SUMMARY
Over the past decade, structures have been determined for broadly neutralizing antibodies that recognize all major exposed surfaces of the prefusion-closed HIV-1-envelope (Env) trimer. Here we analyze 206 antibody-HIV-1 Env structures from the Protein Data Bank with resolution suitable to define interaction chemistries and measured antibody neutralization on a 208-strain panel. Those with >25% breadth segregated into almost two dozen classes based on ontogeny and recognition and into 6 epitope categories based on recognized Env residues. For paratope, the number of protruding loops and level of somatic hypermutation were significantly higher for broad HIV-1 neutralizing antibodies than for a comparison set of non-HIV-1 antibodies (P<0.0001). For epitope, the number of independent sequence segments was higher (P<0.0001), as well as the glycan-component surface area (P=0.0005). The unusual characteristics of epitope and paratope revealed here are likely to respectively reflect virus-immune evasion and antibody solutions that allow effective neutralization of HIV-1.
INTRODUCTION
Antibodies against HIV-1 that neutralize a significant fraction of the diverse primary isolates that typify HIV-1 transmission are highly sought. These antibodies target the HIV-1 envelope (Env) trimer, which is comprised of gp120 and gp41 subunits and shielded from immune recognition by extraordinary glycosylation (Wei et al., 2003), sequence variability (Starcich et al., 1986), and conformational masking (Kwong et al., 2002). Potent broadly neutralizing antibodies that arise in natural infection are selected to overcome these barriers, and consequently tend to have extraordinary characteristics such as long protruding loops, high levels of somatic hypermutation (SHM), and direct glycan interactions (Klein et al., 2013a; McLellan et al., 2011) (reviewed in (Kwong and Mascola, 2012)). A decade ago, only four such broadly neutralizing antibodies had been identified. However, in 2009, the identification by B cell culture of antibody PG9 (along with a somatic variant PG16) (Walker et al., 2009) and in 2010 the identification by probe-based sorting of antibody VRC01 (along with somatic variants VRC02 and VRC03) (Wu et al., 2010) initiated an outbreak of discovery, with dozens of broadly neutralizing antibodies identified by culture or probe-based sorting of B cells from HIV-1-infected donors (reviewed in (Burton and Mascola, 2015)).
Importantly, structures for many of these antibodies in complex with HIV-1 Env have been determined, which link antibody function (breadth and potency of neutralization) with molecular features of antibody recognition (paratope) and chemical details of recognized Env site (epitope). For example, structures of PG9 with scaffolded V1V2 regions of Env (McLellan et al., 2011) and also with Env trimer (Julien et al., 2013) have revealed a protruding anionic tyrosine-sulfated loop penetrating the glycan shield to interact with a cationic site on V2; similarly structures of VRC01 with core gp120 (Zhou et al., 2010) or with a fully glycosylated Env trimer (Stewart-Jones et al., 2016) have revealed how SHM-based optimization of interactions – especially those involving glycan – allows for broad recognition of the CD4-binding site. Most recently, the structure of antibody VRC-PG05 in complex with a glycosylated Env core (Zhou et al., 2018) has shown that even the highly glycosylated center of the Env “silent face” could be recognized by broadly neutralizing antibody.
Here we take advantage of these findings to perform a structure-based survey of all antibody-HIV-1 Env structures in the Protein Data Bank (PDB). We limited analyses to structures of human antibodies in complex with epitopes on the prefusion-closed Env trimer with resolutions sufficient to define side chain orientation and interactions (with the exceptions of antibody b12, which induced a structure distinct from the prefusion-closed conformation, and antibody 2G12, for which we solved the cryo-EM structure in complex with Env trimer). The focus on the prefusion-closed conformation of Env alleviated issues of conformational masking and enabled comparisons on a single structural entity. We classified structures by antibody class (B cell ontogeny and structural mode of recognition) and used a bottom-up classification based on recognized Env residues of each antibody to define epitope categories. We measured antibody neutralization to confirm neutralization breadth of greater than 25% on a diverse cross-clade panel of 208 HIV-1 strains. We correlated structural features of epitope and paratope with functional characteristics of neutralization breadth and potency. We also analyzed paratopes for ease of lineage re-elicitation and epitopes for ease of mimicry. Overall, our structural survey of HIV-1-neutralizing antibodies targeting the prefusion-closed Env trimer delineates categories, identifies underlying relationships between structural properties of epitope and paratope and functional properties of neutralization, and suggests favorable candidates for vaccine design.
RESULTS
Broadly neutralizing antibodies that recognize the prefusion-closed Env trimer segregate into six categories
The PDB contains over 200 coordinate sets for human antibodies in complex with various portions of HIV-1 Env, such as fully glycosylated Env trimers, core gp120, scaffolded domains, representative N-linked glycans, and peptide fragments (Dataset S1). We chose to limit our analysis to structures with resolutions sufficient to define chemistry of interaction: for X-ray crystal structures, we used a resolution limit of 3.9 Å, and for cryo-electron microscopy structures, we used a resolution limit of 4.5 Å. We note that in some cases, multiple complex structures of antibody with epitope were available, some with peptides or glycans, some with gp120 domains, and some with Env trimer; depending on the type of analysis, some complex structures did not provide sufficient information to define the desired characteristic and were excluded from these calculations; such exclusions are noted with footnotes to the figure legends where the characteristics are described. Also, we chose to limit our analysis to antibodies that neutralized at 50 μg/ml at least 25% of a cross-clade panel of at least 100 HIV-1 strains. Finally, to alleviate issues of conformation, we limited our analysis to only those antibodies that recognize the prefusion-closed conformation of Env as this allowed us to focus on a single structural entity: the Env trimer in its prefusion-closed state.
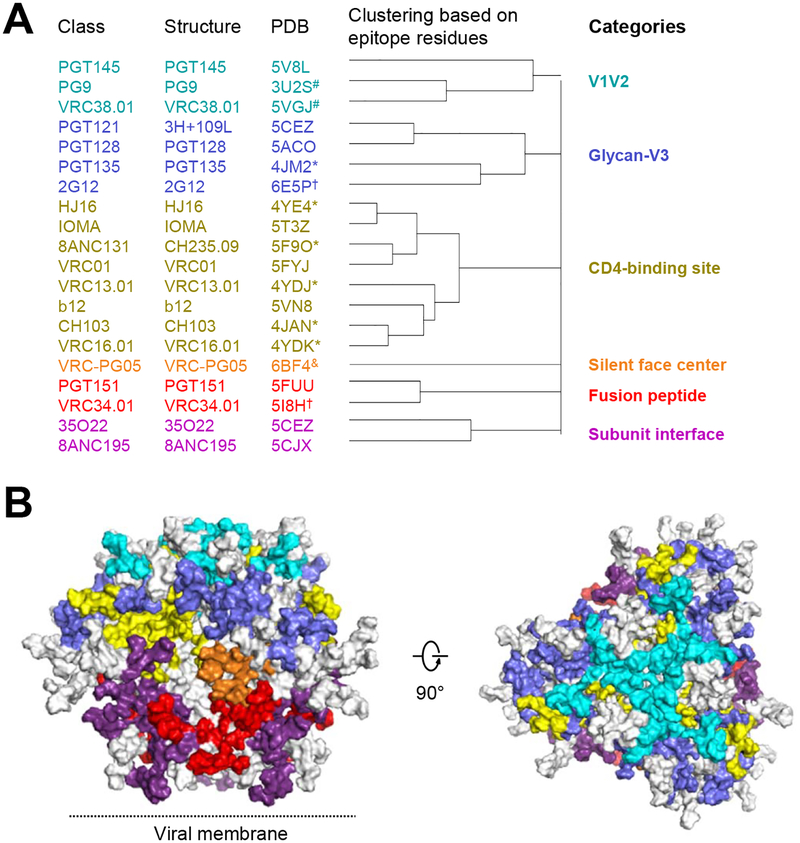
We next used B cell ontogeny and structural mode of recognition to delineate antibody classes (Kwong and Mascola, 2012), and used the name of the first identified antibody in each antibody class as the name of the class (Table 1). While some antibody classes have only been found in a single donor to date (e.g., CH103, VRC38), other classes have been found in more than one donor (e.g. VRC01, 8ANC131). The largest class involved the VRC01 class (Zhou et al., 2013), with 53 PDBs describing 33 antibodies. Antibodies recognizing glycan-V3 derived primarily from classes that had only a single known clonal lineage, although PGT121 and BG18 lineages do share the same D3–3 gene and mode of recognition (Barnes et al., 2018; Mouquet et al., 2012). For antibodies recognizing the spike apex, structural information showed similar parallel strand recognition for PG9 and CH03 indicative of membership in the same class (Gorman et al., 2016); however structures of PGT145 with Env trimer (Lee et al., 2017; Liu et al., 2017) indicated loop insertion into a trimeric hole at the spike apex, and hence a distinct mode of recognition indicative of a separate class for PGT145.
