technical assistance.
Abbreviations
- AMR
Antimicrobials resistance
- CLSI
Clinical & Laboratory Standards Institute
- EMBA
Eosin methylene blue agar
- ESBL
Extended spectrum β-lactamase
- HRM
High resolution melt
- MDR
Multi drug resistance
- MTS
McFarland turbidity standard
- PCR
Polymerase chain reaction
- TMPS
Trimethoprim sulfamethoxazole
Authors’ contributions
TM and CI conducted the 2008 and 2017 cohorts, respectively with assistance from NW, DT, EW, PC and GP. GP, NW, DT, TM and PC. contributed to the study conception and design. TE, EA and PN developed and validated the HRM analysis including pilot samples. GP assisted with the statistical analysis. CI wrote the article and all authors revised the manuscript and approved the final version for submission.
Funding
This work was funded by grants from Horse Trust (Registered Charity No: 231748) (2017 cohort, awarded to Dr. Gina Pinchbeck), the Bransby Home of Rest for Horses (Registered Charity No: 1075601) (2008 cohort, awarded to Prof. Peter Clegg) and the UK Department for Environment, Food and Rural Affairs (Defra). Emma Winward was funded by a Beaufort Cottage Educational Trust summer studentship.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate
Ethical consent was provided by the University of Liverpool Ethical committee (VREC 447) and all horse owners provided written consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 2.Ray S, Anand D, Purwar S, Samanta A, Upadhye KV, Gupta P, et al. Association of high mortality with extended–spectrum β-lactamase (ESBL) positive cultures in community acquired infections. J Crit Care. 2018;44:255–260. doi: 10.1016/j.jcrc.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 3.van Spijk JN, Schmitt S, Schoster A. Infections caused by multidrug-resistant bacteria in an equine hospital (2012-2015). Equine Vet Educ. 2017. 10.1111/eve.12837.
- 4.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 5.McNulty CAM, Lecky DM, Xu-McCrae L, Nakiboneka-Ssenabulya D, Chung K-T, Nichols T, et al. CTX-M ESBL-producing Enterobacteriaceae: estimated prevalence in adults in England in 2014. J Antimicrob Chemother. 2018;73:1368–1388. doi: 10.1093/jac/dky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine M.-H., Blanco J., Leflon-Guibout V., Demarty R., Alonso M. P., Canica M. M., Park Y.-J., Lavigne J.-P., Pitout J., Johnson J. R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. Journal of Antimicrobial Chemotherapy. 2007;61(2):273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SH, Kaufmann ME, Livermore DM, Woodford N, Willshaw GA, Cheasty T, et al. UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J Antimicrob Chemother. 2008;62:1241–1244. doi: 10.1093/jac/dkn380. [DOI] [PubMed] [Google Scholar]
- 9.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 10.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, et al. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother. 2014;69:1224–1230. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 11.Cantón R, Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006. [DOI] [PubMed]
- 12.Manges AR, Johnson JR, Foxman B, O’Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med. 2001;345:1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- 13.Apostolakos I, Franz E, van Hoek AHAMAM, Florijn A, Veenman C, Sloet-van Oldruitenborgh-Oosterbaan MM, et al. Occurrence and molecular characteristics of ESBL/AmpC-producing Escherichia coli in faecal samples from horses in an equine clinic. J Antimicrob Chemother. 2017;72:1915–1921. doi: 10.1093/jac/dkx072. [DOI] [PubMed] [Google Scholar]
- 14.Dolejska M, Duskova E, Rybarikova J, Janoszowska D, Roubalova E, Dibdakova K, et al. Plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding Centre. J Antimicrob Chemother. 2011;66:757–764. doi: 10.1093/jac/dkq500. [DOI] [PubMed] [Google Scholar]
- 15.Walther B, Klein K-S, Barton A-K, Semmler T, Huber C, Wolf SA, et al. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Acinetobacter baumannii among horses entering a veterinary teaching hospital: the contemporary “Trojan horse.”. PLoS One. 2018;13:e0191873. doi: 10.1371/journal.pone.0191873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, Semmler T, et al. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-β-lactamase-producing Escherichia coli among companion animals. J Antimicrob Chemother. 2010. [DOI] [PubMed]
- 17.Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006;57:14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- 18.Edwards T, Sasaki S, Williams C, Hobbs G, Feasey NA, Evans K, et al. Speciation of common gram-negative pathogens using a highly multiplexed high resolution melt curve assay. Sci Rep. 2018;8. 10.1038/s41598-017-18915-5. [DOI] [PMC free article] [PubMed]
