Abstract
Background
P. aeruginosa is considered as one of the most important pathogens, and high antibiotic resistance to P. aeruginosa has become an alarming concern. This study attempts to further improve curcumin solubility and stability by producing the involved nanoparticle and investigate the effect of this nanoparticle on those virulence genes of P. aeruginosa in pathogenicity and biofilm formation.
Methods
In this study, the curcumin nanoparticles were synthesized and characterized, and the antibacterial and antibiofilm effects of Nano-curcumin and curcumin were investigated by microdilution broth and microtiter plate, respectively. In addition, cytotoxic effect of Nano-curcumin on human epithelial cell lines (A549) was determined. The effects of Nano-curcumin on P. aeruginosa virulence genes, mexD, mexB, and mexT (efflux pumps), lecA (adhesion), nfxB (negative regulator of MexCD-OprJ), and rsmZ (biofilm formation) were determined using real-time quantitative PCR.
Results
Synthesized Nano-curcumins were soluble in water, which inhibited the growth of multidrug-resistant (MDR) P. aeruginosa at 128 µg/mL, whereas it was inhibited at 256 µg/mL for soluble curcumin in DMSO. Sub-inhibitory concentrations of Nano-curcumin reduced biofilm formation and, at 64 μg/mL, disrupted 58% of the established bacterial biofilms. In addition, curcumin nanoparticle downregulated the transcription of virulence genes except nfxB and exerted no cytotoxic effect on human epithelial cell lines (A549).
Conclusions
Results suggest that Nano-curcumin could be potentially used to reduce P. aeruginosa virulence and biofilm. However, in vivo studies with respect to an animal model are necessary to validate these results.
Keywords: curcumin, Nano-curcumin, antimicrobial activity, virulence factor, burn wound, biofilm
Introduction
Burn is one of the prevalent and brutal forms of trauma that causes high morbidity rate and is responsible for up to 75% of the mortality rates among burn-induced patients.1,2 Burns destroy skin barrier, decrease systemic and local host cellular defenses, and avascular necrotic tissue and, also, diminish the migration of host immune cells, which provide an ideal environment for microbial colonization; these are important factors that contribute to severe infections in burns injury.3–5 Pseudomonas aeruginosa (P. aeruginosa) is one of the most prevalent etiologic agents of burns infection due to the many virulence factors and antimicrobial resistance property.6–10 It is difficult to treat P. aeruginosa due to its high resistance to antibiotics including fluoroquinolones, β-lactams, and carbapenems.11,12 This resistance is related to the variety of virulence genes such as quorum sensing (QS) and biofilm formation (rsmZ), efflux pumps (mexB, mexD, and mexT), adhesion (lecA), and biofilm.13–15 Biofilm is composed of surface-attached aggregates of bacteria embedded in self-made extracellular polymeric substances, and this barrier reduces the chance of a bacterium to penetrate into immune cells and antibiotics inside the biofilm and acts as an effective defense against the host immune system and antimicrobial agents, resulting in continuous colonization.16–18 Therefore, high antimicrobial resistance to P. aeruginosa appears to affect any risk of a serious wound infection in the future. Antibiotic resistance is a serious challenge to the treatment of burn wound bacterial pathogens, and we should find new approaches to the reduction of death rates associated with bacterial infections in burn injuries. Curcumin or diferuloylmethane is a major phytochemical derived from Curcuma longa (Zingiberaceae family), which is a naturally occurring yellow pigment commonly known as turmeric.19 Curcumin usually exhibits low to no toxicity at active doses and with oral doses as high as 8–12 g per day, which are safe and well tolerated.20 This compound has anticancer and antioxidant properties when combined with other drugs.21 Nevertheless, curcumin therapeutic uses have been hindered by insolubility in water, low oral bioavailability, and stability at physiological pH, rapid degradation and metabolism, limited absorption from gut, and poor blood plasma level, which impaired its clinical uses.22,23 To overcome these problems, we used the wet-milling technique to produce nano-sized particles of curcumin that have increased surface area, and this method increases the rate of dissolution of curcumin in water and enhances chemical and physical stability.23 Therefore, this study attempts to further improve curcumin solubility and stability by producing the involved nanoparticle and investigate the effect of this nanoparticle on those virulence genes of P. aeruginosa in pathogenicity and biofilm formation.
Methods
Ethics statement
The Central Laboratory of Shahid Beheshti University of Medical Sciences provided the P. aeruginosa isolates for this study. The study protocol was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1396.105).
Chemicals and apparatus
Curcumin (purity >65%) was acquired from Sigma-Aldrich (St. Louis, MO, USA). Other reagents including dichloromethane, Dimethyl Sulfoxide (DMSO), and culture medium were purchased from Merck (Germany), and antibacterial disks were purchased from Mast (Diagnostics, UK). All chemicals were of analytical reagent grade and used as received with no further purification. The aqueous solutions were prepared by deionized water (DI, 18.2 MΩ, Millipore). The images of curcumin nanoparticles obtained by transmission electron microscopy (TEM) were captured with a Zeiss (EM10C-80KV) TEM instrument. The atomic force microscopy (AFM) images were captured by a Thermo Microscopes Autoprobe, CP Research (Veeco) in a contact mode. Dynamic light scattering (DLS) analysis was carried out using a DynaPro NanoStar (Wyatt Technology, Santa Barbara, CA, USA) instrument. UV–Vis spectroscopy was performed on a Lambda25 (Perkin-Elmer, Waltham, MA, USA) spectrophotometer using a 1.0 cm quartz cell.
Bacterial strains
This study used a series of 100 clinical strains of P. aeruginosa. They were isolated between October 2014 and March 2016 from those patients who have been admitted to the burns unit of Shahid Motahari Hospital at Tehran, Iran. The location of the burn wound was first cleaned by normal saline and, then, surface culture swabs were collected from all burn patients suspected of problematic burn wound infection and each collected sample was added to Stuart transport medium and inoculated into Tryptic Soy Broth (TSB), blood, and MacConkey agar for 24 hrs. All positive cultures were recognized by their biochemical tests including Gram stain, oxidase, catalase, oxidation-fermentation (OF) test, the ability to grow at 42°C, pigment production in Mueller-Hinton agar, and the Kligler Iron Agar (KIA) tests.24 The reference strain used as positive controls was P. aeruginosa PAO1, and all the strains were maintained in TSB supplemented with 20% glycerol at −70°C until further processing.
