Abstract
The biologist Gordon Walls proposed his “transmutation” theory through the 1930s and the 1940s to explain cone-like morphology of rods (and vice versa) in the duplex retinas of modern-day reptiles, with snakes regarded as the epitome of his hypothesis. Despite Walls' interest, the visual system of reptiles, and in particular snakes, has been widely neglected in favor of studies of fishes and mammals. By analyzing the visual pigments of two henophidian snakes, Xenopeltis unicolor and Python regius, we show that both species express two cone opsins, an ultraviolet-sensitive short-wavelength-sensitive 1 (SWS1) (λmax = 361 nm) pigment and a long-wavelength-sensitive (LWS) (λmax = 550 nm) pigment, providing the potential for dichromatic color vision. They also possess rod photoreceptors which express the usual rod opsin (Rh1) pigment with a λmax at 497 nm. This is the first molecular study of the visual pigments expressed in the photoreceptors of any snake species. The presence of a duplex retina and the characterization of LWS, SWS1, and Rh1 visual pigments in henophidian snakes implies that “lower” snakes do not provide support for Walls' transmutation theory, unlike some “higher” (caenophidian) snakes and other reptiles, such as geckos. More data from other snake lineages will be required to test this hypothesis further.
Introduction
The distinguished comparative biologist Gordon Lynn Walls proposed that the duplex retinas of modern-day reptiles arose by the conversion of cones into rods and cited the visual photoreceptors of snakes and geckos as prime examples of his transmutation theory (Walls, 1934, 1942). In this theory, Walls suggested that the duplex retinas of early terrestrial tetrapods lost all rods in response to a complete diurnal lifestyle with a sole dependency on sunlight. With the rise of birds and mammals and the ensuing complex prey–predator relationships, many reptiles became “secondarily” nocturnal and their cones “transmutated” into photoreceptors with rod-like characteristics of a lower activation threshold and an increased sensitivity to light (Pedier and Tilly, 1964; Tansley, 1964; Kleinschmidt and Dowling, 1975; Rispoli et al., 1993).
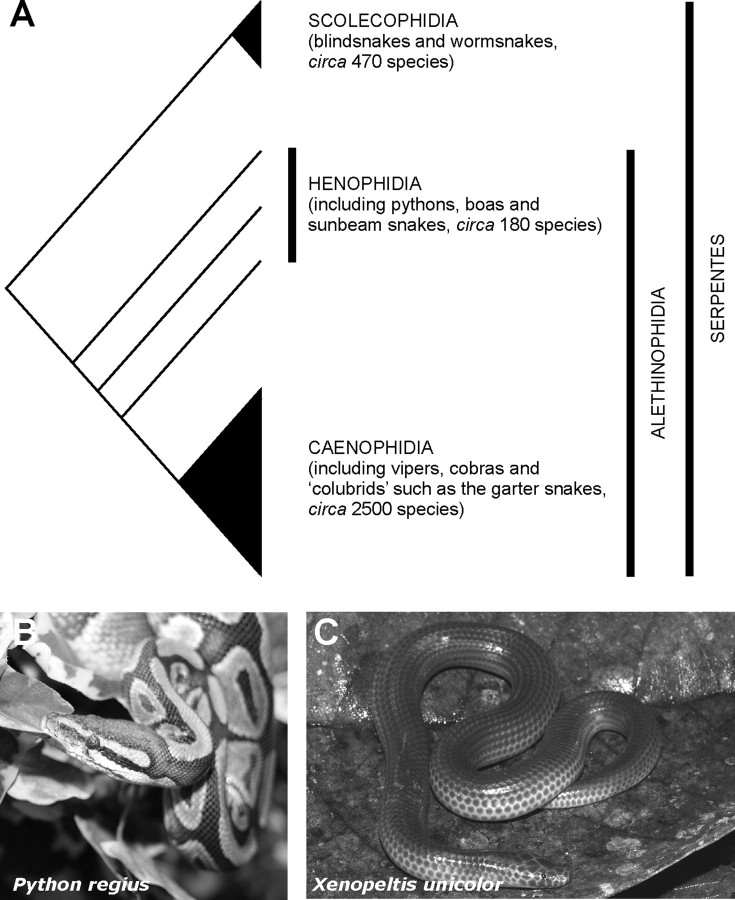
Whether photoreceptor transmutation has occurred within the snakes (Reptilia: Serpentes) remains an important question as yet unanswered. With ∼3000 extant species, snakes are highly adaptive with many having well developed sensory systems such as vision, chemoreception, and infrared detection (the latter being restricted to viperids and boids). Snakes are found on every continent except Antarctica and fill many different ecological niches (Greene, 1997). Indeed, it was this diversity of snake species and a multiplicity in their visual cell types that impressed Walls to make this group a key component of his transmutation theory (Walls, 1934, 1942). A pure-cone retina, with distinct morphological cell types, is present in the essentially diurnal venomous species (e.g., the garter snake Thamnophis sirtalis) (Sillman et al., 1997) that are members of the Caenophidia, a group of phylogenetically deeply nested higher snakes (Fig. 1A). The Henophidia lie outside the Caenophidia and include many nonvenomous, crepuscular/nocturnal and/or burrowing snakes. Among the most familiar henophidians are the python, Python regius, and boa, Boa constrictor. Although both species are partly nocturnal/crepuscular, they nevertheless possess duplex retinas, with rods accounting for in excess of 90% of the total photoreceptors (Sillman et al., 1999, 2001). It does not necessarily follow that such rods express the usual rod (or Rh1) opsin gene, as demonstrated by the expression of the middle-wavelength-sensitive (or Rh2) cone opsin gene in the middle-wavelength-sensitive rod-like photoreceptors of the nocturnal Tokay gecko, Gecko gecko (Yokoyama and Blow, 2001). The latter taxon, therefore, represents an example of the acquisition of rod-like characteristics by a photoreceptor expressing a cone pigment. The transmutation theory of Walls is further supported in this instance by the presence of cone but not rod isoforms of a number of phototransduction genes in the Tokay gecko (Zhang et al., 2006). Rod-like behavior of these photoreceptors is achieved, therefore, with a cone visual pigment linked to cone phototransduction machinery.
