Abstract
A pathological hallmark of the Huntington's disease (HD) is intracellular inclusions containing a huntingtin (Htt) protein with an elongated polyglutamine tract. Aggregation of mutant Htt causes abnormal protein–protein interactions, and the functional dysregulation of aggregate-interacting proteins (AIPs) has been proposed as a pathomechanism of HD. Despite this, a molecular mechanism remains unknown how Htt aggregates sequester AIPs. We note an RNA-binding protein, TIA-1, as a model of AIPs containing a Q/N-rich sequence and suggest that in vitro and in vivo Htt fibrillar aggregates function as a structural template for inducing insoluble fibrillation of TIA-1. It is also plausible that such a cross-seeding activity of Htt aggregates represses the physiological function of TIA-1. We thus propose that Htt aggregates act as an intracellular hub for the cross-seeded fibrillation of Q/N-rich AIPs and that a cross-seeding reaction is a molecular origin to cause diverse pathologies in a polyglutamine disease.
Introduction
Huntington's disease (HD) is a neurodegenerative disorder caused by an abnormal expansion of CAG repeats located in exon I of the HD gene (Zoghbi and Orr, 2000). The CAG repeat encodes a polyglutamine [poly(Q)] stretch in the HD gene product, huntingtin (Htt), and is highly polymorphic; 6–34 repeats in unaffected individuals, but >36 repeats in HD patients (Zoghbi and Orr, 2000). Compared with other neurodegenerative diseases such as Alzheimer's disease, a disease pathology seem more diverse in HD: namely, a wider variety of cellular systems including protein degradation, transcription, and RNA metabolism become dysfunctional (Zoghbi and Orr, 2000).
Several pathomechanisms of HD have been proposed based on a high propensity of an expanded poly(Q) for aggregation, which is evidenced by the observation that mutant Htt is deposited as inclusion bodies inside affected neurons (Davies et al., 1997). Although it remains unclear whether Htt aggregates themselves exert neurotoxicity, mutant Htt has been proposed to sequester many other proteins [aggregate-interacting proteins (AIPs)] including glutamine (Q)/asparagine (N)-rich proteins, leading to cellular dysfunction (Nucifora et al., 2001; Schaffar et al., 2004). Given some of these AIPs remain entrapped in Htt aggregates even after harsh treatment with a strong ionic detergent (Mitsui et al., 2002), irreversible interactions with mutant Htt will be detrimental to physiological functions of AIPs. It is thus tempting to speculate that dysfunction of various AIPs on coaggregation with Htt produces pathological diversity of HD. Despite this, a molecular mechanism underlying coaggregation of AIPs with mutant Htt remains an open question.
Htt with an expanded poly(Q) tract forms insoluble, amyloid-like fibrillar aggregates (Scherzinger et al., 1997). A preformed fibril can act as a seed that significantly accelerates the transition from a soluble protein into an insoluble fibril, and a seed accelerates fibrillation of the same protein molecule with a seed constituent (Harper and Lansbury, 1997). Seeding reactions also occur between different kinds of protein molecules with low sequence identity, albeit with low efficiency in general, which is called as a cross-seeding reaction (O'Nuallain et al., 2004). Unlike a transient association with aggregates, we hypothesize that a possible cross-seeding reaction of Htt integrates some of AIPs into very rigid structure of Htt fibrils, thereby resulting in the functional inactivation of integrated AIPs. To understand the coaggregation process at a molecular level, we note T-cell intracellular antigen-1 (TIA-1) as a model of AIPs in poly(Q) diseases (Waelter et al., 2001).
In this study, we have revealed that TIA-1 is sequestered in inclusions in a HD model mouse. Although TIA-1 contains no poly(Q) tract (at most three consecutive glutamines) in its primary sequence, our results show that in vitro Htt aggregates efficiently seed the fibrillation of TIA-1 through a C-terminal Q/N-rich domain of TIA-1. Using a cultured cell model, we also show that the coaggregation of TIA-1 with mutant Htt decreases amounts of soluble TIA-1 and represses the TIA-1 function as a translational silencer. We thus propose that Htt aggregates act as a hub for recruiting cellular components through a cross-seeding mechanism.
