Abstract
Tumor-stroma interactions significantly influence cancer cell metastasis and disease progression. These interactions partly comprise crosstalk between tumor and stromal fibroblasts, but the key molecular mechanisms within the crosstalk that govern cancer invasion are still unclear. Here we adapted our previously developed microfluidic device as a 3D in vitro organotypic model to mechanistically study tumor-stroma interactions by mimicking the spatial organization of the tumor microenvironment on a chip. We co-cultured breast cancer and patient-derived fibroblast cells in 3D tumor and stroma regions, respectively, and combined functional assessments, including cancer cell migration, with transcriptome profiling to unveil the molecular influence of tumor-stroma crosstalk on invasion. This led to the observation that cancer-associated fibroblasts (CAF) enhanced invasion in 3D by inducing expression of a novel gene of interest, GPNMB, in breast cancer cells, resulting in increased migration speed. Importantly, knockdown of GPNMB blunted the influence of CAF on enhanced cancer invasion. Overall, these results demonstrate the ability of our model to recapitulate patient-specific tumor microenvironments to investigate the cellular and molecular consequences of tumor-stroma interactions.
INTRODUCTION
Tumor-stroma interactions significantly influence cancer cell metastasis and disease progression[1]. These interactions in part make up heterotypic crosstalk between tumor and stromal cells[1]. While conventional thinking has emphasized the importance of epithelial cancer cells, there has been a shift toward understanding the influence of stromal components on tumor progression. Cancer-associated fibroblasts (CAFs) stand out as the most abundant non-cancer cell type within the tumor microenvironment, which allows them a unique position to significantly influence invasion[1, 2]. Recent studies have implicated CAFs as key components in cancer initiation, promotion, and therapeutic responses of different cancers, such as breast, prostate, ovarian, colon, and non-small cell lung cancer[1]. For instance, Orimo et al. demonstrated that CAFs promoted tumor growth and angiogenesis through secreted factors[3]. A separate study found that exosomes secreted by CAFs enhanced the metastatic potential of breast cancer cells[4]. CAFs have also been implicated in altering therapeutic response by activating possible compensatory signaling pathways[5]. On a similar note, triple negative breast cancers (TNBCs), an aggressive form of breast cancer, still lack effective targeted therapies, but it has been hypothesized that interactions with CAFs are crucial for TNBC disease progression presenting a possible area to therapeutically target[2, 3]. However, the mechanism and functional consequences of tumor-stroma interactions on cancer invasion are still not completely understood[1]. As such, understanding and targeting the interaction between CAFs and cancer cells within the tumor microenvironment could provide a potential novel treatment strategy for breast cancer, shifting away from the neoplastic cell-centric toward a tumor-stroma paradigm.
To study the cellular and molecular basis of cancer invasion in response to CAFs, a significant effort has been devoted to recapitulating tumor-stroma interactions[6]. In vivo models play a crucially important role in studying the cellular and molecular basis of disease progression but they suffer from lack of high resolution observation and precise analysis of cell-cell interactions by manipulating stromal cells within the tumor microenvironment[6]. This lack of precise control has led to challenges for determining the cause and effect relationships within the heterotypic dialogues between cancer and stromal cells like CAFs[6]. Furthermore, there are crucial molecular and cellular differences between humans and mice limiting the scope for animal models to fully recapitulate disease progression in humans. To overcome some of these problems, conventional co-culture in vitro platforms, including transwell assays and 3D spheroid-based models, have been utilized for biological studies on invasion[6, 7]. However, these models are often oversimplified and do not replicate proper organotypic arrangement of the tumor-stroma architecture due to random mixing of cells. The scope of analyses within such models are limited to proliferation, morphology, and protein expression as opposed to precise spatial organization of cells which could enable assessment of invasion metrics (i.e. distance, speed, persistence)[3, 7]. Importantly, these models are often end-point assays that do not allow real-time observations of dynamic tumor-stroma interactions at cellular and molecular levels.
Recently, there has been a significant thrust to use microfluidic platforms to develop complex 3D tumor models, with precise control over cell-cell, cell-matrix and cell-soluble factor interactions[7, 8]. Microfluidic models integrated with hydrogel-based 3D matrices allow the study of different steps of the metastatic cascade such as invasion, intravasation, and extravasation[7–11]. Our group recently developed a tumor invasion model of breast cancer on the premise of utilizing and understanding chemoattractants and paracrine signaling[8–10]. We studied the effects of EGF on breast cancer cell invasion, providing quantitative data on real-time invasion in a 3D hydrogel at a single-cell level, cancer cell phenotype, and EGF receptor activation[8]. However, the analyses were limited to cell-based functional assessments mainly built on migration within a 3D tumor microenvironment. The impact of similar studies using complex in vitro models in the context of breast cancer have also been limited specifically due to lack of CAF co-culture or the use of non-mammary cells, such as 3T3 or dermal fibroblasts[12–15]. Most importantly, neither our study nor many others have integrated transcription profiling to better inform the molecular influence of CAFs on invasion[8–10, 16]. Notably, CAFs derived from cancer patients are phenotypically different from normal fibroblasts (NFs) and myofibroblasts due to being influenced and educated by the tumor. As a result, CAFs lack the ability to reverse their phenotype remaining in a perpetually activated state[1]. Therefore, patient-derived CAFs preserve the molecular characteristics and phenotype when taken from tumor tissues. Thus, there is a crucial need to develop a physiologic tumor-stroma model incorporated with patient-derived CAFs to understand the extent of their molecular and cellular influence invasion to better discover relevant therapeutic targets.
In this present study, we adapted our previously developed microfluidic device as an organotypic tumor model incorporated with patient-derived CAFs and NFs to assess the molecular and cellular basis for tumor-stroma interactions on breast cancer invasion (Fig. 1A and B). We found that the interplay between invasive SUM-159 breast cancer cells and mammary fibroblasts had distinct consequences on cellular invasion depending on the fibroblast phenotype (i.e., CAFs or NFs). Notably, we paired our functional assessments with transcriptional profiling to evaluate the molecular changes during cancer invasion. We uncovered a novel gene of interest, glycoprotein non-metastatic B (GPNMB), and unveiled that CAFs enhanced breast cancer invasion through up-regulation of GPNMB on breast cancer cells. Knockdown of GPNMB resulted in attenuating the invasive-promoting effect that CAFs had on cancer invasion providing important insight on molecular mechanisms in tumor-stroma interactions during invasion.
Fig. 1. 3D organotypic co-culture invasion assay.
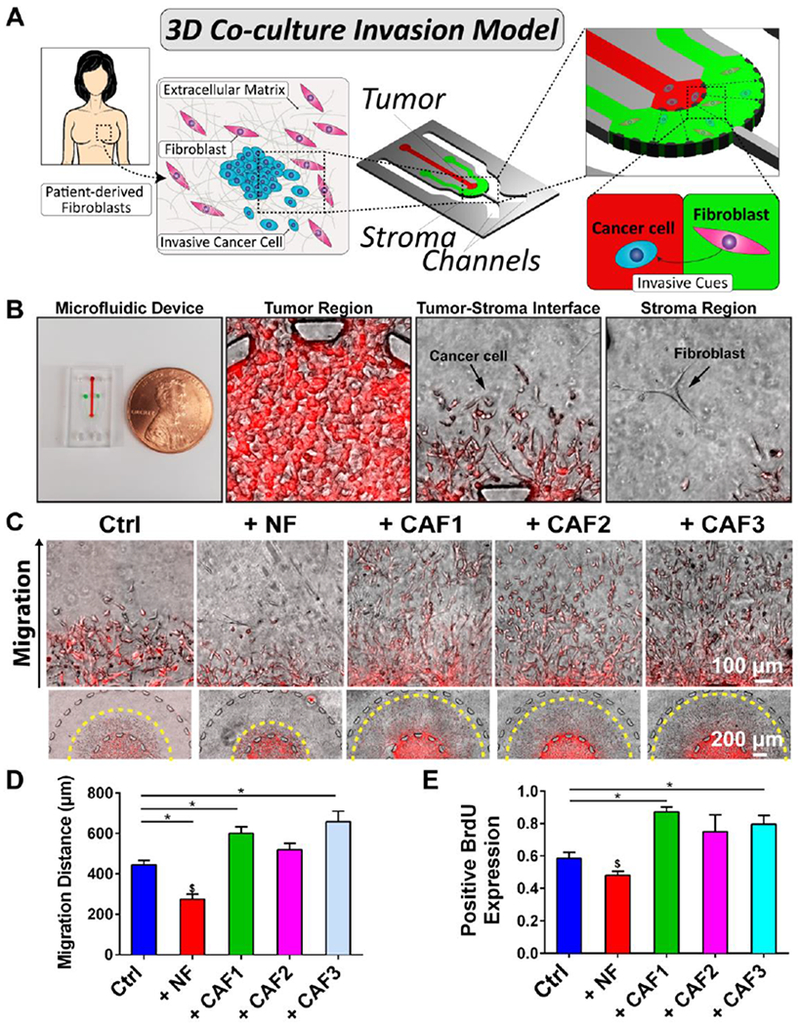
(A) Fibroblasts were derived from patient biopsies. SUM-159 breast cancer cells were cultured in the tumor (red) region while fibroblasts (NFs or CAFs) were cultured in the stroma (green). (B) Microfluidic device shown next to a penny depicting the size of the platform. The tumor region of the platform demonstrates a dense amount of cancer cells. The tumor-stroma interface within the device shows migrating cancer cells. Fibroblasts are shown within the stroma region. (C) Magnified images of cancer migration in the presence of fibroblasts alongside images of the tumor-stroma regions. (D) + CAF1 and + CAF3 migration was significantly higher than Ctrl while + NF was lower than all groups. (E) + CAF1 and + CAF3 had increased expression of BrdU compared to Ctrl. + NF was significantly lower than CAF co-cultures. * denotes significant difference and $ denotes a significantly different group for p < 0.05.