Figure 1.
A, A schematic cladogram showing the relationships between blindsnakes and wormsnakes (Scolecophidia), lower snakes (Henophidia) (e.g., pythons, boas, and sunbeam snakes), and higher snakes (Caenophidia) (e.g., vipers, cobras, and colubrids). B, C, Photograph of the head and upper body of two henophidian snakes, the royal python (Python regius) (B) and the Old World sunbeam snake (Xenopeltis unicolor) (C). Photographs courtesy of Patrick Jean, Muséum d'histoire naturelle de Nantes, France (P. regius) and Jodi Rowley, James Cook University, Australia (X. unicolor).
To determine the molecular basis for the spectral sensitivities of henophidian snake photoreceptors, we investigated the classes of visual pigments present in the retinas of two species of closely related lower snakes (Dong and Kumazawa, 2005; Yan et al., 2008): the partly nocturnal royal python, Python regius (Fig. 1B) and the mainly nocturnal and semifossorial Old World sunbeam snake, Xenopeltis unicolor (Fig. 1C).
Materials and Methods
Animals.
Sunbeam (X. unicolor) and python (P. regius) snakes were obtained from a licensed source as approved by the local Animal Ethics Committee and were killed using an approved procedure.
Microspectrophotometry.
All procedures were performed under dim red light. Eyes were enucleated from a single dark-adapted sunbeam snake (X. unicolor), the globes hemisected, and the outer portion discarded. The retina was placed on a coverslip and mounted in saline containing 10% (w/v) dextran, before being compressed with a second coverslip, which was sealed with wax. Under the microspectrophotometer, rods were easily recognized, but few cones were identified. Recordings from these cells were made in the conventional manner using a Liebman dual-beam microspectrophotometer (Liebman and Entine, 1964; Mollon et al., 1984; Bowmaker et al., 1991). Spectra were recorded at 2 nm intervals from 750 to 350 nm and from 351 to 749 nm on the return scan. The outward and return scans were averaged. A baseline spectrum was measured for each cell, with both beams in an unoccupied area close to the cell, and this was subtracted from the intracellular scan to derive the final spectrum. Two baseline scans were recorded for each cell and averaged. All cells were fully bleached with white light and postbleach spectra recorded. For rods, selection criteria were used to discard records that either had very low absorbance or were clearly distorted, but all cone records were analyzed. The wavelength of maximum absorbance (λmax) of both the absorbance spectra and difference spectra were determined by a standard computer program that best fits a visual pigment template to the right-hand limb of the spectra (Liebman and Entine, 1964; Mollon et al., 1984; Bowmaker et al., 1991).
mRNA source and isolation.
Eyes for mRNA were dissected and preserved in RNAlater (Ambion) at 4°C. Messenger RNA was extracted from homogenized whole eyes using the illustra QuickPrep Micro mRNA Purification Kit (GE Healthcare), according to the manufacturer's instructions.
cDNA synthesis.
A 1 in 10 dilution of ocular mRNA was primed with 500 ng of oligo-dT primer (12-mer) and converted to single-stranded complementary DNA (cDNA) using Superscript III reverse transcriptase (Invitrogen), following the protocol provided by the manufacturer.
PCR amplification and RACE.
“All-opsin, all-species” degenerate PCR primers (AOASF1, AOASF2, AOASR1, AOASR2) (Table 1) were designed to amplify long-wavelength-sensitive (LWS), short-wavelength-sensitive 1 (SWS1), SWS2, RhB/Rh2, and RhA/Rh1 partial sequences based on an alignment of representative opsin sequences from all classes of a number of vertebrate species. A further set of primers were designed that were specific for opsin sequences found in Diapsida (extant reptiles and birds) only (DIAPLMF1, DIAPLMF2, DIAPLMR1, DIAPLMR2 for LWS/MWS opsins; DIAPS1F1, DIAPS1F2, DIAPS1R1, DIAPS1R2 for SWS1 opsins; and DIAPR1F1, DIAPR1F2, DIAPR1R1, DIAPR1R2 for Rh1 opsins) (Table 1). In all cases, negative (no DNA template) controls were included among the reactions, and where appropriate, a positive control was included, such as retinal cDNA derived from the green anole (Anolis carolinensis), with a primer pair designed to amplify a known opsin sequence. First round PCR products were generated by using AOASF1 and AOASR2 primers under the following conditions: an initial denaturation at 94°C for 10 min; 50 cycles at 94°C for 30 s, 45°C for 1 min, 72°C for 1.5 min; and a final extension at 72°C for 10 min. Resulting PCR products, including the negative controls, were diluted 1 in 10 and used as template in a second round heminested PCR using primers AOASF1 and AOASR1, AOASF2 and AOASR2, and AOASF2 and AOASR1. Conditions for the second round PCR were similar to the first round, except for the use of an annealing temperature at 50°C. A similar set up was used for the “Diapsida-specific” experiments, except extension temperatures at 50°C and 55°C were included for first and second round PCRs, respectively. The 5′ and 3′ ends of each opsin sequence were obtained by 5′- and 3′-RACE using the FirstChoice RACE-Ready cDNA kit (Ambion), according to the manufacturer's instructions. PCR conditions were similar to those detailed above, except for the inclusion of an annealing temperature at 60°C for 1.5 min. First and second round PCR and RACE products were visualized by agarose gel electrophoresis and cloned into a pGEM-T easy cloning vector (Promega). Colonies were screened by blue/white selection and a test digestion with endonuclease restriction enzyme EcoRI to confirm the presence of inserts. Selected positive clones were subsequently sequenced using either T7 or SP6 primers and a Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Samples were run on an Applied Biosystems PRISM Genetic Analyser.