Materials and Methods
Preparation of recombinant proteins.
For facilitation of protein purification, a 6× His tag was introduced at the N terminus of glutathione-S-transferase (GST) (a pGEX6P-2 vector; GE Healthcare). GST-Htt42Q was expressed in Escherichia coli (Rosetta) by 0.1 mm isopropyl β-d-thiogalactoside (IPTG) at 20°C for 2 d and purified with Proteus Midi IMAC (Pro-Chem) and glutathione Sepharose (GE Healthcare). TIA-1C (Met 310 to Gln 406) was also N-terminally fused with a His-tagged GST protein, and its expression in E. coli (Rosetta) was induced by 1 mm IPTG at 37°C for 6 h. GST-TIA-1C was obtained as inclusion bodies and purified with Proteus Midi IMAC in the presence of 8 m urea. Refolding of GST-TIA-1C was performed by 60-fold dilution in 20 mm Tris/100 mm NaCl/5 mm DTT/1 mm EDTA, pH 9, with stirring overnight at 4°C. TIA-1FL and TIA-1N (Met 1 to Asp 309) were cloned using a vector, pET15b (Novagen), and the expression in E. coli (Rosetta) was induced by 0.4 mm IPTG and 0.4% glucose at 30°C for 3 h. Both proteins remain in a soluble fraction and were purified with Proteus Midi IMAC. Protein concentration was determined spectroscopically: 42,860 cm−1 m−1 (GST-Htt42Q), 83,770 cm−1 m−1 (GST-TIA-1C), 80,330 cm−1 m−1 (TIA-1FL), and 39,420 cm−1 m−1 (TIA-1N) of an extinction coefficient at 280 nm.
For modification with thiol-specific Alexa dyes, all four Cys residues in GST are mutated to Ser (GSTnoCys), and a unique Cys residue was further introduced at the N terminus of Htt42Q or TIA-1C. In 50 mm Tris/8 m urea/1 mm tris(2-carboxyethyl)phosphine (TCEP)/1 mm EDTA, pH 7.0, 250 μm GSTnoCys-Cys-Htt42Q or GSTnoCys-Cys-TIA-1C was incubated with 1 mm Alexa 555 or Alexa 488 maleimide (Invitrogen) for 1 h at room temperature, respectively. Modified proteins were refolded with 60-fold dilution and overnight stirring in 20 mm Tris/5 mm DTT/1 mm EDTA, pH 9, at 4°C. Followed by buffer exchange to 50 mm Tris/1 mm EDTA, pH 7.0, additional purification was performed using glutathione Sepharose.
Biochemical characterization of protein fibrillation.
Kinetics of protein fibrillation was monitored by thioflavin T fluorescence using SpectraMax M2 (Molecular Devices). In a 96-well plate, 150 μl of the sample solution containing 5 μm proteins in 100 mm Na-Pi/100 mm NaCl/16.7 μm thioflavin T, pH 8.0, were set per well. A fibrillation reaction starts by adding 2 U of a HRV3C protease (Novagen), which specifically cleaves the site between GST and Htt42Q/TIA-1C. A fluorescence signal was monitored at 20°C at intervals of 2 min with 442 and 485 nm of excitation and emission wavelength, respectively. The plate was shaken for 5 s before each fluorescence reading. For a seeding reaction, no plate shaking was performed before each measurement.