MATERIAL AND METHODS
Microfluidic design and fabrication
The microfluidic platform design and fabrication was established in our previous studies[8]. The design of the device consisted of an inner chamber (tumor region) bordered by an outer chamber (stromal region) (Fig 1A). The diameter and height of these concentric chambers were 3 mm and 200 μm, respectively. The distance between the edge of the inner chamber and outer chamber was 1 mm which is the maximum distance of migration from the tumor region. The chambers were bounded by trapezoidal micro-posts to separate the two regions. These trapezoids were spaced at 100 μm apart to create to allow diffusion of biomolecules between the two regions as well as enable interaction of cells. The micro-post design prevented leakage of hydrogel used in the tumor region due to the angle of the trapezoid being supplementary to the contact angle of the gel solution and PDMS. Importantly, this microfluidic system had two inputs used to insert cell culture media. For details on microfluidic fabrication, please see Supplementary Information.
Cell culture
In order to model migration in a 3D microenvironment, SUM-159 breast cancer cells was chosen as a suitable cell type due to their readiness to invade in 3D hydrogels[17]. The mCherry-labeled SUM-159 breast cancer cells, provided by Mouneimne lab, were cultured in SUM specific media (Ham’s F-12 with L-glutamine and supplemented with 5% heat inactivated fetal bovine serum (FBS), 1% penicillin-streptomycin, 1 μg/ml hydrocortisone, and 5 μg/ml insulin). NFs were obtained from ZenBio. CAFs were isolated from breast tumor tissue samples of three different patients with variation in hormone and Her2 receptor statuses from the Mayo Clinic (Phoenix, AZ) (Table S1). Patient studies were conducted in accordance with the ethical guidelines set by Declaration of Helsinki U.S. Common Rule, Belmont Report, and CIOMS. The studies were performed after approval by an Institutional Review Board (IRB) and was deemed minimal risk so no consent was necessary from the patients. For details on fibroblast isolation and characterization, please see Supplementary Information. mCherry-labeled MDA-MB-231 and MCF7 were provided by the Mouneimne Lab. The cells were maintained in complete Dulbecco’s Modified Eagle Media (DMEM supplemented with 10% heat inactivated fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin-streptomycin). Mycoplasma testing performed by nuclear DNA staining and cell line identification of all cells used was confirmed using Human Cell Authentication Service (ATCC).
3D Co-culture Microfluidic Invasion assay
The invasion assay was based on our previous publications[8]. Briefly, a 1:1 mixture of Matrigel® to Collagen I (2.0 mg/mL) was added to the cells to create a mixed hydrogel cell solution (final concentration of Collagen I at 1 mg/mL) with a density of 15 million cells/mL. The mixed hydrogel cell solution was injected into the tumor region of the microfluidic chip. These devices were flipped every minute to maintain homogenous distribution of cells in 3D hydrogel during injection of several devices (n >1–2). Hydrogel was placed within the cell culture incubator at 37 °C to polymerize. After 2 min within the incubator, the devices were taken out and subsequently a 2.0 mg/mL collagen type I solution was injected into the stroma region. For co-culture with CAFs, 50,000 cells/mL were encapsulated in the collagen type I solution. The collagen was polymerized within the humidified incubator at 37 °C for 8 min and flipped every min when fibroblasts were encapsulated. SUM-159 cell media was added into the channels and the devices were put in the cell culture incubator. Media was exchanged daily. For a detailed description of proliferation assay, immunofluorescence staining, western blotting, qPCR, gene expression profiling, and imaging analysis, see Supplementary Information.
Time-lapse imaging
To perform time-lapse imaging, mCherry-labeled SUM-159 cells were mixed together with normal SUM-159 at a ratio of 1:9 prior to the invasion assay. The invasion assay was performed as described above. On day 2 of the invasion assay, the devices were placed inside a custom miniature incubator (TC-MWP, Bioscience Tools) at 37 °C and 5% CO2. Fluorescent time-lapse imaging was performed using a fluorescent microscope (Zeiss Axio Observer Z1, Zeiss) equipped with the Apotome.2 and a 10x objective. The Apotome.2 utilized structural illumination technique to create optical sections of our devices to reduce scattered light and to generate high-resolution Z-stacked fluorescent 3D images. The time interval was set to 45 min. Time-lapse images were taken on day 2 of the invasion assay for 12 h overnight followed by migration metric quantifications.
Statistical analysis
All measurements were compiled from three or more independent devices or replicates for each experimental condition and repeated at least three times. Reported measurements are shown as average ± standard error of mean. The data were compared using unpaired t-test, paired t-test, multiple comparisons test with corrections, and correlation analysis as appropriate within the GraphPad Prism software (GraphPad Software). Volcano plots and heatmaps were generated using R.
RESULTS
Isolation and characterization of patient-derived CAFs
We isolated CAFs from breast tumor tissue samples of three different patients varying in hormone and Her2 receptor status from the Mayo Clinic (Phoenix, AZ) (Table S1). We obtained NFs from reduction mammoplasty. Next, CAFs are routinely identified through expression of alpha smooth muscle actin (αSMA), contractile stress fibers, and vimentin[18]. Immunofluorescent (IF) images of both NFs and CAFs showed positive staining for vimentin and negative expression of cytokeratin demonstrating the overall purity of the fibroblast populations (Fig. S1). CAFs expressed significantly higher αSMA levels (Fig. S1B). We further corroborated the IF results with western blotting (Fig. S1C and D). We characterized the CAF isolates using fibroblast-activation protein (FAP) and fibroblast specific protein-1 (FSP-1 or S100A4)[18] in addition to αSMA. We determined a significant decrease in expression levels FSP-1 with a non-significant increase in αSMA and FAP for CAF isolates compared to NFs (Fig. S1E)[18].
Differences between CAFs and NFs also extended to morphology[1]. Generally, NFs were smaller and more spindle shaped, while CAFs were larger and polygonal with actin stress fibers (Fig. S2A)[19]. Based on morphometric analyses, all three CAFs demonstrated larger cell spreading area and aspect ratio compared to NFs, where the differences for CAF2 and CAF3 but not CAF1 were statistically significant (Fig. S2B and C), but still agreeing with previous work[19]. Next, we analyzed the 2D migration of the fibroblasts using a scratch wound healing assay (Fig. S2D), and found that NFs and CAFs migrated similarly (Fig. S2E). Overall, these results indicated that CAFs shared general mesenchymal markers (i.e. vimentin) with NF in 2D culture, but were heterogeneous in their morphology and αSMA expression.
Stromal CAF and NF behavior within a 3D matrix
To examine the cell morphology and behavior of NF and CAFs in 3D culture, the fibroblasts were cultured in the stroma region of the 3D microfluidic platform. All fibroblast populations showed elongated morphology (Fig. S3A and B). Time-lapse image analysis demonstrated that fibroblasts were mostly stationary for 3 days, and no statistical difference was observed between the migratory activities of NF and CAF similar to the 2D wound healing assay (Fig. S3C). Both NF and CAF1 exhibited significantly lower cell area than CAF2, while CAF3 was not statistically different to any other fibroblasts (Fig. S3B and D). Interestingly, no difference was found for cell aspect ratio in 3D suggesting less morphological heterogeneity in 3D culture (Fig. S3E).