Table 1. Structural details for 20 classes of antibodies in complex with HIV-1 Env.
See also Dataset S1.
| Category | Class | Antibody with most informative structure | Most informative PDB | Experiment method | Resolution (Å) | Env component | Epitope Residues |
|---|---|---|---|---|---|---|---|
| V1V2 | PGT145 | PGT145 | 5V8L | Cryo-EM | 4.3 | Trimer | K121, T123, P124, V127, M161, T162, R166, D167, K169, I309, Q315, NAG608, T123, V127, N160, M161, T162, R166, D167, K168, K169, NAG606, NAG607, BMA608, K121, T123, P124, V127, N160, M161, T162, R166, D167, K168, K169, glycan 160 |
| PG9 | PG9 | 3U2S | X-ray | 1.8 | Scaffolded V1V2 | C157, S158, N160, T162, T163, K166, D167, R168, K169, Q170, K171, V172, N173, glycan 160, glycan 173 | |
| VRC38.01 | VRC38.01 | 5VGJ | X-ray | 3.5 | Scaffolded V1V2 region | H130, T132, N133, V134, T135, I136, N156, C157, S158, F159, N160, K168, K171, E172, Y173, glycan 133, glycan 156, glycan 160 | |
| Glycan-V3 | PGT121 | 3H+109L | 5CEZ | X-ray | 3.0 | Trimer | V134, T135, N136, A137, T139, D140, D321, I322, I323, G324, D325, I326, R327, Q328, H330, T415, P417, glycan 156, glycan 301, glycan 332 |
| PGT128 | PGT128 | 5ACO | Cryo-EM | 4.4 | Trimer | V134, T135, N136, N137, I138, L175, T297, P299, N301, D321, I322, I323, G324, D325, I326, R327, V442, R444, C445, glycan 156, glycan 301, glycan 332 | |
| PGT135 | PGT135 | 4JM2 | X-ray | 3.1 | gp120 | P299, I323, G324, Q328, A329, H330, R335, M373, Y384, N386, Q389, T408, E409, T415, P417, C418, R419, K421, glycan 332, glycan 392, glycan 386 | |
| 2G12 | 2G12 | 6E5P | Cryo-EM | 8.5 | Trimer | N411, Glycans 295, 332, 339, and 392 | |
| CD4-binding site | HJ16 | HJ16 | 4YE4 | X-ray | 2.7 | gp120 | T278, N279, N280, A281, T283, V360, Q362, P363, S364, S365, G366, G367, I371, T455, R456, D457, G458, G459, N460, N461, N462, K463, I469, R471, P472, G473, G474, D476, glycan 276 |
| IOMA | IOMA | 5T3Z | X-ray | 3.5 | Trimer | K97, E102, T198, T257, T278, N279, N280, A281, K282, R360, S365, G366, G367, D368, E370, V371, M426, W427, Q428, I430, T455, R456, D457, G458, G459, S460, T461, N462, S463, E466, T467, R469, G472, G473, D474, M475, R476, glycan 197, glycan 276, glycan 363, K65, H66, R308, glycan 262, glycan 301 | |
| 8ANC131 | CH235.09 | 5F9O | X-ray | 1.9 | gp120 | W96, K97, E275, N276, L277, T278, N279, N280, A281, K282, T283, S365, G366, G367, D368, I371, M426, G429, G431, Q432, T455, R456, D457, G458, G459, A460, N461, R469, P470, G471, G472, N474, D477, R480, glycan 276 | |
| VRC01 | VRC01 | 5FYJ | X-ray | 3.1 | Trimer | W96, K97, V198, E275, N276, T278, D279, N280, A281, K282, T283, I360, S365, G366, G367, D368, I371, R425, Q428, R429, V455, R456, D457, G458, C459, K460, S461, T465, E466, T467, R469, A471, G472, G473, D474, R476, glycan 197, glycan 276, glycan 386, Y 61, S 62, E 64, glycan 301 | |
| VRC13.01 | VRC13.01 | 4YDJ | X-ray | 2.3 | gp120 | W96, K97, E275, N279, N280, A281, K282, T283, R327, P364, S365, G366, G367, D368, L369, I371, T372, M373, Y384, K419, I420, K421, I423, T455, G458, G459, R469, P470, G471, G472, G473, N474, D477, R480, glycan 386 | |
| b12 | b12 | 5VN8 | Cryo-EM | 3.6 | Trimer | K178, L179, V181, V182, P183, L184, E185, N185, K185, N185, N187, I188, T189, R192, I194, N195, N197, T198, S199, T257, N280, A281, H363, S364, S365, G366, G367, D368, P369, E370, I371, V372, T373, Y384, N386, Q417, C418, R419, K421, N425, M426, W427, Q428, G429, V430, G431, T455, R456, D457, R460, R469, G471, G472, G473, N474, M475, glycan 197, glycan 362, glycan 386 | |
| CH103 | CH103 | 4JAN | X-ray | 3.2 | gp120 | S256, D279, N280, A281, E362, H364, S365, G366, G367, D368, L369, E370, I371, T455, R456, D457, G458, G459, N460, D461, D462, N463, R469, P470, G471, G472 | |
| VRC16.01 | VRC16.01 | 4YDK | X-ray | 2.1 | gp120 | Q105, L122, T123, G124, G198, S199, T257, E275, N280, A281, K282, T283, P364, S365, G366, G367, D368, L369, E370, I371, H375, N425, M426, W427, Q428, G429, G431, Q432, T455, R456, D457, G458, G459, A460, T463, T467, R469, P470, G471, G472, G473, N474, D477 | |
| Silent face center | VRC-PG05 | VRC-PG05 | 6BF4 | X-ray | 2.4 | gp120 | N262, K290, S291, V292, E293, D334, E337, L446, S447, N448, glycan 262, glycan 295, glycan 448 |
| Fusion peptide | PGT151 | PGT151 | 5FUU | Cryo-EM | 4.2 | Trimer | A58, P79, N80, Q82, E83, V84, V85, A224, T244, V245, Q246, glycan 241, glycan 262, glycan 295, glycan 448, A512, V513, G514, I515, G516, A517, V518, F519, G521, F522, R542, L543, S546, G547, V549, Q550, Q551, Q552, N553, N554, L592, N637, Y638, S640, E641, Y643, T644, E647, glycan 611, glycan 637 |
| VRC34 | VRC34.01 | 5I8H | X-ray | 4.3 | Trimer | N80, Q82, E83, I84, H85, E87, K229, D230, K231, S241, glycan 88, A512, V513, G514, I515, G516, A517, V518, F519, L520, G521, E648 | |
| Subunit interface | 35O22 | 35O22 | 5CEZ | X-ray | 3.0 | Trimer | G527, S528, T529, A532, R617, N618, S620, E621, D624, N625, M626, T627, L629, Q630, K633, glycan 618, E87, N88, T90, E91, E92, P238, P240, glycan 234, glycan 88 |
| 8ANC195 | 8ANC195 | 5CJX | X-ray | 3.6 | Trimer | S612, S613, W614, S615, N616, R617, M626, L629, Q630, D632, K633, E634, S636, N637, Y638, glycan 637, V44, W45, K46, D47, V89, T90, E91, E92, F93, N94, K97, T236, G237, P238, E275, N276, I277, T278, K351, H352, F353, R456, K487, E492, glycan 234, glycan 276 |
We selected a structural representative antibody for each class based on the most informative structure (for example, choosing an antibody-Env trimer complex over the same antibody with a deglycosylated core gp120) (Figure 1A, Table 1). Based on the 20 representative antibody-antigen complex structures, we defined epitope residues on the prefusion-closed Env trimer by buried surface area calculations (see methods). For 2G12, the most complete antibody-Env complexes deposited in the PDB defined interactions with only a few N-linked glycans (Calarese et al., 2003); therefore, we determined a Cryo-EM structure of 2G12 in complex with BG505 DS-SOSIP, defining interactions with glycans 295, 332, 339, and 392 (Figure S1, Table 2). Next, we carried out hierarchical clustering of epitope residues, which segregated the 20 antibody classes into 6 categories (Figure 1A,B and Table S1). Three of the categories had been previously described: V1V2, glycan-V3 and CD4-binding site, and each of these categories encompassed at least three different antibody classes. Three additional categories were identified, including “silent face center” (with antibody VRC-PG05), “fusion peptide” (with antibodies PGT151 and VRC34), and “subunit interface” (with antibodies 35O22 and 8ANC195). Beyond shared epitope residues, each of the categories did appear to have specific characteristics. For example, single-molecule fluorescence energy transfer (smFRET) measurements define three prefusion states (states 1–3) of the Env trimer and found broadly neutralizing antibodies to preferentially recognize state 1 (Munro et al., 2014); only two antibodies, VRC34.01 (Kong et al., 2016) and PGT151 (personal communications, Walther Mothes), have been found to shift the smFRET population to state 2, and both were assigned to the same epitope category. Thus, our structure-based epitope analysis defined six categories of broadly neutralizing antibodies, and antibodies in each category generally shared similar characteristics.
