Abstract
The sesquiterpenoid deoxynivalenol (DON) is an important trichothecene mycotoxin produced by the cereal pathogen Fusarium graminearum. DON is synthesized in specialized subcellular structures called toxisomes. The first step in DON synthesis is catalyzed by the sesquiterpene synthase (STS), Tri5 (trichodiene synthase), resulting in the cyclization of farnesyl diphosphate (FPP) to produce the sesquiterpene trichodiene. Tri5 is one of eight putative STSs in the F. graminearum genome. To better understand the F. graminearum terpenome, the volatile and soluble fractions of fungal cultures were sampled. Stringent regulation of sesquiterpene accumulation was observed. When grown in trichothecene induction medium, the fungus produces trichothecenes as well as several volatile non-trichothecene related sesquiterpenes, whereas no volatile terpenes were detected when grown in non-inducing medium. Surprisingly, a Δtri5 deletion strain grown in inducing conditions not only ceased accumulation of trichothecenes, but also failed to produce the non-trichothecene related sesquiterpenes. To test whether Tri5 from F. graminearum may be a promiscuous STS directly producing all observed sesquiterpenes, Tri5 was cloned and expressed in E. coli and shown to produce primarily trichodiene in addition to minor, related cyclization products. Therefore, while Tri5 expression in F. graminearum is necessary for non-trichothecene sesquiterpene biosynthesis, direct catalysis by Tri5 does not explain the sesquiterpene deficient phenotype observed in the Δtri5 strain. To test whether Tri5 protein, separate from its enzymatic activity, may be required for non-trichothecene synthesis, the Tri5 locus was replaced with an enzymatically inactive, but structurally unaffected tri5N225D S229T allele. This allele restores non-trichothecene synthesis but not trichothecene synthesis. The tri5N225D S229T allele also restores toxisome structure which is lacking in the Δtri5 deletion strain. Our results indicate that the Tri5 protein, but not its enzymatic activity, is also required for the synthesis of non-trichothecene related sesquiterpenes and the formation of toxisomes. Toxisomes thus not only may be important for DON synthesis, but also for the synthesis of other sesquiterpene mycotoxins such as culmorin by F. graminearum.
1. Introduction
Fusarium graminearum sensu stricto (1) is a fungal pathogen of major cereal crops causing the disease Fusarium head blight (FHB) (2). FHB is an increasingly damaging disease worldwide, recently causing billions of dollars in crop losses, with regional losses of more than 50% occurring with regularity (3). Thus, global wheat and barley production are vulnerable to FHB, raising the cost of production, and sometimes necessitating widespread use of fungicides (4). As a result, understanding the physiology and pathogenesis of F. graminearum is important for combatting FHB infection, as well as for breeding resistant cereal crops.
Fusarium and other fungi such as Myrothecium and Stachybotrys produce a complex suite of natural products collectively known as trichothecenes (5). Trichothecenes encompass over 200 sesquiterpenoids, all derived from the unmodified sesquiterpene trichodiene. This shared precursor is produced by the cyclization of farnesyl pyrophosphate (FPP) by a sesquiterpene synthase (STS) called trichodiene synthase or Tri5 (6–8) (Fig. 1). Trichothecenes may accumulate to high levels during infection of wheat and barley by F. graminearum (9,10).
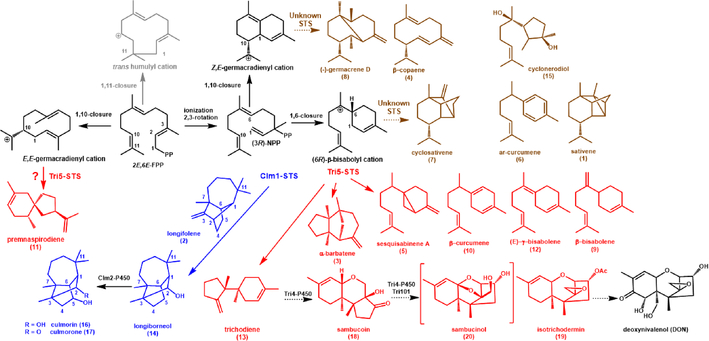
Figure 1. Sesquiterpene cyclization pathways in F. graminearum.
Product profiling shows that F. graminearium possess STS activities that catalyze the initial 1,10 and 1,6 cyclization of FPP or NPP (nerolidyl diphosphate) to the corresponding germacradienyl or bisabolyl cations shown in black. Subsequent cyclization and/or rearrangement reactions followed by a final quenching reaction of the cation gives rise to the characteristic products of a STS. No sesquiterpenes derived from a trans-humulyl cation intermediate appear to be produced by F. graminearum. Tri5-STS cyclizes FPP via a (6R)-β-bisabolyl cation primarily into trichodiene 13, which is modified by additional enzymes into the trichothecene sesquiterpenoids (red compounds). Tri5-STS produces additional, minor sesquiterpenoids (shown in red) via this 1,6-cyclization pathway; although premnaspirodiene 11 may be derived from a different cation intermediate. Clm1-STS cyclizes the same bisabolyl cation intermediate via a different route (blue compounds) primarily to longiborneol 14 with longifolene 2 as an alternative product. Longiborneol becomes the precursor to culmorin 16 and culomorone 17. Several sesquiterpenes (brown) produced by F. graminearum are not associated with Tri5 and made by yet to be identified STS activities.
Trichothecene biosynthesis is the best characterized sesquiterpenoid biosynthetic pathway in fungi, and has been described in detail (5,11). Mutant F. graminearum strains deficient in Tri5, and thus lacking production of the trichothecene deoxynivalenol (DON), are unable to spread within an infected wheat spike (9,12). Trichothecenes not only potentiate FHB disease in wheat, where they inhibit protein synthesis (13), but also are toxic to humans and other animals who consume them from contaminated food, rendering the grain inedible (14).
Fusarium species also are known to produce the non-trichothecene related sesquiterpenoids (NTS) culmorin and cyclonerodiol (15,16) (Fig. 1). The mycotoxin culmorin is derived from a sesquiterpenoid synthesized by the STS longiborneol synthase (Clm1), whereas the enzymatic basis for synthesis of cyclonerodiol is currently unknown. In addition to the characterized genes Tri5 and Clm1, the genome of F. graminearum contains six additional predicted STS genes with currently unknown function (Fig. 2, Table S1). Two of the unknown STS genes, designated FGSG_08181 and FGSG_16873 (corresponding Fusgr1|10122 and Fusgr1|8874 in Fig. 2), are co-expressed under conditions where Tri5 is induced (17). There is ample evidence that the wide variety of sesquiterpenes produced by an organism can have significant effect on chemical signaling and toxicity (18). However, the effect of these NTS on FHB, and whether additional, yet uncharacterized NTS are produced by F. graminearum, need further investigation.
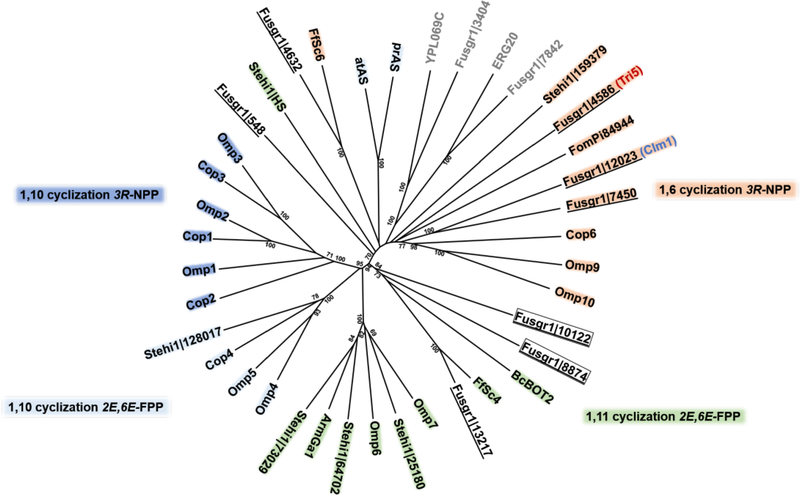
Figure 2. The terpenome of F. graminearum.