Fibroblasts differentially influence breast cancer cell invasion
To investigate the influence of patient-derived CAFs on migration and proliferation of invasive breast cancer cells, we utilized a 3D tumor-stroma microfluidic as a co-culture system (Fig. 1B)[8]. The microfluidic platform was designed to organize the cancer cells into a central tumor region surrounded by a stroma to simulate the invasion of cancer cells away from the primary ‘tumor’. SUM-159 breast cancer cells, which were derived from invasive ductal carcinoma of a TNBC patient, were chosen for their propensity to migrate within a 3D hydrogel[8, 10]. SUM-159 cells in the tumor region were observed over 3 days invading into the stroma region of the microfluidic either with or without fibroblasts (Fig. 1C, Fig. S4). We found a significant increase in migration distance for CAF1 (+ CAF1) and CAF3 (+ CAF3) but not with CAF2 (+ CAF2) when compared to the mono-culture condition (Ctrl) (Fig. 1D). Notably, the NF co-culture (+ NF) had significantly lower migration compared to all CAF co-cultures and the Ctrl (Fig. 1D). In parallel, we investigated cancer cell proliferation influenced by fibroblasts. The + NF condition had a lower fraction of BrdU-positive SUM-159 cells compared to the three CAF co-cultures indicating less proliferation in NF co-culture (Fig. 1E). On the other hand, we observed a significant increase in BrdU expression for + CAF1 and + CAF3 versus the Ctrl, and for + CAF2 to a lesser extent. In summary, NFs demonstrated a suppressive effect on cancer cell invasion and proliferation, while CAF1 and 3 exhibited an invasion- and proliferation-promoting activities. Taken together, this suggests that fibroblasts of different microenvironmental origin (i.e. patients) and phenotype, although all were mammary derived, could exert distinct effects on invasion and proliferation of cancer cells in 3D culture conditions.
We also investigated MDA-MB-231, another TNBC cell line, but derived from pleural effusion, and MCF7 cells, which are also pleural effusion-derived but less invasive. We asked if CAFs had a similar influence on migration for these breast cancer cell lines. The CAF3 population was utilized as it demonstrated the highest increase in SUM-159 cell migration (Fig. 1D). We observed that both MCF7 and MDA-MB-231 cells showed propensity to migrate into the 3D stroma but MCF7 had far less migration capacity. Importantly, adding CAFs into the stroma influenced both MCF7 and MDA-MB-231 cells by significantly enhancing their 3D invasive capacity into the stroma similar to SUM-159 cells (Fig. S5A and B). Consistent with the literature, MDA-MB-231 had less invasive capacity as compared to SUM-159 cells. Taken together, this suggests that patient-derived CAFs promoted invasion of broad types of breast cancer cell lines.
Real-time analysis of cell migration reveals bi-directional cancer-fibroblast interaction
To investigate the migration of cancer cells in real-time, at a single cell level, within our co-culture system, we measured the influence of CAFs on SUM-159 migration speed using CAF3. We traced cell migration tracks and quantified the migration speed observing that the presence of CAFs significantly enhanced migration speed of SUM-159 cells (Fig. 2A–C, Movie S1 and S2). However, there was no statistical difference for persistence of migration (Fig. 2D). Surprisingly, when we analyzed the migration of CAFs, which were largely stationary in the absence of cancer cells (Fig. S3A), we instead found enhanced migration behavior when co-cultured with migrating cancer cells (Fig. 2E and F, Movie S3 and S4). Quantification indeed showed that presence of SUM-159 cells significantly promoted both migration speed and persistence of CAFs (Fig. 2G and H). Taken together, these analyses demonstrated the presence of a bi-directional influence between cancer and fibroblast cells mutually affecting their migratory behaviors of both SUM-159 cells and CAFs.
Fig. 2. Real-time analysis of cell migration.
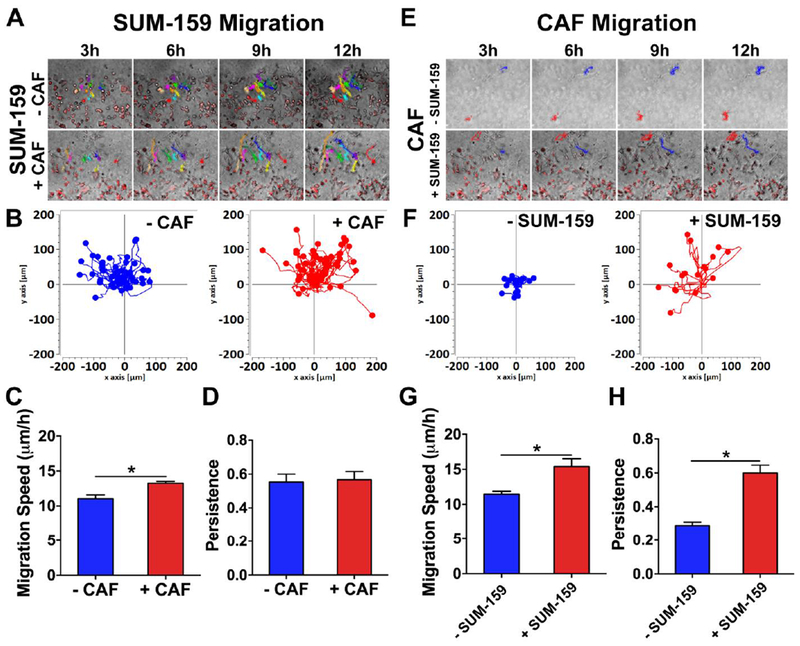
(A) SUM-159 migration tracked over 12 h in presence of absence of CAFs. (B) Overlay of all migration tracks showed SUM-159 cells migrated further in presence of CAFs. (C, D) CAFs significantly enhanced migration speed of SUM-159 cells but not for persistence. * denotes significant difference for p < 0.05. (E) CAF migration tracked over 12 h in presence of absence of SUM-159 cells. (F) Overlay of all migration tracks showed CAFs migrated further in presence of SUM-159 cells. Presence of SUM-159 cells significantly enhanced both (G) migration speed and (H) persistence of CAFs. * denotes significant difference for p < 0.05.
Morphometric analysis of SUM-159 cells co-cultured with fibroblasts
As morphology is closely linked to cell migratory behavior, we utilized shape descriptors to analyze the morphology of cancer cells by assessing the cell area, circularity, aspect ratio, and protrusiveness of SUM-159 cells that have migrated into the stroma (Fig. 3A and B)[20]. Only CAF3 but no other fibroblast population significantly increased cell area and decreased circularity of the SUM-159 cells (Fig. 3C and D). We found significant lowered aspect ratio between + NF vs. + CAF1 and + NF vs. + CAF2 (Fig. 3E). Protrusiveness was measured using the inverse of solidity and we found that SUM-159 cells showed significantly higher levels of protrusion in CAF co-cultures than the Ctrl and + NF (Fig. 3F). To summarize the phenotypic influence (i.e. changes in morphology, proliferation, and migration) of the heterogeneous fibroblast population and SUM-159 cells, we performed a hierarchical clustering on the morphological and behavioral measurements (Fig. 3G). The analysis showed that CAF co-culture conditions clustered together and that + CAF3 was separated from + CAF1 and + CAF2, agreeing with the data showing that + CAF3 exerted the largest influence in cell migration compared to the other two CAF isolates.
Fig. 3. Morphometric analysis of SUM-159 cells after co-culture.
