Abstract
The molecular diversity of many gene products functioning in the nervous system is enhanced by alternative splicing and adenosine-to-inosine editing of pre-mRNA. Using RDL, a Drosophila melanogaster GABA-gated ion channel, we examined the functional impact of RNA editing at several sites along with alternative splicing of more than one exon. We show that alternative splicing and RNA editing have a combined influence on the potency of the neurotransmitter GABA, and the editing isoforms detected in vivo span the entire functional range of potencies seen for all possible edit variants expressed in Xenopus laevis oocytes. The extent of RNA editing is developmentally regulated and can also be linked to the choice of alternative exons. These results provide insights into how the rich diversity of signaling necessary for complex brain function can be achieved by relatively few genes.
Introduction
Alternative splicing (Stetefeld and Ruegg, 2005) and adenosine (A)-to-inosine (I) editing of pre-mRNA (Keegan et al., 2001) lead to the generation from a single gene of multiple protein isoforms and may contribute to achieving complexity in highly evolved organisms that have a surprisingly low total gene count (Hahn and Wray, 2002). Whereas alternative splicing introduces, excises, or substitutes peptide regions of a protein, A-to-I editing alters individual nucleotides through adenosine deaminases acting on RNA (ADARs) (Keegan et al., 2004) converting select adenosine residues in pre-mRNA to inosine. Because inosine is interpreted by cellular machineries as guanosine (G), A-to-I editing generates transcripts with a nucleotide composition distinct from the corresponding genomic DNA.
Many neuronal gene products are diversified by alternative splicing (Li et al., 2007), defects of which are implicated in neurological diseases (Licatalosi and Darnell, 2006). The genome recoding by RNA editing occurs particularly in proteins involved in neuronal signaling (Hoopengardner et al., 2003), which is consistent with the neurological phenotypes observed for ADAR-deficient Caenorhabditis elegans (Tonkin et al., 2002), Drosophila melanogaster (Palladino et al., 2000), and mice (Higuchi et al., 2000). Ligand-gated ion channels (LGICs) play central roles in mediating neuronal signaling, and an increasing number of LGICs are found to undergo RNA A-to-I editing which alters amino acid residues in functionally significant regions (Hoopengardner et al., 2003). The association of deficient RNA editing of the glutamate AMPA receptor subunit GluR2 with amyotrophic lateral sclerosis highlights the importance of RNA editing of LGICs (Kawahara et al., 2004).
Gene products can undergo both alternative splicing and RNA editing, and several reports of these processes being coregulated have been published (Laurencikiene et al., 2006; Schoft et al., 2007). For example, RNA editing leading to an arginine-to-glycine switch may regulate alternative splicing which generates Flip and Flop variants in glutamate receptors (Schoft et al., 2007). RNA editing can also alter several nucleotides of a transcript without affecting splicing. However, the combined functional effects of such editing and multiple alternative splicing events of LGICs remain to be determined. We have, therefore, addressed this question using RDL, a homomer-forming GABA receptor subunit (Hosie et al., 1997). RDL mediates fast synaptic inhibition in the nervous system of D. melanogaster, where it is an important modulator of olfactory associative learning (Liu et al., 2007) and sleep latency (Agosto et al., 2008). Splicing of Rdl yields two variants each for exons 3 (variants a and b) and 6 (variants c and d) (Buckingham et al., 2005) (Fig. 1), and all four possible alternative isoforms (ac, ad, bc, and bd) are transcribed (ffrench-Constant and Rocheleau, 1993). In addition, Rdl undergoes A-to-I editing (Hoopengardner et al., 2003), which leads to the substitution of four amino acid residues. Thus, the combination of alternative splicing and RNA editing has the potential to generate considerable variation in the RDL receptor.
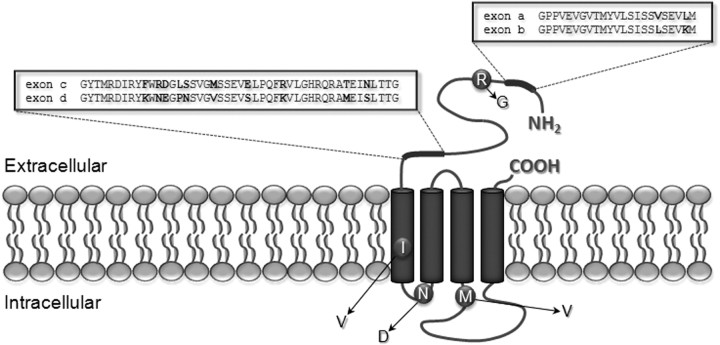
Figure 1.