Figure 1. 20 classes of broadly neutralizing antibodies recognize the prefusion-closed Env trimer and segregate into 6 categories based on the Env residues with which they interact.
(A) All HIV-1 Env-antibody complexes structures in the PDB (Table 1) were assigned to classes (leftmost column, listed by the name of first reported antibody of the class) based on similarities in B cell ontogeny and mode of recognition, with a representative structure and PDB for the class (2nd and 3rd columns from left), which were chosen based on resolution and degree to which Env in the structure resembled prefusion-closed trimer; “*” indicate structures determined in deglycosylated gp120-core context; “&” indicates structures determined in partially glycosylated gp120-core context; “#” indicates structures determined with V1/V2 scaffold; “†”indicates a structure with high resolution peptide- or glycan-antibody complex but lower resolution Env trimer-antibody complex structure; “@” indicates glycan N295, N332, and N392 used for 2G12 epitope (see methods). (B) Prefusion-closed Env trimer with molecular surface colored by categories defined in (A). Epitope residues shared by antibodies in separate classes were colored according to surface area and requirements for antibody binding. For example, glycan N276 has interactions with antibody 8ANC195 of the subunit interface category and with multiple antibodies of the CD4-binding site category; however, because glycan N276 is required for 8ANC195, and generally only accommodated by antibodies that target the CD4-binding site, we colored glycan N276 to be part of the subunit interface category. Left image is shown with viral membrane at bottom; right image is rotated 90° to look down on the trimer apex. See also Figures S1 and S5, Tables S1 and S3, and Dataset S1.
Table 2.
Cryo-EM data collection and map refinement statistics.
| 2G12-BG505 DS-SOSIP-VRC03 | |
|---|---|
| EMD-8981; PDB 6E5P | |
| Data collection and processing | |
| Magnification | 105000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 63.84 |
| Defocus range (μm) | −1.2 – −2.2 |
| Pixel size (Å) | 1.1 |
| Symmetry imposed | C3 |
| Initial particle images (no.) | 463,860 |
| Final particle images (no.) | 5245 |
| Map resolution (Å) | 8.5@ (8.8#) |
| FSC threshold | 0.143 |
as reported by crypoSparc
as reported by RELION
Neutralization characteristics for identified antibody classes
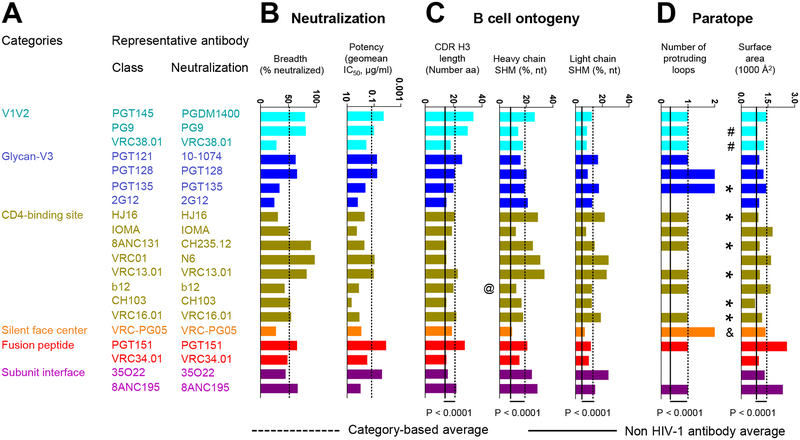
As the most important functional property of the uniquely identified antibody classes is their neutralization of diverse HIV-1 strains, we felt it important to accurately measure this feature in a way that allowed comparison between different antibodies. We thus measured both neutralization breadth and potency for the representative member of each class on a diverse cross-clade panel of 208 HIV-1 strains (Seaman et al., 2010) (Figure 2A,B, Dataset S2). For functional representative, we chose the antibody-class member with highest neutralization breadth. For the PGT121 class, we chose antibody 10–1074 (Mouquet et al., 2012); for the VRC01 class, we chose antibody N6 (Huang et al., 2016); and for the 8ANC131 class we chose antibody CH235.12 (Bonsignori et al., 2016). Notably, the newly measured median neutralization IC50 generally differed by less than 2 fold from prior reported values (Alam et al., 2017; Bonsignori et al., 2017; Bonsignori et al., 2016; Cale et al., 2017; Chuang et al., 2013; Doria-Rose et al., 2012; Huang et al., 2016; Huang et al., 2012; Julg et al., 2017; Kong et al., 2016; Kwon et al., 2015; Liao et al., 2013; McLellan et al., 2011; Pancera et al., 2017; Pancera et al., 2013; Rudicell et al., 2014; Williams et al., 2017; Wu et al., 2010; Xu et al., 2017; Zhou et al., 2015; Zhou et al., 2018), with all 20 classes averaging 56.7% breadth with an average geometric mean IC50 of 0.27 μg/ml.
Figure 2. Neutralization, B cell ontogeny, and paratope characteristics for 20 classes of broadly neutralizing antibodies.
(A) The broadest neutralizing antibody from each class was chosen as representative. (B) Neutralization properties. Bar graphs show breadth and potency of each representative antibody on the 208-isolate panel, with dotted line showing averages. (C) Features of B cell ontogeny. CDR H3 lengths are defined based on Kabat nomenclature. @ indicates that antibody b12 is derived from a phage library. (D) Paratope properties. “*” indicate structures determined in deglycosylated gp120-core context; “&” indicates structures determined in partially glycosylated gp120-core context; “#” indicates structures determined with V1/V2 scaffold. Structures denoted with “*”, “&”, or “#” were included in calculations of the category averages for protruding loop numbers, but not for paratope surface area. P-values were determined using two-tailed Mann-Whitney test. See also Figure S1, Table S2 and Datasets S2–S4.
B cell ontogenies, paratopes, and epitopes of antibodies recognizing the prefusion-closed Env trimer
B cell ontogenies involve the recombination events that lead to creation of the ancestor B cell for each lineage, as well as SHM that leads to mature antibody clones. To estimate elicitation likelihoods related to recombination and SHM, we extracted CDR H3 length and V-gene SHM (Figure 2C). We observed that the average CDR H3 length and heavy chain SHM of the 20 HIV-1 broadly neutralizing antibodies were higher than those of next generation sequencing (NGS) reads from three healthy donors (P<0.0001) (Bonsignori et al., 2016).
We also extracted critical features of the paratope (Figure 2D). We selected a measure related to recognition mode (number of protruding antibody loops) and a second measure related to recognition extent (the surface area of the interacting paratope). Fifteen of the 20 antibodies utilized protruding loops (predominantly CDR H3), although this feature was less common in non-HIV-1 antibodies (as defined on a set of 81 non-HIV-1 antibody-antigen structures, see Datasets S3 and S4). The paratope surface areas were almost twice as high as observed with non-HIV-1 antibodies (P<0.0001), and this may relate to glycan interactions, as antibodies in complex with deglycosylated gp120 cores generally showed surface areas similar to non-HIV antibodies.
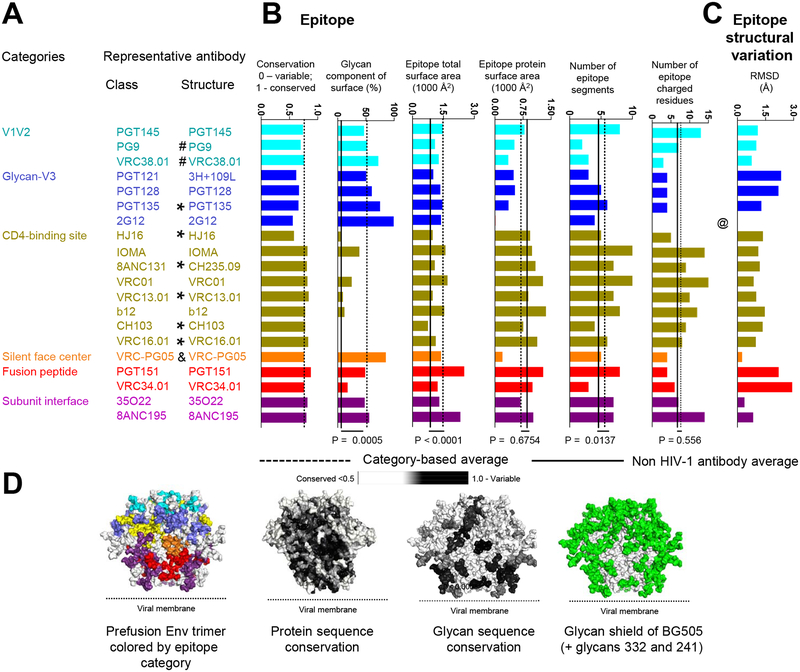
For epitope, we measured several features including sequence conservation, extent of glycan contribution, number of independent sequence segments in each epitope, and the structural variability of the epitope (Figure 3A–D). For sequence conservation, epitopes of broadly neutralizing antibodies (average conservation of 0.75) were similar to the average conservation of the closed trimer (0.69). Further analyses of entropy score at individual residue level revealed that most of the entropy score distributions were right skewed, but distribution for 3H+109L and PGT128 were bimodal (Figure S3). The standard deviation of the entropy scores within each epitope ranged from 0.13 (for 2G12 epitope) to 0.34 (for PGT128 epitope). For glycan, we observed the average contribution to epitope surface area (45%) was similar to the surface area contribution of MAN5 glycans to the overall Env trimer surface area (53%). Also, we observed that the average epitope glycan composition (P=0.0005), total epitope surface area (P<0.0001), and number of epitope segments (P=0.0137) of the 20 HIV-1 broadly neutralizing antibodies were higher than those of non-HIV-1 antibodies (Datasets S3 and S4; for glycan composition specifically, the comparison was done with a non-HIV-1 glycoprotein set with 16 antibody antigen complexes, see Methods and Dataset S5). Features common in HIV-1 neutralizing antibodies, but generally rare, may provide insight into requirements for recognition: in this case identifying features of broadly neutralizing antibodies selected to overcome Env sequence variability and glycosylation.