Antimicrobial susceptibility
To evaluate the profiles associated with susceptibility to antibiotic disks of the 100 strains of P. aeruginosa isolated from Iranian adults with burns infection, the Kirby–Bauer disk diffusion, and broth microdilution methods were used in accordance with the guidelines established by the Clinical and Laboratory Standards Institute (CLSI).25,26
All the strains were grown in Mueller-Hinton agar and dissolved in sterile saline solution to achieve turbidity equal to that of a No. 0.5 McFarland opacity standard. The following antibiotic disks were tested: ticarcillin (TC), piperacillin (PRL), piperacillin/tazobactam (PTZ), ceftazidime (CAZ), doripenem (DOR), meropenem (MEM), imipenem (IMI), cefepime (CMP), aztreonam (ATM), amikacin (AK), gentamicin (GM), and ciprofloxacin (CIP). P. aeruginosa ATCC27853 and Escherichia coli ATCC 25922 were applied as a control for susceptibility testing. The MICs of ceftazidime, ciprofloxacin, imipenem, meropenem, and colistin were obtained by CLSI broth microdilution method (MIC range, 0.5–256 μg/ml). P. aeruginosa ATCC 27853 was applied as a control to ensure the test quality.
Synthesis of curcumin nanoparticles
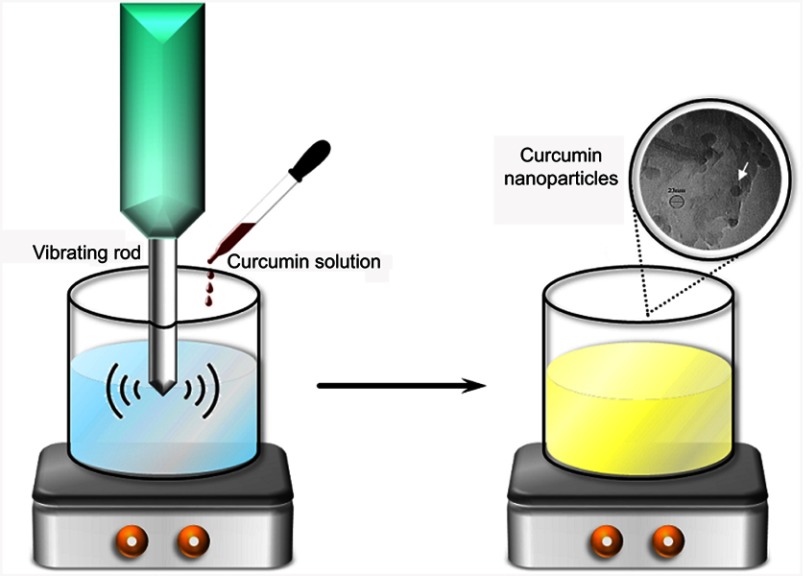
Curcumin powder was stored under dry conditions at −20°C when not in use and was allowed to warm the space to room temperature (≈25°C) before the synthesis of nanoparticle and antibacterial tests. Colloids of curcumin nanoparticle with an average diameter of 20–40 nm were prepared in accordance with the method described previously.23,27 Curcumin nanoparticles were synthesized by the aid of sonication method. For this purpose, an ultrasonic homogenizer (Hielscher, Germany) was used. One milliliter of curcumin solution was sprayed dropwise into 50 mL of boiling water with a flow rate of 0.2 mL/min, while the solution was vigorously stirred using a probe sonicator. In order to achieve nano-sized curcumin particles, the highest ultrasonic condition, ie, power of 100 W with a frequency of 30 KHz, was used. The solution was then sonicated for another 10 mins followed by stirring at 500 rpm for 20 mins until a clear yellow solution was obtained (Figure 1). The solution was concentrated and, finally, freeze-dried to obtain the curcumin nanoparticles powder. The lyophilized Nano-curcumin was re-dispersed in DI water and used for further characterizations such as TEM, DLS, and AFM analyses.
Figure 1.
Synthesize procedure of curcumin nanoparticles.
Note: The white arrow shows the Nano-curcumin produced.
Cytotoxicity assay
Cytotoxicity assay on human epithelial cell lines (A549 ATCC CCL-185TM purchased from Pasteur Institute of Iran) was carried out to study the percentage of cells left alive in Nano-curcumin. To perform the cytotoxicity assay, A549 cell lines were kept in RPMI 1640 medium (Biosera, USA) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamic acid, 1% non-essential amino acid, and 1% penicillin-streptomycin and incubated at 37°C in CO2 humidified atmosphere. For morphological and viability studies, when the cells reached a confluence level of 80%, they were inserted in 100 μL of complete medium on 96-well plates with 50,000 cells per well and, then, incubated for 24 hrs in a humidified atmosphere at 37°C and 5% CO2 to allow for attachment of cells on the surface. Subsequently, selected concentrations of Nano-curcumin were added to the wells and, after 24 hrs of incubation, cell morphology was evaluated with an Olympus IX70 Inverted microscope. On the other hand, 24 hrs after seeding the cells, different concentrations of Nano-curcumin were added to the microtiter wells to evaluate mitochondrial functions of the cells. Then, 20 µL of MTT purchased from Roche (Mannheim, Germany) was directly added to the wells and, after 4 hrs incubation in dark conditions, the absorbance at 570 nm was measured with a standard microplate reader (Anthos Labtec instruments).28
Antimicrobial activity of curcumin and Nano-curcumin
In this section, at first, five MDR strains of P. aeruginosa with the highest antibiotic resistance (Table 4) were selected and PAO1 was used as a standard strain. Then, the minimum inhibitory concentrations (MICs) of curcumin and Nano-curcumin were measured by Broth microdilution methods according to the CLSI.8,15 To prepare Nano-curcumin and curcumin stock solution, 1.024 mg of each substance was dissolved in 1 mL water and DMSO (4%), respectively, because curcumin is completely insoluble in water, unlike Nano-curcumin. The stock solution was serially diluted (in the range of 64–1024 μg/mL); then, serial two-fold dilutions of curcumin and nanoparticle initiating at 64 μg/mL were added to 96-well plates, and each well was inoculated with 10 μL (5×106 CFU/mL) of bacteria in Mueller-Hinton broth to a final concentration (5×105 CFU/mL). The wells with P. aeruginosa PAO1 and media alone served as positive and negative controls, respectively. The lowest concentration of curcumin and Nano-curcumin that showed no visible growth after 24 hrs of incubation at 37°C was determined as the MIC.
Table 4.
Phenotypic and genotypic characteristics of P. aeruginosa MDR strains were evaluated in this study
| Strain | Resistance pattern MIC (µg/ml) | ESBL genes | Biofilm | Amonoglycosid e-Modifying enzyme genes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GEN | AK | IMP | MEM | CAZ | CIP | COL | ||||
| PA11 | 256 | 64 | 8 | 64 | 128 | 16 | 0 | blaTEM | Strong |
aadA- aadB- rmtA- armA |
| PA17 | 128 | 32 | 4 | 128 | 128 | 32 | 0 | blaTEM | Strong |
aadA- aadB rmtA |
| PA22 | 32 | 32 | 256 | 64 | 128 | 16 | 0 | blaTEM | Strong |
aadA- aadB- rmtA- armA |
| PA31 | 32 | 32 | 256 | 128 | 128 | 128 | 0 | blaTEM | Strong |
aadA- aadB- armA |
| PA42 | 256 | 256 | 256 | 128 | 64 | 32 | 0 | blaTEM | Strong |
aadA- aadB- rmtA- armA |
Abbreviations: AK, amikacin; GM, gentamicin; CAZ, ceftazidime; IMI, imipenem; MEM, meropenem; COL, colistin; MIC, minimum inhibitory concentration; ESBL, extended-spectrum beta-lactamases; P. aeruginosa, Pseudomonas aeruginosa; MDR, multi-drug resistant.