Phylogenetic analysis comparing the eight predicted F. graminearum sesquiterpene synthases (STSs) (underlined) to characterized Basidiomycota and Ascomycota STS showed that three of the uncharacterized STS clade with 1,6-cyclizing STS, including the known longiborneol synthase (Clm1) and trichodiene synthase (Tri5). The remaining three clades with known enzymes that follow an 1,11 initial cyclization mechanism shown in Fig. 1. Transcriptome analysis comparing STS gene expression under a variety of conditions (data not shown) identified STS (boxed) (see Table S1 for gene model designators) as being co-expressed with Clm1 and Tri5. Accession numbers and references for STS sequences are listed in the Materials and methods. The tree was rooted with FPP synthases (F. graminearum Fusgr1|7842 and S. cerevisiae ERG20) and GGPP synthases (F. graminearum Fusgr1|3404 and S. cerevisiae YPL069C) as outgroup. The tree was generated in MEGA7 using the Neighbor-Joining method with bootstrap values of >70 shown for 500 replicates.
Numerous cellular changes occur upon induction of trichothecene biosynthesis in vitro and in planta including hyphal thickening, subapical swelling, and increased vacuole size (19,20). Additionally, oxygenase enzymes of the trichothecene biosynthetic pathway co-localize to induced and highly remodeled, organized smooth endoplasmic reticulum (OSER) structures referred to as “toxisomes” (21,22). The remodeled ER appears to be important for high level trichothecene production because treatments that prevent the formation of toxisomes diminish the capacity of cells to accumulate trichothecenes (23).
The initial aim of this study was to determine the spectrum of NTS produced by F. graminearum cultures induced to produce trichothecenes. Previous studies determined that the genes for culmorin synthesis (Clm1 and Clm2 (corresponding to Fusgr1|1203, Fusgr1|7450 in Fig. 2)) are co-expressed with genes for trichothecene pathway enzymes under the conditions studied here. Moreover, deletion of either Clm1 or Clm2 was shown to increase levels of trichothecene that accumulate in culture (15,24,25). These results could be explained by an increased pool of the shared isoprenoid pathway precursor, farnesyl pyrophosphate, available for trichothecene synthesis in the culmorin minus mutants. To characterize additional NTS in F. graminearum cultures, we sought to maximize NTS synthesis and we reasoned that their concentration may be similarly increased in strains lacking trichothecene production by deleting Tri5. However, unexpectedly we found that NTS synthesis was actually reduced in a Δtri5 mutant, and that the Tri5 protein itself may be a key player in the synthesis of both trichothecenes and NTS.
2. Materials and methods
2.1. Strains, media, materials and cultivation conditions
A list of all F. graminearum and Escherichia coli strains used or generated in this study is provided in Table S2. All primers used to create F. graminearum strains, construct plasmids and Tri5 constructs are listed in Table S3.
E. coli JM109 was used for cloning and plasmid maintenance, E. coli BL21 (DE3) (New England Biolabs (NEB), Ipswich, MA) for sesquiterpene production and E. coli Rosetta™ cells (Millipore Sigma, Burlington, MA) for protein expression. All E. coli strains were grown at 200 rpm in LB medium supplemented with appropriate antibiotics (kanamycin, chloramphenicol) at 37°C for cloning and plasmid amplification and at 30°C to express Tri5 and Tri5N255D S229T for product profiling and for protein purification.
For Tri5 and Tri5N255D S229T sesquiterpene profiling in E. coli, 50 mL cultures (250 mL shake flasks, covered with double-layered aluminum foil) were inoculated with 0.5 mL of overnight cultures of recombinant E. coli BL21 (DE3) (New England Biolabs (NEB), Ipswich, MA) cells transformed with the corresponding pET28a expression plasmids. Recombinant cultures were grown to an OD600 ~0.3 and induced with 0.5 mM IPTG. Sesquiterpenoids were analyzed 4 h post induction.
For Tri5 and Tri5N255D S229T protein purification, 500 mL cultures (2.5 L shake flasks) were inoculated with 5 mL overnight cultures, grown to an OD600 ~0.3 and induced with 0.5 mM IPTG. After three hours of cultivation, cells were harvested at 6000 rpm for 10 minutes in a Beckman (Brea, CA) J2-HS floor centrifuge and stored at −80°C until used for purification.
For fungal sesquiterpenoid profiling, fifty milliliter F. graminearum cultures (250 mL shake flasks, covered in double-layered aluminum foil) were cultivated as described in (19) as adapted from (26). Briefly, conidia were resuspended at a concentration of 104 mL−1 in either Toxin Induction Medium (TBI) supplemented with 5 mM putrescene at pH 4.5, or Non-Induction Medium (NIM) containing 10 mM sodium nitrate at pH 4.5. All other ingredients were identical for both media (30 g sucrose, 1 g KH2PO4, 0.5 g MgSO4*7H2O, 0.5 g KCl, 2 mL FeSO4*7H2O (5 mg mL−1 stock), and 200 μl trace elements (5 g citric acid, 5 g ZnSO4 *7H2O, 0.25 g CuSO4*5H2O, 50 mg of MnSO4* H2O, 50 mg of H3BO3, and 50 mg NaMoO4 *2H2O per 100 ml) per L). Culture media were sterile filtered. Growth was at 25 °C and 150 rpm shaking in the dark for 3 days.
2.2. Sesquiterpene profiling
Volatile sesquiterpenoids were extracted from E. coli and fungal culture headspace with a 100 μm polydimethylsiloxane-divinylbenzene (PDMS) Solid Phase MicroExtraction fiber (SPME, Supelco, Bellfonte, PA) inserted through aluminum foil seal for 10 min as described previously (27). Likewise, non-volatile, soluble sesquiterpenoids were extracted by SPME from 50 mL centrifuged culture media (2655 × g, 10 mins, Eppendorf 5810R swinging-bucket centrifuge) that was clarified by passage through a HT Tuffryn low protein binding 0.45 μm syringe filter (Pall, Port Washington, NY).
Extracted terpenoid products were separated and analyzed by GC-MS on an HP GC 7890A gas chromatography coupled to an anion-trap mass spectrometer equipped with an HP MSD triple axis detector (Agilent Technologies, Santa Clara, CA) and using an HP-5MS capillary column (30 m × 0.25 mm × 1.0 μm) with an injection port temperature of 250 °C and helium as a carrier gas. The oven temperature started at 60 °C and was increased at 6 °C min−1 to a final temperature of 250 °C with a 38 min cycle time. Terpenoids were identified by comparing mass spectra and retention indices to MassFinder’s (Software v.4) terpene library (28) and the NIST chemical database as described previously (29).
2.3. Plasmid construction
F. graminearum cDNA was prepared as described previously (29) and used for Tri5 cloning. The Tri5 ORF was initially amplified with its single intron and cloned in frame with an N-terminal 6xHis-Tag into plasmid pET28a (Millipore Sigma) using primers F_tri5_NdeI and R_tri5_NotI. The intron was subsequently removed with these primers and primers Tri5-Exon1-R and Tri5-Exon2-F by overlapextension PCR followed by religation into pET28a. The resulting plasmid was Sanger sequenced and tested for trichodiene synthase activity upon expression in E. coli. The intron was identified by comparison to the well-characterized F. sporotrichioides Tri5, which was also initially cloned with this intron and was similarly removed prior to obtaining a functional Tri5 enzyme (8). All PCR utilized Phusion™ high-fidelity polymerase (New England Biolabs, Ipswich, MA). The Tri5N225D S229T mutant was subsequently created via Q5 Site-directed Mutagenesis™ (New England Biolabs, Ipswich, MA) using the primers Tri5N225D S229T Q5 1 and Tri5N225D S229T Q5 2 following the manufacturer-recommended conditions to create the pET28a-Tri5N225D S229T plasmid.