Figure 3. Epitope features for 20 classes of broadly neutralizing antibodies.
(A) Categories, classes, and representative structures. “*” indicate structures determined in deglycosylated gp120-core context; “&” indicates structures determined in partially glycosylated gp120-core context; “#” indicates structures determined with V1/V2 scaffold; @ indicates that the epitope structural variation of antibody 2G12 was not calculated due to the lack of protein epitope. Structures denoted with “*”, “&”, or “#” were not included in calculations of the category averages for number of epitope segments and epitope total surface. Structures denoted with “*” were not included in calculations of the category average for epitope glycan composition. (B) Epitope properties. Bar graphs show conservation and glycan-interactive surface for each representative antibody, with dotted line showing averages. “*” indicate structures determined in gp120-core context, where glycan surface areas may not reflect trimer interactions. (C) Number of epitope segments, shown for segments with >25 Å2 or > 100 Å2 surface area. VRC34 has low number epitope segments and low glycan context (black arrow), and this may explain why fusion peptide immunization are yielding promising results from epitope-based vaccine design. (D) Observed variation in epitope structure, as calculated from RMSD of recognized amino acid residues, computed pairwise among class members of each category and unliganded trimer. (E) Prefusion-closed Env trimer colored by epitope and by various epitope features. P-values were determined using two-tailed Mann-Whitney test. See also Figures S1–S4 and Datasets S3–S5.
Underlying relationships between neutralization, paratope and epitope
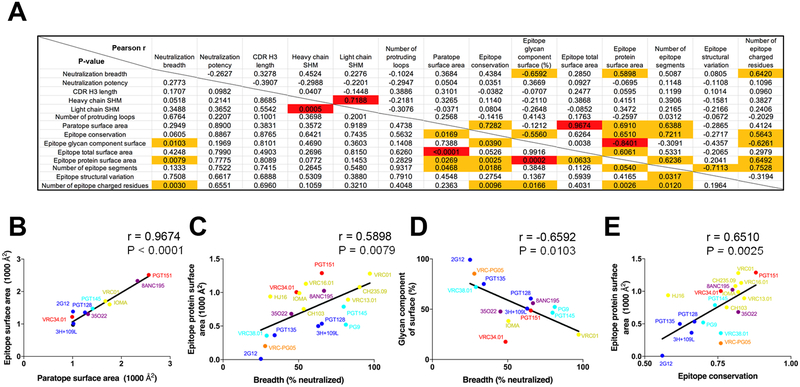
To provide insight into other features specific to these HIV-1 neutralizing antibodies, we correlated neutralization with epitope and paratope features to reveal underlying relationships (Figure 4A). Although a number of features correlated with p-values of less than 0.05, when corrected for multiple comparison, few correlations were significant. Significant correlations included the expected positive correlation between surface area of paratope and surface area of epitope (P < 0.0001, r = 0.9674) (Figure 4B) and a positive correlation between heavy chain and light chain SHM (P = 0.0005, r = 0.7188). Other correlations did not achieve statistical significance, but were potentially revealing. For example, we observed antibody neutralization breadth to correlate positively with buried protein-epitope surface area (P = 0.0079, r = 0.5898) (Figure 4C) and negatively with degree of epitope glycan (P = 0.0103, r = −0.6592) (Figure 4D), indicating that while antibody recognition can include glycan, this nevertheless reduced breadth. Meanwhile sequence conservation correlated positively with the size of the protein epitope (P = 0.0025, r = 0.6510) (Figure 4E) and with the size of the paratope (P = 0.0169, r = 0.7282), suggesting that one way to overcome variation is by reducing the area of recognized surface. We also noticed that the broadest HIV-1-neutralizing antibodies had high levels of SHM and the most potent had unusual CDR H3s (Figure 2). Overall, these observations suggested both the frequency and the extent that neutralizing antibodies targeting the prefusion-closed Env trimer utilize protruding loops, unusual recombination, and extensive SHM to overcome immune evasion mechanism of extraordinary glycosylation and high sequence variability.
Figure 4. Correlations of neutralization and features of epitope and paratope reveal underlying relationships.
Correlation matrices are shown for properties of neutralizing antibodies that recognize the prefusion closed Env trimer. (A) Pearson correlations (r) are provided in the upper right; P-values (not adjusted for multiple comparisons) are provided in the lower left. Correlations with P-value of less than 0.05 were highlighted with orange fill, and correlations with P-value of less than 0.05 after Bonferroni correction were highlighted with red fill. (B) Correlation between paratope surface area and epitope surface area. (C) Correlation between Neutralization breadth and epitope protein surface area. (D) Correlation between neutralizing breadth and glycan component of surface. (E) Correlation between epitope conservation and epitope protein surface area. Note that labels in (B-E) correspond to antibodies in structures analyzed and that not all antibodies were included in the correlation analysis, as some of the antibody structures did not provide information sufficient for the analysis. See Methods and Dataset S2.
Antibody-based vaccine design
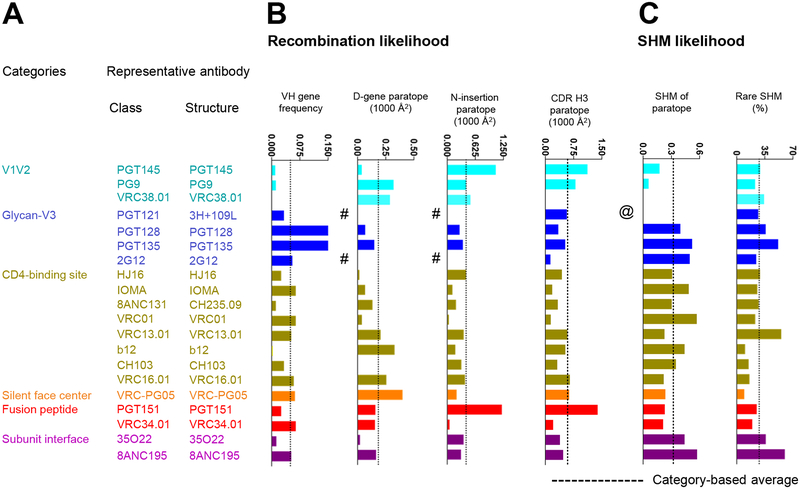
We analyzed antibodies to identify which B cell ontogenies might be most easily re-elicited by antibody-lineage based vaccine design (Kwong and Mascola, 2018). Such design is premised on the re-elicitation of antibodies with similar ontogenies, through priming of ancestor B cells and induction of their maturation (Crowe, 2016; Gorman et al., 2016; Jardine et al., 2013). Two factors affect re-elicitation: the likelihood that a recombination event produces an appropriate unmutated common ancestor (UCA) and the likelihood that this UCA will mature through processes of SHM to achieve similar functional properties of neutralization. The former is affected by prevalence of appropriate V-genes and by the contribution to the paratope of CDR H3 features such as D-gene and N-nucleotide addition, which we calculated for each of the 20 classes of neutralizing antibodies (Figure 5). We also calculated features of SHM, such as the percent of the V gene-contributed paratope surface altered by SHM or the prevalence of rare mutations (Sheng et al., 2017). Although some of the broadest and most potent antibodies had features that lowered the probability of their re-elicitation, there were antibody classes with less rare SHM and CDR H3 characteristics such as IOMA, as well as antibodies like PG9 with extensive D-gene contributions to antigen recognition that might be re-elicited with greater ease.
Figure 5. Factors affecting re-elicitation.