Schematic illustration showing alternative splicing and RNA editing in the RDL subunit. Alternative splicing of Rdl yields two variants each for exons 3 (variants a and b) and 6 (variants c and d) changing amino acid sequence in the N-terminal extracellular domain where ligand binding occurs. Residues that are different in alternative exons are shown in bold. RNA editing leads to the substitution of arginine to glycine (R122G) in the N-terminal extracellular domain and of isoleucine to valine (I283V) in the first transmembrane domain, whereas asparagine and methionine residues in intracellular regions are converted to aspartic acid (N294D) and valine (M360V), respectively.
Here, we report an analysis of all possible editing combinations in two splice variant backgrounds. We show that editing and the choice of alternative exons are linked and that both processes act in concert to alter GABA potency suggesting a key role for both processes in fine-tuning neuronal signaling.
Materials and Methods
Drosophila stocks.
Oregon-R flies were used in this study.
RNA editing analysis.
Embryo total RNA was obtained from Stratagene UK, and total RNA was extracted from CNSs dissected from L3 larvae or adult (mixed gender) using Trizol (Invitrogen). cDNA was synthesized from the total RNA using Expand reverse transcriptase (Roche) and an oligo(dT)18 primer to generate template for PCR using Pfu Turbo DNA polymerase (Stratagene). The extent of RNA editing shown in Figure 2 was estimated by ratiometric A/G measurement from sequence chromatograms (Jepson and Reenan, 2007) of PCR products amplified using primers specific to alternative exons (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). To determine RNA editing profiles shown in Figure 3, Rdl PCR products amplified from adult CNS using Pfu Turbo DNA polymerase (Stratagene), and primers listed in supplemental Figure 1, available at www.jneurosci.org as supplemental material, were cloned into pCR-Blunt (Invitrogen) and sequenced.
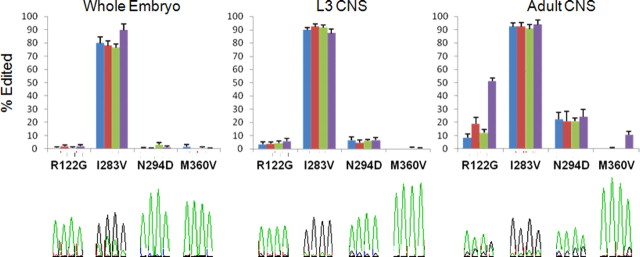
Figure 2.
The extent of RNA editing in Rdl is related to developmental stage and alternative splicing. Percentage editing at the four sites (R122G, I283V, N294D, and M360V) is shown for each of the four possible splice variants, ac (blue), ad (red), bc (green), and bd (purple), at three developmental stages of Drosophila (whole embryo, L3 CNS, and adult CNS). Values are shown as mean + SEM (n = 5). Also shown are example traces from cDNA sequence chromatograms corresponding to each of the editing sites in the four splice variants. The presence of black peaks (guanosine) with the green peaks (adenosine) indicates RNA A-to-I editing.
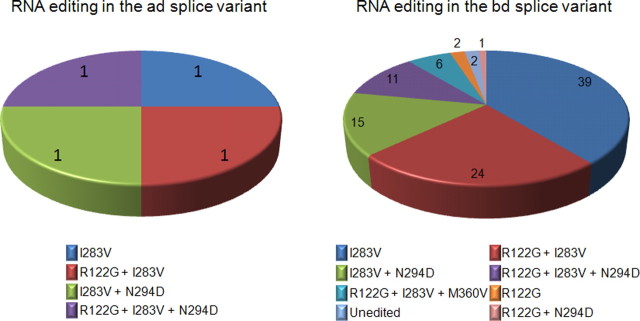
Figure 3.
Particular RNA editing profiles are highly represented in vivo. The pie charts show the RNA editing isoforms found in the 104 clones analyzed. Four of the clones have ad alternative exons, whereas the remaining 100 have bd.
SYBR Green PCR.
To determine relative transcript levels of Rdl splice variants, SYBR Green PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and the ABI Prism 7000 Sequence Detection System. Primers listed in supplemental Figure 1, available at www.jneurosci.org as supplemental material, were used on adult CNS cDNA template. The comparative cycle threshold method (User Bulletin 2, 1997; Applied Biosystems) was used to calculate relative values of the Rdl splice variant cDNA normalized against the calibrator gene rp49.
Two-electrode voltage-clamp electrophysiology.