Table 1.
Oligonucleotide sequences used in nested-PCR to generate partial sequences for X. unicolor and P. regius LWS, SWS1, and Rh1 cone opsin genes and full-length sequences for in vitro regeneration of sunbeam LWS, SWS1, and Rh1 visual pigments
| Primer | Sequence |
|---|---|
| AOASF1 | 5′-CGCGAGAGATACATNGTNRTNTGYAARCC-3′ |
| AOASF2 | 5′-ATTTTAGAAGGTCTGCCRGWSNTCNTGYGG-3′ |
| AOASR1 | 5′-ATTGGTCACCTCCTTYTCNGCYYTYTGNGT-3′ |
| AOASR2 | 5′-CCCGGAAGACGTAGATGANNGGRTTRWANA-3′ |
| DIAPLMF1 | 5′-AAGCGTATTYAYTTAYACCRACASCAACAA-3′ |
| DIAPLMF2 | 5′-AGTGTCATCAACCAGWTCTYBGGSTAYTTC-3′ |
| DIAPLMR1 | 5′-CATCCTBGACACYTCCYTCTCVGCCTTCTG-3′ |
| DIAPLMR2 | 5′-CATCATCCACTTTYTTSCCRAASAGCTGCA-3′ |
| DIAPS1F1 | 5′-TCCCATGTCCGGAGAVGAVGABTTYTACCT-3′ |
| DIAPS1F2 | 5′-GGCCTTCGARCGHTACATYGTYATCTGCAA-3′ |
| DIAPS1R1 | 5′-CACCACSACCATSCGVGASACCTCCCGCTC-3′ |
| DIAPS1R2 | 5′-TTAGCTGGGGCYGACYTGRCTGGAGGACAC-3′ |
| DIAPR1F1 | 5′-GTCAAAATTTCTAYRTBCCCWTKTCCAACA-3′ |
| DIAPR1F2 | 5′-AATAGGATGCWRCWTYGARGGCTTCTTTGC-3′ |
| DIAPR1R1 | 5′-ACAGTGCAGACAAGRYKYCCRTAGCAGAAG-3′ |
| DIAPR1R2 | 5′-ATTCTTTCCACARCARAGRGTBRTGATCAT-3′ |
| PESBLWSF | 5′-GCGCGAATTCCACCATGCAAACAAGGGGGAAAGAA-3′ |
| PESBLWSR | 5′-CGGCGTCGACGCTGCTGGCGATACAGAAGAGTT-3′ |
| PESBSWS1F | 5′-GCGCGAATTCCACCATGTCCGGAGAAGAGGACTTC-3′ |
| PESBSWS1R | 5′-CGGCGTCGACGCGCTCCTTGGAGAACCCGTGGT-3′ |
| PESBRH1F | 5′-GCGCGAATTCCACCATGAATGGAACAGAAGGGCTT-3′ |
| PESBRH1R | 5′- CGGCGTCGACGCGGCAGGGGAAACCTGACTTGT-3′ |
Phylogenetic analysis.
Phylogenetic analysis was performed on a codon-match nucleotide alignment of a core region of the opsin sequence that contained all seven transmembrane domains [nucleotide sequence encoding amino acids 20–319, based on bovine rod opsin (NM001014860) numbering] (see supplemental material, available at www.jneurosci.org), where LWS, SWS1, and Rh1 retinal opsin sequences expressed in the eyes of the sunbeam and python snakes were compared with the coding sequences of LWS, SWS1, SWS2, RhB/Rh2, and RhA/Rh1 opsin classes of other vertebrate species. A Bayesian probabilistic inference method was applied with a Metropolis Markov chain Monte Carlo algorithm using MrBayes 3.1.2 software (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). A general time-reversal model (Lanav et al., 1984) was used, applying a gamma-distributed rate of variation across all sites with a proportion of invariable sites. Two simultaneous and independent runs starting from different random trees were performed for 1,400,000 generations with a chain frequency of 1000 generations until stationarity was reached. The first 25% (350) of all trees generated were discarded as burnin to allow for tree convergence and a low (close to 0.01) SD of split frequencies. Resultant trees were visualized with Treeview 1.6.6 (Page, 1996). The ciliary opsin of Dumeril's clam worm, Platynereis dumerilii (GenBank accession number AY692353), was used as an outgroup.
Expression of recombinant opsins.
The full-length coding sequences for the sunbeam LWS, SWS1, and Rh1 opsins were generated as a single contiguous amplicon from retinal cDNA, using Thermococcus kodakaraensis DNA polymerase (Novogen), according to the manufacturer's recommendations and forward/reverse opsin-specific primers (PESBLWSF and PESBLWSR for the LWS opsin; PESBSWS1F and PESBSWS1R for the SWS1 opsin; PESBRH1F and PESBRH1R for the Rh1 opsin) (Table 1). The primer pairs designed to the 5′- and 3′- ends of the opsin sequences included EcoRI and SalI restriction sites. The resulting products were then cloned via these restriction sites into the expression vector pMT4 (Franke et al., 1988) and used to transfect HEK-293T cells using GeneJuice (Invitrogen) according to manufacturer's instructions. Twelve 140 mm plates were used per transfection, and the cells were harvested 48 h after transfection and washed with 1× PBS. The visual pigments were regenerated in 1× PBS with 40 μm 11-cis-retinal in the dark. The pigments were isolated by incubating with 1% (w/v) dodecyl-maltoside and 20 mg/ml phenylmethylsulphonylfluoride before passage over a cyanogen bromide-activated Sepharose-binding column coupled to an anti-1D4 monoclonal antibody.