(A) Categories, classes, and representative structures. (B) Recombination properties. Bar graphs show average VH gene frequency and paratope properties of D gene, N addition and CDR H3 for each representative antibody. Average VH gene frequencies report from healthy donors in PRJNA304166. D gene and N additions occurring at the junctions are from IMGT V-quest (http://www.imgt.org/IMGT_vquest/vquest). # indicates that antibody missing nucleotide sequences at junction. (C) SHM likelihood. Bar graphs show the paratope contributed from SHM (1st amino acid to the 2nd conserved Cys) and rare SHM. @ indicates that antibody missing nucleotide sequence. See also Table S2.
We also analyzed epitopes for ease of structural mimicry, as related to the development of immunogens for epitope-based vaccine design (Kwong and Mascola, 2018). We observed antibody VRC34.01 to have few epitope segments, low epitope-glycan content, and high epitope-conformational variability. Few epitope segments enable easy creation of epitope mimics; low epitope-glycan content reduces barriers to immune recognition (glycan masking is a strategy used broadly by pathogens (Park and Williamson, 2015)); and high epitope-conformational variability permits epitope recognition in multiple structural contexts (Tainer et al., 1984). Thus, despite VRC34-class antibodies only being identified in a single donor and only with 50% breadth, its characteristics suggest that vaccine design based on the epitope of antibody VRC34.01 could be especially promising.
DISCUSSION
Structural characterization of the recognition by broadly neutralizing antibodies of all major exposed surfaces on the prefusion-closed HIV-1 Env trimer enables both comprehensive and aggregate-level analyses: Relative to paratope, we could address which characteristics allow for broad recognition of the glycosylated and sequence-variable Env trimer, and relative to the Env trimer surface, we could address which features were recognized and which were avoided. For paratope, our results indicated protruding loops and extensive SHM to dominate recognition (extending observations previously reported by Klein and colleagues (Klein et al., 2013b) utilizing a less complete set of HIV-1-directed antibodies), although antibodies with more common features were also observed. For epitope, our results indicate both glycan and sequence variation to be recognized by antibody, which may relate to the extent that these features dominate the surface of the prefusion-closed Env trimer. As additional broadly neutralizing antibodies emerge, the epitope-paratope knowledge base can be further expanded using the analysis framework outlined in this study. We note that a critical aspect of the knowledge base is its grounding in standardized functional assessment – in our case comprising a cross-clade panel of 208 Env-pseudovirus strains of HIV-1.
While our focus on the prefusion-closed trimer allowed us to relate properties of recognized epitopes to average properties of the Env trimer in a specific conformation, this focus also led to the omission from our analysis of an important class of broadly neutralizing antibodies, those that target the membrane-proximal external region (MPER) (Figure S5 and Table S3). MPER antibodies generally show low levels of recognition of the native Env trimer (Chakrabarti et al., 2011) and have thus been proposed to recognize Env trimer in a different conformation (Ruprecht et al., 2011). In addition to differences in conformation of Env recognized, MPER antibodies also target a region of Env with substantially lower glycan and sequence variability. Thus, the immune-evasion mechanisms that shield the MPER appear to differ from those shielding the rest of the prefusion-closed Env trimer, and the antibody mechanisms that allow for recognition of the MPER likely differ as well from those that allow recognition of the closed Env trimer.
In terms of the most promising epitopes on the Env trimer, the features described here allow for the selection of favorable candidates for vaccine design. For antibody lineage-based design, the impact of immunological frequencies of recombination and SHM on antibody elicitation is only now being delineated (Abbott et al., 2018; Wu et al., 2015; Zhou et al., 2015). For epitope-based vaccine design, however, the criteria for elicitation related to particular epitope features has been more extensively studied, and as noted above the epitope of antibody VRC34.01 may be especially promising. Indeed, we recently achieved the elicitation of antibodies in mice, guinea pigs and rhesus macaques that neutralized diverse HIV-1 Env isolates through immunogen design based on the epitope of VRC34.01 (Xu et al., 2018). Thus, in addition to revealing features of paratope and epitope that allow for immune recognition of the prefusion closed spike, the structural survey also provides insight into which antibody templates are most suitable for vaccine design.
ACCESSION NUMBERS
Cryo-EM electron density map of 2G12 in complex with BG505 DS-SOSIP and VRC03 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession number EMD-8981, and the fitted coordinates were deposited in the Protein Data Bank (PDB) under accession code 6E5P.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter D. Kwong (pdkwong@nih.gov)
METHOD DETAILS
Structural Dataset and Selection of Structural Representative for Each of the 20 Unique HIV-1 Antibody Classes.
All the antibody-antigen complex structures were collected from Protein Data Bank (PDB) (Berman et al., 2000) using the key words “HIV-1 antibody” on 28 February 2018. Structures that contain antibody alone were excluded. Non-human and synthetic antibodies were also excluded. The final comprehensive list of current deposited HIV-1 human antibody-antigen complex structures is provided in Dataset S1. Only the antibodies that targeted the HIV-1 Env trimer and neutralized Tier-2 HIV-1 isolates were considered. To focus on broadly neutralizing HIV-1 antibodies, a neutralizing breadth cutoff of >25% was also used. Based on ontogenies in separate donors, we defined 20 unique classes that recognize the prefusion-closed Env trimer (Figure 1A). For each antibody class, we selected the most informative structure based on the following criteria: 1) Resolution cutoff for X-ray crystal structures < 3.9 Å and < 4.5 Å for cryo-EM structures. 2) The complex structure of the trimer with highest resolution was selected, followed by glycosylated monomer, by deglycosylated monomer, or by scaffolded domain with highest resolution. We note that epitope properties were calculated based on only one antigen-antibody complex for each of the 20 antibody classes, so some of the observed contacts (or their absence) could be strain specific. This was especially true for glycan contacts. For example, VRC34.01 was solved in complex with the envelope trimer from strain BG505 where glycan 241 is missing, but this antibody might make contact with this glycan in another HIV-1 strain. The final PDB IDs we selected included 5V8L (Lee et al., 2017) (for PGT145), 3U2S (McLellan et al., 2011) (for PG9), 5VGJ (Cale et al., 2017) (for VRC38.01), 5CEZ (Garces et al., 2015) (for 3H+109L and 35O22), 5ACO (Lee et al., 2015) (for PGT128), 4JM2 (Kong et al., 2013) (for PGT135), 4YE4 (for HJ16), 5T3Z (Gristick et al., 2016) (for IOMA), 5F9O (Bonsignori et al., 2016) (for CH235.09), 5FYJ (Stewart-Jones et al., 2016) (for VRC01), 4YDJ (Zhou et al., 2015) (for VRC13.01), 5VN8 (Ozorowski et al., 2017) (for b12), 4JAN (Liao et al., 2013) (for CH103), (Zhou et al., 2015) 4YDK (for VRC16.01), 6BF4 (Zhou et al., 2018) (for VRC-PG05), 5FUU (Lee et al., 2016) (for PGT151), 5I8H (Kong et al., 2016) (for VRC34.01), and 5CJX (Scharf et al., 2015) (for 8ANC195). For 2G12, we solved a cryo-EM structure of 2G12 in complex with BG505 DS-SOSIP for this purpose (see “Cryo-EM structure of 2G12 in complex with HIV-1 Env trimer” for details).
Cryo-EM structure of 2G12 in complex with HIV-1 Env trimer.
HIV-1 BG505 DS-SOSIP Env (Kwon et al., 2015) at a final concentration of 0.5 mg/mL was incubated with 3–fold molar excess each of VRC03 Fab and 2G12 Fab’2 for 4 hours. To prevent aggregation during vitrification, the sample was incubated in 0.085 mM dodecyl-maltoside (DDM), followed by vitrification using a semi-automated Spotiton V1.0 robot (Dandey et al., 2018; Razinkov et al., 2016) The grids used were nanowire self-blotting grids with a lacey carbon support substrate (Wei et al., 2018). Sample was dispensed onto the nanowire grids using a picoliter piezo dispensing head. A total of ~5 nL sample, dispensed as 50 pL droplets, was applied in a stripe across each grid, followed by a pause of a few milliseconds, before the grid was plunged into liquid ethane.
Data was acquired using the Leginon system (Suloway et al., 2005) installed on a Titan Krios electron microscope operating at 300kV, fitted with Gatan K2 Summit direct detection device and an energy filter. The dose was fractionated over 50 raw frames and collected over a 10-s exposure time. Individual frames were aligned and dose-weighted (Zheng et al., 2017). CTF was estimated using the GCTF package (Zhang, 2016). Particles were picked using DoG Picker (Voss et al., 2009) within the Appion pipeline (Lander et al., 2009). Particles were extracted from the micrographs using RELION (Scheres, 2012). 2D classification, ab initio reconstruction, heterogeneous refinement, and final map refinement were performed using cryoSparc (Punjani et al., 2017). Map sharpening was performed in RELION.