The vectors pNB14.1 encoding Rdlac (Hosie et al., 2001) or Rdlbc (constructed in this study) as well as pcDNA3.1/Zeo+ (Invitrogen) with Rdlbd (constructed in this study) were used. Point mutations corresponding to all possible combinations of edited and unedited residues were generated in Rdlac and Rdlbd splice variants using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). pNB14.1 Rdlac or Rdlbc and pcDNA3.1/Zeo+ Rdlbd were linearized to be used as template for cRNA synthesis with the SP6 and T7 mMESSAGE mMACHINE kits (Ambion), respectively. Xenopus laevis oocytes, which were injected with combinations of cRNAs to a total of 50 nl cRNA (1 ng/nl), were incubated and voltage clamped at −60 mV 2–4 d after injection as described previously (Hosie et al., 2001; Buckingham et al., 2006). Concentration–response curves were constructed by challenging oocytes with increasing concentrations of GABA at a flow rate of 5 ml/min and with at least 2 min between challenges to reduce the effects of desensitization. Peak current amplitudes were measured as the maximum deflection of the current trace from the baseline using Clampfit v8.1. EC50s were determined by normalizing the concentration–response curve obtained from each oocyte to the maximum current obtained from that oocyte and fitting the normalized data to the following expression:
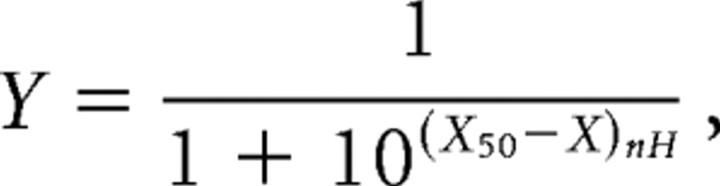 |
where Y is the normalized response amplitude, X50 is the log10 of the EC50, X is the log10 of the GABA concentration (m), and nH is the Hill coefficient. Fits to the data were performed using GraphPad Prism.
Statistical analysis.
Data were analyzed by either one-way or two-way ANOVA followed, if significant, by the Tukey post-test using GraphPad Prism 4.
Results
RNA editing of Rdl is developmentally regulated and can be dependent on the choice of alternative exons
We analyzed sequence traces of cDNA PCR products to investigate the extent of RNA editing in each of the splice variants at three stages of Drosophila development (whole embryo, larval L3, and adult nervous system). Consistent with findings that RNA A-to-I editing is primarily involved in the adult nervous system (Palladino et al., 2000), it was here that we found the greatest extent of RNA editing (Fig. 2). Our analysis also revealed that the implementation of editing at each site varied with developmental stage. For example, we found only I283V editing in the embryo, whereas R122G and N294D editing was also detected in larval nervous system, and M360V editing was seen only in adults. Editing levels vary between each site, as shown by our discovery that isoleucine is predominantly edited, whereas the other three sites are mostly unedited. When considering different splice variants of a given edit site, editing levels can be either similar (e.g., I283V or N294D in the adult nervous system) or different, as shown by significantly higher R122G editing levels in the bd splice variant (p < 0.001). The inclusion of certain alternative exons and the decision to edit might in some cases be linked, as we saw M360V editing only in the bd splice variant. Thus, the extent of RNA editing is dependent on developmental stage and can differ between alternative splice variants.
Particular editing profiles are favored in vivo
To determine RNA editing profiles present in vivo, we sequenced cloned Rdl subunits amplified from adult nervous system cDNA. Of the 104 clones analyzed, four were of the ad splice isoform, whereas the remaining 100 were bd. SYBR Green PCR, using primers shown in supplemental Figure 1, available at www.jneurosci.org as supplemental material, was used to confirm that the bd isoform is clearly the predominant splice variant, with transcript levels 26 times greater than ad, which is the second most abundant variant (Table 1). Each of the four ad clones has a different combination of editing, which corresponds to the four most abundant isoforms seen in the bd transcripts (Fig. 3). As shown in Figure 3, only half of the 16 possible isoforms that could arise from RNA editing were detected in the bd clones, indicating that certain edit isoforms are preferentially adopted and that editing at distinct sites is linked. Editing combinations in sequenced clones were present in varying proportions ranging from 1 to 39%. A small proportion (2%) of sequenced subunits was completely unedited, and fully edited isoforms were not detected. M360V editing, which was only seen in the bd splice variant (Fig. 2), was observed only in combination with R122G and I283V editing. In contrast, R122G, I283V, and N294D editing was seen in various combinations. The proportion of editing at each site of bd transcripts shows a reasonable correlation with editing levels estimated by measuring A/G peak height ratios (Table 2) showing a consistency between two techniques used to quantify RNA editing (Jepson and Reenan, 2007).