Detection of virulence and resistance genes
Specific primers were used based on PCR-based assays to investigate virulence genes (rsmZ, mexT, mexD, mexB, lecA, and nfxB) (Table 1), beta-lactamase resistance gene (blaCTXM, blaSHV, and blaTEM), and Aminoglycoside Modifying Enzyme Genes (aadA, aadB, rmtA, and armA) that have important roles in antibiotic resistance, as shown in our previous study.3,29
Table 1.
DNA sequences used in PCR and real-time RT-PCR
| gene | Sequence (5′→3′) | Expected size of amplicon (bp) | Reference |
|---|---|---|---|
| mexB | CAAGGGCGTCGGTGACTTCCAG ACCTGGGAACCGTCGGGATTGA |
297 | Krausz et al38 |
| mexD | CGACCAGGCCGTGAGCAAGCAGC GGAGACCTTCGCCGCGTTGTCGC |
79 | Oh et al39 |
| mexT | GAAGGCGTTTATGTTCATAC GTATGTTTCAAGAGTGATGC |
165 | This study |
| rsmZ | CGCCGACAAGAAGAACTAGC GTCGATTACAGGATCGACAG |
102 | This study |
| nfxB | TGATTTCCCATGACGAGCGACTCA AGGCCTGGATGATCTGGTTCAGTA |
198 | This study |
| lecA | GTTTGGTCGCATATCGCAAC AATGCGCAGCACCAGGATA | 51 | This study |
| rpsL | GGCGTGCGTTACCACACCGT GGACGCTTGGCGCCGTACTT |
92 | Oh et al39 |
RNA extraction and quantitative real-time PCR
This study used real-time PCR to evaluate and determine the effect of subinhibitory concentration of curcumin and Nano-curcumin on the expression of rsmZ, mexD, mexB, mexT, nfxB, and lecA in five MDR P. aeruginosa strains and PAO1 using the primers listed in Table 1.
Total RNA was extracted in the middle of the exponential growth phase from untreated and treated bacteria cultivated in LB medium in the presence or absence of subinhibitory concentration of curcumin and Nano-curcumin. Total RNA was purified using an RNeasy Mini Kit with 1 hr on-column DNase digestion (Qiagen NV, Venlo, The Netherlands), and quantified using a spectrophotometer (WPA Biowave II Nanospectrophotometer, USA). The complementary DNA (cDNA) synthesis was carried out in 20 μL reaction volumes using the Applied Biosystems High-Capacity cDNA Reverse Transcriptase Kit (Takara kit Japan). Then, 1 μg of cDNA and 50 nM (final concentration) of each primer were mixed with 10 μL 2× SYBR Green PCR Master Mix (Takara kit Japan). Assays were performed in triplicate with Corbett Rotor-Gene 6000 system (Corbett Life Science, Australia). All of the data in gene expression were calculated through Equation 2−ΔΔCT, and all values were normalized with respect to the internal standard rpsL (encoding ribosomal subunit).30 Melting curve analysis demonstrated that SYBR Green-bound DNA was accumulated in a target gene-specific manner. The negative and positive control (P. aeruginosa PAO1) was included in all of the experiments, and the results are expressed as means±standard deviation.
Biofilm formation assay and quantification
The biofilm formation assay was performed in 96-well polystyrene plate in which the strains (PAO1 and five MDR P. aeruginosa strain) were grown simultaneously with subinhibitory concentration of curcumin (128 and 64 µg/mL) and Nano-curcumin (64 µg/mL). P. aeruginosa grew overnight either in a tryptic soy broth (TSB) medium at 37°C, shaken at 250 rpm, and diluted at 100-fold to 106 CFU/mL (determined by OD and plate count assay) with TSB media. A 100 μL aliquot of the culture was placed into each well followed by the addition of 100 μL curcumin and Nano-curcumin at varying concentrations. The plates were incubated without shaking at 37°C. After 24 hrs, the wells were washed three times with normal saline solution, and the remaining attached biofilm was fixed with 150 μL of 96% (v/v) ethanol and stained with crystal violet (1.5% w/v), left for 15 mins at room temperature, and then rinsed and de-stained with ethanol-acetone. The OD was measured using an Anthos Labtec microtiter plate reader at a wavelength of 570 nm. Biofilm-forming capacity (BFC) was calculated as follows:
In another method, at first, bacteria are allowed to form biofilms; then, 100 mL aliquot of re-suspended curcumin and Nano-curcumin at varying concentrations was added to the wells containing biofilm and incubated for 4 hrs at 37°C. The wells were washed extensively with PBS and the process continued according to the method described earlier and the biomass of biofilm corresponded to the absorbance at 570 nm. The percentage of biofilm detachments was measured through the following equation.31
Statistical analysis
This study features a completely randomized design. Data of ODs and comparisons for the gene expression study before and after treatment with Nano-curcumin and curcumin were investigated by paired t-test and Wilcoxon Signed Ranks test. Differences between the two were found quite considerable when the P-value was <0.05.
Results
Antimicrobial susceptibility profile
The profiles of the susceptibility of 100 isolates to antimicrobial agents were obtained by disk diffusion agar method (Table 2). The result of disk diffusion showed that 100% of the strains exhibited susceptibility to colistin, while the highest resistance rate was reported against IMI, MEM, and GM (95%). The result of MIC indicated that 100% of the strains were susceptible to colistin, while most of the strains were resistant to IMI, MEM, and CIP (Table 3). Moreover, 95% (n=95/100) of the P. aeruginosa was MDR. The five MDR isolates were selected for further study. The genotypic and phenotypic characteristics of the five selected MDR P. aeruginosa isolates are shown in Table 4.
Table 2.
Antibiotic susceptibility of the 100 P. aeruginosa isolates by disk diffusion method
| Antibiotic | N (%) of isolates (n=100) | ||
|---|---|---|---|
| R | I | S | |
| Amikacin (30 μg) | 91 (91%) | 4 (4%) | 5 (5%) |
| Aztreonam (10 μg) | 90 (90%) | 7 (7%) | 3 (3%) |
| Ceftazidime (30 μg) | 75 (75%) | 9 (9%) | 16 (16%) |
| Cefepime (30 μg) | 93 (93%) | 3 (3%) | 4 (4%) |
| Ciprofloxacin (30 μg) | 94 (94%) | 2 (2%) | 4 (4%) |
| Doripenem (10 μg) | 94 (94%) | 0 (0%) | 6 (6%) |
| Gentamicin (10 μg) | 95 (95%) | 0 (0%) | 5 (5%) |
| Imipenem (10 μg) | 95 (95%) | 1 (1%) | 4 (4%) |
| Meropenem (10 μg) | 90 (90%) | 5 (5%) | 5 (5%) |
| Piperacillin (100 μg) | 90 (90%) | 3 (3%) | 7 (7%) |
| Piperacillin/Tazobactam (100/10 μg) | 82 (82%) | 10 (10%) | 8 (8%) |
| Ticarcillin (75 μg) | 98 (98%) | 1 (1%) | 1 (1%) |
| Colisitin | 0 (0%) | 0 (0%) | 100 (100%) |
Abbreviations: S, susceptible; I, intermediate; R, resistant; P. aeruginosa, Pseudomonas aeruginosa.