Protein aggregation was also examined in a 96-well plate using a BioShaker MBR-024 (TAITEC). One-hundred fifty microliters of 10 μm proteins in 100 mm Na-Pi/100 mm NaCl/1 mm EDTA/5 mm TCEP, pH 7.0, were set per well, which was shaken overnight at 1200 rpm, 37°C. For a measurement of thioflavin T fluorescence, 10 μl of the sample solution was mixed with 90 μl of 100 mm Na-Pi/100 mm NaCl/1 mm EDTA/25 μm thioflavin T, pH 7.0. A fluorescence signal was obtained using a plate reader, ARVO MX (PerkinElmer), with a CW-lamp filter (440 nm cutoff) and an emission filter (486 nm cutoff). For a Congo red binding assay, a sample solution (150 μl; 10 μm) was first ultracentrifuged (TLA55; Beckman Coulter) at 50,000 rpm for 30 min at 4°C, and the protein precipitate was then resuspended in 50 μl of 100 mm Na-Pi/100 mm NaCl/1 mm EDTA, pH 7.0. Ten microliters of the resuspended solution was mixed with 90 μl of 100 mm Na-Pi/100 mm NaCl/1 mm EDTA/5 μm Congo red, pH 7.0, and electronic absorption spectra were recorded using Shimadzu UV-2400PC.
To observe aggregate morphologies using an electron microscope, protein aggregates were first adsorbed on 400-mesh grids coated by a glow-charged supporting membrane. For an immunoelectron microscopic observation, the aggregates adsorbed on the grids were further incubated with mouse monoclonal anti-Htt (EM48; Millipore Bioscience Research Reagents) and rabbit polyclonal anti-hemagglutinin (HA) (Y-11; Santa Cruz Biotechnology) antibodies in 1:250 dilution. After being washed with 0.1 m phosphate buffer, the aggregates were then incubated with colloidal gold-conjugated secondary antibodies (anti-mouse with 5 nm gold particle and anti-rabbit with 10 nm gold particle; British Biocell) in 1:250 dilution. After washing with 0.1 m phosphate buffer and pure water, negative staining with neutralized 2% sodium phosphotungstic acid was performed. Images were obtained using an electron microscope (1200EX; JEOL).
Images of atomic force microscopy were acquired on a Digital Instruments Multimode Nanoscope IIIa scanning microscope using a J-Scanner. Solution containing TIA-1C aggregates (10 μl) were deposited on freshly cleaved mica substrate and incubated for 1 min. Unbound proteins and salt in the buffer solution were washed away with 50 μl of H2O. Imaging was performed in tapping mode with RTESP (Veeco) cantilevers and acquired at a scan rate of 1.0 Hz with 512 lines per image.
Transgenic mice.
The mouse experiments were approved by the animal experiment committee of the RIKEN Brain Science Institute. Details of transgenic mice, R6/2, have been published previously (Davies et al., 1997). Brain homogenates were fractionated as previously described (Díaz-Hernández et al., 2004), and an outline of the fractionation is also shown in Figure 1D. Slight modifications of the fractionation were performed as follows; after treatment with 1% Sarkosyl in PBS containing 1% β-mercaptoethanol, the pellet was washed with 4% Sarkosyl in PBS. Additional wash of the pellet was done with 2% SDS in PBS. The pellet was then dissolved by incubation in 100% formic acid at 37°C for 1 h, dried up using SpeedVac, and redissolved in PBS containing 2% SDS. Protein concentration was determined by a BCA assay using BSA as a standard.
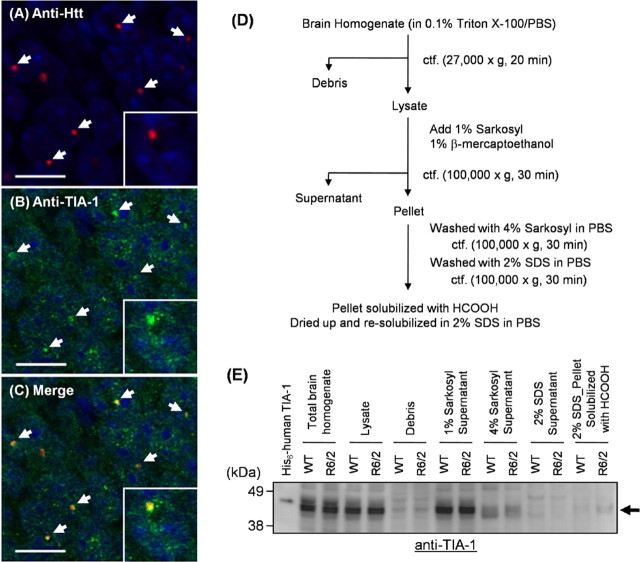
Figure 1.