Table 1.
Rdl splice variants show differential transcript levels and GABA potency
| Splice variant | Transcript levels relative to Rdlac | GABA EC50 (μm) |
|---|---|---|
| ac | 1 ± 0.5 | 27 ± 1* |
| ad | 11.9 ± 2.8 | 58 ± 4* |
| bc | 7.5 ± 1.6 | 20 ± 0.2 |
| bd | 311 ± 1.1 | 152 ± 10* |
Relative transcript levels are given as mean ± SEM (n = 3). GABA EC50 values marked by an asterisk are previously published (Hosie et al., 2001), while the concentration–response curve for RDLbc is shown in supplemental Figure 2, available at www.jneurosci.org as supplemental material. Note that all splice variants used to generate the EC50s have I283V editing. EC50s are shown as mean ± SEM (n ≥ 3).
Table 2.
Editing levels obtained by clone counting (Fig. 3) are consistent with levels determined by measuring A/G peak height ratios (Fig. 2)
| Edit site | A/G peak height ratio | Clone counting |
|---|---|---|
| R122G | 51 ± 2.1 | 44 |
| I283V | 94 ± 3.4 | 95 |
| N294D | 24 ± 5.4 | 27 |
| M360V | 10 ± 2.5 | 6 |
Values are indicated as percentages edited in the Rdlbd splice variant from adult CNS. A/G peak height ratios are shown as mean ± SEM (n = 5).
Alternative splicing of Rdl affects GABA potency
Previous work with three RDL variants (ac, ad, and bd) has shown that alternative splicing affects agonist potency (Hosie et al., 2001). Using the two-electrode voltage-clamp technique (Buckingham et al., 2006), we report the GABA EC50 for the fourth splice variant, bc, which is similar to that of RDLac (Table 1; supplemental Fig. 2, available at www.jneurosci.org as supplemental material). RDLbd shows the most strikingly different EC50 indicating that GABA acts with less potency on this isoform than the other three splice variants. Interestingly, RDLbd is distinct in other respects because we found that when compared with other splice variants, bd transcripts are by far the most abundant (Table 1) and that bd is the only splice variant observed to be edited at all four sites (Fig. 2).
Alternative splicing and RNA editing have a combined effect on GABA potency
To study the impact of alternative splicing and RNA editing of a ligand-gated ion channel on agonist potency, we introduced point mutations to generate isoforms reflecting all 16 possible combinations of nonediting and editing in both Rdlac and Rdlbd. These two splice variants cover all four alternative exons and are also the least and most abundant, respectively, of the four splice isoforms (Table 1). Capped RNA of the 32 variants were injected into Xenopus laevis oocytes for functional analysis. Application of GABA (1 μm–3 mm) to oocytes expressing Rdl resulted in concentration-dependent inward currents for all editing isoforms in both splice backgrounds tested. The relatively slow delivery of GABA in the Xenopus oocyte expression system does not permit the accurate measurement of current kinetics. Nevertheless, oocytes give highly robust and reproducible responses, and the expression of insect LGICs in other systems have often proved difficult. We determined the GABA EC50s which varied up to 60-fold between different edit and splice variants (range, 3.2–192.9 μm) (Fig. 4, Table 3). Reponses to high concentrations of GABA relative to the EC50 declined in amplitude, as evidenced by a fall in I/Imax at the right hand of the concentration response curves (Fig. 4b) (RDLbd I283v plus N294D). This is likely attributable to the large current fluxes involved, because such large responses were accompanied by a change in reversal potential (data not shown).