2.4. Tri5 and Tri5N225D S229T purification
Frozen E. coli cells from 500 mL cultures were resuspended in 30 mL STS purification buffer (50 mM Tris, pH 8, 250 mM NaCl, 5 mM imidazole, 10 mM MgCl2, 1 mM PMSF) supplemented with cOmplete protease inhibitor (Sigma-Aldrich) and sonicated for 4 min (1 sec pulse on, 2 sec pulse off at 30% power), followed by 30 min centrifugation at 12,000 rpm at 4 °C in a Beckman J2-HS centrifuge. Soluble protein was filtered through a 0.45 μm syringe and purified by FPLC (GE Healthcare, Chicago, IL) using a 5 mL His-trap™ FF Ni2+ affinity column (GE Healthcare, Chicago, IL) equilibrated with STS purification buffer. After washing, bound protein was eluted with STS elution buffer (50 mM Tris, pH 8, 250 mM NaCl, 250 mM imidazole, 10 mM MgCl2, 1 mM PMSF) collecting 4 mL fractions. Eluted fractions were then concentrated using Amicon (EMD Millipore, Burlington, MA) Ultra-15 3,000 NMWL centrifugation filter units and dialyzed overnight into size exclusion buffer (5% glycerol, 20 mM Tris, pH 7.5, 50 mM NaCl, 0.22 μm filtered) for Superdex 200 10/300 (GE Healthcare) purification with 1 mL fractionation. Protein fractions were combined and concentrated using a 3000 NMWL Amicon filter to a concentration of approximately 1 mg mL−1. All protein containing fractions obtained after chromatography and concentration, were assessed by 12% SDS-PAGE analysis with Coomassie staining for Tri5 protein purity. In addition, WT Tri5 samples were analyzed for trichodiene synthase activity as described previously with 2 μM FPP as substrate (29). Purified and concentrated proteins were then dialyzed overnight into 10 mM potassium phosphate buffer, pH 8.0 for circular dichroism analysis as described (30).
2.5. Circular dichroism
Purified and concentrated Tri5 and Tri5N225D S229T were diluted to a concentration of 0.1 mg mL−1 (2.146 and 2.168 μmol L−1 for WT and mutant, respectively) and analyzed by circular dichroism (CD) from 260 to 185 nm with single nanometer step resolution. CD analysis was performed on a JASCO J-815 spectropolarimeter (Ishikawacho, Japan) (31) with appropriate air and solvent-only controls. CD spectra comparing WT Tri5 and Tri5 N225D S229T molar ellipticity (Θ) from 185–260 nm were overlaid to identify any loss of α-helical structure resulting from the active site mutations performed.
2.6. OSCAR deletion
The Tri5 deletion strain for sesquiterpenoid profiling was made using a modified OSCAR method (Paz et al., 2011). Marker vector pBLUE-HPH-GFP was constructed by excising the hph-GFP fragment from plasmid pPK2-hphGFP (32) using BamHI and PstI and ligated into plasmid pBlue digested with BamHI and NsiI. Tri5 upstream flank and downstream flank were amplified from F. graminearum PH-1 genomic DNA using primers FGSG03537-del-LF1F, FGSG03537-del-LF2F and FGSG03537-del-RF3F, FGSG03537-del-RF4R, respectively. Invitrogen Gateway system (Thermo-Fisher, Waltham, MA) was used to combine flanks and HPH-GFP fragment into T-DNA vector plasmid pOSCAR (GenBank: HM623914) to create the final transformation construct. Primers OSC-F, Hyg-R(210) and Hyg-F(850), OSC-R were used to verify the transformation construct. Agrobacterium-mediated transformations of F. graminearum PH-1 were performed as described previously (33). Strains were checked for the predicted deletion of TRI5 by PCR.
2.7. Fluorescent protein tagging and Tri5 allele mutation in F. graminearum
A fusion PCR-based method (34) was used to synthesize the constructs for generating a full-length Tri5 (FGSG_03537; FGRAMPH1_01T13111) protein tagged with GFP and a full-length Hmr1 (FGSG_09197; FGRAMPH1_01G27691) protein tagged with RFP. The Neurospora knock-in vector pGFP::hph::loxP (GenBank: FJ457011.1) (35) was used as a template for the synthesis of the GFP::hph portion of the fusion construct for GFP tagging. Vector pAL12-Lifeact (36) was used as a template for synthesis of the RFP::nat1 cassette for the fusion construct for RFP tagging. Oligonucleotides used to amplify the upstream and downstream regions flanking the Tri5 stop codon and GFP::hph as well as the primers used to amplify Hmr1 flanks and the RFP::nat1 cassette are listed in Table S3. Transformation of F. graminearum protoplasts was performed using gel purified fusion constructs as described (21). V8 juice agar supplemented with 250 μg ml−1 hygromycin B was used for isolation of Tri5-GFP transformants. Half-strength potato dextrose agar supplemented with 50 μg mL−1 nourseothricin was used to isolate Hmr1-RFP transformants. Integration of GFP-tagged and RFP tagged constructs were confirmed via amplification of DNA with gene specific oligonucleotides.
A Tri5 deletion containing a neomycin gene replacement (ΔTri5-NEO) was constructed in order to facilitate making F. graminearum strains expressing Tri5N225D S229T. Split-marker recombination mutagenesis (37,38) was used to replace Tri5 with the neomycin resistance cassette amplified from plasmid pSM334 (39). Tri5 upstream and downstream flanks were at the same loci as used for the Tri5 OSCAR deletion.
F. graminearum strains expressing Tri5N225D S229T were made using the Tri5N225D S229T cassette amplified from plasmid pET28a-Tri5N225D S229T with primers FGSG03537-delNEO-LF1F and FGSG03537-delNEO-RF4R. The amplified cassette was then used to transform protoplasts of the ΔTri5-NEO strain as described above. Transformants were isolated on V8 juice containing 250 μg mL−1 geneticin and confirmed using amplification with gene specific primers. Integration at the Tri5 locus was verified using PCR, Sanger sequencing, and Southern blot analysis.
2.7. Crossing strains
Dual tagged strains expressing both Tri5-GFP/Hmr-RFP, Tri5N225D S229T/Hmr1-RFP, and ΔTri5-NEO/Hmr1RFP were created by crossing the individual strains on carrot agar as per Boenisch et al., 2017 with minor modifications. Ascospores from the crosses were plated on half strength PDA containing 150 μg ml−1 and 50 μg ml−1 nourseothricin (Tri5-GFP/Hmr-RFP, Tri5N225D S229T/Hmr1-RFP) or 150 μg ml−1 geneticin and 50 μg ml−1 nourseothricin (ΔTri5-NEO/Hmr1-RFP). Strains were verified using amplification with gene specific primers. Tri5N225D S229T/Hmr1-RFP Tri5 locus was also verified with DNA sequencing.
2.8. Microscopy
To observe expression of fluorescently tagged proteins in vivo, conidia were suspended in 5 ml TBI cultures at a final concentration of 104 conidia mL−1 and grown at 25°C on an orbital shaker at 150 rpm in the dark. Cultures were sampled at 72 h following inoculation. Wet mounts of tissue were viewed using a Nikon Eclipse 90i upright epifluorescence microscope. NIS-Elements AR software was used for image generation and analysis (Nikon Instruments Inc., Melville, NY, USA).
2.9. Phylogenetic analysis and STS sequences
Sequence alignments and phylogenetic analysis of Fungal STS sequences were carried out in MEGA 7 (40) with default parameters: MUSCLE (41) was used for sequence alignments and the Neighbor-Joining Method (42) was used to build a phylogenetic tree. Accession numbers and/or references for protein sequences used are as follows: Coprinus cinereus (Cop1–4, Cop6) [XP 001832573, XP 001836556, XP 01832925, XP 01836356, XP 01832548] (43), Omphalotus olearius (Omp1–10) (44), Fomitopsis pinicola [FomPi84944] (44), Stereum hirsutum [Stehi1|159379, 128017, 25180, 64702, 73029] (29,45), Armillaria gallica (ArmGa1) [P0DL13] (46); Botrytis cinerea (BcBOT2) [AAQ16575.1] (47), Fusarium fujikori (Ffsc4) [HF563560.1] and Ffsc6 [HF563561.1](48) Fusarium gramineareum (Clm1) [GU123140]) (48–50) and (Tri5) [AAM48886] (50,51), Aspergillus terreus (atAS) [Q9UR08) (52), Penicillium roqueforti (prAS) [W6Q4Q9] (53). Table S1 lists gene model designators for F. graminearum STS sequences.