Table 3.
Antibiotic susceptibility of the 100 P. aeruginosa isolates by Broth microdilution method
| Antibiotics | MIC | ||
|---|---|---|---|
| R | I | S | |
| Meropenem | 90% | 7% | 3% |
| Imipenem | 90% | 5% | 5% |
| Ceftazidime | 72% | 3% | 25% |
| Ciprofloxacin | 96% | 1% | 3% |
| Colistin | 0% | 0% | 100% |
Abbreviations: S, susceptible; I, intermediate; R, resistant; P. aeruginosa, Pseudomonas aeruginosa.
Characterization of curcumin nanoparticles
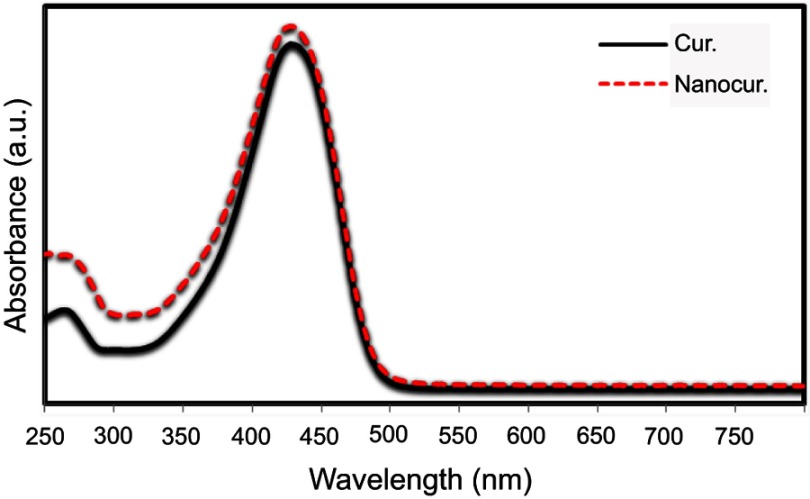
In order to study the effect of the ultrasonic condition during the preparation of curcumin nanoparticles, UV-Vis spectroscopy was performed. As observed, there is no change observed in the UV-Vis spectrum of Nano-curcumin, which indicates that the structure does not change after the sonication process (Figure 2).
Figure 2.
UV-Vis spectrum of curcumin (solid line) and curcumin nanoparticles (dashed line).
Abbreviations: Cur., curcumin; Nanocur., Nano-curcumin.
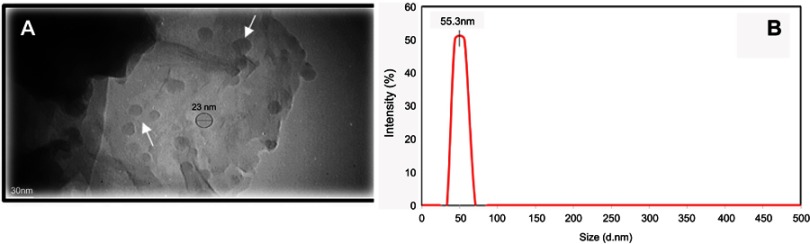
As mentioned previously, the size distribution and morphology of synthesized curcumin nanoparticles were evaluated by DLS and TEM techniques, respectively. Based on the TEM image shown in Figure 3A, curcumin nanoparticles with a well-spherical shape and an average diameter of 20–40 nm were synthesized. Furthermore, dynamic light scattering (DLS) was also used to obtain the size distribution of curcumin nanoparticles. The results showed that the prepared Nano-curcumin exhibited very narrow-size distribution with an average particle size of ca. 55 nm, which is slightly bigger than the average size measured from TEM images due to the hydrodynamic radius of solvated nanoparticles (Figure 3B).
Figure 3.
(A) TEM image and (B) dynamic light scattering analysis of the as-prepared curcumin nanoparticles which exhibit very narrow size distribution.
Note: (A) Arrows show synthesized curcumin nanoparticles.
Abbreviation: TEM, transmission electron microscopy.
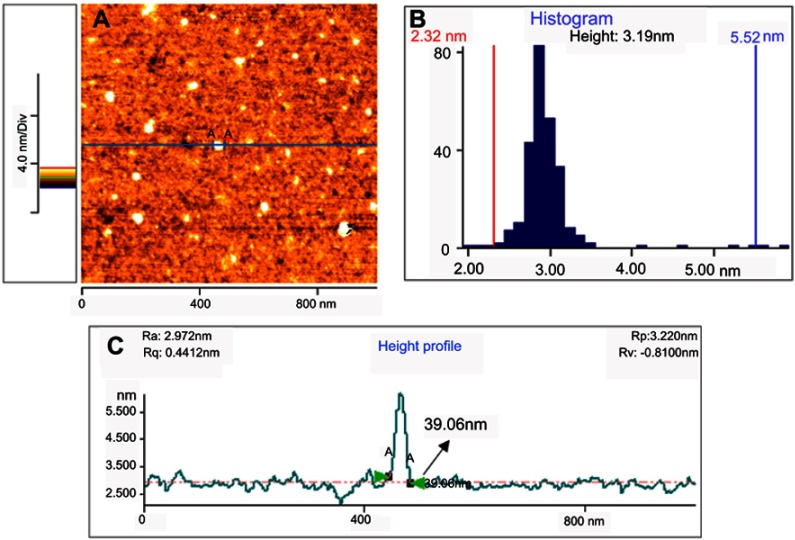
Figure 4 presents typical atomic force microscopy (AFM) images of the curcumin nanoparticles. From the AFM image of the Nano-curcumin deposited on a mica substrate, it can be clearly observed that the diameters of nanoparticles are mainly distributed in the range of 23–42 nm, and their topographic heights are mostly between 2.32 and 5.52 nm with an average height of 3.19 nm.
Figure 4.
AFM image (A), height distribution (B) and lateral size distribution (C) of the curcumin nanoparticles deposited on freshly cleaved mica substrate.
Abbreviation: AFM, atomic force microscope.
Frequency of virulence genes
PCR was applied to evaluate the strong presence of virulence genes in 100 P. aeruginosa clinical strains. The results showed that mexB, mexD, mexT, nfxB, and rsmZ were detected in 100% of the P. aeruginosa strains, while lecA gene was detected in 93% of the isolates. Only seven strains were lecA negative.