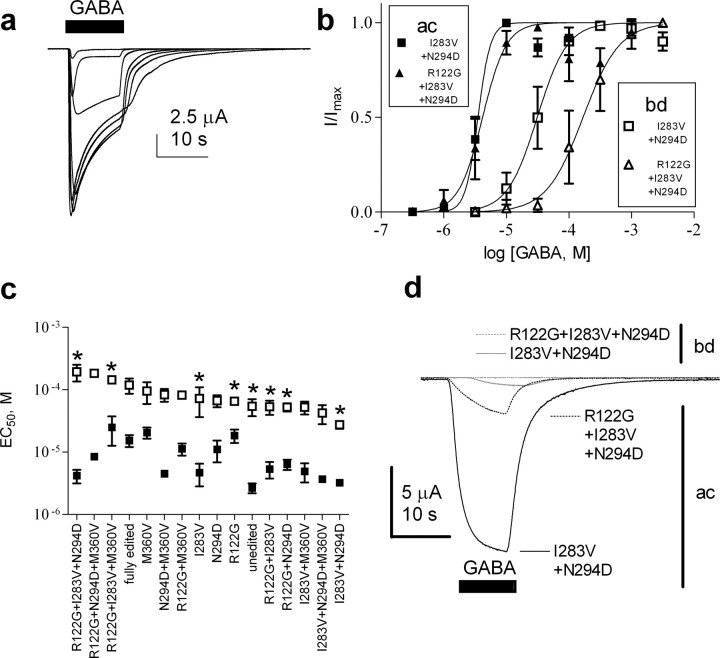
Figure 4.
Alternative splicing and RNA editing of Rdl affect GABA potency. a, Representative current responses of RDL expressed in Xenopus oocytes to GABA at concentrations from 1 μm to 3 mm. b, Example concentration–response curves of two RDL edit isoforms in ac and bd splice backgrounds. c, The graph shows GABA EC50 values of all possible editing isoforms as mean ± SEM (n = 3–7 oocytes, each oocyte from a different animal), ordered from left to right according to the EC50 values for the bd splice variant. All the actual EC50 values are listed in Table 3, and the two concentration–response curves giving the extreme EC50 values in the bd splice variant are shown in Figure 4b. EC50 values of edit isoforms found to exist in vivo through clone analysis (Fig. 3) are marked by an asterisk. Closed squares, ac variant; open squares, bd variant. d, Splice/edit isoforms can show variation in current response to the same GABA concentration (4 mm).
Table 3.
The EC50 values for all the splicing and editing combinations performed in the present study
| ac |
bd |
|||||
|---|---|---|---|---|---|---|
| EC50, μm | SEM | n | EC50, μm | SEM | n | |
| R122G plus I283V plus N294D | 4.2 | 1 | 4 | 192.9 | 57.3 | 4 |
| R122G plus N294D plus M360V | 8.5 | 0.8 | 4 | 182.8 | 19.2 | 4 |
| R122G plus I283V plus M360V | 24.9 | 12.3 | 5 | 143.5 | 13.4 | 4 |
| Fully edited | 15.4 | 3.3 | 5 | 118.6 | 32.2 | 7 |
| M360V | 20.6 | 4.2 | 5 | 95 | 36.1 | 3 |
| N294D plus M360V | 4.5 | 0.6 | 5 | 83.6 | 19.4 | 4 |
| R122G plus M360V | 11.3 | 2.5 | 5 | 81.8 | 8.4 | 5 |
| I283V | 4.7 | 1.9 | 5 | 72.6 | 36.5 | 4 |
| N294D | 11.1 | 4.3 | 5 | 66.9 | 14.7 | 4 |
| R122G | 18.5 | 4.5 | 3 | 65.6 | 7.4 | 4 |
| Unedited | 2.7 | 0.5 | 5 | 54 | 16.6 | 4 |
| R122G plus I283V | 5.3 | 1.6 | 5 | 53.2 | 13.8 | 5 |
| R122G plus N294D | 6.4 | 1.2 | 4 | 52.6 | 8.2 | 6 |
| I283V plus M360V | 4.9 | 1.7 | 4 | 52.3 | 12.2 | 3 |
| I283V plus N294D plus M360V | 3.7 | 0.4 | 5 | 42.3 | 14.4 | 5 |
| I283V plus N294D | 3.2 | 0.2 | 5 | 27.4 | 3.9 | 6 |
The SEM and number of experiments from different batches of oocytes are also given. The EC50 values are arranged in descending order for the bd splice variant.
Two-way ANOVA for the effects of editing and splicing on the EC50 using unweighted means indicated that effects of editing and splicing on EC50s were both highly significant (p < 0.0001). In addition, there was a significant interaction between editing and splicing (p < 0.0001), indicating that the effects of editing these residues differs between the two splice backgrounds. The ANOVA also indicated that splicing contributed an estimated 38% percent to the variance, with editing contributing 19% and the interaction between editing and splicing accounting for 16%. In one striking example, I283V plus N294D and R122G plus I283V plus N294D edit isoforms in the bd splice variant showed significantly different (p < 0.001) GABA EC50s of 27.4 μm and 192.9 μm, respectively (Fig. 4b,c, Table 3), which are the lowest and highest values among all of the possible edit isoforms. For the ac splice variant, this is not the case as the GABA EC50s of these two edit isoforms are similar with 3.2 and 4.2 μm (Table 3). Thus, editing and splicing can have a combined effect on agonist potency. Interestingly, the EC50s of the splice/edit forms identified in vivo (Fig. 3) span the whole range of values including the two extremes (Fig. 4c), suggesting that RNA editing adds precision modulation to the function of proteins involved in rapid neurotransmission.