3. Results
3.1. F. graminearum produces a suite of complex sesquiterpenoids
To produce trichothecenes in culture, F. graminearum requires particular molecular signals. Among the most potent inducers of trichothecene biosynthesis are polyamines, including putrescine and agmatine (26), that may be the signals recognized by the fungus during the infection of plants (54). With these strong inducers at hand, our initial goal was to profile the full repertoire of sesquiterpenes produced by F. graminearum under trichothecene inducing and non-inducing cultivation conditions.
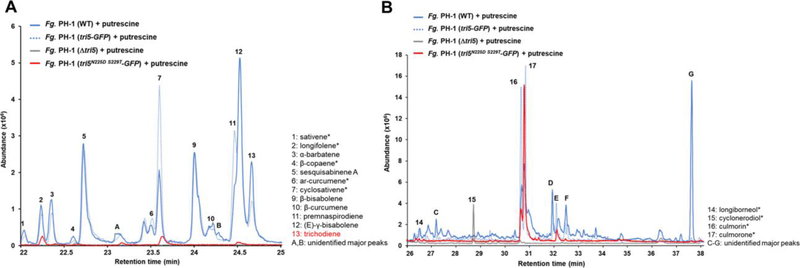
During growth in minimal medium without 5 mM putrescine as inducer, no volatile sesquiterpenes were detected in the culture headspace (Fig. 3A). In cultures grown in the same medium but with putrescine instead of sodium nitrate as the primary nitrogen source, 13 identifiable sesquiterpenes and two major unknown sesquiterpenes (peaks A and B) were found in the volatile fraction. One of the sesquiterpene peaks is trichodiene 13 (red), the major product of the trichodiene synthase Tri5 (55). Additionally, both α-barbatene 3 and β-bisabolene 9 were produced by induced cultures, which have previously been identified as minor 1,6-cyclization products of trichodiene synthase catalyzed reactions (56,57). Other major 1,6-cyclization pathway derived sesquiterpenoids that could be Tri5 products are sesquiabinene A 5 and (E)-γ-bisabolene 12. The STS and cyclization mechanism that yields another major product, premnaspirodiene 11, is unknown (see Fig. 1 for structures).
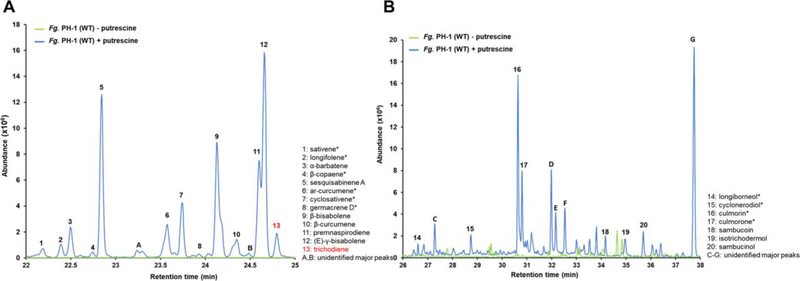
Figure 3. Sesquiterpene production in F. graminearum in response to putrescine induction.
A: SPME GC/MS analysis of volatile sesquiterpenes produced in culture headspace. Wild-type F. graminearum grown on non-inducing medium ( − putrescine) produces no volatile sesquiterpenes. The same strain grown with putrescine ( + putrescine) produced 13 identifiable sesquiterpenes and two major sesquiterpenes that could not be identified (A, B). Accumulation of trichodiene is presumably depressed because of conversion to trichothecenes by Tri4 and other enzymes (Fig. 1). B: SPME GC/MS analysis of soluble sesquiterpenes in liquid cultures. Seven identifiable non-volatile sesquiterpenoids are similarly produced in putrescine-induced F. graminearum cultures, most notably culmorin 16 and its derivative culmorone 17, as well as five significant but unidentified sesquiterpenoids (C-G). GC/MS spectra for all labelled compounds are shown in Fig. S1. Numbered compounds structures are shown in Fig. 1. Unlabeled minor peaks are not sesquiterpene derived compounds.
Upon induction with putrescine, non-volatile sesquiterpenoids also accumulated and could be identified in the aqueous phase of F. graminearum cultures. These include the expected trichodiene-derived trichothecene pathway intermediates isotrichodermin 19, sambucoin 18 and sambucinol 20, as well as the longiborneol synthase Clm1-derived longiborneol 14 and its derivatives culmorin 16 and culmorone 17 (Fig. 3B). Additionally, the sesquiterpene cyclonerodiol 15 was present, produced by an unknown STS. Five additional, modified sesquiterpenoid products (peaks C-G) were also produced that could not be unambiguously identified. In contrast, the non-induced wild type control accumulated no detectable non-volatile sesquiterpenoids which corresponds to the absence of volatile sesquiterpenes in the head space of uninduced cultures (Fig. 3A).
In-depth biochemical characterization, including mechanistic and structural studies, of Tri5 have been performed with the trichodiene synthase cloned from Fusarium sporotrichioides (58,59). Therefore, to more accurately determine which sesquiterpenes may have arisen from cyclization reactions catalyzed by Tri5 in F. graminearum (and which are NTS), we cloned the Tri5 gene for expression in E. coli and comparison of volatile sesquiterpene product profiles between the recombinant E. coli cultures and putrescine induced F. graminearum cultures (Fig. 4). Of the 13 previously identified sesquiterpene compounds made by the induced fungal culture, six sesquiterpenes labeled with a star in Fig. 4 are not made by recombinant Tri5 in E. coli. Among those are the Clm1 product longifolene 2 and the Z,E-germacradienyl cation derived germacrene D 8 and β-copaene 4 that are the likely products of separate, as-yet unidentified STS activities. Sativene 1, cyclosativene 7 and ar-curcumene 8, which could be derived via a 1,6-cyclization reaction, are also not made by Tri5 expressed in E. coli, suggesting that they may be the product of another STS activity in F. graminearum (note: the mass fragmentation spectrum of the E. coli Tri5 peak eluting slightly before the F. graminearum cyclosativene 7 in Fig. 4 does not match cyclosativene, and has the characteristic MS spectrum of a siloxane column component). Interestingly, premnaspirodiene 11 is also a significant E. coli Tri5 product which may not be synthesized via the common 1,6-cyclization pathway of the other Tri5 products based on the 1,10-cyclization mechanism proposed for a characterized plant premnaspirodiene synthase (60). We noticed that the levels of trichodiene were much higher in the E. coli culture than in the F. graminearum culture presumably because in F. graminearum, trichodiene would be further metabolized to other, non-volatile oxygenated trichothecene compounds.
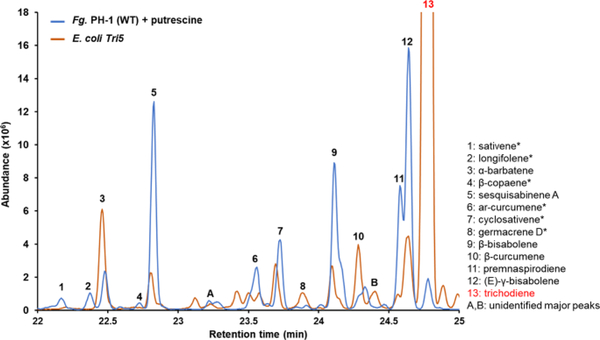
Figure 4: Identification of non-Tri5 derived sesquiterpenes.