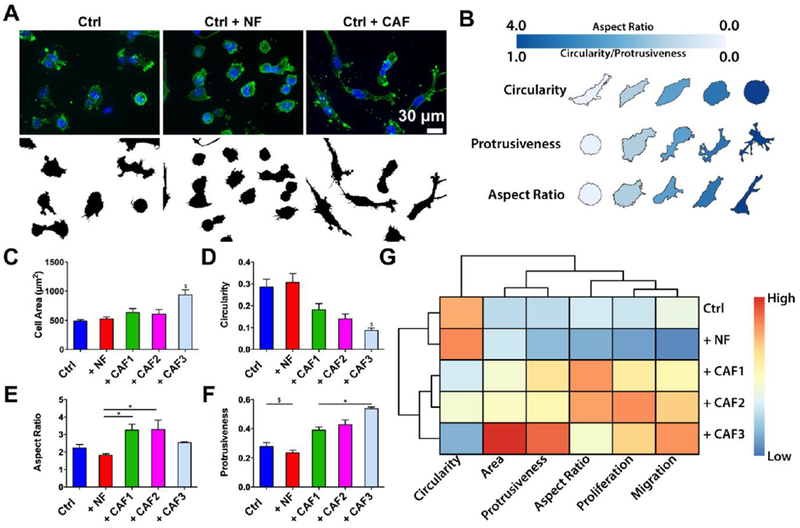
(A) Depiction of cancer cell shape during co-culture with fibroblasts. (B) Morphometric key for the analysis circularity, protrusiveness, and aspect ratio. (C-F) Graph comparing different cell shape descriptors. * denotes significant difference for p < 0.05. $ denotes significantly different groups for p < 0.05. (G) Unsupervised clustering revealed Ctrl and NF co-culture as more similar than the CAF co-cultures.
Gene expression profiles of SUM-159 cells upon tumor-stroma interaction
To couple our functional assessments (i.e. migration, proliferation, morphology) with molecular changes, we performed RNA-seq on sorted populations of SUM-159 cells extracted from the microfluidic device after co-culture with fibroblasts. Multi-dimensional scaling (MDS) analysis on total expression profiles showed that SUM-159 cells in NF and CAF co-cultured conditions separated away from the mono-cultured cells (Ctrl, Fig. 4A). Furthermore, + CAF1, 2, and 3 clustered together indicating that SUM-159 cells in CAF co-cultures share a similar transcriptional profile distinct from the + NF condition, concordant with our functional assessments (Fig. 3G). By an ANOVA-like test, we identified 280 differentially expressed genes (DEGs, fold change ≥ 1.5 and FDR < 0.05 in any pairwise comparison) across all conditions. Hierarchical clustering on the expression profiles (Fig. 4B) resulted in an identical grouping pattern as the MDS analysis (Fig. 4A). We then compared Ctrl vs. all fibroblast co-culture conditions, and identified 149 DEGs (92 up and 57 down, fold change ≥ 1.5 and FDR < 0.05). These genes represented a common set of transcriptional changes elicited by any fibroblasts, and were associated with Pathways in Cancer, Focal Adhesion, and PI3K-Akt signaling pathway (Table S2 and S3). We next compared the expression profiles of Ctrl vs. NF as well as Ctrl vs. CAF co-cultures to identify DEGs uniquely regulated by either NFs or CAFs, and found 108 DEGs in the NF co-culture (70 up and 38 down, fold change ≥ 1.5 and FDR < 0.05, Fig. 4C and D, Table S4) and 175 DEGs in CAF co-cultures compared to mono-culture (118 up and 57 down, fold change ≥ 1.5 and FDR < 0.05, Fig. 4 and D, Table S5). KEGG pathway analysis revealed enrichment for cell adhesion, collagen fibril organization, and extracellular matrix organization-related terms for Ctrl vs. CAFs, while inflammatory response, response to liposaccharide, and leukocyte migration-related terms were enriched in Ctrl vs. NFs (Fig. 4E, Table S6 and S7). Finally, we identified 22 unique DEGs between NF and CAF co-cultures that could potentially play a role in cancer invasion (FDR < 0.05, Table S8), including VAMP1, GPNMB, BGN, and CEND1, which were up-regulated in the CAF co-cultures, and RGS16, SOCS3, CXCL8, SAA2, and IL6, which were expressed higher in NF co-culture. Among the identified genes, glycoprotein non-metastatic B (GPNMB) encodes for a cell surface transmembrane glycoprotein that is highly expressed among many cancers, including breast cancer. Prior literature had associated GPNMB with poor prognosis within basal/triple-negative subtype of breast cancers[21]. GPNMB has also been implicated in tumor invasion, angiogenesis, cell adhesion, and immune suppression[21]. Previous studies have shown GPNMB to be a mediator in cell migration through transwell assays but without the presence of CAFs[22]. In that regard, we aimed to discover the role of GPNMB during cancer migration due to tumor-stroma interactions.
Fig. 4. Gene expression profiling of SUM-159 breast cancer cells after interacting with fibroblasts.
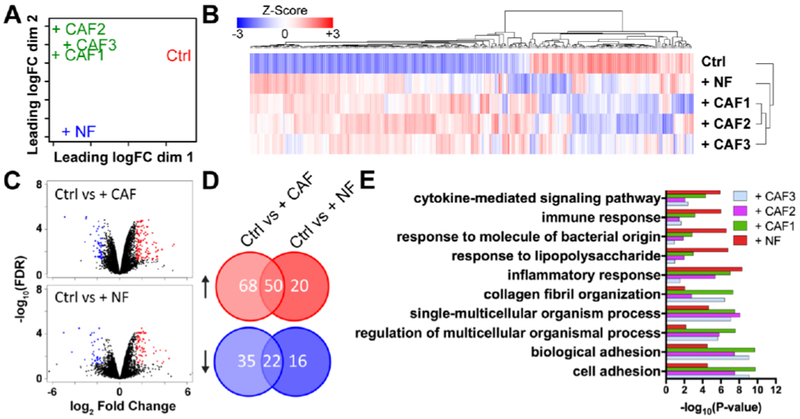
(A) Multi-dimensional scaling of gene expression profile revealed that cancer cells in CAF co-cultures shared a similar transcriptional profile distinct from the + NF condition (B) Heatmap of 280 differentially expressed genes (DEGs, fold change ≥ 1.5 and FDR < 0.05 in any pairwise comparison) across all conditions. (C and D) Volcano plots and Venn diagrams show Ctrl vs + CAF (118 up and 57 down, fold change ≥ 1.5 and FDR < 0.05) and Ctrl vs + NF (70 up and 38 down, fold change ≥ 1.5 and FDR < 0.05). (E) Top 5 GO terms for Ctrl vs NF co-culture combined with the Top 5 GO terms for Ctrl vs CAF co-cultures.
Clinical relevance of glycoprotein non-metastatic B in cancer
As we found striking differences in CAF co-cultures compared to Ctrl (mono-culture of SUM-159 cells), we asked whether GPNMB may play a tumor-promoting role in breast cancer (Fig. 5A, Table S8). Therefore, we investigated GPNMB as one of the possible mediator of promoting invasion. GPNMB encodes for a cell surface transmembrane glycoprotein that is highly expressed among many cancers, including breast cancer. It has also been implicated in tumor invasion, angiogenesis, cell adhesion, and immune suppression[21]. To determine whether GPNMB may play a tumor-promoting role in breast cancer, we evaluated two large publically available datasets (METABRIC and TCGA) for mutations, copy-number variations and changes in expression[23–25]. We found that alterations occurred in 10 and 16% of breast cancer patients, respectively, in two data sets (Fig. 5B)[23–27]. mRNA overexpression in breast carcinoma was significantly increased in TCGA and other data sets, when compared to the normal tissue controls (Fig. 5C) [28–30]. Additionally, the TNBC-related subtypes (i.e. basal-like and claudin-low) expressed higher levels of GPNMB than other subtypes (Fig. S6A). Next, expression of GPNMB also positively correlated with tumor stage (Fig. 5D). To determine the impact of GPNMB overexpression, we next investigated the effect on patient survival. Analysis of the public breast cancer data sets from TCGA-BRCA and web-based GeneAnalytics revealed that high expression of GPNMB correlated with poorer metastasis-free and overall survival (Fig. 5E and Fig. S6B and C). Taken together, these data provided important evidence for the potential clinical relevance of GPNMB in breast cancer.
Fig. 5. Clinical relevance for GPNMB.