Two-way ANOVA using unweighted means showed that alternative splicing had a significant effect on the maximum currents (p < 0.0001) (supplemental Table 1, available at www.jneurosci.org as supplemental material), whereas RNA editing did not. This is highlighted in Figure 4d, where I283V plus N294D and R122G plus I283V plus N294D edit isoforms in the ac and bd splice variants show considerably different current amplitudes in response to the same GABA concentration. The different currents indicate that alternative splicing of Rdl may influence receptor trafficking, assembly, and/or channel conductance or kinetics.
Discussion
The diversity of LGICs can be broadened considerably by the presence of more than one alternatively spliced exon along with RNA editing at multiple sites. To investigate the functional consequences of such complexity, we expressed in Xenopus oocytes all 16 possible editing isoforms of Rdl in two splice variants and measured their effect on GABA potency. Our study is the first functional analysis encompassing both RNA editing and alternative splicing in a cys-loop LGIC, which is a member of a superfamily of important signaling molecules including nicotinic acetylcholine receptors (Kalamida et al., 2007), serotonin type 3 receptors (Reeves and Lummis, 2002), and GABA receptors (Bormann, 2000). Previous studies have shown that alternative splicing of Rdl affects agonist potency (Hosie et al., 2001). In line with this, splice variants have different amino acids in functionally significant regions. Thus, alternative splicing of exon 3 effectively introduces amino acid diversity near a highly conserved valine, which is proposed to form a contact between the ligand-binding and transmembrane domains (Miyazawa et al., 2003), suggesting that Rdl exon 3 is involved in the coupling of ligand binding to pore opening (Buckingham et al., 2005). Alternative splicing of exon 6 is likely to directly affect GABA potency, because it generates diversity in loops C and F which contribute to the ligand-binding site (Buckingham et al., 2005). As with exon 6 splicing (Hosie et al., 2001), R122G RNA editing may also directly affect agonist potency, because it alters a residue located between ligand-binding loops D and A. This was shown to be the case where the replacement of arginine by glycine slightly but significantly increases GABA EC50 in the bd splice variant (Es-Salah et al., 2008). In our study, we also found that R122G, when compared with the unedited isoform, shows a slightly increased EC50 for GABA (Table 3), although our analysis considering all bd edit isoforms indicated this increase was not significantly different. However, R122G editing in the bd splice variant had a dramatic affect on GABA potency when in combination with I283V plus N294D editing, because R122G plus I283V plus N294D and I283V plus N294D have significantly different (p < 0.001) EC50s (Table 3). I283 and N294 are located, respectively, in the intracellular end of TM1 and the linker between TM1 and TM2 (Fig. 1), both of which regions are proposed to contribute to the pathway of the permeating ion (Filippova et al., 2004). Substitution of the equivalent residues with cysteine in the human ρ1 GABA subunit significantly alters GABA EC50 (Filippova et al., 2004). We found that individually, or in combination, I283V and N294D editing had no significant effect on RDLbd GABA EC50 (Table 3), although the triple edit isoform R122G plus I283V plus N294D had a significantly different GABA EC50 to both R122G plus I283V and R122G plus N294D (p < 0.01) (Table 3), indicating that I283V and N294D editing influences GABA sensitivity when in combination with editing at other sites. The finding that R122G, I283V, and N294D editing alone or as double edit (any two from three) isoforms has no significant effect on GABA EC50 (Table 3) further points to a combinatorial effect of RNA editing at several sites which is relevant in vivo, because we sequenced clones with almost all of these editing profiles (Fig. 3). The combinatorial effect of the R122G plus I283V plus N294D triple edit isoform seen in bd was not mirrored in the ac splice variant (Table 3, Fig. 4b), showing that alternative splicing presents another factor in determining the effectiveness of RNA editing in modulating agonist potency. Our results, therefore, highlight that RNA editing at several sites can act in concert and with alternative splicing to broaden, considerably, the spectrum of LGIC sensitivity to its neurotransmitter.