Endogenous TIA-1 is colocalized with Htt inclusions in R6/2 mouse brain. A–C, Immunohistochemistry of hippocampus CA1 of an R6/2 brain was performed by double staining with mouse anti-Htt [EM48; red (A)] and goat anti-TIA-1 [C-20; green (B)] followed with Alexa 546-anti-mouse and Alexa 488-anti-goat antibodies (scale bar, 10 μm). Images were obtained by a confocal microscopy. A merged image is shown in C, and colocalization of endogenous TIA-1 with Htt inclusions is shown by white arrows. Nuclei were counterstained with TOTO-3 (blue). A magnified image is also shown in the inset of each panel. Brain homogenates of R6/2 and WT mice at 12 weeks of age were prepared as shown in D, and each fraction was analyzed by Western blot using anti-TIA-1 antibody (E). Recombinant 6× His-tagged human TIA-1 (molecular weight, 45,090) was loaded as a positive control and exhibits slower mobility than endogenous mouse TIA-1 (molecular weight, 42,800).
Plasmid construction for cell transfection.
All plasmids including TIA-1 variants, Htt60Q-green fluorescent protein (GFP), Htt18Q-yellow fluorescent protein (YFP), Htt62Q-YFP, Htt62Q-cyan fluorescent protein (CFP), and Htt150Q-CFP were constructed using either pIRESneo3 (Clontech) or pcDNA3.1 (Invitrogen). YFP used in this study is an improved version of YFP called Venus (Nagai et al., 2002). In some experiments, coexpression of Htt18Q/62Q-YFP and TIA-1FL-HA was performed by using a modified pIRESneo3 vector (pIRES-TIA-1), in which a neomycin phosphotransferase coding sequence (SmaI/XbaI) is replaced with TIA-1FL-HA, and Htt18Q/62Q-YFP was inserted in the multiple cloning site (AgeI/BamHI) of pIRES-TIA-1.
A pre-micro-RNA (miRNA) sequence targeting a human TIA-1 gene was cloned into a pcDNA6.2-GW/EmGFP-miR vector (Invitrogen). A forward sequence is as follows: 5′-TGCTGATGTGCTGCACTT-TCATGGGAGTTTTGGCCACTGACTGACTCCCATGAGTGCAGC-ACAT-3′. An miR-LacZ provided by Invitrogen was used as a negative control for miRNA experiments.
Cell culture and sample preparations.
Neuro2a cells as well as HEK293T cells were maintained in DMEM containing 10% FBS and penicillin/streptomycin and transfected with plasmid(s) indicated in each experiment using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
For preparation of Htt/TIA-1 coaggregates formed in cells, Neuro2a cells in a six-well plate were transfected per well with 2 μg of pIRESneo3 containing Htt62Q-YFP together with 2 μg of pIRESneo3 containing either TIA-1FL-HA or TIA-1C-HA. After 4 h of transfection, a medium was changed to DMEM containing 10% FBS and 5 mm N6,2′-O-dibutyryl cAMP (dbcAMP) (Nacalai Tesque). The cells were further incubated for a day and then lysed with 2% SDS in PBS with a Complete protease inhibitor mixture (Roche), sonicated, and ultracentrifuged with 50,000 rpm for 30 min at 4°C (TLA55; Beckman Coulter). Pellets were washed once with 2% SDS/PBS and twice with PBS. After being resuspended in PBS, the pellets were immunodecorated by anti-Htt and anti-HA antibodies with gold-conjugated secondary antibodies (5 and 10 nm, respectively) and observed by an electron microscope.