SPME GC/MS analysis of volatile sesquiterpenoids in the culture headspace produced by putrescine induced F. graminearum and by E. coli expressing Tri5. Overlay of GC chromatographs identifies six sesquiterpenes (sativene 1, cyclosativene 7, longifolene 2, β-copaene 4, ar-Curcumene 6, and germacrene D 8 and two unidentified sesquiterpenes (A and B) that are not produced directly by Tri5. GC/MS spectra for all labelled compounds are shown in Fig. S1. Numbered compounds structures are shown in Fig. 1. Non-Tri5 pathway derived compounds are labeled with a star. Unlabeled minor peaks are not sesquiterpene derived compounds.
To confirm that the NTS are produced by STS other than Tri5, and also attempt to increase the levels of minor sesquiterpenes synthesized by F. graminearum cultures, we created a tri5 deletion (Δtri5) strain hypothesizing this would redirect FPP flux towards NTS biosynthesis similar to the increase of trichothecene production previously observed in a longiborneol synthase clm1 deletion strain (15,24,25). However, contrary to expectations, all volatile sesquiterpenes except for minor quantities of the non-Tri5 product β-copaene 4 were missing from the tri5 deletion strain culture headspace grown under inducing conditions (Fig. 5A). Absent any sesquiterpene precursors, this strain therefore also ceased to produce almost all modified, soluble sesquiterpenoids, including culmorin 16 and culmorone 17, which are derived from the Clm1 product longiborneol 14; only cyclonerodiol was still produced at a significant level (Fig. 5B). This meant our simple model of FPP availability determining sesquiterpene levels was not correct and warranted further investigations.
Figure 5. Sesquiterpenoid product profiles of F. graminearum strains with different tri5 alleles.
A: SPME GC/MS analysis of volatile sesquiterpenes in the headspace of different, putrescine induced cultures. Overlay of GC/MS chromatograms show that the Δtri5 deletion strains ceased production of all volatile sesquiterpenoids, except for minor quantities of β–copaene 4, compared to the WT strain. The Tri5N225D S229T-GFP strain retained production of longifolene 2 and cyclosativene 7 compared to the Tri5-GFP control strain. B: SPME GC/MS analysis non-volatile sesquiterpenes in liquid cultures produced by different, putrescine induced strains. The Δtri5 strain accumulates cyclonerodiol 15 as the only major sesquiterpenoid compared to the WT, which produces both trichodiene-derived and non-trichodiene related (NTS) compounds. The Tri5N225D S229T-GFP strain restored production of the NTS longiborneol 14, culmorin 16 and culmorone 17, but is deficient in the production of trichodiene-derived compounds compared to the Tri5-GFP control strain. GC/MS spectra for all labelled compounds are shown in Fig. S1. Numbered compounds structures are shown in Fig. 1. Non-Tri5 pathway derived compounds are labeled with a star. Unlabeled minor peaks are not sesquiterpene derived compounds.
3.2. The Tri5 protein is important for NTS synthesis
The loss of most non-trichothecene sesquiterpenes in the Δtri5 mutant suggested that Tri5 must be involved in the regulation of global sesquiterpenoid biosynthesis. We posited that either the Tri5 protein itself (hypothesis 1) or Tri5-derived pathway products such as trichodiene 13, trichothecene intermediates or DON (hypothesis 2) are required for production of the full spectrum of sesquiterpenoids by F. graminearum, including some, like longifolene, whose synthesis is not directly catalyzed by Tri5.
We chose to address first the role of the Tri5 protein in overall sesquiterpene regulation as the most straightforward approach to test our hypothesis, as this would not require laborious isolation and testing of many possible Tri5 pathway products. For this, we planned to create a Tri5 protein without enzymatic activity but retaining its overall secondary structure. Complementation of the inactive Tri5 mutant into the F. graminearum tri5 locus, and subsequent analysis of its effect on the sesquiterpenoid product profile would therefore directly test both hypotheses, if mutually exclusive. If culmorin 16, culmorone 17 and other NTS production is restored in strains expressing the inactive Tri5 protein, then co-regulation must be occurring via direct protein interaction, while lack of restoration indicates trichodiene or downstream reaction pathway products may be required for co-production of NTS.
To create an inactive Tri5 protein, we mutated two key residues (N225D and S229T) in the conserved NSE/DTE active site motif of sesquiterpene synthases required for the coordination of Mg2+. Previously, it was shown that a mutated Tri5N225D S229T from F. sporotrichioides completely lacked activity while retaining three dimensional structure of the wild-type enzyme (56). Expression of the F. graminearum Tri5N225D S229T mutant in E. coli confirmed that the F. graminearum enzyme also was inactive and did not produce any sequiterpenes (Fig. S2). Overnight incubation of the purified Tri5N225D S229T with FPP also validated complete loss of enzyme activity. Conservation of secondary structure was then confirmed by comparing the Circular Dichroism (CD) spectra of purified wild-type and mutant Tri5 (Fig. S3), with both proteins exhibiting the expected minima at 208 and 222 nm for correctly formed, primarily alpha-helical proteins such as the STS (31).
Next, we inserted the enzymatically inactive Tri5N225D S229 but structurally intact protein into the deleted tri5 locus of the previously analyzed Δtri5 strain of F. graminearum. To allow for eventual visualization of cellular localization of the Tri5 protein, we first created a Tri5N225D S229T-GFP fusion construct along with a wild-type Tri5-GFP fusion as a control. Successful generation of the corresponding F. graminearum tri5N225D S229T-GFP and tri5-GFP strains was confirmed by DNA sequencing, PCR, and Southern blot analysis (Fig. S4). To ensure that the GFP fusion would have no or only a minimal effect on Tri5 function in F. graminearum, we compared the sesquiterpenoid product profile of the transgenic strain with the wild-type strain (Fig. 5A and 5B). The GFP fusion did not substantially perturb the function of Tri5, resulting in the production of the same compounds mostly at similar levels as the wild-type strain; although the unknown compound G was not produced as a major, nonvolatile compound by the Tri5GFP strain.
Remarkably, when Tri5N225D S229T-GFP replaced the Δtri5 allele, it partially restored the ability to produce the volatile sesquiterpenes longifolene 2 and cyclosativene 7 (Fig. 5A) and fully restored the ability to produce the non-volatile Clm1- derived sesquiterpenoids culmorin 16 and culmorone 17 (Fig. 5B), which are products of a separate NTS pathway (15,25) (Fig. 1). As anticipated, the Tri5N225D S229T-GFP did not restore the Tri5-GFP major product, trichodiene, or other Tri5 products in the volatile fraction (Fig. 5A) or trichothecene peaks D and F in the non-volatile fraction (Fig. 5B). These results demonstrated that the catalytically inactive Tri5N225D S229T but structurally intact protein is both necessary and sufficient to restore NTS production in the Δtri5 deletion strain. Therefore, the Tri5 protein itself may play a role in synthesis of NTS that is independent of its enzymatic activity. One possible explanation for this may be due to the ability of Tri5, a cytosolic protein, to interact with the modified organized smooth endoplasmic reticulum (OSER) of the toxisome formed in F. graminearum (61), which we investigated next.
3.3. Tri5 protein is important for toxisome formation
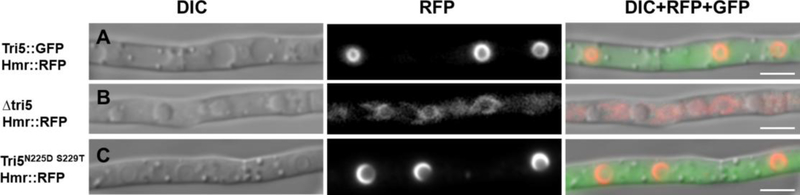
To visualize the potential effect of Tri5 on toxisome structure and function, we created a strain of F. graminearum with fluorescently tagged ER by labelling the ER integral membrane protein, HMG CoA reductase, with the red fluorescent protein RFP (Hmr1-RFP) via a translational fusion. To this strain, we then introduced the three different Tri5 alleles created in this study to produce the following three strains: (Tri5-GFP, Hmr1-RFP); (Δtri5, Hmr1-RFP); and (Tri5N225D S229T-GFP, Hmr1-RFP). The formation of toxisomes under inducing conditions was then visualized as described previously (22,61).