Virulence-gene expression in response to Nano-curcumin and curcumin treatment
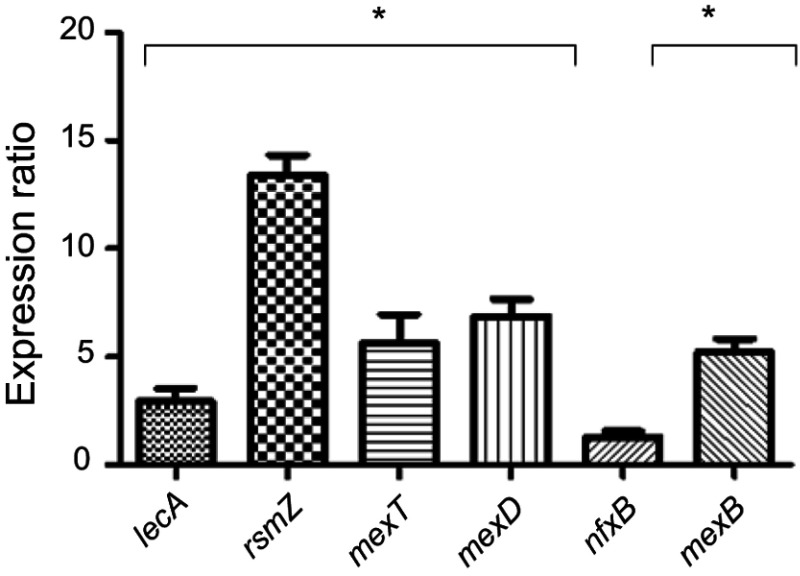
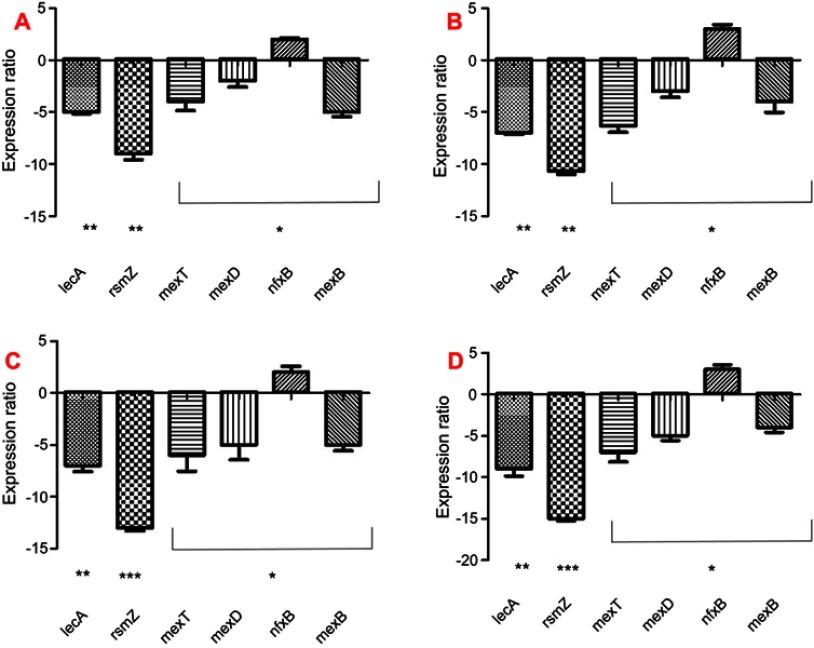
Expression of virulence genes involved in resistance, stress response, pathogenicity, attachment, and biofilm formation was performed for PAO1 and five MDR P. aeruginosa clinical strains. The results showed a significant increase in the virulence genes in clinical strains compared to PAO1 except for nfxB gene, which did not significantly increase (P<0.05) (Figure 5). To study the molecular effects of Nano-curcumin and curcumin against the MDR P. aeruginosa strains and PAO1, virulence genes involved in pathogenicity were selected for quantification of their transcriptional expression using quantitative real-time PCR and, then, compared to untreated strains. Average relative amounts of target genes were normalized to the average relative amount of the rpsL reference gene in the same sample. As expected, the presence of Nano-curcumin significantly downregulated the expression of the biofilm formation gene rsmZ by 16- and 13-fold in the PAO1 and MDR strain, respectively (P<0.001). The lecA gene expression (adhesion and biofilm formation) was also significantly downregulated by 9- and 7-fold for the PAO1 and MDR strains, respectively (P<0.01). Other virulence factors that were downregulated in both strains included mexD, mexB, and mexT (encoding efflux pump); yet, nfxB (negative regulator of MexCD-OprJ) showed upregulation of gene-expression patterns in MDR strains and PAO1 (P<0.05). All of the virulence genes were inhibited more in PAO1 as expected; however, mexB is downregulated more in five MDR strains. According to our results, the curcumin also downregulated the expression of virulence genes; however, its effect was weaker than Nano-curcumin (P<0.001) (Figure 6).
Figure 5.
Expression of virulence genes in 5 MDR clinical strains compared to PAO1 (*P<0.05).
Abbreviation: MDR, multi-drug resistant.
Figure 6.
Relative gene expression levels of curcumin and Nano-curcumin-treated PAO1 and 5 MDR clinical strains of P. aeruginosa.
Notes: *P<0.05; **P<0.01; ***P<0.001. (A) and (B) are curcumin-treated MDR strain of P. aeruginosa and PAO1, respectively; (C) and (D) are Nano-curcumin treated MDR strains of P. aeruginosa and PAO1, respectively.
Abbreviations: MDR, mult-drug resistant; P. aeruginosa, Pseudomonas aeruginosa.