M360V editing in vivo was only seen in the bd splice variant (Fig. 2) and only in combination with R122G and I283V editing (Fig. 3), indicating that particular RNA editing profiles are preferentially adopted in certain splice variants. M360 is located in the large intracellular loop between TM3 and TM4 (Fig. 1), which has been shown to influence GABAA desensitization kinetics and GABA EC50 (Fisher, 2004). We found that the EC50 of R122G plus I283V plus M360V is among the highest of all possible RDLbd edit isoforms (Table 3, Fig. 4c). As can be seen in Figure 4c, the EC50 of R122G plus I283V plus M360V, along with the other seven edit isoforms observed in vivo, span the entire functional range seen for all possible edit isoforms, suggesting a role for RNA editing in fine-tuning LGIC function.
In addition to its actions on EC50, we also noted that splicing affected the maximum amplitude of GABA-evoked currents (Fig. 4d, supplemental Table 1, available at www.jneurosci.org as supplemental material). This may reflect changes in the receptors, such as alterations in single-channel conductance or open time, or it could point to changes in the efficiency of receptor trafficking or assembly. Thus, in concert, alternative splicing and RNA A-to-I editing have the potential to profoundly influence GABA-mediated inhibitory signaling in the insect CNS.
It has been suggested that in glutamate receptors, arginine-to-glycine editing influences Flip Flop alternative splicing, because the edit site is located only one nucleotide away from the 5′ splice site of intron 13 (Barbon et al., 2003; Schoft et al., 2007). Editing could, thereby, reduce the efficiency of a 5′ consensus splice site or introduce a splice silencer consensus sequence. A similar process is unlikely to account for the links between alternative splicing and RNA editing we observed for Rdl, because the four editing sites are not located adjacent to exon boundaries. The mechanism of RNA editing involves ADARs targeting the site to be edited which has formed a duplex by base pairing with a complementary RNA strand known as the editing site complementary sequence (ECS) (Jepson and Reenan, 2007). The ECS, which is commonly located in the intron downstream of the exon containing the editing site, forms a hairpin structure with the editing site (Hanrahan et al., 2000), although a more complex pseudoknot structure has been described where intronic elements pair with separate exonic regions to create two ADAR target sites (Reenan, 2005). Our findings suggesting that alternative splicing of exons is coordinated with preferential editing at several sites may add to the complexity of RNA structures recognized by ADARs.
Thus, the RDL receptor should be thought of in terms of a rich array of splice and edit variants. Interestingly, a study on Drosophila neurons demonstrated heterogeneity in GABA receptors containing RDL (Lee et al., 2003), which, in light of this report, may be due to the differential expression of various splice/edit isoforms. A future challenge, therefore, is to determine the precise stoichiometry of subunit isoforms which make up native RDL receptors in vivo. Similar or even greater complexity may be achieved in vertebrate GABA-gated ion channels because there are more subunit-encoding genes, several products of which are alternatively spliced (Whiting et al., 1990; Poulsen et al., 2000; Tian et al., 2005), and, recently, A-to-I editing has been to shown to modulate function of the GABAA α3 subunit (Ohlson et al., 2007; Rula et al., 2008). By examining all possible edit combinations in the most abundant and least abundant splice variants, we have uncovered a profound repertoire of neurotransmitter responses with major implications for timing in inhibitory circuits and the consequent impact on behavior.
Footnotes
This work was supported by The Integrative Physiology Initiative in Ion Channels and Diseases of Electrically Excitable Cells. We thank Drs. Christopher Mee and Laurence Brown for their assistance and discussions as well as Dr. Alastair Hosie for providing the Rdlac clone.
The authors declare no conflicts of interest.