- 19.Merino Irene, Hernández-García Marta, Turrientes María-Carmen, Pérez-Viso Blanca, López-Fresneña Nieves, Diaz-Agero Cristina, Maechler Friederike, Fankhauser-Rodriguez Carolina, Kola Axel, Schrenzel Jacques, Harbarth Stephan, Bonten Marc, Gastmeier Petra, Canton R, Ruiz-Garbajosa P, Desilets Marc, Dul Stéphanie, Scherrer-Muller Fabienne, Huttner Benedikt, Uçkay Ilker, Prendki Virginie, Renzi Gesuele. Emergence of ESBL-producing Escherichia coli ST131-C1-M27 clade colonizing patients in Europe. Journal of Antimicrobial Chemotherapy. 2018;73(11):2973–2980. doi: 10.1093/jac/dky296. [DOI] [PubMed] [Google Scholar]
- 20.Timofte D, Maciuca IE, Williams NJ, Wattret A, Schmidt V. Veterinary hospital dissemination of CTX-M-15 extended-Spectrum Beta-lactamase–producing Escherichia coli ST410 in the United Kingdom. Microb Drug Resist. 2016;22:609–615. doi: 10.1089/mdr.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pomba C, López-Cerero L, Bellido M, Serrano L, Belas A, Couto N, et al. Within-lineage variability of ST131 Escherichia coli isolates from humans and companion animals in the south of Europe. J Antimicrob Chemother. 2014;69:271–273. doi: 10.1093/jac/dkt343. [DOI] [PubMed] [Google Scholar]
- 22.Pomba C, da Fonseca JD, Baptista BC, Correia JD, Martínez-Martínez L. Detection of the pandemic O25-ST131 human virulent Escherichia coli CTX-M-15-producing clone harboring the qnrB2 and aac(6′)-Ib-cr genes in a dog. Antimicrob Agents Chemother. 2009;53:327–328. doi: 10.1128/AAC.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timofte D, Maciuca IE, Kemmett K, Wattret A, Williams NJ. Detection of the human pandemic Escherichia coli B2-O25b-ST131 in UK dogs. Vet Rec. 2014;174:352. doi: 10.1136/vr.101893. [DOI] [PubMed] [Google Scholar]
- 24.Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018. [DOI] [PMC free article] [PubMed]
- 25.Guenther S, Grobbel M, Beutlich J, Bethe A, Friedrich ND, Goedecke A, et al. CTX-M-15-type extended-spectrum beta-lactamases-producing Escherichia coli from wild birds in Germany. Environ Microbiol Rep. 2010;2:641–645. doi: 10.1111/j.1758-2229.2010.00148.x. [DOI] [PubMed] [Google Scholar]
- 26.Zong Z, Yu R. Escherichia coli carrying the blaCTX-M-15gene of ST648. J Med Microbiol. 2010;59:1536–1537. doi: 10.1099/jmm.0.022459-0. [DOI] [PubMed] [Google Scholar]
- 27.Critically important antimicrobials for human medicine, 6th revision. 2018. https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/.
- 28.Hughes LA, Pinchbeck G, Callaby R, Dawson S, Clegg P, Williams N. Antimicrobial prescribing practice in UK equine veterinary practice. Equine Vet J. 2013;45:141–147. doi: 10.1111/j.2042-3306.2012.00602.x. [DOI] [PubMed] [Google Scholar]
- 29.Maddox TW, Williams NJ, Clegg PD, OA J, Dawson S, Pinchbeck GL, et al. Longitudinal study of antimicrobial-resistant commensal Escherichia coli in the faeces of horses in an equine hospital. Prev Vet Med. 2011;100:134–145. doi: 10.1016/j.prevetmed.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, CLSI, 5th Edition. https://clsi.org/standards/products/veterinary-medicine/documents/vet01/.
- 31.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 32.McDaniels AE, Rice EW, Reyes AL, Johnson CH, Haugland RA, Stelma Jr. GN. Confirmational identification of Escherichia coli , a comparison of genotypic and phenotypic assays for glutamate decarboxylase and Confirmational identification of Escherichia coli , a comparison of genotypic and phenotypic assays for glutamate Decarboxy. Appl Enviromental Microbiol 1996;62:3350–3354. http://aem.asm.org/content/62/9/3350.short. Accessed 25 Sept 2018. [DOI] [PMC free article] [PubMed]
- 33.Batchelor M, Hopkins K, Threlfall EJ, Stallwood AD, Davies RH, Liebana E. bla CTX-M Genes in Clinical Salmonella Isolates Recovered from Humans in England and Wales from 1992 to 2003 bla CTX-M Genes in Clinical Salmonella Isolates Recovered from Humans in England and Wales from 1992 to 2003. Am Soc Microbiol. 2005;49:1319–1322. doi: 10.1128/AAC.49.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins KL, Batchelor MJ, Liebana E, Deheer-Graham AP, Threlfall EJ. Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int J Antimicrob Agents. 2006;28:180–192. doi: 10.1016/j.ijantimicag.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 35.Dallenne C, da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important Beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 36.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006. [DOI] [PMC free article] [PubMed]
- 37.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, et al. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 38.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother. 2009. [DOI] [PubMed]
- 39.Sergeant E. Epitools epidemiological calculators: Ausvet Pty Ltd; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding authors on reasonable request.