The Tri5-GFP, Hmr1-RFP dual-tagged strain formed toxisomes (Fig. 6A) as we expected from its expression of a wild-type like Tri5 enzyme. In contrast, the Δtri5, Hmr1-RFP dual-tagged strain showed only undifferentiated ER, similar to that seen in non-induced strains (22) (Fig. 6B). This phenotype was coupled to the loss of production of trichodiene-derived and most other NTS (Fig. 5A and 5B). As posited from its restoration of NTS but not trichodiene-derived sesquiterpene production, the Tri5N225D S229T-GFP, Hmr1-RFP strain also restored toxisome structure to the ER (Fig. 6C).
Figure 6. Toxisome structure in F. graminearum cells having different Tri5 genotypes.
The Tri5-GFP allele (A), a Δtri5 deletion (B), or a Tri5N225D S229T-GFP allele (C) were introduced into the Tri5 locus in the same genetic background having the ER tagged with Hmr1-RFP. Strains were treated with 5 mM putrescine and cells visualized by epifluorescent microscopy 72 h after induction. Tri5N225D S229T complementation of the Δtri5 allele (B) restores toxisome structure to cells (C) comparable to structures seen in the wild-type like Tri5-GFP control cells (A). Columns illustrate differential interference contrast (DIC) images, red channel fluorescence (RFP), and a merged image of green (GFP) and red (RFP) channel fluorescence with DIC. Exposure time for RFP was 3s for A and C, 5s for B. Scale bar = 5 μm.
4. Discussion
We have demonstrated that F. graminearum produces several non-trichothecene sesquiterpenes (NTS) in response to induction by putrescine. No sesquiterpene compounds were detected in cultures grown with nitrate as the sole source of nitrogen. Putrescine induced cultures produced the Tri5 product trichodiene 13 and previously reported trichothecene derivatives (Fig. 1) including sambucoin 18 (16) and pathway intermediate isotrichodermin 19 (62,63). The sesquiterpene cyclonerodiol 15 also was observed in putrescine induced cultures. Cyclonerodiol 15 is not a product of trichodiene synthase Tri5 and has been previously reported in cultures of F. graminearum grown in a medium containing ammonium as the sole carbon source (16,64). Culmorin 16 and culmorone 17 were also observed in putrescine induced cultures, and are known to be oxygenated products of longiborneol 14, and thus a product arising from longiborneol synthase Clm1 (15,25). Longifolene 2, found in the volatile fraction of cultures, has previously been determined to be a minor product of Clm1 (15).
The volatile sesquiterpenes sativene 1, β-copaene 4, ar-curcumene 6, cyclosativene 7 and germacrene D 8 are produced by F. graminearum in induced cultures, but are not produced by Tri5 expressed in E. coli, nor have they been previously reported to result from Clm1 (15). Therefore, these compounds likely arise by cyclization of FPP by enzymes encoded by other as-yet uncharacterized STSs, such as FGSG_08181/Fusgr1|10122 or FGSG_16873/Fusgr1|8874, that are co-expressed with Tri5 and Clm1 (Fig. 2, boxed). FGSG_08181/Fusgr1|10122 has been reported to be expressed during infection of barley and wheat, but not maize, whereas FGSG_16873/Fusgr1|8874 is expressed in wheat and maize but not barley (17). FGSG_08181/Fusgr1|1012 appears to be part of the F. graminearum accessory genome and is likely is part of a five gene cluster that includes a predicted transcription factor and three cytochrome P450s (65). Both STS are closely related to two characterized 1,11-cyclizing enzymes; koraiol synthase Ffsc4 from rice pathogenic fungus Fusarium fujikori (Ffsc4) (48), presilphiperfolan-8β-ol synthase BcBOT2 from the gray mold causing plant pathogen Botrytis cinerea (66) (Fig. 2). No sesquiterpenes derived via a 1,11-cyclization pathway were produced by F. graminearum cultures and it remains to be seen if these two STS may have switched cyclization mechanism to produce the non-Tri5 products which could be derived via 1,10- and 1,6-cyclization of NPP (Fig. 1). Cycloneridiol 5 is another intriguing sesquiterpenoid produced by F. graminearum which could be the product of a stand-alone STS that produces a sesquiterpene alcohol like Clm1 or Ffsc4 and BcBOT2. Its production was also not affected by the deletion of Tri5 like the production of most of the other volatile and non-volatile sesquiterpenoids by induced cultures.
Even though at least four putative STSs are expressed, and a complex mixture of volatile and non-volatile sesquiterpenes were produced by F. graminearum in induced cultures, deletion of Tri5 led to the loss of all compounds except the minor product β-copaene 4 and the aforementioned cycloneridiol 15 (Fig. 5). It is unclear how the Tri5 protein could control synthesis of most sesquiterpenes produced in culture, including those known to be synthesized via separate enzymes, such as longiborneol 14, longifolene 2, culmorin 16 and culmorone 17 that require Clm1. One formal possibility was that Tri5 plays a role in the cellular localization of sesquiterpene synthesis apart from its enzymatic activity.
Recently we have shown that many enzymes of the trichothecene pathway, including the three cytochrome P-450 enzymes Tri1, Tri11 and Tri4, are co-localized in approximately three-micron structures organized as stacks of smooth ER cisternae (21,22). These structures, called toxisomes, localize and incorporate sequential enzymes in the trichothecene pathway that may promote pathway efficiency. Assembly of toxisomes involves interactions with the actin cytoskeleton mediated by the protein myosin 1 (Myo1) (23). Myosin 1 proteins previously have been reported to interact with cellular endomembranes, allowing for stabilization of ER sheets (67). Disruption of toxisome structure by phenamacril, a small molecule that interferes with the motor domain of Myo1 (68,69), or by inhibition of F-actin formation using latrunculin b (23), greatly reduces the concentration trichothecenes that accumulate in cultures. These results suggest that toxisome structures, dependent upon the actin cytoskeleton for formation and persistence, are required for maximum trichothecene synthesis, and that treatments that prevent toxisome formation may also inhibit not only synthesis of trichothecenes, but also potentially other sesquiterpenes.
To our surprise, toxisomes did not form in the Δtri5 mutant (Fig. 6). Nevertheless, addition of trichodiene, the product of the Tri5-mediated reaction, to the Δtri5 mutant did lead to the formation of isotrichodermin, indicating that Tri4 and presumably other downstream enzymes in the trichothecene pathway are present and remain active in this mutant (data not shown). Cells inhibited in formation of toxisomes by phenamacril also produce reduced amounts of trichothecenes, suggesting that while toxisomes are needed for high levels of trichothecene production they are not absolutely necessary (23). What is remarkable is that the Δtri5 mutant is deficient in accumulation of many other NTS, most notably the Clm1-derived pathway compounds culmorin 16 and culmorone 17 that are produced at significant levels by wild-type cultures. This suggests that the toxisome also may be essential for synthesis of these two mycotoxins as well as other, less characterized sesquiterpenoids in addition to its known role in trichothecene production. The role of culmorin in the pathogenicity of F. graminearum toward plants is unclear (24,25). Nevertheless culmorin is considered an “emerging mycotoxin” whose synthesis and toxicology will be of greater interest for food safety consideration in the future (70). As such, understanding its pathway regulation and interconnection with trichothecene mycotoxin biosynthesis will therefore become an important piece in characterizing the terpenome of F. graminearum.