Absorbance spectra were recorded in the dark using a dual-path spectrophotometer (Spectronic Unicam). Pigments were either bleached by exposure to broad-spectrum white fluorescent light at a distance of 50 cm for 60 min (for LWS and Rh1 visual pigments) or acid-denatured with 27.3 mm HCl in the dark for 30 min (for the SWS1 visual pigment). The λmax values for each pigment were determined by subtracting the bleached or acid-denatured spectrum from the dark spectrum to generate a difference spectrum. This was then fitted to a standard Govardovskii rhodopsin A1 template (Govardovskii et al., 2000) to determine the λmax.
Results
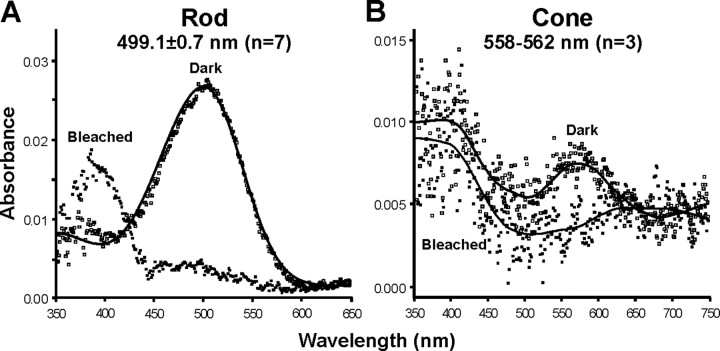
Previous microspectrophotometric (MSP) analysis of the visual pigments of the rods, and the large and small single cones of P. regius, gave spectral peaks at 494, 551, and 360 nm, respectively (Sillman et al., 1999). Our MSP analysis of the photoreceptors of X. unicolor identified a predominance of rods in the retina with a spectral peak of absorbance (λmax) at 499.1 ± 0.7 nm (Fig. 2A) and a smaller number of cones with λmax values between 558 and 562 nm (Fig. 2B). Using cDNA derived from X. unicolor and P. regius ocular mRNA, partial sequences for three visual pigments encoded by the rod Rh1 and cone LWS and SWS1 opsin genes were successfully amplified using multiple sets of degenerate opsin primer pairs designed to amplify either all vertebrate opsin sequences or each opsin class within the Diapsida (Table 1). The failure to identify any short-wavelength-sensitive cones by MSP suggests that cones expressing the SWS1 pigment are present at low frequency. We failed to detect either SWS2 or Rh2 opsin cDNA sequences in either snake species despite all five opsins being readily amplified from retinal cDNA derived from the green anole lizard, A. carolinensis, and conclude that these pigments are not expressed in the python or sunbeam snake retina. Overlapping cDNA sequences were obtained by 5′- and 3′-RACE, and full-length opsin sequences were generated as one contiguous amplicon. Alignments of the three opsin coding sequences for both P. regius and X. unicolor are shown in Figure 3 and in the supplemental data (available at www.jneurosci.org as supplemental material).
Figure 2.
Mean absorbance spectra from microspectrophotometry of rods (A) and cones (B) of X. unicolor under dark and light-bleached conditions. The prebleach rod spectrum (open squares) is overlaid with a vitamin-A1 (rhodopsin) visual pigment template (line); cone spectra are fitted with variable point unweighted running averages.
Figure 3.
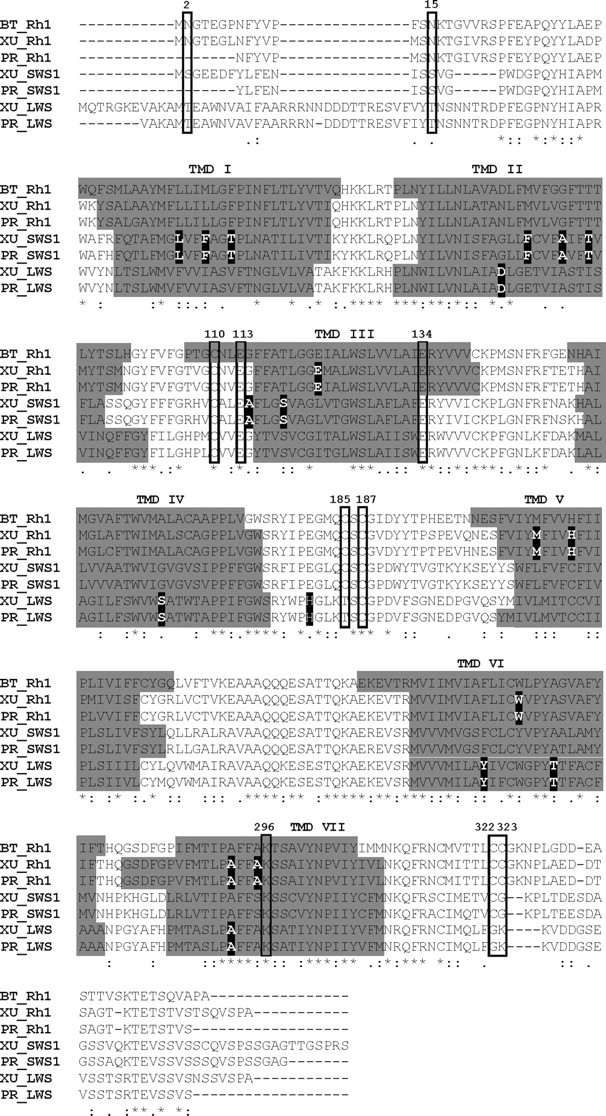
Alignment of the amino acid sequences of visual pigments expressed in the retina of the sunbeam (X. unicolor) and python (P. regius) snakes. Amino acid numbering is based on the Rh1 pigment of the common cow (Bos taurus, BT). Asterisks denote an identical consensus residue between all snake opsin sequences and bovine Rh1. “:” and “.” indicate a conservative or semiconservative amino substitution, respectively, with the codon-matched protein alignment. Gaps inserted to maintain a high degree of identity present between the opsin sequences derived from both sunbeam and python snake retina and bovine Rh1 are indicated by dashes (−). Seven putative transmembrane domains (TMDs) are indicated by gray shading. The TMDs shown for bovine rhodopsin were determined by crystallography (Palczewski et al., 2000). The putative positions of the