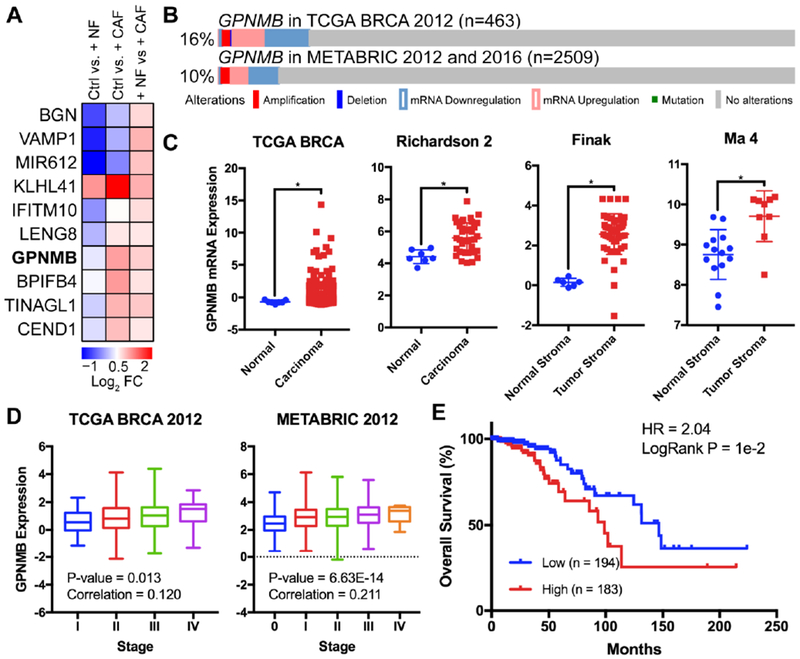
(A) Heat map of top 10 DEGs when comparing NF and CAF co-cultures. (B) Incidence of GPNMB alterations in two different cohorts from public datasets[23–27]. (C) GPNMB expression from TCGA-BRCA in breast cancer compared to normal. * denotes significant difference for p < 0.05; unpaired t-test[28–30]. (D) GPNMB expression positively correlated with tumor stage[24, 27]. (E) Kaplan–Meier analysis of overall survival from TCGA-BRCA data sets. HR, 2.04; P = 1e-2. High expression of GPNMB was correlated to poorer patient prognosis using TCGA data [24].
Role of GPNMB in tumor-stroma interaction and SUM-159 cancer cell migration
We first validated for expression of GPNMB in our cell lines. Western blotting demonstrated that GPNMB was indeed expressed highly in SUM-159 and MDA-MB-231 cells and lowly in MCF7 and MCF10A cells corroborating prior studies (Fig. S7A)[31]. Next, to study the role of GPNMB in the context of tumor microenvironment, we assessed the expression of GPNMB in SUM-159 cells in mono- or co-culture with CAFs (CAF3). IF imaging revealed punctate expression throughout the cell body of SUM-159 cells while quantification of the IF signal demonstrated a significant increase in GPNMB expression for SUM-159 cells within the CAF co-culture agreeing with the RNA-seq data (Fig. S7B). Next, we knocked down GPNMB using two independent short hairpin RNAs (shRNA) on SUM-159 breast cancer cells and characterized the resulting invasive phenotype of GPNMB knockdown lines (shG). The shRNA plasmid consisted of a psi-LVRU6MP vector with a control shRNA (shCtrl) containing a scrambled sequence. Successful knockdown was confirmed by a western blot (Fig. S8A), qPCR (Fig. S8B), and IF images of 2D cultured GPNMB knockdown lines which demonstrated qualitatively less punctuates compared to the control cells (Fig. S8C). Proliferation of shG lines were significantly attenuated as compared to the shCtrl line in 2D cultures (Fig. S8D). We also observed reduced cell spreading area and higher circularity on a 2D substrate, which was correlated to reduced invasive capacity (Fig. S8E and F), but no difference among the knockdown lines was found in adhesion to collagen substrate (Fig. S8G). Then, we asked whether GPNMB is crucial for breast cancer cell migration in a 3D hydrogel-based stroma using our microfluidic model. It was observed that GPNMB knockdown significantly decreased migration of SUM-159 breast cancer cells in a 3D microenvironment (Fig. 6A). To further confirm the role of GPNMB in migration of another cancer cell line, we produced stable knockdown lines for MDA-MB-231 cells (Fig. S9A and B) and observed a similar trend in decreased cell migration (Fig. S9C). Furthermore, as the MCF7 cell line did not have significant GPNMB expression based on literature data as well as our data (Fig. S7A), we used an overexpression vector to overexpress GPNMB and observed its effect on migration within our model (Fig. S10A and B). Interestingly, we found that increased GPNMB expression in MCF7 cells promoted invasion but not to the extent we found for SUM-159 or MDA-MB-231 wildtypes (Fig. S10C). Next, we co-cultured CAFs (CAF3) with the SUM-159 knockdown lines, and we found a significant increase in migration into the stroma for only the shCtrl line but not for the shG knockdown lines (Fig. 6A). Next, we examined expression change of the GPNMB protein in SUM-159 cells by IF when in presence of CAFs and found an increase in expression for the shCtrl line but found no significant difference for the shG lines (Fig. 6B). Then we asked if the slower migration of shG cells was due to either reduced cell proliferation, cell migration speed, or both. EdU assay was used to measure cell proliferation, and we found no significant difference between the shCtrl and shG lines (Fig. 6C). On the other hand, time-lapse imaging of cell migration in 3D revealed that GPNMB knockdown significantly attenuated migration speed by nearly half and migration persistence to a lesser extent when compared to the control knockdown (Fig. 6D and E, Movie S5 and S6). This suggested that the decreased migration distance into the 3D stroma for the GPNMB knockdown lines was likely due to attenuated cell migration speed. Taken together, these data demonstrated that GPNMB plays a key role in enhancing cancer invasion in 3D by affecting cell migration speed and that co-cultured CAFs induced GPNMB expression in cancer cells.
Fig. 6. Functional study of GPNMB in tumor-stroma interactions.
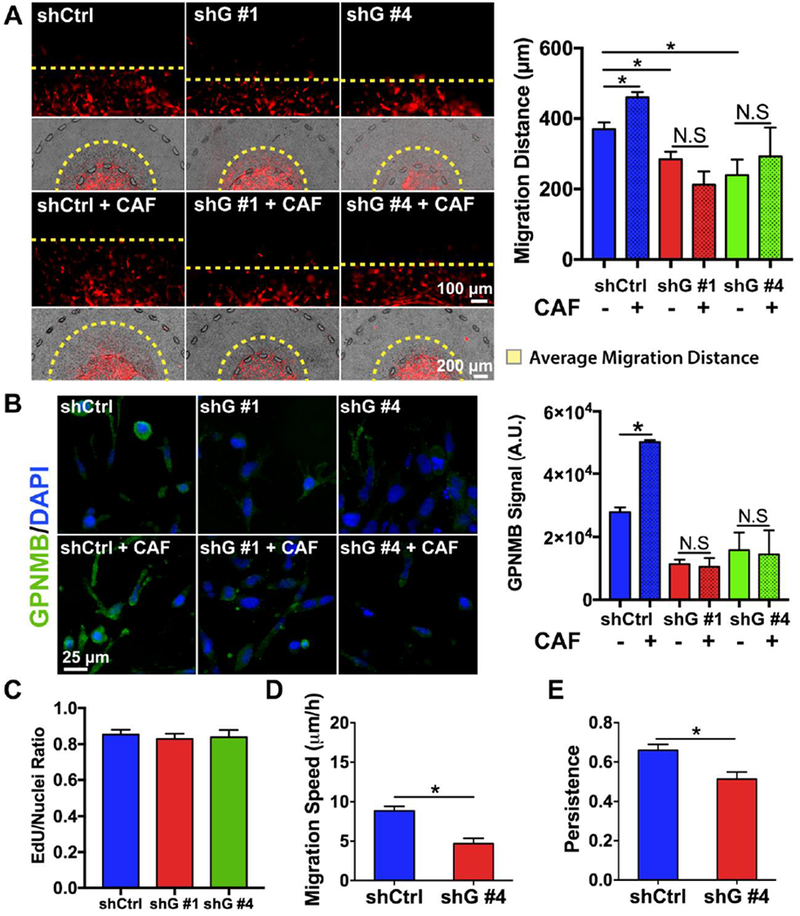
(A) Migration distance was lowered in GPNMB knockdown lines compared to the scrambled shRNA control. CAF co-culture increased migration of control knockdown, but did not increase migration of GPNMB knockdowns. * denotes significant difference for p < 0.05.(B) CAF co-culture increased IF expression of GPNMB in shCtrl cells, but did not increase IF expression of GPNMB in shG knockdown lines. * denotes significant difference for p < 0.05. (C) No difference was found in proliferation using EdU assay. (D, E) GPNMB knockdown significantly reduced migration speed and persistence of SUM-159 cells. * denotes significant difference for p < 0.05.