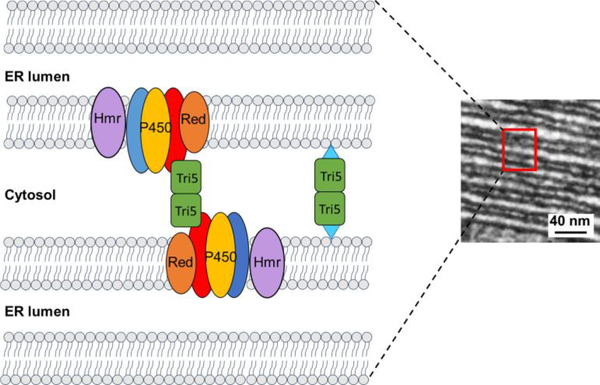
Finally, the mechanism by which Tri5 actually promotes toxisome formation and stability remains unknown. While it is established that Tri5 is a cytosolic enzyme (22,55,71) it also is enriched in the vicinity of the toxisome, seemingly within the layers of cytosol separating stacks of ER cisternae (61). At this position within the toxisome, the cytosolic regions between adjacent ER lamellae are 10 nm or less (22) and so Tri5, whose active form exists as an elongate homodimer (55), would be in a position to physically interact with adjacent membranes themselves, or with the cytosolic-facing surfaces of the ER membrane-anchored proteins such as Tri1, Tri4 and Tri11 as proposed in the model shown in Fig. 7. Current work, is therefore focused on examining the potential role of post-translational modification of Tri5 and lipid content of the toxisome in defining the dependence of toxisome structure on the presence of Tri5. Additional inquiries will aim at completing the characterization of the F. graminearum terpenome along with investigating the interactions of other STS’s with the Tri5-toxisome complex.
Figure 7. Proposed model for Tri5 interaction with the toxisome.
The cytochrome P450 oxygenases Tri1, Tri4 and Tri11 (blue, yellow, red ovals) involved in DON biosynthesis are clustered and anchored to the smooth ER membrane of the toxisome along with P450 reductase (“Red” in orange) and HMG-CoA reductase (Hmr in violet). Sheets of ER within the toxisome are stacked together and separated by thin layers of cytosol as shown in the transmission electron micrograph to the right. Active Tri5 (green), which exists as a homodimer in the cytosol, may allow for formation of stacked ER cisternae by directly interacting with the cytosolic surface of proteins positioned in the ER membrane or by anchoring adjacent membranes by attachment through post-translationally added lipophilic adducts (blue triangles).
Supplementary Material
Highlights.
Induced F. graminearum produces trichothecene and unrelated sesquiterpenes.
Trichodiene synthase (Tri5) cyclizes FPP into minor, related products besides trichodiene.
Δtri5 strain fails to produce trichodiene and most other unrelated sesquiterpenes.
Inactive but structurally intact Tri5N225D S229T mutant restores unrelated sesquiterpene synthesis.
The Tri5 protein appears essential for formation of reorganized ER toxisomes.
Acknowledgements
We thank Burçu Yordem for helpful conversations leading to the initiation of this project. Funding for this work was provided by award 2018-67013-28512 from the Agriculture and Food Research Initiative of the National Institute of Food and Agriculture, United States Department of Agriculture to HCK. CMF was supported from a Doctoral Dissertation Fellowship by the University of Minnesota and with funds from a National Institutes of Health Grant GM080299 (to C.S.-D.). USDA is an equal opportunity provider and employer.
Footnotes
Conflict of interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O’Donnell K, Ward TJ, Geiser DM, Kistler HC, and Aoki T (2004) Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol 41, 600–623 [DOI] [PubMed] [Google Scholar]
- 2.Goswami RS, and Kistler HC (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5, 515–525 [DOI] [PubMed] [Google Scholar]
- 3.McMullen M, Bergstrom G, De Wolf E, Dill-Macky R, Hershman D, Shaner G, and Van Sanford D (2012) A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis 96, 1712–1728 [DOI] [PubMed] [Google Scholar]
- 4.Wegulo SN, Baenziger PS, Nopsa JH, Bockus WW, and Hallen-Adams H (2015) Management of Fusarium head blight of wheat and barley. Crop Protection 73, 100–107 [Google Scholar]
- 5.Proctor RH, McCormick SP, Kim H-S, Cardoza RE, Stanley AM, Lindo L, Kelly A, Brown DW, Lee T, Vaughan MM, Alexander NJ, Busman M, and Gutiérrez S (2018) Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. Plos Pathog 14, e1006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormick SP, Stanley AM, Stover NA, and Alexander NJ (2011) Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grovey J (2007) The trichothecenes and their biosynthesis in Progress in the Chemistry of Organic Natural Products, Springer; pp 63–130 [PubMed] [Google Scholar]
- 8.Hohn TM, and Beremand PD (1989) Isolation and nucleotide-sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79, 131–138 [DOI] [PubMed] [Google Scholar]
- 9.Jansen C, von Wettstein D, Schafer W, Kogel KH, Felk A, and Maier FJ (2005) Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. P Natl Acad Sci USA 102, 16892–16897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goswami RS, and Kistler HC (2005) Pathogenicity and in planta mycotoxin accumulation among members of the Fusarium graminearum species complex on wheat and rice. Phytopathology 95, 1397–1404 [DOI] [PubMed] [Google Scholar]
- 11.Alexander NJ, Proctor RH, and McCormick SP (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Reviews 28, 198–215 [Google Scholar]
- 12.Proctor RH, Hohn TM, and McCormick SP (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe In 8, 593–601 [DOI] [PubMed] [Google Scholar]
- 13.Varga E, Wiesenberger G, Hametner C, Ward TJ, Dong YH, Schofbeck D, McCormick S, Broz K, Stuckler R, Schuhmacher R, Krska R, Kistler HC, Berthiller F, and Adam G (2015) New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ Microbiol 17, 2588–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escriva L, Font G, and Manyes L (2015) In vivo toxicity studies of fusarium mycotoxins in the last decade: A review. Food and Chemical Toxicology 78, 185–206 [DOI] [PubMed] [Google Scholar]
- 15.McCormick S, Alexander N, and Harris L (2010) CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl Environ Microb 76, 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauren DR, Sayer ST, and di Menna ME (1992) Trichothecene production by Fusarium species isolated from grain and pasture throughout New Zealand. Mycopathologia 120, 167–176 [Google Scholar]
- 17.Harris LJ, Balcerzak M, Johnston A, Schneiderman D, and Ouellet T (2016) Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biology 120, 111–123 [DOI] [PubMed] [Google Scholar]
- 18.Gershenzon J, and Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3, 408–414 [DOI] [PubMed] [Google Scholar]
- 19.Menke J, Dong YH, and Kistler HC (2012) Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol Plant Microbe In 25, 1408–1418 [DOI] [PubMed] [Google Scholar]
- 20.Boenisch MJ, and Schafer W (2011) Fusarium graminearum forms mycotoxin producing infection structures on wheat. Bmc Plant Biology 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menke J, Weber J, Broz K, and Kistler HC (2013) Cellular development associated with induced mycotoxin synthesis in the filamentous fungus Fusarium graminearum. PLoS One 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boenisch MJ, Broz KL, Purvine SO, Chrisler WB, Nicora CD, Connolly LR, Freitag M, Baker SE, and Kistler HC (2017) Structural reorganization of the fungal endoplasmic reticulum upon induction of mycotoxin biosynthesis. Scientific Reports 7, 44296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang G, Chen Y, Xu J-R, Kistler HC, and Ma Z (2018) The fungal myosin I is essential for Fusarium toxisome formation. Plos Pathog 14, e1006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner DM, Kazan K, and Manners JM (2009) Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol Plant Microbe In 22, 1588–1600 [DOI] [PubMed] [Google Scholar]
- 25.Bahadoor A, Schneiderman D, Gemmill L, Bosnich W, Blackwell B, Melanson JE, McRae G, and Harris LJ (2016) Hydroxylation of longiborneol by a Clm2-encoded CYP450 monooxygenase to produce culmorin in Fusarium graminearum. Journal of Natural Products 79, 81–88 [DOI] [PubMed] [Google Scholar]
- 26.Gardiner DM, Kazan K, and Manners JM (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet Biol 46, 604–613 [DOI] [PubMed] [Google Scholar]
- 27.Agger S, Lopez‐Gallego F, and Schmidt‐Dannert C (2009) Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72, 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joulain D, and König WA (1998) The atlas of spectral data of sesquiterpene hydrocarbons, EB-Verlag [Google Scholar]