For fitting into the cryo-EM reconstructions we used complex of BG505 DS-SOSIP and VRC03 from PDB ID 6CUI (after removing vFP16.02 and PGT122 from the coordinates) and the crystal structure of 2G12 Fab’2 bound to two glycans (PDB ID 1OP5) (Calarese et al., 2003). Fits of the HIV-1 Env trimer, VRC03 Fab and 2G12 Fab’2 coordinates to the cryo-EM reconstructed maps were performed using UCSF Chimera (Pettersen et al., 2004). The two glycans bound to 2G12 in the docked crystal structure overlay well with glycans 295 and 392 of the docked trimer. The electron density showed contacts of 2G12 at its domain swap interface with two additional glycans, 332 and 339. The positions of the glycans were optimized to the cryo-EM electron density using rigid body fitting in Coot (Emsley and Cowtan, 2004), followed by geometry minimization in Phenix (Adams et al., 2010). Figures were generated in UCSF Chimera and Pymol (www.pymol.org). Map-fitting cross correlations were calculated using Fit-in-Map feature in UCSF Chimera. For depositing co-ordinates to the RCSB database the side chains of all amino acid residues were truncated down to their Cβ atoms.
Epitope and Paratope Buried Surface Area Calculations.
The buried surface area between antibody and antigen was calculated using program NACCESS (Hubbard and Thornton, 1993). The epitope residues and paratope residues for each antibody were defined as residues with non-zero buried surface area. For 2G12, the epitope residues were defined as residue 411 and glycans N295, N332, N339, and N392, based on a cryo-EM reconstruction (Figure S1).
Structural Definition of HIV-1 Env Broadly Neutralizing Antibody Categories.
The structural representative of the 20 unique antibodies were clustered using hierarchical clustering, with the distance between each of the antibody is defined as , where no is the number of overlapping epitope residues, and Ns is the number of epitope residues for the antibody with the least number of epitope residues of the two.
Display of Epitope Residues on Prefusion-Closed HIV-1 Env.
The BG505 SOSIP trimer structure (PDB:5FYL) was used as the template with each glycan (with the addition of glycans 241 and 332) modeled as MAN5. Amino acid and glycan epitope residues were colored based on the structurally defined antibody category. Epitope residues with more than 5 Å C-alpha deviation from PDB:5FYL were excluded. For residue positions shared in epitopes from multiple antibody categories, the color of the antibody category with the antibody that has the largest epitope surface area was used.
Antibody Expression and Purification.
The 20 representative antibody heavy and light chain expression constructs were synthesized (Gene Universal Inc., Newark, DE) and cloned into pVRC8400 expressing vector. For antibody protein expression, 1.5 ml of Turbo293 transfection reagent (Speed BioSystems) were mixed into 25 ml Opti-MEM medium (Life Technology) and incubated for 5 min at room temperature. 500 μg of plasmid DNAs (250 heavy chain and 250 μg of light chain) were mixed into 25 ml of Opti-MEM medium in another tube. Then the diluted Turbo293 were added into Opti-MEM medium containing plasmid DNAs. Transfection reagent and DNA mixture was incubated for 15 min at room temperature, and added to 400 ml of Expi293 cells (Life Technology) at 2.5 million cells/ml. The transfected cells were cultured in shaker incubator at 120 rpm, 37 °C, 9% CO2 overnight. On the next day of transfection, 40 ml of AbBooster medium (ABI scientific) were added to each flask of transfected cells and the flasks were transferred to shaker incubators at 120 rpm, 33 °C, 9% CO2 for additional 5 days. At 6 days post transfection, antibodies in clarified supernatants were purified over 3 ml Protein A (GE Health Science) resin in columns. Antibody was eluted with IgG elution buffer (Pierce), immediately neutralized with one tenth volume of 1M Tris-HCL pH 8.0. The antibodies were then buffer exchanged in PBS by dialysis, adjusted concentration to 1.0 mg/ml and filtered (0.22 μm) for neutralization assays (Kwon et al., 2018) (45).
Virus Neutralization.
Single-round-of-replication Env pseudoviruses were prepared, titers were determined, and the pseudoviruses were used to infect TZM-bl target cells as described previously in an optimized and qualified automated 384-well format (Sarzotti-Kelsoe et al., 2014). Briefly, antibodies were serially diluted, a constant amount of pseudovirus added, and plates incubated for 60 minutes; followed by addition of TZMbl cells which express luciferase upon viral infection. The plates were incubated for 48 hours and then lysed, and luciferase activity measured. Percent neutralization was determined by the equation: (virus only)-(virus+antibody)/(virus only) multiplied by 100. Data are expressed as the antibody concentration required to achieve 50% neutralization (IC50) and calculated using a dose-response curve fit with a 5-parameter nonlinear function. We used a previously described panel (Cale et al., 2017; Georgiev et al., 2013; Wu et al., 2010) of 208 geographically and genetically diverse Env pseudoviruses representing the major subtypes and circulating recombinant forms. The IC50 values reported here are from the complete set of 208 viruses run at the VRC. In some cases, multiple runs were averaged. The values used here differed slightly, generally within 2 fold of previous median IC50, from previously reported values in references (Alam et al., 2017; Bonsignori et al., 2017; Bonsignori et al., 2016; Cale et al., 2017; Chuang et al., 2013; Doria-Rose et al., 2012; Huang et al., 2016; Huang et al., 2012; Julg et al., 2017; Kong et al., 2016; Kwon et al., 2015; Liao et al., 2013; McLellan et al., 2011; Pancera et al., 2017; Pancera et al., 2013; Rudicell et al., 2014; Williams et al., 2017; Wu et al., 2010; Xu et al., 2017; Zhou et al., 2015; Zhou et al., 2018). The differences have two sources: in earlier publications, the panels contained between 178 and 198 of the 208 viruses; and because the neutralization IC50 values are known to vary up to 3-fold between repeat assays, as documented in (Sarzotti-Kelsoe et al., 2014).
CDR H3 Length, Germline Gene Assignment, N-Insertion, and SHM Calculations.
Antibody CDR H3 length, germline gene assignment, N-insertion, and SHM Calculation was performed using the IMGT/HighV-QUEST webserver (http://www.imgt.org/HighV-QUEST/).
Number of Protruding Loops.
A CDR loop was considered as a protruding loop based on the following procedure. First, we defined the long axial vector of the Fab based on two points: (i) average coordinates of the Cα atoms for heavy chain C22 and light chain C23, and (ii) average coordinates of the Cα atoms for heavy chain G106 and light chain G101. Second, we defined a plane using the Cα atom of heavy chain C22 and the long axial vector defined previously as normal vector. Finally, if the maximum distance of a CDR loop (based on Cα atoms) to the defined plane was at least 5Å longer than the average of maximum distances for all six CDR loops, we consider it as a protruding loop. The definition of the CDR loops were calculated using the IMGT/HighV-QUEST webserver (http://www.imgt.org/HighV-QUEST/). Due to its unique VH domain-exchanged structure, visual inspection was performed to determine the number of protruding loop for 2G12.
Epitope Conservation Calculations.
The conservation of each HIV-1 Env residue was calculated using the entropy function of the R package bio3d (H.norm column), with the addition of glycan as residue type. The calculation was based on the filtered web alignment of the year 2016 from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/). The epitope conservation was defined as the average conservation for each residue, weighted by epitope surface area. The average Env surface conservation was defined as the average conservation for each Env residue, weighted by accessible surface area as determined by NACCESS (Hubbard and Thornton, 1993). Alignment gap and putative N-linked glycosylation sequon were treated as additional residue types.
Average Glycan Surface Calculations.
The glycan surface area of the HIV-1 Env prefusion-closed trimer was calculated applying a two-step procedure. First, we approximated the accessible surface area (ASA), using NACCESS program (Hubbard and Thornton, 1993) with the default radius of 1.4 Å, for all protein as well as glycan residues based on a molecular dynamics simulation with a 500-nanosecond timescale (Stewart-Jones et al., 2016). Second, we determined the percentage of the glycan surface area, by dividing the ASA of glycan residues by the sum of the ASAs of protein and glycan residues.
Number of Epitope Segment Calculations.
The number of epitope segment is defined as number of continuous stretches on epitope residues, allowing skipping of one to two residues, which in total have an epitope surface area of greater than 25Å2.
Epitope Structural Variation Calculations.
The epitope structural variation for each antibody class was determined by the averaged root-mean-square-deviation of recognized epitope amino acid residues computed pairwise among category members and the unliganded structure (PDB ID: 4ZMJ) (Kwon et al., 2015) using PyMOL software. Antibody 2G12 conformational variability was not calculated due to lack of amino acid epitope residues.
Non-HIV-1 Antibody Sequence Dataset and V-gene Frequency Estimation.
Heavy and light chain NGS samples (Sheng et al., 2017) from 454 pyrosequencing downloaded from Short Read Archive and processed to calculate average SHM (Table S3A – S3C) and HV germline frequency (Table S3D). The next-generation sequencing (NGS) data from healthy donor performed with 5’ primer-adapters should avoid bias of germline gene usage. All heavy chain reads shorter than 350 nucleotides were removed, and all lambda and kappa chain reads longer than 300 nucleotides were kept. Germline genes were assigned to all filtered reads using IgBLAST (Ye et al., 2013). After assigned all reads, an in-house python script applied to process IgBLAST output, and non-IG sequences were removed. The variation between alleles were ignored, and we treated those alleles as identical VH gene. The VH gene frequency was calculated by the number of reads of VH germline divided by the total good IG sequences.