TMDs for each snake visual pigment were determined online using TMHMM Server Version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Residues identified as being critical for correct opsin protein conformation are boxed. Comparison of the opsin amino acid sequences of X unicolor, P. regius, and bovine rod opsin demonstrated that the critical residues involved in the maintenance of the tertiary structure of the opsin molecule are present. Using the conventional numbering system of the bovine rod opsin polypeptide sequence, these key sites include (1) three conserved cysteine (C) residues at positions 110 (TMD3), 185 [extracellular domain (ECD2)], and 187 (ECD2) that are involved in disulfide bond formation (Karnik and Khorana, 1990), except for a Thr (T) residue at position 185 in the LWS opsin of both snakes, which is also conserved throughout the rest of the vertebrate LWS opsin class; (2) a conserved glutamate (E) at position 113 (TMD3) that provides the negative counterion to the proton of the Schiff base (Sakmar et al., 1989); (3) a conserved glutamate (E) at position 134 (TM3) that provides a negative charge to stabilize the inactive opsin molecule (Cohen et al., 1992); (4) a conserved lysine (K) at position 296 (TM7) that is covalently linked to the chromophore via a Schiff base (Dratz and Hargrave, 1983); (5) conservation of two cysteine (C) residues at putative palmitoylation positions 322 and 323 (Ovchinnikov et al., 1988) in both sunbeam and python snake Rh1 opsins but not LWS and SWS1 opsins; (6) the presence of a number of Ser (S) and Thr (T) residues in the carboxy terminus, which are potential targets for phosphorylation by rhodopsin kinases in the deactivation of metarhodopsin II (Palczewski et al., 1993; Zhao et al., 1997); and (7) the conserved glycosylation sites at positions 2 and 15 (Kaushal et al., 1994) in the Rh1 opsin identified in the retina of X. unicolor and P. regius. Amino acids important for the spectral tuning of LWS, SWS1, and Rh1 visual pigments are indicated as black highlighted boxes with white text.
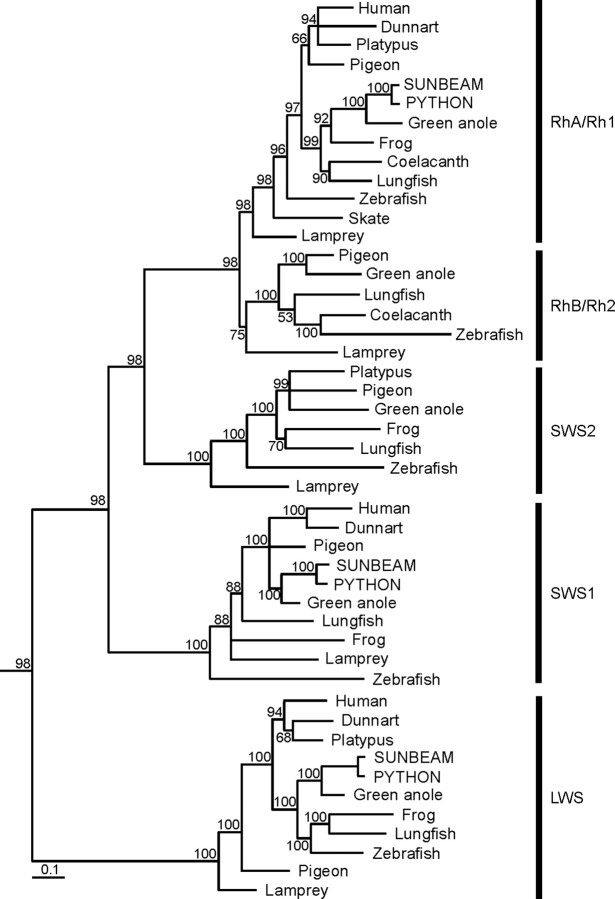
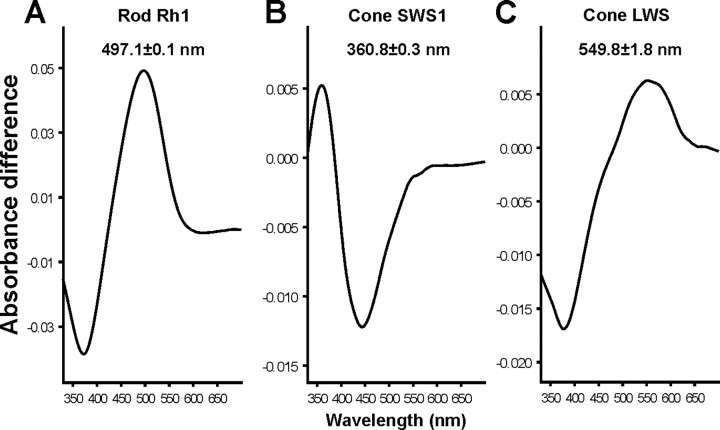
Phylogenetic analysis of the three snake opsin genes confirms that they are orthologues of the opsins of other vertebrates (Fig. 4). In all three cases, the snake gene sequences robustly cluster together as the sister group to the only other incorporated reptilian species, the green anole. A complete set of spectra obtained by MSP is already available for P. regius photoreceptors (Sillman et al., 1999) but less complete for X. unicolor, based on our MSP analysis. Spectra for the three X. unicolor pigments were therefore obtained in vitro by transfection of mammalian cells with the cDNAs sequences, isolation of expressed opsins by immunoaffinity chromatography, and reconstitution with 11-cis-retinal. The spectrum for the Rh1 pigment gave an absorbance peak at 497.1 ± 0.1 nm (Fig. 5A), similar to the λmax of 499.1 ± 0.7 nm obtained by MSP. Seven amino acids within the retinal-binding pocket have been shown to “tune” vertebrate Rh1 pigments to a spectral peak at 500 nm (D83, E122, M207, H211, W265, A292, A295; numbering based on the bovine rod opsin sequence, NM001014860) (Yokoyama, 2000). These sites in the Rh1 opsins of P. regius and X. unicolor show a profile of NEMHWAA, differing only at site 83 from the former sequence, which may explain the small blue-shift in the spectral peaks obtained for both species of snake.