For functional analysis of TIA-1 in a cell, HEK293T cells in a six-well plate were transfected with 2 μg of pIRESneo3 or pIRES-TIA-1 containing Htt18/62Q-YFP cDNA and 2 μg of pPUR (Clontech). After 24 h of transfection, 1.5 μg/ml puromycin was added.
TIA-1 knockdown experiments were performed by transfecting HEK293T cells with 4 μg of pcDNA6.2-GW/EmGFP-miR containing miR-LacZ (negative control) or miR-TIA-1. After 24 h transfection, 10 μg/ml blasticidin was added.
In the functional analysis of TIA-1 as well as TIA-1 knockdown experiments, the cells were further incubated for 2 d after addition of antibiotics and then lysed in 150 μl of a radioimmunoprecipitation assay (RIPA) buffer containing a Complete protease inhibitor mixture with sonication. After ultracentrifugation (TLA55; Beckman Coulter) at 50,000 rpm for 30 min at 4°C, a supernatant was saved as a RIPA-soluble fraction. The pellet was washed once with a RIPA buffer, sonicated, and incubated in 100 μl of formic acid at 37°C for 1 h. After being dried up using a Speed-Vac, the pellet fraction was redissolved/sonicated in 150 μl of 100 mm Na-Pi/100 mm NaCl/2% SDS, pH 7.0 and saved as an RIPA-insoluble fraction. Protein concentration was determined by a BCA assay using BSA as a standard. Amounts of TIA-1 and Htt in both RIPA-soluble and -insoluble fractions were estimated by Western blotting.
Immunostaining of cultured cells and mouse tissues.
Neuro2a cells seeded on a four-well chamber slide were cotransfected with 0.72 μg each of Htt60Q-GFP in pcDNA3.1 and TIA-1 variants C-terminally tagged with HA in pIRESneo3. After 4 h of transfection, cells were differentiated by exchanging the culture medium to DMEM containing 10% FBS and 5 mm dbcAMP. After 2 d incubation at 37°C, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The antibody dilutions were 1:300 for rabbit polyclonal anti-HA (Y-11) and 1:1000 for anti-rabbit IgG conjugated with Alexa 555 (Invitrogen). Nuclei were counterstained with Hoechst 33342. Images were obtained by using a fluorescence microscope (Olympus IX70, LCPlan Fl ×40).
For immunostaining of mouse brain tissues, mice were anesthetized and immediately fixed by perfusion through the left ventricle with 4% paraformaldehyde in PBS. A brain was collected and further fixed with the same fixative overnight, followed by treatment with 30% sucrose in PBS for 2 d and then with 20% sucrose solution overnight. The brains of R6/2 mice (12 weeks of age) mounted in Tissue mount (Chiba Medical) were cut into 10 μm sections with cryostat. For immunostaining, the primary antibody dilutions were used as follows: 1:500 for mouse monoclonal anti-Htt (EM48), 1:1000 for goat polyclonal anti-TIA-1 (C-20; Santa Cruz Biotechnology). The secondary antibody dilutions were 1:300 for goat anti-mouse IgG conjugated with Alexa 546 (Invitrogen) and donkey anti-goat IgG conjugated with Alexa 488 (Invitrogen). Nuclei were counterstained with TOTO-3. After being mounted using Immu-mount (Thermo Fisher Scientific), the tissues were observed under a confocal microscope (TCS SP2; Leica Microsystems).
Electrophoresis.