SHM Rarity.
Rarity score of a somatic hypermutation was calculated as 1-frequency of the SHM observed in gene-specific substitution profiles (Sheng et al., 2017). SHMs with rarity score lower than 0.5% were counted as rare SHMs.
Non-HIV Antibody-Antigen Structural Dataset.
A non-redundant non-HIV-1 Env antibody/antigen structural dataset was obtained from SAbDab (Dunbar et al., 2014), using the “Non-redundant search” function (Antibody identity = 90%, Antigen identity = 99%, for protein antigens, Resolution = 3.9 Å). The list was further filtered down by keeping only the human antibodies, and removing all HIV antibodies, resulting a total of 81 antibody/antigen complexes (Dataset S2).
Non-HIV Glycoprotein Structural Dataset.
To understand the difference of epitope glycan composition between HIV-1 antibodies and non-HIV-1 glycoprotein, a non-HIV-1 glycoprotein structural dataset was assembled with 16 antibodies complexed with one of the following glycoproteins: Ebola virus glycoprotein, Herpes simplex viruses −2 gD, Human Metapneumovirus fusion protein, Hepatitis C virus envelope glycoprotein E2 core, dengue virus type 2 envelope glycoprotein, Human cytomegalovirus glycoprotein B, Respiratory syncytial F, Human cytomegalovirus pentamer, and influenza hemagglutinin (Dataset S5). To avoid the dataset over-represented by influenza hemagglutinin, only four structures (the number of structures for the next abundant antigen) were selected. Resolution cutoff for X-ray crystal structures < 3.9 Å and < 4.5 Å for cryo-EM structures.
QUANTIFICATION AND STATISTICAL ANALYSIS
Two tailed Mann-Whitney test was used to calculated the statistical difference of different properties between HIV-1 (N=20) and non-HIV-1 antibodies (N=81) (Figures 2 and 3). Pearson correlation for every pair of antibody properties listed in Figure 2 and 3 were calculated (N=19) and displayed in Figure 4. b12 class was not included in the correlation calculation as b12 was derived from phage display. For correlations involving total epitope surface, paratope surface, and number of epitope segment calculations, PG9, VRC38.01, PGT135, H16, CH235, VRC13.01, CH103, VRC16.01, and VRC-PG05 were excluded as their representative structures were determined with partial Env domain. For correlations involving epitope glycan composition, HJ16, CH235, VRC13.01, CH103, and VRC16.01 were excluded as their representative structures were determined with deglycosylated gp120 monomers. All statistical analyses were performed using GraphPad Prism 7.
Supplementary Material
Dataset S1. List of human HIV-1 antibody-Env complexes. Structures used in the analyses are highlighted in red. Related to Figure 1 and Table 1.
Dataset S2. Antibody neutralization evaluated on 208 HIV-1 isolates. Related to Figures 2 and 4.
Dataset S3. 81 non-HIV-1 antibody-antigen complexes obtained from SAbDab. Related to Figures 2 and 3.
Dataset S4. Calculated properties for 81 non-HIV-1 antibody-antigen complexes obtained from SAbDab. Related to Figures 2 and 3.
Dataset S5. Non-HIV-1 glycoprotein-anibody complexes obtained from Protein Data Bank. Related to Figure 3.
Figure S1. Details of cryo-EM structure of HIV-1 BG505 DS-SOSIP in complex with antibodies VRC03 and 2G12. Related to Figures 1–3. (A) Representative micrograph, (B) Representative 2D class averages (C) Ab initio model generated using cryoSparc showing HIV-1 Env in green VRC03 in grey and 2G12 in magenta. (D) Refined map fitted with model of BG505 DS-SOSIP (green) VRC03 (grey) and 2G12 (magenta) (left) Fourier shell correlation plotted as a function of resolution with resolution reported according to the gold standard FSC0.143 criterion (FSC0.143 shown as dotted line) (right). (E) Details of HIV-1 Env glycan interactions with 2G12. Left. Domain swapped 2G12 shown in surface representation (magenta and pink) bound to HIV-1 Env (green ribbon with glycans shown as sticks). Right. Details of the interactions of 2G12 with the four glycans in its binding site. For each pair of figures for the 4 glycans, the left image shows the Env and 2G12 as ribbons, glycans as sticks, and electron density as grey mesh. The right images show 2G12 as surface, Env as ribbon, and glycans as sticks.
Figure S2. Conservation score for individual epitope residues. Related to Figure 3. Conservation score 0 indicates the most variable, whereas score 1 indicates the most conserved.
Figure S3. Histogram distribution of conservation score for individual epitope residues. Related to Figure 3.
Figure S4. WebLogo Plots for individual epitope residues. Related to Figure 3. X denotes putative N-linked glycosylation sequon. The plots were generated using https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html.
Figure S5. MPER antibodies segregate into two additional categories, N-term MPER and C-term MPER. Related to Figure 1. Representative antibody-Env structures and PDBs for MPER-directed HIV-1 neutralizing antibodies (10E8, 2F5, 4E10, DH511 and Z13e1) were analyzed with representative antibodies recognizing the prefusion-closed Env trimer by neighbor-joining analysis based on epitope-residue overlap (dendrogram). The MPER antibodies segregated into two additional categories, N-term MPER (containing the single antibody 2F5, which recognizes primarily residues 661–670) and C-term MPER (containing antibodies 10E8, 4DE10, DH5511 and Z13e1, which recognize primarily residues 671–683).
Table S1. Matrix of epitope distances among 20 unique antibodies. Epitope distance between each of the antibody is defined as , where no is the number of overlapping epitope residues, and Ns is the number of epitope residues for the antibody with the least number of epitope residues of the two. Related to Figure 1.
Table S2. NGS reads statistics. Related to Figures 2 and 5.
Table S3. Structural details for MPER antibodies in complex with HIV-1 Env. Related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PGDM31400, PG9, VRC38.01, 10–1074, PGT128 | NIH/VRC | N/A |
| PGT135, 2G12, HJ16, IOMA, CH235.12, N6 | NIH/VRC | N/A |
| VRC13.01, b12, CH103, VRC16.01, VRC-PG05 | NIH/VRC | N/A |
| PGT151, VRC34.01, 35O22, 8ANC195, VRC03 | NIH/VRC | N/A |
| Virus Strains | ||
| VRC 208 virus panel | NIH/VRC | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BG505 DS-SOSIP | NIH/VRC | N/A |
| Deposited Data | ||
| Cryo-EM electron density map of 2G12 in complex with BG505 DS-SOSIP and VRC03 | This Paper | EMD-8981 |
| Fitted coordinates for cryo-EM electron density map of 2G12 in complex with BG505 DS-SOSIP and VRC03 | This Paper | PDB ID: 6E5P |
| Experimental Models: Cell Lines | ||
| Human: TZM-bl cells | NIH AIDS Reagent Program | Cat# 8129; RRID: CVCL_B478 |
| Human: Expi293F cells | Thermo Fisher | Cat# A14527; RRID: CVCL_D615 |
| Recombinant DNA | ||
| pVRC8400 vector | NIH/VRC | N/A |
| Software and Algorithms | ||
| PRISM 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| IMGT V-quest | (Lefranc et al., 2015) | http://www.imgt.org/IMGT_vquest/vquest |
| Pymol | Schrödinger | https://pymol.org |
| R | The R Foundation | https://www.r-project.org |
| Other | ||
| Los Alamos HIV sequence database | LANL | http://www.hiv.lanl.gov/ |
ACKNOWLEDGEMENTS
We thank J. Stuckey for assistance with figures, and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions or comments on the manuscript. We thank P. J. Bjorkman and A. P. West, Jr., for discussions on IOMA and H. Mouquet and M. Nussenzweig for the nucleotide sequences of antibody 10–1074. We also thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panel. Some of this work was performed at the Simons Electron Microscopy Center (SEMC) and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310). Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, and by IAVI Neutralizing Antibody Consortium (NAC).
Footnotes
DECLARATION OF INTERESTS: The authors declare no competing interests.