Figure 4.
Phylogenetic analysis of P. regius and X. unicolor LWS, SWS1, and Rh1 retinal opsins (GenBank accession numbers FJ497233-FJ497238) with LWS, SWS1, SWS2, RhB/Rh2, and RhA/Rh1 visual pigment coding sequences of other vertebrate species. Posterior probability values (represented as a percentage) are indicated for each resolved node. The scale bar indicates the number of nucleotide substitutions per site. The ciliary opsin of Dumeril's clam worm, Platynereis dumerilii, was used as an outgroup (data not shown). The sequences used for generating the tree are as follows: (1) RhA/Rh1 opsin class: human (Homo sapiens), NM000539; fat-tailed dunnart (Sminthopsis crassicaudata), AY159786; platypus (Ornithorhynchus anatinus), EF050076; pigeon (Columba livia), AH007730; sunbeam snake (Xenopeltis unicolor), FJ497233; python (Python regius), FJ497236; green anole (Anolis carolinensis), AOIRHODOPS; clawed frog (Xenopus laevis), NM001087048; Comoran coelacanth (Latimeria chalumnae), AF131253; Australian lungfish (Neoceratodus forsteri), EF526295; zebrafish (Danio rerio), NM131084; little skate (Raja erinacea), U81514; pouched lamprey (Geotria australis) (RhA), AY366493; (2) RhB/Rh2 opsin class: pigeon (Columba livia), AH007731; green anole (Anolis carolinensis), AH004781; Australian lungfish (Neoceratodus forsteri), EF526296; Comoran coelacanth (Latimeria chalumnae), AF131258; zebrafish (Danio rerio), NM131253 (Rh2.1); pouched lamprey (Geotria australis) (RhB), AY366494; (3) SWS2 opsin class; platypus (Ornithorhynchus anatinus), EF050077; pigeon (Columba livia), AH007799; green anole (Anolis carolinensis), AF133907; clawed frog (Xenopus laevis), BC080123; Australian lungfish (Neoceratodus forsteri), EF526299; zebrafish (Danio rerio), NM131192; pouched lamprey (Geotria australis), AY366492; (4) SWS1 opsin class: human (Homo sapiens), NM001708; fat-tailed dunnart (Sminthopsis crassicaudata), AY442173; pigeon (Columba livia), AH007798; sunbeam snake (Xenopeltis unicolor), FJ497234; python (Python regius), FJ497237; green anole (Anolis carolinensis), AH007736; Australian lungfish (Neoceratodus forsteri), EF526298; clawed frog (Xenopus laevis), XLU23463; pouched lamprey (Geotria australis), AY366495; zebrafish (Danio rerio), NM131319; (5) LWS opsin class: human (Homo sapiens), NM020061; fat-tailed dunnart (Sminthopsis crassicaudata), AY430816; platypus (Ornithorhynchus anatinus), EF050078; sunbeam snake (Xenopeltis unicolor), FJ497235; python (Python regius), FJ497238; green anole (Anolis carolinensis), ACU08131; clawed frog (Xenopus laevis), XLU90895; Australian lungfish (Neoceratodus forsteri), EF526297; zebrafish (Danio rerio), NM131175; pigeon (Columba livia), AH007800; pouched lamprey (Geotria australis), AY366491.
Figure 5.
Difference absorbance spectra for the Rh1 (A), SWS1 (B), and LWS (C) visual pigments of X. unicolor, regenerated in vitro with 11-cis-retinal.
Regeneration of the sunbeam LWS opsin with 11-cis-retinal gave a λmax of 549.8 ± 1.8 nm (Fig. 5C). The spectral tuning of all vertebrate LWS pigments is influenced by residues present at five critical sites (164, 181, 261, 269, and 292), and the combination of SHYTA at these sites yields a spectral peak at 560 nm (Yokoyama, 2000). An identical complement of “tuning” residues is found in P. regius and X. unicolor LWS pigments, although, in both cases, the λmax is blue-shifted close to 550 nm, suggesting that other presently unidentified residues are responsible for the short-wavelength shift of snake LWS visual pigments.
An SWS1 opsin gene is expressed in the retina of both species, although we were unable to identify, by MSP, any short-wavelength-sensitive single cones in X. unicolor. An ultraviolet (UV)-sensitive cone class is present in P. regius (Sillman et al., 1999), and the in vitro expression of the X. unicolor SWS1 coding sequence showed that this pigment was also UV-sensitive with a peak at 360.8 ± 0.3 nm (Fig. 5B). The residue present at site 86 has been shown to be critical in determining the peak sensitivity in all nonavian pigments (Hunt et al., 2007). Both snake SWS1 pigments have F86, which is known to confer UV sensitivity (Cowing et al., 2002; Fasick et al., 2002).