Three or 10 μg of total proteins indicated in each experiment were boiled in an SDS-PAGE sample buffer containing β-mercaptoethanol and loaded on a 5–20% gradient or a 15% SDS-PAGE gel (PAGEL; ATTO). After electrophoresis, proteins were electroblotted on a 0.2 μm PVDF (polyvinylidene difluoride) membrane (Bio-Rad) and analyzed by Western blotting using either mouse monoclonal anti-poly(Q) (1C2; Millipore Bioscience Research Reagents; 1:1000), goat polyclonal anti-TIA-1 (C-20; 1:1000), mouse anti-GFP (Roche; 1:1000), or mouse monoclonal anti-cytochrome c (Cytc) (7H8.2C12; BD Biosciences Pharmingen; 1:1000) as a primary antibody. Corresponding secondary antibodies that are conjugated with HRP were used: anti-goat (1:3000) and anti-mouse (1:1000) antibodies. Blots were developed with the SuperSignal West Dura (Pierce), and images were obtained using LAS1000 (FUJI FILM).
Time-lapsed live-cell imaging.
Neuro2a cells were cotransfected with a pair of Htt62Q-CFP and TIA-1FL-YFP or Htt150Q-CFP and TIA-1C-red fluorescent protein (RFP) on a glass-bottom dish (MatTeck). After 24 h of transfection, an observation of cells is started using a time-lapsed confocal microscopy. During the experiment, cells were maintained at 37°C with 5% CO2 in a live-cell incubation chamber. Imaging was performed on an Olympus FV1000 confocal microscope equipped with a 60× UPlanSApo oil objective lens, a CO2 live-cell incubator (MI-IBC-IF), a motorized XY stage, and an IR autofocus system (ZDC-IMAGE). Images at 10 different sites were acquired at intervals of 5 min for 10 h. For each site, five different Z planes (1 μm) in 110 μm pinhole were acquired to account for the aggregate location in the Z plane. CFP, YFP, and RFP were excited by 440 nm light-emitting diode, 515 nm argon, and 547 nm He–Ne laser, respectively. Fluorescence from CFP, YFP, and RFP is observed between 460 and 500 nm, 530 and 630 nm, and 600 and 700 nm emission wavelength, respectively.
Results
TIA-1 colocalizes with Htt inclusions in transgenic mice expressing Htt exon I with abnormal length of poly(Q)
Htt constitutes intranuclear inclusions when the length of consecutive glutamine residues in exon I exceeds a pathogenic threshold (∼36 glutamines). An R6/2 transgenic mouse, in which human Htt exon I with ∼130 glutamines is expressed, mimics pathological processes of HD including the formation of neuronal intranuclear inclusions (Davies et al., 1997). In R6/2 mice, mutant Htt starts to form inclusions at an early stage of the disease (4 weeks), and accumulates until the disease endstage (12 weeks). Although TIA-1 has been implied to colocalize with inclusions of mutant Htt in a cultured cell model (Waelter et al., 2001), we first examined whether TIA-1 is a constituent of pathological inclusions in an R6/2 mouse brain. Double immunostaining of a brain section of an R6/2 mouse at 12 weeks of age has been performed using anti-Htt (EM48) and anti-TIA-1 (C-20) antibodies. We confirmed intranuclear Htt-positive inclusions in cortex, hippocampus, and striatum (data not shown) and found that extensive colocalization of TIA-1 with Htt-positive inclusions especially in hippocampus (Fig. 1A–C). These immunohistochemical observations suggest that TIA-1 is recruited into pathological HD inclusions in vivo.
Aggregates of mutant Htt remain insoluble in 2% SDS but can be solubilized after treatment with 100% formic acid (Hazeki et al., 2000). To examine biochemical alterations of TIA-1 on its recruitment into pathological inclusions, a fractionation was performed using homogenates of wild-type (WT) and R6/2 mouse brain at 12 weeks of age (Díaz-Hernández et al., 2004) (Fig. 1D). As expected, Htt aggregates solubilized with formic acid are detected in a 2% SDS-resistant fraction of an R6/2 mouse brain homogenate (Fig. 2). Although current experimental conditions do not detect difference in Sarkosyl-soluble amounts of endogenous TIA-1 between WT and R6/2 mice at 12 weeks of age, TIA-1 remains in a final SDS-resistant pellet fraction in R6/2 but not WT mice (Fig. 1D,E). Furthermore, the insolubilization of TIA-1 associated with the inclusion formation is found to be age dependent (Fig. 2). At 4 weeks of age, significant amounts of Htt aggregates are already found in an SDS-resistant pellet, whereas insoluble TIA-1 is not yet detected. TIA-1 starts to accumulate in SDS-resistant pellets between 4 and 8 weeks of age, and amounts of SDS-insoluble TIA-1 increase at 12 weeks of age in R6/2 mice. Together, a recruitment of TIA-1 into pathological inclusions occurs after Htt aggregation and leads to decrease the solubility of a TIA-1 protein.