Antibacterial and biofilms eradication assay
Susceptibility of P. aeruginosa to curcumin and Nano-curcumin was determined by the microdilution broth assay. The Nano-curcumin solution was prepared with distilled water, while the curcumin solution was prepared with 4% DMSO. The MIC of Nano-curcumin for the five MDR P. aeruginosa strains and PAO1 was 128 μg/mL, while it was 256 μg/mL for curcumin. This is because Nano-curcumin is more soluble (water) than the parent curcumin (DMSO).27
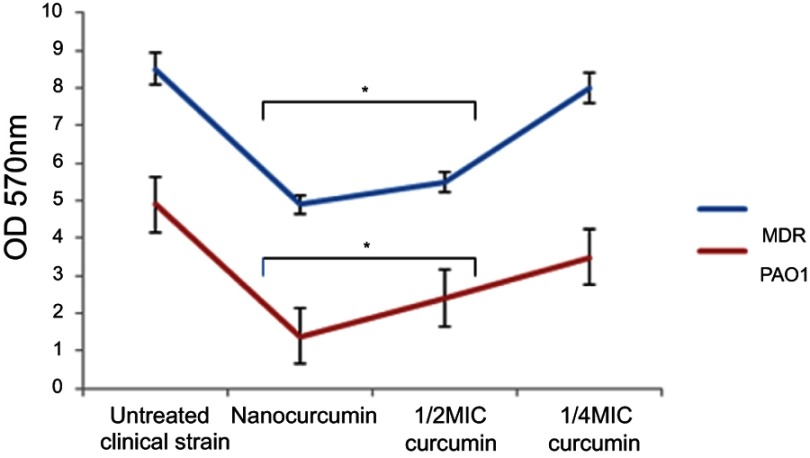
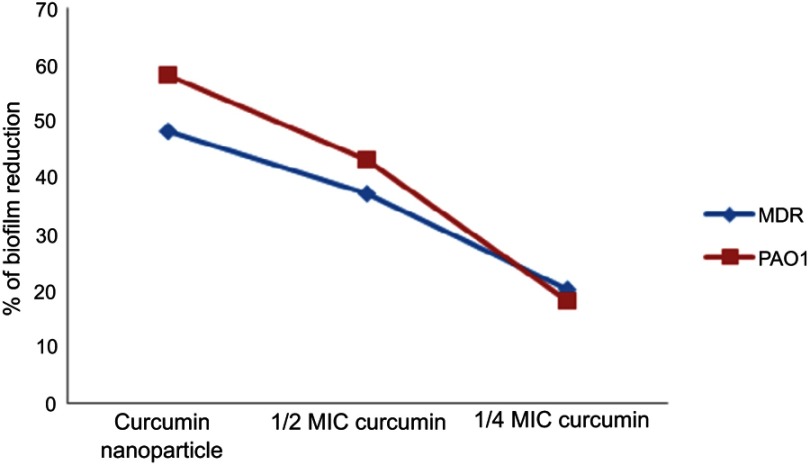
Quantitative biofilm determination using the microtiter assay revealed that 96 strains were biofilm producers, 30% of which produced strong biofilms, and 47% and 23% produced medium and weak biofilms, respectively. Only 4 strains were non-producers. Based on the results, it was concluded that Nano-curcumin (64 µg/mL) and 1/2 MIC curcumin (128 µg/mL) reduced the ODs of the biofilms; however, a considerable decrease in biofilm formation was not detected when strains were grown with 1/4 MIC of curcumin (64 µg/mL) (P<0.05) (Figure 7). Therefore, it was concluded that Nano-curcumin inhibited P. aeruginosa biofilm formation. In another method, the susceptibility of established biofilms to different concentrations of components was studied by spectrophotometric analysis, and the results revealed that Nano-curcumin resulted in a rapid decrease of biofilm amounting to 58% and 46% detachments in PAO1 and MDR strains, respectively (Figure 8).
Figure 7.
The effects of Nano-curcumin and curcumin on the biofilm of P. aeruginosa MDR strains and PAO1 (*P<0.05).
Abbreviations: MDR, mult-drug resistant; P. aeruginosa, Pseudomonas aeruginosa.
Figure 8.
The effect of Nano-curcumin and curcumin on the reduction of pre-formed P. aeruginosa biofilm.
Abbreviation: P. aeruginosa, Pseudomonas aeruginosa.
Cytotoxicity assay
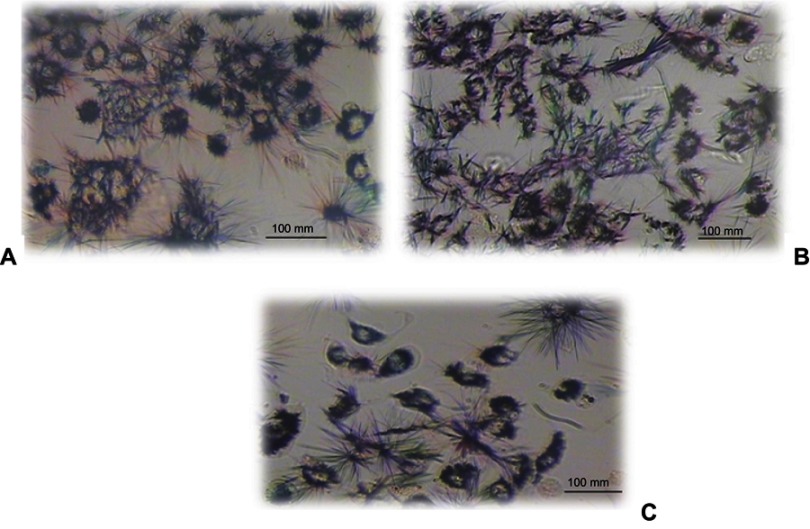
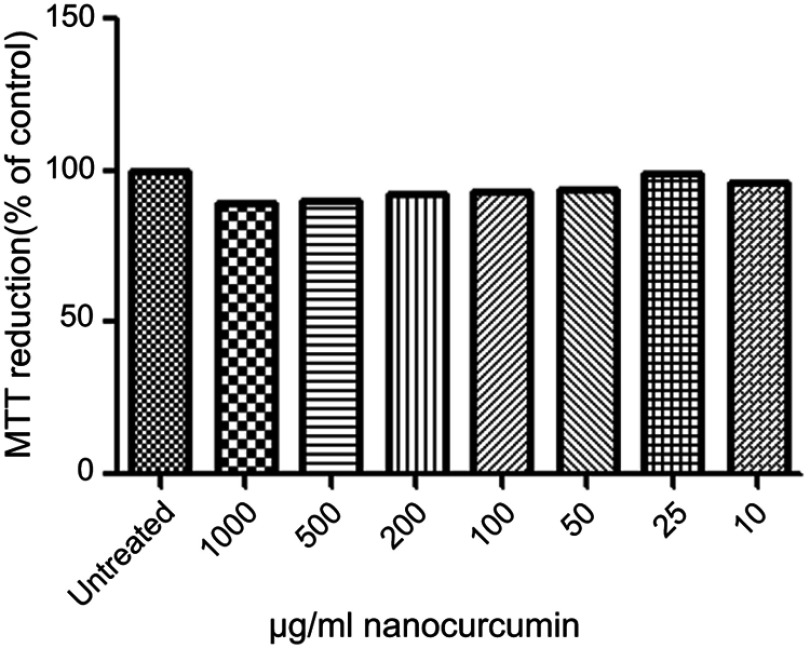
The morphology of the A549 cell lines after 24 hrs of incubation with different concentrations (25–1,000 µg/mL) of Nano-curcumin in phase-contrast microscopy is shown in Figure 9. The result showed that the human alveolar epithelial cell well spread, and there was no distinct change in morphology after 24 hrs of incubation with any concentration of curcumin nanoparticles relative to control cells (Figure 10). Nevertheless, dramatic changes (becoming irregular, necrotic, and detached from the culture dishes) occur with absolute DMSO, which is considered as positive control for cytotoxicity in this report. Our data from MTT assay indicate that Nano-curcumin exerted no significant effect on mitochondrial activity.
Figure 9.
Morphology of the human epithelial cell lines (A549) after incubation with Nano-curcumin for 24 hrs (original magnification, 100×). (A) Control, (B) Nano-curcumin 1,000 µg/mL, (C) Nano-curcumin 200 µg/mL.
Figure 10.
Influence of the different concentrations of Nano-curcumin on the viability of human epithelial cell lines (A549) after 24-hr incubation. The relative cell viability (%) was computed by this formula: [A]test/[A]control ×100. The experiment was performed 2 times in duplicates.