Discussion
Phylogenetically, the three visual pigments present in the two species of henophidian snakes, P. regius and X. unicolor, studied here are identified as belonging to the Rh1 rod class and the SWS1 and LWS cone classes. Thus, these lower snakes have retained two of the four classes of ancestral vertebrate cone pigments, a reflection perhaps of the partial nature of their nocturnality/crepuscularity/fossoriality. A similar situation exists in the pure rod retinas of nocturnal Tokay geckos (Crescitelli et al., 1977), where three of the cone pigment classes, SWS1, Rh2, and LWS, have been retained (Kojima et al., 1992; Yokoyama and Blow, 2001). In these geckos, the middle-wavelength-sensitive photoreceptors which are described as rod-like on the basis of morphology and sensitivity to light (Pedier and Tilly, 1964; Tansley, 1964; Kleinschmidt and Dowling, 1975; Rispoli et al., 1993) express a “cone” Rh2 gene (Yokoyama and Blow, 2001), consistent with Walls' transmutation theory (Walls, 1934, 1942). In contrast, P. regius and X. unicolor have retained the Rh1 gene in their middle-wavelength-sensitive rod photoreceptors. Both snake species presumably possess dichromatic color vision based on the two cone pigments, although this will be restricted by the limited degree of spectral overlap at longer wavelengths between the 360 and 550 nm pigments. Many henophidian snakes are at least partly fossorial and/or crepuscular/nocturnal, which may explain the presence of only a small population of cone photoreceptors (Conant and Collins, 1998). Under such mesopic conditions, rod pigments may also be active (Reitner et al., 1991; Stabell and Stabell, 1994; Buck, 1997) and contribute to color vision (Sillman et al., 1999), a form of conditional trichromacy.
Interestingly, the garter snake, Thamnophis sirtalis (a higher or caenophidian snake), possesses a pure-cone retina with four cone classes. Two small single cone classes are spectrally tuned to either 360 or 482 nm, whereas the large single and double cones both have a spectral peak at 554 nm (Sillman et al., 1997). If the loss of the Rh2 pigment occurred in the ancestor of all extant snakes, then it is possible that in addition to cones expressing SWS1 and LWS pigments with spectral sensitivities of 360 and ∼555 nm, respectively (Sillman et al., 1997), the garter snake middle-wavelength-sensitive cone (with a λmax of 482 nm) contains an Rh1 “rod” visual pigment with a blue-shifted spectral peak such as has occurred in some deep-sea teleosts (Hunt et al., 2001), the coelacanth (Yokoyama et al., 1999), and in some marine mammals (Fasick and Robinson, 2000). Therefore, in higher snakes, the Rh1-expressing rod photoreceptor of the ancestral snake may have transmutated into a cone, a change that may be restricted to caenophidian or phylogenetically more deeply nested snakes and occurring in the reverse direction to that proposed by Walls' theory. In henophidian snakes, however, with their morphologically distinct rods and cones that express rod and cone pigments, respectively, it would appear that the transmutation of photoreceptors as proposed by Walls has not occurred and that unlike in geckos, nocturnality/crepuscularity/fossoriality may be an ancestral state. However, more data from other snake lineages will be required to test this hypothesis further.
Footnotes
This work was supported by grants awarded by the United Kingdom Biotechnological and Biological Sciences Research Council and the Leverhulme Trust. We thank Rosalie Crouch for supplying the 11-cis-retinal and Patrick Jean and Jodi Rowley for the photographs of P. regius and X. unicolor, respectively.
References
- Bowmaker JK, Astell S, Hunt DM, Mollon JD. Photosensitive and photostable pigments in the retinae of Old World monkeys. J Exp Biol. 1991;156:1–19. doi: 10.1242/jeb.156.1.1. [DOI] [PubMed] [Google Scholar]
- Buck SL. Influence of rod signals on hue perception: evidence from successive scotopic contrast. Vision Res. 1997;37:1295–1301. doi: 10.1016/s0042-6989(96)00276-3. [DOI] [PubMed] [Google Scholar]
- Cohen GB, Oprian DD, Robinson PR. Mechanism of activation and inactivation of opsin: role of Glu113 and Lys296. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- Conant R, Collins J. Boston: Houghton Mifflin; 1998. A field guide to reptiles and amphibians of eastern and central North America (Peterson field guide series) [Google Scholar]
- Cowing JA, Poopalasundaram S, Wilkie SE, Robinson PR, Bowmaker JK, Hunt DM. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem J. 2002;367:129–135. doi: 10.1042/BJ20020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli F, Dartnall H, Loew E. The visual pigments of the gecko and other vertebrate eyes. In: Dartnall H, editor. Handbook of sensory physiology. Berlin: Springer; 1977. pp. 391–450. [Google Scholar]
- Dong S, Kumazawa Y. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 2005;61:12–22. doi: 10.1007/s00239-004-0190-9. [DOI] [PubMed] [Google Scholar]
- Dratz EA, Hargrave PA. The structure of rhodopsin and the outer segment disc membrane. TIBS. 1983;8:128–131. [Google Scholar]
- Fasick JI, Robinson PR. Spectral-tuning mechanisms of marine mammal rhodopsins and correlations with foraging depth. Vis Neurosci. 2000;17:781–788. doi: 10.1017/s095252380017511x. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6965. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- Franke RR, Sakmar TP, Oprian DD, Khorana HG. A single amino acid substitution in rhodopsin (Lys248Leu) prevents activation of transducin. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. In search of the visual pigment template. Vis Neurosci. 2000;17:509–528. doi: 10.1017/s0952523800174036. [DOI] [PubMed] [Google Scholar]