REFERENCES
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Aussedat B, Vohra Y, Meyerhoff RR, Cale EM, Walkowicz WE, Radakovich NA, Anasti K, Armand L, Parks R, et al. (2017). Mimicry of an HIV broadly neutralizing antibody epitope with a synthetic glycopeptide. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CO, Gristick HB, Freund NT, Escolano A, Lyubimov AY, Hartweger H, West AP Jr., Cohen AE, Nussenzweig MC, and Bjorkman PJ (2018). Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat Commun 9, 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, and Bourne PE (2000). The Protein Data Bank. Nucleic Acids Res 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Kreider EF, Fera D, Meyerhoff RR, Bradley T, Wiehe K, Alam SM, Aussedat B, Walkowicz WE, Hwang KK, et al. (2017). Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. (2016). Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell 165, 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, and Mascola JR (2015). Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, et al. (2003). Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071. [DOI] [PubMed] [Google Scholar]
- Cale EM, Gorman J, Radakovich NA, Crooks ET, Osawa K, Tong T, Li J, Nagarajan R, Ozorowski G, Ambrozak DR, et al. (2017). Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop. Immunity 46, 777–791 e710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, and Wyatt RT (2011). Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. Journal of virology 85, 8217–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Acharya P, Schmidt SD, Yang Y, Louder MK, Zhou T, Kwon YD, Pancera M, Bailer RT, Doria-Rose NA, et al. (2013). Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. J Virol 87, 10047–10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JE Jr. (2016). Teaching a Clone to Walk, One Step at a Time. Cell 166, 1360–1361. [DOI] [PubMed] [Google Scholar]
- Dandey VP, Wei H, Zhang Z, Tan YZ, Acharya P, Eng ET, Rice WJ, Kahn PA, Potter CS, and Carragher B (2018). Spotiton: New features and applications. J Struct Biol 202, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Louder MK, Yang Z, O’Dell S, Nason M, Schmidt SD, McKee K, Seaman MS, Bailer RT, and Mascola JR (2012). HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol 86, 3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J, Krawczyk K, Leem J, Baker T, Fuchs A, Georges G, Shi J, and Deane CM (2014). SAbDab: the structural antibody database. Nucleic Acids Res 42, D1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. (2015). Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity 43, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. (2013). Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756. [DOI] [PubMed] [Google Scholar]
- Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, et al. (2016). Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol 23, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick HB, von Boehmer L, West AP Jr., Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, et al. (2016). Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 23, 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. (2016). Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 45, 1108–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. (2012). Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, and Thornton JM (1993). ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology, University College London. [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julg B, Tartaglia LJ, Keele BF, Wagh K, Pegu A, Sok D, Abbink P, Schmidt SD, Wang K, Chen X, et al. (2017). Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. (2013). Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev I, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. (2013a). Somatic mutations of the immunoglobulin framework are generaly required for broad and potent HIV-1 neutralizing activity. Cell in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. (2013b). Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. (2013). Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20, 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Chuang GY, Zhang B, Bailer RT, Doria-Rose NA, and Gindin TS (2018). Surface-Matrix Screening Identifies Semi-specific Interactions that Improve Potency of a Near Pan-reactive HIV-1-Neutralizing Antibody. Cell Rep 22, 1798–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. (2002). HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682. [DOI] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2012). Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 48, 855–871. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Andrabi R, Su CY, Yasmeen A, Julien JP, Kong L, Wu NC, McBride R, Sok D, Pauthner M, et al. (2017). A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure. Immunity 46, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, de Val N, Lyumkis D, and Ward AB (2015). Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy. Structure 23, 1943–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, et al. (2017). Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol 24, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. (2011). Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. (2012). Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109, E3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB 3rd, Kwong PD, et al. (2014). Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, et al. (2017). Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Lai YT, Bylund T, Druz A, Narpala S, O’Dell S, Schon A, Bailer RT, Chuang GY, Geng H, et al. (2017). Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 13, 1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, et al. (2013). Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nat Struct Mol Biol 20, 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YD, and Williamson PR (2015). Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts. J Fungi (Basel) 1, 397–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Razinkov I, Dandey VP, Wei H, Zhang Z, Melnekoff D, Rice WJ, Wigge C, Potter CS, and Carragher B (2016). A new method for vitrifying samples for cryoEM. J Struct Biol 195, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. (2014). Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol 88, 12669–12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht CR, Krarup A, Reynell L, Mann AM, Brandenberg OF, Berlinger L, Abela IA, Regoes RR, Gunthard HF, Rusert P, et al. (2011). MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J Exp Med 208, 439–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. (2014). Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409, 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, Wang H, Gao H, Chen S, McDowall AW, and Bjorkman PJ (2015). Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell 162, 1379–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. (2010). Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Schramm CA, Kong R, Program NCS, Mullikin JC, Mascola JR, Kwong PD, and Shapiro L (2017). Gene-Specific Substitution Profiles Describe the Types and Frequencies of Amino Acid Changes during Antibody Somatic Hypermutation. Front Immunol 8, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, et al. (1986). Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. (2016). Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 165, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, and Hendrickson WA (1984). The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature 312, 127–134. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B (2009). DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. (2009). Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Dandey VP, Zhang Z, Raczkowski A, Rice WJ, Carragher B, and Potter CS (2018). Optimizing “self-wicking” nanowire grids. J Struct Biol 202, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422, 307–312. [DOI] [PubMed] [Google Scholar]
- Williams WB, Zhang J, Jiang C, Nicely NI, Fera D, Luo K, Moody MA, Liao HX, Alam SM, Kepler TB, et al. (2017). Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat Commun 8, 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. (2015). Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell 161, 470–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang GY, and Kwong PD (2018). Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. bioRxiv 10.1101/306282/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Pegu A, Rao E, Doria-Rose N, Beninga J, McKee K, Lord DM, Wei RR, Deng G, Louder M, et al. (2017). Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science 358, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41, W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, et al. (2010). Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. (2015). Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zheng A, Baxa U, Chuang GY, Georgiev IS, Kong R, O’Dell S, Shahzad-Ul-Hussan S, Shen CH, Tsybovsky Y, et al. (2018). A Neutralizing Antibody Recognizing Primarily N-Linked Glycan Targets the Silent Face of the HIV Envelope. Immunity 48, 500–513 e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. (2013). Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1. List of human HIV-1 antibody-Env complexes. Structures used in the analyses are highlighted in red. Related to Figure 1 and Table 1.
Dataset S2. Antibody neutralization evaluated on 208 HIV-1 isolates. Related to Figures 2 and 4.
Dataset S3. 81 non-HIV-1 antibody-antigen complexes obtained from SAbDab. Related to Figures 2 and 3.
Dataset S4. Calculated properties for 81 non-HIV-1 antibody-antigen complexes obtained from SAbDab. Related to Figures 2 and 3.
Dataset S5. Non-HIV-1 glycoprotein-anibody complexes obtained from Protein Data Bank. Related to Figure 3.
Figure S1. Details of cryo-EM structure of HIV-1 BG505 DS-SOSIP in complex with antibodies VRC03 and 2G12. Related to Figures 1–3. (A) Representative micrograph, (B) Representative 2D class averages (C) Ab initio model generated using cryoSparc showing HIV-1 Env in green VRC03 in grey and 2G12 in magenta. (D) Refined map fitted with model of BG505 DS-SOSIP (green) VRC03 (grey) and 2G12 (magenta) (left) Fourier shell correlation plotted as a function of resolution with resolution reported according to the gold standard FSC0.143 criterion (FSC0.143 shown as dotted line) (right). (E) Details of HIV-1 Env glycan interactions with 2G12. Left. Domain swapped 2G12 shown in surface representation (magenta and pink) bound to HIV-1 Env (green ribbon with glycans shown as sticks). Right. Details of the interactions of 2G12 with the four glycans in its binding site. For each pair of figures for the 4 glycans, the left image shows the Env and 2G12 as ribbons, glycans as sticks, and electron density as grey mesh. The right images show 2G12 as surface, Env as ribbon, and glycans as sticks.
Figure S2. Conservation score for individual epitope residues. Related to Figure 3. Conservation score 0 indicates the most variable, whereas score 1 indicates the most conserved.
Figure S3. Histogram distribution of conservation score for individual epitope residues. Related to Figure 3.
Figure S4. WebLogo Plots for individual epitope residues. Related to Figure 3. X denotes putative N-linked glycosylation sequon. The plots were generated using https://www.hiv.lanl.gov/content/sequence/ANALYZEALIGN/analyze_align.html.
Figure S5. MPER antibodies segregate into two additional categories, N-term MPER and C-term MPER. Related to Figure 1. Representative antibody-Env structures and PDBs for MPER-directed HIV-1 neutralizing antibodies (10E8, 2F5, 4E10, DH511 and Z13e1) were analyzed with representative antibodies recognizing the prefusion-closed Env trimer by neighbor-joining analysis based on epitope-residue overlap (dendrogram). The MPER antibodies segregated into two additional categories, N-term MPER (containing the single antibody 2F5, which recognizes primarily residues 661–670) and C-term MPER (containing antibodies 10E8, 4DE10, DH5511 and Z13e1, which recognize primarily residues 671–683).
Table S1. Matrix of epitope distances among 20 unique antibodies. Epitope distance between each of the antibody is defined as , where no is the number of overlapping epitope residues, and Ns is the number of epitope residues for the antibody with the least number of epitope residues of the two. Related to Figure 1.
Table S2. NGS reads statistics. Related to Figures 2 and 5.
Table S3. Structural details for MPER antibodies in complex with HIV-1 Env. Related to Figure 1.