Discussion
P. aeruginosa is one of the important bacteria involved in infections in burn patients worldwide, and infections caused by MDR isolates of P. aeruginosa represent an important problem in the burn units that consequently reduce treatment process. In the present study, more than 94% of our isolates were resistant to ciprofloxacin, gentamicin, imipenem, and ticarcillin, while the isolates showed high sensitivity to colistin followed by ceftazidime (100% and 16%, respectively). Another study reported similar variations in the pattern of antibiotic resistance among their P. aeruginosa isolates; however, the resistance to imipenem was lower than our study.32 In the investigation conducted by Anvarinejad et al, the rates of resistance to ceftazidime and imipenem were 72% and 98%, respectively, similar to our study.33 Other researchers reported the most prevalent rate of resistance to ceftazidime (90.5%) and gentamicin (88.5%) and showed that colistin and imipenem were the most effective antibiotics against P. aeruginosa.34 Biofilm is one of the main causes of antibiotic resistance and, in the present study, 96% of the isolates produced biofilm, among which 30% showed strong biofilm formation, 47% and 23% formed moderate and weak biofilm, respectively. This result is similar to those of Jabalameli et al24 and Banar et al,32 indicating the high rate of biofilm formation by P. aeruginosa in burn infections and one of the important factors that decrease the treatment process. This study investigates the antibiofilm and antimicrobial activity of curcumin and Nano-curcumin against PAO1 and MDR strains of P. aeruginosa, which is one of the important nosocomial pathogens.
We used the wet-milling technique to synthesize nanoparticles of curcumin, and characterization and distribution of nanoparticle were performed by DLS, TEM, and AFM analyses. The result of TEM indicated the nanoparticle size to be in the range of 2–40 nm. Of note, the synthesized curcumin nanoparticles were found to have good bioavailability and solubility, and this enhanced aqueous solubility could be attributed to their larger surface area, which promotes dissolution (Figure 11). Similar results were reported by Bhawana et al23 and Kesisoglou et al35 studies, which indicated that a reduction in size of nanoparticles caused improvement in their bioavailability and solubility. Our result showed a broad-spectrum inhibitory effect of Nano-curcumin against MDR P. aeruginosa isolates; however, curcumin also had an inhibitory effect, yet MIC value was higher than the MIC range (128 μg/ml) for the Nano-curcumin. Previous study indicated that Nano-curcumin destroyed the cell wall, and the disruption of the structure of bacteria caused cell lysis and death.23 It was revealed that a significant reduction in the number of genes enabled P. aeruginosa pathogenicity and biofilm formation by Nano-curcumin. Although curcumin inhibited the growth of P. aeruginosa and led to a decrease in expression of virulence gene and biofilm formation, its effect was much lower than Nano-curcumin, because curcumin did not have enough stability, bioavailability, and solubility. In the investigation conducted by Roudashti et al,36 curcumin decreases QS-related virulence traits and biofilm formation at 1/4 and 1/16 MIC (128 μg/mL) alone and in combination with antibiotics. In another study by Ching-Yee Loo et al,37 it was reported that curcumin nanoparticles in combination with silver nanoparticles at 100 μg/ml disrupted 50% of the established bacterial biofilms. Aimee E. Krausz et al38 reported that their curcumin-encapsulated nanoparticles inhibited in vitro growth of P. aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA) in a dose-dependent manner, inhibited bacterial growth, and enhanced wound healing in an in vivo murine. Therefore, it was concluded that Nano-curcumin had a very good antibacterial effect on gram-positive and gram-negative bacteria. Toxicity of nanoparticle is a common concern, which led us to investigate the toxicity effect of synthesized Nano-curcumin on human epithelial cell; the results consistently showed no change in cell morphology and mitochondrial activity of the cells; therefore, the nanoparticles were not toxic.
Figure 11.
Solubility of Nano-curcumin (left) and curcumin (right) in water.
Conclusion
In summary, Nano-curcumin represents a significant advance as an antimicrobial agent against MDR P. aeruginosa strains and burn infections. Our synthesized nanoparticles destroyed biofilm, making them good candidates for future studies in terms of antibiofilm agents, disinfecting surfaces, and deregulation of virulence-gene expression. Curcumin nanoparticles have good stability and solubility; therefore, the difficulties inherent in curcumin administration can be circumvented.
Acknowledgments
The present study was financially supported by the research Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences (grant No 9526).
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev. 2006;19(2):403–434. doi: 10.1128/CMR.19.2.403-434.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vindenes H, Bjerknes R. Microbial colonization of large wounds. Burns. 1995;21(8):575–579. [DOI] [PubMed] [Google Scholar]
- 3.Fallah F, Borhan RS, Hashemi A. Detection of bla(IMP) and bla(VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int J Burns Trauma. 2013;3(2):122–124. [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander JW. Mechanism of immunologic suppression in burn injury. J Trauma Acute Care Surg. 1990;30:70–74. doi: 10.1097/00005373-199012001-00017 [DOI] [PubMed] [Google Scholar]
- 5.Grogan JB. Altered neutrophil phagocytic function in burn patients. J Trauma. 1976;16(9):734–738. [DOI] [PubMed] [Google Scholar]
- 6.Clark NM, Patterson J, Lynch JP. Antimicrobial resistance among gram-negative organisms in the intensive care unit. Curr Opin Crit Care. 2003;9(5):413–423. [DOI] [PubMed] [Google Scholar]
- 7.Dalamaga M, Karmaniolas K, Chavelas C, Liatis S, Matekovits H, Migdalis I. Stenotrophomonas maltophilia: a serious and rare complication in patients suffering from burns. Burns. 2003;29(7):711–713. [DOI] [PubMed] [Google Scholar]
- 8.Bahramian A, Khoshnood S, Shariati A, Doustdar F, Chirani AS, Heidary M. Molecular characterization of the pilS2 gene and its association with the frequency of Pseudomonas aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island. Infect Drug Resist. 2019;12:221. doi: 10.2147/IDR.S188527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shariati A, Fallah F, Pormohammad A, et al. The possible role of bacteria, viruses, and parasites in initiation and exacerbation of irritable bowel syndrome. J Cell Physiol. 2019;234(6):8550–8569. doi: 10.1002/jcp.27828 [DOI] [PubMed] [Google Scholar]