- 29.Flynn CM, and Schmidt-Dannert C (2018) Sesquiterpene Synthase-3-Hydroxy-3-Methylglutaryl Coenzyme A Synthase Fusion Protein Responsible for Hirsutene Biosynthesis in Stereum hirsutum. Appl Environ Microbiol 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quin MB, Michel SN, and Schmidt-Dannert C (2015) Moonlighting Metals: Insights into Regulation of Cyclization Pathways in Fungal Delta(6)-Protoilludene Sesquiterpene Synthases. Chembiochem 16, 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield NJ (2006) Using circular dichroism spectra to estimate protein secondary structure. Nature protocols 1, 2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michielse CB, van Wijk R, Reijnen L, Manders EMM, Boas S, Olivain C, Alabouvette C, and Rep M (2009) The Nuclear Protein Sge1 of Fusarium oxysporum Is Required for Parasitic Growth. Plos Pathog 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonkers W, Dong YH, Broz K, and Kistler HC (2012) The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. Plos Pathog 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, and Oakley BR (2006) Fusion PCR and gene targeting in Aspergillus nidulans. Nature protocols 1, 3111. [DOI] [PubMed] [Google Scholar]
- 35.Honda S, and Selker EU (2009) Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182, 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berepiki A, Lichius A, Shoji J-Y, Tilsner J, and Read ND (2010) F-actin dynamics in Neurospora crassa. Eukaryot Cell 9, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catlett NL, Lee B-N, Yoder O, and Turgeon BG (2003) Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genetics Reports 50, 9–11 [Google Scholar]
- 38.Goswami RS, Xu JR, Trail F, Hilburn K, and Kistler HC (2006) Genomic analysis of host-pathogen interaction between Fusarium graminearum and wheat during early stages of disease development. Microbiol-Sgm 152, 1877–1890 [DOI] [PubMed] [Google Scholar]
- 39.Fuchs U, Czymmek KJ, and Sweigard JA (2004) Five hydrophobin genes in Fusarium verticillioides include two required for microconidial chain formation. Fungal Genet Biol 41, 852–864 [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, and Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitou N, and Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 43.Agger S, Lopez-Gallego F, and Schmidt-Dannert C (2009) Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Molecular microbiology 72, 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, and Schmidt-Dannert C (2012) Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem Biol 19, 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quin MB, Flynn CM, Wawrzyn GT, Choudhary S, and Schmidt-Dannert C (2013) Mushroom hunting by using bioinformatics: application of a predictive framework facilitates the selective identification of sesquiterpene synthases in basidiomycota. Chembiochem 14, 2480–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engels B, Heinig U, Grothe T, Stadler M, and Jennewein S (2011) Cloning and characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. The Journal of biological chemistry 286, 6871–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgant G, Collado IG, Cane DE, and Viaud M (2008) Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS chemical biology 3, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brock NL, Huss K, Tudzynski B, and Dickschat JS (2013) Genetic dissection of sesquiterpene biosynthesis by Fusarium fujikuroi. Chembiochem 14, 311–315 [DOI] [PubMed] [Google Scholar]
- 49.McCormick SP, Alexander NJ, and Harris LJ (2010) CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl Environ Microbiol 76, 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hohn TM, and Beremand PD (1989) Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79, 131–138 [DOI] [PubMed] [Google Scholar]
- 51.Ward TJ, Bielawski JP, Kistler HC, Sullivan E, and O’Donnell K (2002) Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci U S A 99, 9278–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shishova EY, Costanzo LD, Cane DE, and Christianson DW (2007) X-ray Crystal Structure of Aristolochene Synthase from Aspergillus terreus and Evolution of Templates for the Cyclization of Farnesyl Diphosphate. Biochemistry 46, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caruthers JM, Kang I, Rynkiewicz MJ, Cane DE, and Christianson DW (2000) Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti. J Biol Chem 275, 25533–25539 [DOI] [PubMed] [Google Scholar]
- 54.Gardiner DM, Kazan K, Praud S, Torney FJ, Rusu A, and Manners JM (2010) Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. Bmc Plant Biology 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rynkiewicz MJ, Cane DE, and Christianson DW (2001) Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proceedings of the National Academy of Sciences 98, 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vedula LS, Jiang J, Zakharian T, Cane DE, and Christianson DW (2008) Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: Probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233–motif. Archives of biochemistry and biophysics 469, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixit M, Weitman M, Gao J, and Major DT (2017) Chemical control in the battle against fidelity in promiscuous natural product biosynthesis: The case of trichodiene synthase. ACS Catalysis 7, 812–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rynkiewicz MJ, Cane DE, and Christianson DW (2001) Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci U S A 98, 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cane DE, Xue Q, and Fitzsimons BC (1996) Trichodiene synthase. Probing the role of the highly conserved aspartate-rich region by site-directed mutagenesis. Biochemistry 35, 12369–12376 [DOI] [PubMed] [Google Scholar]
- 60.Koo HJ, Vickery CR, Xu Y, Louie GV, O’Maille PE, Bowman M, Nartey CM, Burkart MD, and Noel JP (2016) Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants. J Antibiot (Tokyo) 69, 524–533 [DOI] [PubMed] [Google Scholar]
- 61.Boenisch MJ, Broz K, Blum A, Gardiner DM, and Kistler HC (201x) Nanoscale enrichment of the cytosolic enzyme trichodiene synthase near reorganized endoplasmic reticulum in Fusarium graminearum. Fungal Genetics and Biology In press. [DOI] [PubMed] [Google Scholar]
- 62.Alexander NJ, Hohn TM, and McCormick SP (1998) The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl Environ Microb 64, 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maeda K, Tanaka A, Sugiura R, Koshino H, Tokai T, Sato M, Nakajima Y, Tanahashi Y, Kanamaru K, Kobayashi T, Nishiuchi T, Fujimura M, Takahashi-Ando N, and Kimura M (2016) Hydroxylations of trichothecene rings in the biosynthesis of Fusarium trichothecenes: evolution of alternative pathways in the nivalenol chemotype. Environ Microbiol, n/a–n/a [DOI] [PubMed] [Google Scholar]
- 64.Lauren D, Di Menna M, Greenhalgh R, Miller J, Neish G, and Burgess L (1988) Toxin-producing potential of some Fusarium species from a New Zealand pasture. New Zealand journal of agricultural research 31, 219–225 [Google Scholar]
- 65.Walkowiak S, Rowland O, Rodrigue N, and Subramaniam R (2016) Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genomics 17, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C-M, Hopson R, Lin X, and Cane DE (2009) Biosynthesis of the Sesquiterpene Botrydial in Botrytis cinerea. Mechanism and Stereochemistry of the Enzymatic Formation of Presilphiperfolan-8-ol. J Am Chem Soc 131, 8360–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joensuu M, Belevich I, Rämö O, Nevzorov I, Vihinen H, Puhka M, Witkos TM, Lowe M, Vartiainen MK, and Jokitalo E (2014) ER sheet persistence is coupled to myosin 1c–regulated dynamic actin filament arrays. Molecular Biology of the Cell 25, 1111–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Chen Y, Yin Y, Ji HH, Shim WB, Hou Y, Zhou M, Li X. d., and Ma Z (2015) A small molecule species specifically inhibits Fusarium myosin I. Environ Microbiol 17, 2735–2746 [DOI] [PubMed] [Google Scholar]
- 69.Zheng Z, Hou Y, Cai Y, Zhang Y, Li Y, and Zhou M (2015) Whole-genome sequencing reveals that mutations in myosin-5 confer resistance to the fungicide phenamacril in Fusarium graminearum. Scientific Reports 5, 8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gruber-Dorninger C, Novak B, Nagl V, and Berthiller F (2017) Emerging Mycotoxins: Beyond Traditionally Determined Food Contaminants. Journal of Agricultural and Food Chemistry 65, 7052–7070 [DOI] [PubMed] [Google Scholar]
- 71.Blum A, Benfield AH, Stiller J, Kazan K, Batley J, and Gardiner DM (2016) High-throughput FACS-based mutant screen identifies a gain-of-function allele of the Fusarium graminearum adenylyl cyclase causing deoxynivalenol over-production. Fungal Genet Biol 90, 1–11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.