DISCUSSION
Despite the significance of tumor-stroma interactions in early steps of cancer metastasis, not many models have studied and paired the functional assessments to molecular changes in cancer cell invasion in a suitable and relevant tumor microenvironment[1, 6]. In this work, we aimed to take a significant step forward by developing an innovative organotypic tumor microenvironment model with configurable tumor and stroma regions coupled with integrated molecular and cellular studies on the influences of patient-derived fibroblasts on breast cancer invasion. The 3D microfluidic co-culture system contained side-by-side tumor and stroma regions to resemble the architecture of the early tumor microenvironment. This spatial organization enabled bi-directional crosstalk between the cancer cells and stromal cells, while still allowing imaging-based quantifications such as migration and proliferation. Most importantly, our model also addressed the limitations of previous systems, which only used functional assessments (i.e. cell migration and proliferation), by incorporating RNA-seq to profile the transcriptome of the cancer cells in co-culture with patient-derived fibroblasts to better dissect the molecular mechanisms and provide insights in tumor-stroma crosstalk during invasion.
In this work, NFs demonstrated lower expression of αSMA and spreading area when compared to CAFs with higher levels of αSMA and larger spreading area. The differences in spreading area corroborated with earlier work[19]. Despite that patient-derived CAFs showed no difference in αSMA expression among themselves in 2D, only CAF1 and CAF3 significantly promoted migration and proliferation and CAF2 to a lesser extent suggesting further CAF heterogeneity beyond αSMA levels[1]. Alongside α–SMA, we utilized two additional common molecular markers of CAFs, FAP and FSP-1[18]. These markers have been used to describe CAFs in several works that have isolated fibroblasts from patient tumors[32]. However, we found that there were varying levels of these markers among the CAF isolates. Furthermore, several studies have pointed out that although these markers usually identify CAFs, there is no defined single or set of molecular markers that exclusively defines CAFs owing to their heterogeneity. Moreover, gene expression profile studies using an mRNA microarray on CAFs isolated from different breast cancer subtypes (ER+, HER2+, and TNBC) showed that there exist subtype specific gene expression profiles for CAFs[32]. This again provides further evidence of the heterogeneity of CAFs. Due to the complexity in fibroblast heterogeneity, previous studies have also proposed that fibroblasts could be subdivided into categories based on their functional heterogeneity, like tumor-restraining or tumor-promoting[1, 18]. The NFs that we examined showed tumor-restraining properties which indeed corroborated with prior studies[33]. On the other hand, we demonstrated that two of three patient-derived CAFs, including CAFs from a TNBC patient, exhibited tumor-promoting behavior. Therefore, we envision that the changes in SUM-159 behavior when in co-culture with fibroblasts were due to the differences in fibroblast phenotype and their microenvironmental origin.
One interesting consequence of tumor-stroma interactions that was revealed through our model was the possibility of a bi-directional effect between SUM-159 cells and fibroblasts influencing migration of both cell types. Specifically, real-time analysis of CAF migration demonstrated increased cell motility and persistence in presence of SUM-159. In reverse, SUM-159 cells also expressed enhanced migration speed in presence of CAFs. Previous studies have added cancer-conditioned media to fibroblast cultures resulting in enhanced elongation and migration of these cells in scratch assays[34]. Building upon these studies, our work incorporated both cancer cells and fibroblasts in a 3D matrix to reveal this reciprocal effect on cell motility. This suggests that there may be mechanisms that cancer cells use to promote CAF migration or recruitment. For instance, during normal wound healing, different stromal cells, like fibroblasts and mesenchymal stem cells (MSCs), are recruited and induced to migrate to the wound site[35]. In the context of tumors, these stromal cells promote survival, proliferation, and invasion of cancer cells. Interestingly CAFs and MSCs share similar behavior and molecular markers and it is even debatable if MSCs could be a possible origin for CAFs. For instance, McGrail et al. designed an experiment by incubating MSCs and 3T3 fibroblasts in tumor-conditioned media. They found increased migration speed overtime for both cell types similar to what we observed in our primary CAFs[36]. Therefore, we suspect that there may be conserved cell-cell signaling mechanisms during wound healing, and we propose that this could be the same mechanism for CAF migration we observed in our work. Alternatively, Mauney et al. suggested that matrix remodeling by degradation can promote recruitment of stem cells[37]. However, there still does not exist, to our knowledge, literature data to support tumor-driven matrix degradation promoting CAF migration. Perhaps a future direction of research could involve studying the possible molecular changes in fibroblasts overtime due to interactions with cancer cells either from cell-cell signaling or tumor-driven matrix degradation
Many studies have indicated that the shape of the cell carries information determining the cell behavior, such as invasiveness[20]. Here we showed that SUM-159 cells alone or when in co-culture with NFs demonstrated higher circularity as well as lower protrusiveness, area, and aspect ratio compared to when co-cultured with CAF populations. We connected the shape changes of the cancer cells to invasive behavior by demonstrating that the increase in migration and proliferation of SUM-159 cells correlated with increases in aspect ratio and protrusiveness. These results build upon the growing body of literature that have previously reported the relationship between protrusive activity and invasion[17]. Taken together, this suggested that the CAF isolates within our system promoted invasive morphology and migratory behavior of breast cancer cells[1, 20].
To screen for molecular influences on cancer migration due to the stromal fibroblasts (i.e. both CAFs and NFs), we profiled the transcriptome of the cancer cells and performed bioinformatics analyses. Within the NF co-culture, we found higher expression of genes related to inflammatory responses, such as CXCL8, ILR2, and IL6. A recent paper by Camp et al. co-cultured basal-like cancer cells (i.e. SUM-159) with immortalized fibroblasts obtained from reduction mammoplasty similar to NFs in our study[38]. Consistently, they showed up-regulated immune responses, such as IL6, CXCL8, STAT1, CXCL1, and ILR2, corroborating our results. In addition, we found over-expression of Regulator of G protein signaling 16 (RGS16) within NF co-culture. RGS16 is a possible tumor suppressor gene down-regulating cancer migration and proliferation[39]. On the other hand, CAF co-culture over-expressed genes related to cell adhesion, like BGN, GPNMB, and IFITM10. The gene BGN encodes for biglycan, a small leucine-rich repeat proteoglycan found within the ECM, which has been demonstrated to be up-regulated within the breast tumor stroma[40]. GPNMB has been associated with poor prognosis within basal/triple-negative subtype of breast cancers[21]. Prior research has implicated GPNMB in tumor invasion, angiogenesis, cell adhesion, and immune suppression for melanoma, breast, prostate, pancreatic, and lung cancers[22, 41]. However, previous studies have mainly utilized transwell and colony forming assays to assess the influence of GPNMB in tumor invasion in absence of CAFs or the tumor microenvironment[22]. Therefore, of the genes we screened for, we intended to discover role of GPNMB in cancer migration within a 3D tumor microenvironment with tumor fibroblast interactions.
Using our 3D model, we confirmed that GPNMB knockdown reduced breast cancer cell invasion within a 3D hydrogel-based stroma, suggesting its requirement for efficient migration in a 3D microenvironment. We showed through time-lapse imaging that the decrease in migration distance was due to attenuated cell migration speed in the 3D matrix. Our work is the first time that it was shown that GPNMB contributed to breast cancer cell migration speed in 3D culture. In prior works, GPNMB has been demonstrated to engage integrins and MMPs which are crucial for migration in a 3D microenvironment[22, 41]. We did indeed observe similar association between GPNMB and integrins and MMPs in our RNA-seq data (Table S9). GPNMB interaction with integrin was also shown to activate SRC and FAK signaling, which regulate cell migration[31, 42]. Our results taken together with the current findings suggested that GPNMB plays a crucial role in cancer invasion in 3D microenvironment.