- 10.Jafari M, Fallah F, Borhan RS, et al. The first report of CMY, aac (6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran. Archiv Pediatric Infect Dis. 2013;1(3):109–112. doi: 10.5812/pedinfect.11392 [DOI] [Google Scholar]
- 11.Taherpour A, Hashemi A, Erfanimanesh S, Taki E. Efficacy of methanolic extract of green and black teas against extended-spectrum β-Lactamase-producing Pseudomonas aeruginosa. Pak J Pharm Sci. 2016;29(4):1257–1261. [PubMed] [Google Scholar]
- 12.Li X-Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39(9):1948–1953. doi: 10.1128/aac.39.9.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balasubramanian D, Schneper L, Merighi M, et al. The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7(3):e34067. doi: 10.1371/journal.pone.0034067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida H, Ishida Y, Kurosaka Y, Otani T, Sato K, Kobayashi H. In vitro and in vivo activities of levofloxacin against biofilm-producing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42(7):1641–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ardehali SH, Azimi T, Owrang M, Aghamohammadi N, Azimi L. Role of efflux pumps in reduced susceptibility to tigecycline in Acinetobacter baumannii. New Microbes New Infecti. 2019;100547. doi: 10.1016/j.nmni.2019.100547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoodley P, Sauer K, Davies D, Costerton JW. Biofilms as complex differentiated communities. Ann Rev Microbiol. 2002;56(1):187–209. doi: 10.1146/annurev.micro.56.012302.160705 [DOI] [PubMed] [Google Scholar]
- 17.Shigeta M, Tanaka G, Komatsuzawa H, Sugai M, Suginaka H, Usui T. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy. 1997;43(5):340–345. doi: 10.1159/000239587 [DOI] [PubMed] [Google Scholar]
- 18.Sharahi JY, Azimi T, Shariati A, Safari H, Tehrani MK, Hashemi A. Advanced strategies for combating bacterial biofilms. J Cell Physiol. 2019. doi: 10.1002/jcp.28225 [DOI] [PubMed] [Google Scholar]
- 19.Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as “Curecumin”: from kitchen to clinic. Biochem Pharmacol. 2008;75(4):787–809. doi: 10.1016/j.bcp.2007.08.016 [DOI] [PubMed] [Google Scholar]
- 20.Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. Aaps J. 2013;15(1):195–218. doi: 10.1208/s12248-012-9432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moshe M, Lellouche J, Banin E. Curcumin: a natural antibiofilm agent. In: Mendez-Vilas A, editor. Science and Technology Against Microbial Pathogens Singapore: World Scientific; 2011:89–93. [Google Scholar]
- 22.Flora G, Gupta D, Tiwari A. Nanocurcumin: a promising therapeutic advancement over native curcumin. Crit Rev Ther Drug Carrier Syst. 2013;30(4):331–368. [DOI] [PubMed] [Google Scholar]
- 23.Basniwal RK, Buttar HS, Jain V, Jain N. Curcumin nanoparticles: preparation, characterization, and antimicrobial study. J Agric Food Chem. 2011;59(5):2056–2061. doi: 10.1021/jf104402t [DOI] [PubMed] [Google Scholar]
- 24.Jabalameli F, Mirsalehian A, Khoramian B, et al. Evaluation of biofilm production and characterization of genes encoding type III secretion system among Pseudomonas aeruginosa isolated from burn patients. Burns. 2012;38(8):1192–1197. doi: 10.1016/j.burns.2012.07.030 [DOI] [PubMed] [Google Scholar]
- 25.Americal Association for Clinical Chemistry. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement (M100-S24) Washington, DC: AACC; 2014. Available from: https://www.researchgate.net/file.PostFileLoader.html?id=59202a0696b7e4d462166956&assetKey=AS%3A496054988533760%401495280134033. Accessed June 28, 2019. [Google Scholar]
- 26.Bahramian A, Shariati A, Azimi T, et al. First report of New Delhi metallo-β-lactamase-6 (NDM-6) among Klebsiella pneumoniae ST147 strains isolated from dialysis patients in Iran. Infect Genet Evol. 2019;69:142–145. doi: 10.1016/j.meegid.2019.01.030 [DOI] [PubMed] [Google Scholar]
- 27.Adahoun M, Al-Akhras M-AH, Jaafar MS, Bououdina M. Enhanced anti-cancer and antimicrobial activities of curcumin nanoparticles. Artif Cells, Nanomed Biotechnol. 2017;45(1):98–107. doi: 10.3109/21691401.2015.1129628 [DOI] [PubMed] [Google Scholar]
- 28.Braydich-Stolle L, Hussain S, Schlager JJ, Hofmann M-C. In vitro cytotoxicity of nanoparticles in mammalian germline stem cells. Toxicol Sci. 2005;88(2):412–419. doi: 10.1093/toxsci/kfi256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shariati A, Azimi T, Ardebili A, et al. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microbes New Infecti. 2018;21:75–80. doi: 10.1016/j.nmni.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumas J-L, van Delden C, Perron K, Köhler T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett. 2006;254(2):217–225. doi: 10.1111/j.1574-6968.2005.00008.x [DOI] [PubMed] [Google Scholar]
- 31.Loo C-Y, Young PM, Cavaliere R, Whitchurch CB, Lee W-H, Rohanizadeh R. Silver nanoparticles enhance Pseudomonas aeruginosa PAO1 biofilm detachment. Drug Dev Ind Pharm. 2014;40(6):719–729. doi: 10.3109/03639045.2013.780182 [DOI] [PubMed] [Google Scholar]
- 32.Banar M, Emaneini M, Satarzadeh M, et al. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS One. 2016;11(10):e0164622. doi: 10.1371/journal.pone.0164622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anvarinejad M, Japoni A, Rafaatpour N, et al. Burn Patients Infected With Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa: multidrug-Resistant Strains. Arch Trauma Res. 2014;3(2):e18182–e18182. doi: 10.5812/atr.18182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khosravi AD, Shafie F, Montazeri EA, Rostami S. The frequency of genes encoding exotoxin A and exoenzyme S in Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2016;42(5):1116–1120. doi: 10.1016/j.burns.2016.02.012 [DOI] [PubMed] [Google Scholar]
- 35.Kesisoglou F, Panmai S, Wu Y. Nanosizing—oral formulation development and biopharmaceutical evaluation. Adv Drug Deliv Rev. 2007;59(7):631–644. doi: 10.1016/j.addr.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 36.Roudashti S, Zeighami H, Mirshahabi H, Bahari S, Soltani A, Haghi F. Synergistic activity of sub-inhibitory concentrations of curcumin with ceftazidime and ciprofloxacin against Pseudomonas aeruginosa quorum sensing related genes and virulence traits. World J Microbiol Biotechnol. 2017;33(3):50. doi: 10.1007/s11274-016-2195-0 [DOI] [PubMed] [Google Scholar]
- 37.Loo C-Y, Rohanizadeh R, Young PM, et al. Combination of silver nanoparticles and curcumin nanoparticles for enhanced anti-biofilm activities. J Agric Food Chem. 2015;64(12):2513–2522. doi: 10.1021/acs.jafc.5b04559 [DOI] [PubMed] [Google Scholar]
- 38.Krausz AE, Adler BL, Cabral V, et al. Curcumin-encapsulated nanoparticles as innovative antimicrobial and wound healing agent. Nanomedicine. 2015;11(1):195–206. doi: 10.1016/j.nano.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh H, Stenhoff J, Jalal S, Wretlind B. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist. 2003;9(4):323–328. doi: 10.1089/107662903322762743 [DOI] [PubMed] [Google Scholar]