References
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbon A, Vallini I, La Via L, Marchina E, Barlati S. Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Res Mol Brain Res. 2003;117:168–178. doi: 10.1016/s0169-328x(03)00317-6. [DOI] [PubMed] [Google Scholar]
- Bormann J. The ‘ABC’ of GABA receptors. Trends Pharmacol Sci. 2000;21:16–19. doi: 10.1016/s0165-6147(99)01413-3. [DOI] [PubMed] [Google Scholar]
- Buckingham SD, Biggin PC, Sattelle BM, Brown LA, Sattelle DB. Insect GABA receptors: splicing, editing, and targeting by antiparasitics and insecticides. Mol Pharmacol. 2005;68:942–951. doi: 10.1124/mol.105.015313. [DOI] [PubMed] [Google Scholar]
- Buckingham SD, Pym L, Sattelle DB. Oocytes as an expression system for studying receptor/channel targets of drugs and pesticides. Methods Mol Biol. 2006;322:331–345. doi: 10.1007/978-1-59745-000-3_23. [DOI] [PubMed] [Google Scholar]
- Es-Salah Z, Lapied B, Le Goff G, Hamon A. RNA editing regulates insect gamma-aminobutyric acid receptor function and insecticide sensitivity. Neuroreport. 2008;19:939–943. doi: 10.1097/WNR.0b013e32830216c7. [DOI] [PubMed] [Google Scholar]
- ffrench-Constant RH, Rocheleau TA. Drosophila gamma-aminobutyric acid receptor gene Rdl shows extensive alternative splicing. J Neurochem. 1993;60:2323–2326. doi: 10.1111/j.1471-4159.1993.tb03523.x. [DOI] [PubMed] [Google Scholar]
- Filippova N, Wotring VE, Weiss DS. Evidence that the TM1-TM2 loop contributes to the rho1 GABA receptor pore. J Biol Chem. 2004;279:20906–20914. doi: 10.1074/jbc.M401012200. [DOI] [PubMed] [Google Scholar]
- Fisher JL. The alpha 1 and alpha 6 subunit subtypes of the mammalian GABA(A) receptor confer distinct channel gating kinetics. J Physiol. 2004;561:433–448. doi: 10.1113/jphysiol.2003.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Wray GA. The g-value paradox. Evol Dev. 2002;4:73–75. doi: 10.1046/j.1525-142x.2002.01069.x. [DOI] [PubMed] [Google Scholar]
- Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA. RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–1160. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Hosie AM, Aronstein K, Sattelle DB, ffrench-Constant RH. Molecular biology of insect neuronal GABA receptors. Trends Neurosci. 1997;20:578–583. doi: 10.1016/s0166-2236(97)01127-2. [DOI] [PubMed] [Google Scholar]
- Hosie AM, Buckingham SD, Presnail JK, Sattelle DB. Alternative splicing of a Drosophila GABA receptor subunit gene identifies determinants of agonist potency. Neuroscience. 2001;102:709–714. doi: 10.1016/s0306-4522(00)00483-8. [DOI] [PubMed] [Google Scholar]
- Jepson JE, Reenan RA. Genetic approaches to studying adenosine-to-inosine RNA editing. Methods Enzymol. 2007;424:265–287. doi: 10.1016/S0076-6879(07)24012-1. [DOI] [PubMed] [Google Scholar]
- Kalamida D, Poulas K, Avramopoulou V, Fostieri E, Lagoumintzis G, Lazaridis K, Sideri A, Zouridakis M, Tzartos SJ. Muscle and neuronal nicotinic acetylcholine receptors. Structure, function and pathogenicity. FEBS J. 2007;274:3799–3845. doi: 10.1111/j.1742-4658.2007.05935.x. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Leroy A, Sproul D, O'Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencikiene J, Källman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Su H, O'Dowd DK. GABA receptors containing Rdl subunits mediate fast inhibitory synaptic transmission in Drosophila neurons. J Neurosci. 2003;23:4625–4634. doi: 10.1523/JNEUROSCI.23-11-04625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Liu X, Krause WC, Davis RL. GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron. 2007;56:1090–1102. doi: 10.1016/j.neuron.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Poulsen CF, Christjansen KN, Hastrup S, Hartvig L. Identification and cloning of a gamma 3 subunit splice variant of the human GABA(A) receptor. Brain Res Mol Brain Res. 2000;78:201–203. doi: 10.1016/s0169-328x(00)00085-1. [DOI] [PubMed] [Google Scholar]
- Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- Reeves DC, Lummis SC. The molecular basis of the structure and function of the 5-HT3 receptor: a model ligand-gated ion channel (review) Mol Membr Biol. 2002;19:11–26. doi: 10.1080/09687680110110048. [DOI] [PubMed] [Google Scholar]
- Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABAA receptor function by RNA editing. J Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res. 2007;35:3723–3732. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Tian H, Chen HJ, Cross TH, Edenberg HJ. Alternative splicing and promoter use in the human GABRA2 gene. Brain Res Mol Brain Res. 2005;137:174–183. doi: 10.1016/j.molbrainres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P, McKernan RM, Iversen LL. Another mechanism for creating diversity in gamma-aminobutyrate type A receptors: RNA splicing directs expression of two forms of gamma 2 phosphorylation site. Proc Natl Acad Sci U S A. 1990;87:9966–9970. doi: 10.1073/pnas.87.24.9966. [DOI] [PMC free article] [PubMed] [Google Scholar]