The influences of GPNMB on invasion has mainly been studied using a neoplastic-cell centric approach. To identify the involvement of the tumor microenvironment, we specifically asked whether CAFs promoted invasion of cancer cells through a GPNMB-dependent manner. Using the CAF co-culture model, we showed through IF that patient-derived CAFs residing in the biomimetic stroma enhanced protein expression of GPNMB on breast cancer cells validating the RNA-seq data. Furthermore, knockdown of GPNMB of SUM-159 cells in the co-culture model blunted the effect of CAFs on promoting invasion. To the best of our knowledge, tumor-stroma interaction through GPNMB has not been explored before. In a similar study to ours, Le bras et al. established a 3D organotypic co-culture model using a collagen matrix, human esophageal epithelial cells, and human skin fibroblasts and demonstrated enhanced tumor proliferation and growth[43]. Concordant with our work, gene expression analysis revealed more than a 1.5 fold-increase in GPNMB compared to non-invasive cultures. However, studying the role of GPNMB was not the focus of that study. To that end, our work adds to the growing body of literature implicating GPNMB in promoting tumor invasion and progression, while supporting that GPNMB expression is enhanced through tumor-stroma interactions between breast cancer and CAFs[22, 41, 42]. There are some potential mechanisms by which CAFs could induce GPNMB expression in cancer cells. Smuczek et al. showed that C16, a laminin-111 subunit, could enhance GPNMB expression and promote MDA-MB-231 cell migration[31]. It was suggested that C16 was produced through cell-induced proteolysis of the ECM through MMP9 and MMP9 has been shown to be secreted by CAFs[44, 45]. In a separate study, GPNMB was upregulated in hepatocellular carcinoma by colony-stimulating factor 1, which is a known factor secreted from fibroblasts[46]. Taking these prior findings with our results indicate that either directly through paracrine signaling or indirectly by enzymatic release of bound peptides from ECM, GPNMB could be induced through tumor-stroma interactions.
In future studies, we intend to correlate the changes in gene expression to the molecular differences in fibroblasts to predict how patient heterogeneity within tumor microenvironments could influence cancer progression. We believe that to fully recapitulate patient heterogeneity, there is a need for larger sample sizes to increase the power of the study as well as large-scale screening of multidimensional phenotyping using –omics technologies[18, 32]. Understanding the source of tumor heterogeneity and its implication on tumor progression is important for developing effective cancer therapies. Therefore, our technology could be used to mechanistically understand how stromal cells play a role in promoting tumor heterogeneity. As selective pressure is one of the main hypothesis for tumor heterogeneity, one could devise experiments to understand how cancer-associated fibroblasts drive proliferation or migration of subpopulation of cancer cells followed by single-cell sequencing to classify the different subpopulations[1, 18]. These subpopulations could then be exploited based on their heterogeneic expression of proteins or gene expression.
Furthermore, we would like to expand the usage of this microfluidic model for characterizing the role of the tumor microenvironment on the evolution of non-invasive cancer cells toward malignancy. As we noted, segregation of tumor and stroma enabled measurement of an invasive breast cancer cell migration toward a stroma embedded with CAFs followed by analyzing the transcriptomic changes in the cancer cells. Therefore, the continuation of this work would entail studying the influence of CAFs on less invasive cell lines and analyzing the transcriptomic profile for potential drivers of malignancies. Importantly, much of cancer research is still devoted to utilizing cancer cell lines due to their ease and feasibility for drug screening[47]. For our current work, we needed a stable cell line with known characterizations in terms of robust invasion to conduct our mechanist study[47]. This led us to choosing well-characterized cell lines, such as SUM-159 breast cancer cells. This cell line demonstrated robust invasion in literature as well as in our model providing us a proper baseline for migration experiments[8, 17]. Importantly, prior studies have shown that this specific cell line demonstrated similar genomic features as primary breast tumors from which the cell line was derived from[48]. In our future work, we will expand the capability of our tumor model by incorporating patient-derived tumor cells as to tailor the platform more towards a patient-specific tumor microenvironment.
In conclusion, we engineered a 3D organotypic microfluidic co-culture system to represent the breast tumor microenvironment by juxtaposing 3D tumor and stroma regions in order to study the molecular and cellular influence of patient-derived fibroblasts on breast cancer invasion. We found clear evidence that interactions between invasive SUM-159 breast cancer cells and mammary fibroblasts displayed distinct consequences on cancer migratory behavior depending on the fibroblast phenotype. Particularly, NFs expressed a tumor-suppressive behavior shown by reduction in migration, proliferation, and cell aspect ratio. On the other hand, two of the three CAFs studied demonstrated tumor-promoting behavior while the other to a lesser extent. Through transcriptome profiling of the SUM-159 cells, we found that NF co-cultures were enriched for inflammatory pathways while CAF co-cultures were enriched cell adhesion and ECM. This led to the observation that CAFs enhanced breast cancer invasion within a 3D microenvironment by inducing the expression of a novel gene of interest, GPNMB, in breast cancer cells resulting in increased invasion speed. Importantly, knockdown of GPNMB led to blunting the effect of CAFs on promoting cancer invasion. Overall, we propose that this organotypic microfluidic model has potential to tease out and understand key molecular pathways in tumor-stroma interactions for discovering druggable targets for inhibiting cancer invasion.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
An organotypic model of tumor-stroma interactions on a microfluidic chip reveals that CAFs promote invasion by enhancing expression of GPNMB in breast cancer cells.
ACKNOWLEDGEMENTS
We would like to acknowledge National Science Foundation Award # CBET 1510700 received by M. Nikkhah, the National Institutes of Health R01CA196885 received by G. Mouneimne, funding from Breast Cancer Research Foundation received by J. LaBaer and J.G. Park, the 2017-2018 Achievement Rewards for College Scientists Scholarship received by D.D. Truong, the 2016-2017 and 2017-2018 International Foundation for Ethical Research Fellowship received by D.D. Truong, and funding from ASU Graduate & Professional Student Association and the ASU Graduate College received by D.D. Truong. We also acknowledge Ali Navaei for his input in scientific discussions, Zachary Camacho for his help on microfluidic fabrication, Jaimeson Veldhuizen for her assistance on wafer fabrication, Padhmavathy Yuvaraj and Ian Shoemaker for assistance on western blot imaging, Crystal Willingham for assistance on RNA isolation, Kassondra Hickey for qPCR protocols, and the Smith and Stabenfeldt lab at ASU for equipment usage. Plasmids were obtained from DNASU and Addgene plasmid repositories. Lastly, anti-BrdU was purchased from the Developmental Studies Hybridoma Bank, deposited to the DSHB by Kaufman, S.J., and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- [1].Kalluri R, The biology and function of fibroblasts in cancer, Nat Rev Cancer 16(9) (2016) 582–98. [DOI] [PubMed] [Google Scholar]
- [2].Tchou J, Conejo-Garcia J, Targeting the tumor stroma as a novel treatment strategy for breast cancer: shifting from the neoplastic cell-centric to a stroma-centric paradigm, Advances in pharmacology, Elsevier: 2012, pp. 45–61. [DOI] [PubMed] [Google Scholar]
- [3].Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA, Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion, Cell 121(3) (2005) 335–48. [DOI] [PubMed] [Google Scholar]
- [4].Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL, Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration, Cell 151(7) (2012) 1542–1556. [DOI] [PubMed] [Google Scholar]
- [5].Takai K, Le A, Weaver VM, Werb Z, Targeting the cancer-associated fibroblasts as a treatment in triple-negative breast cancer, Oncotarget 7(50) (2016) 82889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weigelt B, Ghajar CM, Bissell MJ, The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer, Adv Drug Deliv Rev 69–70 (2014) 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Peela N, Truong D, Saini H, Chu H, Mashaghi S, Ham SL, Singh S, Tavana H, Mosadegh B, Nikkhah M, Advanced biomaterials and microengineering technologies to recapitulate the stepwise process of cancer metastasis, Biomaterials 133 (2017) 176–207. [DOI] [PubMed] [Google Scholar]