- Greene H. Berkeley, CA: University of California; 1997. Snakes: the evolution of mystery in nature. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 2001;204:3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Carvalho LS, Cowing JA, Parry JW, Wilkie SE, Davies WL, Bowmaker JK. Spectral tuning of shortwave-sensitive visual pigments in vertebrates. Photochem Photobiol. 2007;83:303–310. doi: 10.1562/2006-06-27-IR-952. [DOI] [PubMed] [Google Scholar]
- Karnik SS, Khorana HG. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J Biol Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- Kaushal S, Ridge KD, Khorana HG. Structure and function of rhodopsin: the role of asparagine-linked glycosylation. Proc Natl Acad Sci U S A. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt J, Dowling JE. Intracellular recordings from gecko photoreceptors during light and dark adaptation. J Gen Physiol. 1975;66:617–648. doi: 10.1085/jgp.66.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima D, Okano T, Fukada Y, Shichida Y, Yoshizawa T, Ebrey TG. Cone visual pigments are present in gecko rod cells. Proc Natl Acad Sci U S A. 1992;89:6841–6845. doi: 10.1073/pnas.89.15.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Liebman PA, Entine G. Sensitive low-light-level microspectrophotometer: detection of photosensitive pigments of retinal cones. J Opt Soc Am. 1964;54:1451–1459. doi: 10.1364/josa.54.001451. [DOI] [PubMed] [Google Scholar]
- Mollon JD, Bowmaker JK, Jacobs GH. Variations of colour vision in a New World primate can be explained by polymorphism of retinal photopigments. Proc R Soc Lond B Biol Sci. 1984;222:373–399. doi: 10.1098/rspb.1984.0071. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Abdulaev NG, Zolotarev AS, Artamonov ID, Bespalov IA, Dergachev AE, Tsuda M. Octopus rhodopsin. Amino acid sequence deduced from cDNA. FEBS Lett. 1988;232:69–72. doi: 10.1016/0014-5793(88)80388-0. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Buczyłko J, Lebioda L, Crabb JW, Polans AS. Identification of the N-terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J Biol Chem. 1993;268:6004–6013. [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Pedler C, Tilly R. The nature of the Gecko visual cell. A light and electron microscopic study. Vision Res. 1964;4:499–510. doi: 10.1016/0042-6989(64)90056-2. [DOI] [PubMed] [Google Scholar]
- Reitner A, Sharpe LT, Zrenner E. Is colour vision possible with only rods and blue-sensitive cones? Nature. 1991;352:798–800. doi: 10.1038/352798a0. [DOI] [PubMed] [Google Scholar]
- Rispoli G, Sather WA, Detwiler PB. Visual transduction in dialysed detached rod outer segments from lizard retina. J Physiol. 1993;465:513–537. doi: 10.1113/jphysiol.1993.sp019691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillman AJ, Govardovskii VI, Röhlich P, Southard JA, Loew ER. The photoreceptors and visual pigments of the garter snake (Thamnophis sirtalis): a microspectrophotometric, scanning electron microscope and immunological study. J Comp Physiol [A] 1997;181:89–101. doi: 10.1007/s003590050096. [DOI] [PubMed] [Google Scholar]
- Sillman AJ, Carver JK, Loew ER. The photoreceptors and visual pigments in the retina of a boid snake, the ball python (Python regius) J Exp Biol. 1999;202:1931–1938. doi: 10.1242/jeb.202.14.1931. [DOI] [PubMed] [Google Scholar]
- Sillman AJ, Johnson JL, Loew ER. Retinal photoreceptors and visual pigments in Boa constrictor imperator. J Exp Zool. 2001;290:359–365. doi: 10.1002/jez.1076. [DOI] [PubMed] [Google Scholar]
- Stabell U, Stabell B. Mechanisms of chromatic rod vision in scotopic illumination. Vision Res. 1994;34:1019–1027. doi: 10.1016/0042-6989(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tansley K. The gecko retina. Vision Res. 1964;4:33–37. doi: 10.1016/0042-6989(64)90029-x. [DOI] [PubMed] [Google Scholar]
- Walls G. The reptilian retina. I. A new concept of visual cell evolution. Am J Ophthalmol. 1934;17:892–915. [Google Scholar]
- Walls G. The vertebrate eye and its adaptive radiation. Bloomfield Hills, MI: Cranbrook Institute of Science; 1942. Duplicity and transmutation; pp. 163–168. [Google Scholar]
- Yan J, Li H, Zhou K. Evolution of the mitochondrial genome in snakes: gene rearrangements and phylogenetic relationships. BMC Genomics. 2008;9:569. doi: 10.1186/1471-2164-9-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Blow NS. Molecular evolution of the cone visual pigments in the pure rod-retina of the nocturnal gecko, Gekko gekko. Gene. 2001;276:117–125. doi: 10.1016/s0378-1119(01)00643-6. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Zhang H, Radlwimmer FB, Blow NS. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae) Proc Natl Acad Sci U S A. 1999;96:6279–6284. doi: 10.1073/pnas.96.11.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wensel TG, Yuan C. Tokay gecko photoreceptors achieve rod-like physiology with cone-like proteins. Photochem Photobiol. 2006;82:1452–1460. doi: 10.1562/2006-01-05-RA-767. [DOI] [PubMed] [Google Scholar]
- Zhao X, Haeseleer F, Fariss RN, Huang J, Baehr W, Milam AH, Palczewski K. Molecular cloning and localization of rhodopsin kinase in the mammalian pineal. Vis Neurosci. 1997;14:225–232. doi: 10.1017/s0952523800011366. [DOI] [PubMed] [Google Scholar]