- [8].Truong D, Puleo J, Llave A, Mouneimne G, Kamm RD, Nikkhah M, Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment, Scientific reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nagaraju S, Truong D, Mouneimne G, Nikkhah M, Microfluidic Tumor-Vascular Model to Study Breast Cancer Cell Invasion and Intravasation, Adv Healthc Mater (2018). [DOI] [PubMed] [Google Scholar]
- [10].Peela N, Barrientos ES, Truong D, Mouneimne G, Nikkhah M, Effect of suberoylanilide hydroxamic acid (SAHA) on breast cancer cells within a tumor-stroma microfluidic model, Integr Biol (Camb) 9(12) (2017) 988–999. [DOI] [PubMed] [Google Scholar]
- [11].Truong D, Fiorelli R, Barrientos ES, Melendez EL, Sanai N, Mehta S, Nikkhah M, A three-dimensional (3D) organotypic microfluidic model for glioma stem cells – Vascular interactions, Biomaterials (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nikkhah M, Strobl JS, Schmelz EM, Roberts PC, Zhou H, Agah M, MCF10A and MDA-MB-231 human breast basal epithelial cell co-culture in silicon micro-arrays, Biomaterials 32(30) (2011) 7625–32. [DOI] [PubMed] [Google Scholar]
- [13].Strobl JS, Nikkhah M, Agah M, Actions of the anti-cancer drug suberoylanilide hydroxamic acid (SAHA) on human breast cancer cytoarchitecture in silicon microstructures, Biomaterials 31(27) (2010) 7043–50. [DOI] [PubMed] [Google Scholar]
- [14].Peela N, Sam FS, Christenson W, Truong D, Watson AW, Mouneimne G, Ros R, Nikkhah M, A three dimensional micropatterned tumor model for breast cancer cell migration studies, Biomaterials 81 (2015) 72–83. [DOI] [PubMed] [Google Scholar]
- [15].Nikkhah M, Strobl JS, De Vita R, Agah M, The cytoskeletal organization of breast carcinoma and fibroblast cells inside three dimensional (3-D) isotropic silicon microstructures, Biomaterials 31(16) (2010) 4552–61. [DOI] [PubMed] [Google Scholar]
- [16].Saini H, Eliato KR, Silva C, Allam M, Mouneimne G, Ros R, Nikkhah M, The Role of Desmoplasia and Stromal Fibroblasts on Anti-cancer Drug Resistance in a Microengineered Tumor Model, Cellular and Molecular Bioengineering 11(5) (2018) 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, Mullins RD, Brugge JS, Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion, Cancer cell 22(5) (2012) 615–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ohlund D, Elyada E, Tuveson D, Fibroblast heterogeneity in the cancer wound, The Journal of experimental medicine 211(8) (2014) 1503–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Evans RA, Tian YC, Steadman R, Phillips AO, TGF-beta1-mediated fibroblast-myofibroblast terminal differentiation-the role of Smad proteins, Exp Cell Res 282(2) (2003) 90–100. [DOI] [PubMed] [Google Scholar]
- [20].Yu H, Lim KP, Xiong S, Tan LP, Shim W, Functional morphometric analysis in cellular behaviors: shape and size matter, Adv Healthc Mater 2(9) (2013) 1188–97. [DOI] [PubMed] [Google Scholar]
- [21].Rose AA, Grosset A-A, Dong Z, Russo C, MacDonald PA, Bertos NR, St-Pierre Y, Simantov R, Hallett M, Park M, Glycoprotein nonmetastatic B is an independent prognostic indicator of recurrence and a novel therapeutic target in breast cancer, Clinical Cancer Research 16(7) (2010) 2147–2156. [DOI] [PubMed] [Google Scholar]
- [22].Rose AA, Pepin F, Russo C, Abou Khalil JE, Hallett M, Siegel PM, Osteoactivin promotes breast cancer metastasis to bone, Molecular cancer research : MCR 5(10) (2007) 1001–14. [DOI] [PubMed] [Google Scholar]
- [23].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov 2(5) (2012) 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].N. Cancer Genome Atlas, Comprehensive molecular portraits of human breast tumours, Nature 490(7418) (2012) 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah S, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Borresen-Dale AL, Earl HM, Pharoah PD, Ross MT, Aparicio S, Caldas C, The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes, Nat Commun 7 (2016) 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Science signaling 6(269) (2013) pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups, Nature 486(7403) (2012) 346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M, Stromal gene expression predicts clinical outcome in breast cancer, Nat Med 14(5) (2008) 518–27. [DOI] [PubMed] [Google Scholar]
- [29].Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC, Gene expression profiling of the tumor microenvironment during breast cancer progression, Breast cancer research : BCR 11(1) (2009) R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S, X chromosomal abnormalities in basal-like human breast cancer, Cancer Cell 9(2) (2006) 121–32. [DOI] [PubMed] [Google Scholar]
- [31].Smuczek B, Santos ES, Siqueira AS, Pinheiro JJV, Freitas VM, Jaeger RG, The laminin-derived peptide C16 regulates GPNMB expression and function in breast cancer, Exp Cell Res 358(2) (2017) 323–334. [DOI] [PubMed] [Google Scholar]
- [32].Tchou J, Kossenkov AV, Chang L, Satija C, Herlyn M, Showe LC, Pure E, Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles, BMC Med Genomics 5 (2012) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sadlonova A, Mukherjee S, Bowe DB, Gault SR, Dumas NA, Van Tine BA, Frolova N, Page GP, Welch DR, Novak L, Frost AR, Human breast fibroblasts inhibit growth of the MCF10AT xenograft model of proliferative breast disease, Am J Pathol 170(3) (2007) 1064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Henriksson ML, Edin S, Dahlin AM, Oldenborg PA, Oberg A, Van Guelpen B, Rutegard J, Stenling R, Palmqvist R, Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion, Am J Pathol 178(3) (2011) 1387–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Arnold KM, Opdenaker LM, Flynn D, Sims-Mourtada J, Wound healing and cancer stem cells: inflammation as a driver of treatment resistance in breast cancer, Cancer growth and metastasis 8 (2015) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McGrail DJ, Ghosh D, Quach ND, Dawson MR, Differential mechanical response of mesenchymal stem cells and fibroblasts to tumor-secreted soluble factors, PLoS One 7(3) (2012) e33248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mauney J, Olsen BR, Volloch V, Matrix remodeling as stem cell recruitment event: a novel in vitro model for homing of human bone marrow stromal cells to the site of injury shows crucial role of extracellular collagen matrix, Matrix Biology 29(8) (2010) 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Camp JT, Elloumi F, Roman-Perez E, Rein J, Stewart DA, Harrell JC, Perou CM, Troester MA, Interactions with fibroblasts are distinct in Basal-like and luminal breast cancers, Mol Cancer Res 9(1) (2011) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Carper MB, Boskovic G, Denvir J, Primerano D, Hardman WE, Claudio PP, Investigation of RGS16 mediated inhibition of pancreatic cancer metastasis, The FASEB Journal 27(1 Supplement) (2013) 611.6–611.6. [Google Scholar]
- [40].Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Weaver D, Muss H, Plaut K, Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer, Breast Cancer Res Treat 114(1) (2009) 47–62. [DOI] [PubMed] [Google Scholar]
- [41].Fiorentini C, Bodei S, Bedussi F, Fragni M, Bonini SA, Simeone C, Zani D, Berruti A, Missale C, Memo M, Spano P, Sigala S, GPNMB/OA protein increases the invasiveness of human metastatic prostate cancer cell lines DU145 and PC3 through MMP-2 and MMP-9 activity, Exp Cell Res 323(1) (2014) 100–11. [DOI] [PubMed] [Google Scholar]
- [42].Maric G, Annis MG, Dong Z, Rose AA, Ng S, Perkins D, MacDonald PA, Ouellet V, Russo C, Siegel PM, GPNMB cooperates with neuropilin-1 to promote mammary tumor growth and engages integrin alpha5beta1 for efficient breast cancer metastasis, Oncogene 34(43) (2015) 5494–504. [DOI] [PubMed] [Google Scholar]
- [43].Le Bras GF, Taylor C, Koumangoye RB, Revetta F, Loomans HA, Andl CD, TGFbeta loss activates ADAMTS-1-mediated EGF-dependent invasion in a model of esophageal cell invasion, Exp Cell Res 330(1) (2015) 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Siqueira AS, Pinto MP, Cruz MC, Smuczek B, Cruz KS, Barbuto JA, Hoshino D, Weaver AM, Freitas VM, Jaeger RG, Laminin-111 peptide C16 regulates invadopodia activity of malignant cells through beta1 integrin, Src and ERK ½, Oncotarget 7(30) (2016) 47904–47917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stuelten CH, DaCosta Byfield S, Arany PR, Karpova TS, Stetler-Stevenson WG, Roberts AB, Breast cancer cells induce stromal fibroblasts to express MMP-9 via secretion of TNF-alpha and TGF-beta, J Cell Sci 118(Pt 10) (2005) 2143–53. [DOI] [PubMed] [Google Scholar]
- [46].Tian F, Liu C, Wu Q, Qu K, Wang R, Wei J, Meng F, Liu S, Chang H, Upregulation of glycoprotein nonmetastatic B by colony-stimulating factor-1 and epithelial cell adhesion molecule in hepatocellular carcinoma cells, Oncol Res 20(8) (2013) 341–50. [DOI] [PubMed] [Google Scholar]
- [47].Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH, Systematic identification of genomic markers of drug sensitivity in cancer cells, Nature 483(7391) (2012) 570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J-P, Tong F, A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes, Cancer cell 10(6) (2006) 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