Figure 2.
Htt aggregation precedes TIA-1 insolubilization in R6/2 mouse brain. The SDS-insoluble fraction redissolved with HCOOH treatment was prepared from WT and R6/2 mouse brains at 4, 8, and 12 weeks of age. Ten micrograms of total proteins were loaded on a gradient gel and analyzed by Western blot using anti-poly(Q) (1C2) or anti-TIA-1 (C-20) antibodies. Calculated molecular weight of exon I of Htt with 130Q is 24,075 Da, but it is known that a long poly(Q) tract retards the electrophoretic mobility and increases apparent molecular weight. In addition, redissolved Htt aggregates exhibit smear signals in the high molecular weight region probably because of the SDS-resistant interactions between long poly(Q) tracts in Htt proteins.
C-terminal domain of TIA-1 is responsible for its recruitment into Htt inclusions
TIA-1 is comprised of three RNA recognition motifs at its N terminus and a Q/N-rich auxiliary domain at its C terminus (Tian et al., 1991) (Fig. 3A). To test which region(s) of TIA-1 is responsible for the recruitment into inclusions containing mutant Htt, exon I of Htt with a pathological (60Q) length of a poly(Q) tract (Htt60Q) was coexpressed with either a full-length (TIA-1FL, 1–406), N-terminal RNA recognition motifs (TIA-1N, 1–309), or a C-terminal Q/N-rich domain (TIA-1C, 310–406) of human TIA-1 in mouse neuroblastoma cells, Neuro2a. Htt60Q and TIA-1 variants were C-terminally fused with GFP and HA, respectively. After incubation for 2 d, Htt60Q-GFP exhibits cytoplasmic inclusions, with which TIA-1FL-HA and TIA-1C-HA are colocalized (Fig. 3B,D). In contrast, TIA-1N-HA is segregated from Htt60Q-GFP inclusions (Fig. 3C). We also attempted to test specificity of the recruitment of TIA-1FL-HA or TIA-1C-HA into Htt inclusions; for that purpose, we note Cu,Zn-superoxide dismutase (SOD1) with a G85R mutation, which is causative for familial form of amyotrophic lateral sclerosis (Rosen et al., 1993). Overexpression of SOD1G85R C-terminally tagged with GFP (SOD1G85R-GFP) in Neuro2a cells leads to formation of cytoplasmic inclusions (Furukawa et al., 2008); however, TIA-1FL-HA or TIA-1C-HA was not recruited into the SOD1 inclusions (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). These immunocytochemical results thus show that interaction of TIA-1 with Htt inclusions does not appear to be nonspecific and that TIA-1 interacts with Htt inclusions through a C-terminal Q/N-rich domain.
Figure 3.
C-terminal Q/N-rich domain of TIA-1 is responsible for coaggregation with Htt. A, Schematic representation of domain structure of TIA-1. Thirty-one percent of a C-terminal domain corresponds to glutamine (Q) or asparagine (N) residues. Neuro2a cells were doubly transfected with Htt60Q-GFP and TIA-1FL-HA (B), TIA-1N-HA (C), or TIA-1C-HA (D). Two days after transfection, cells were fixed, stained with rabbit anti-HA antibody and Alexa 555-anti-rabit antibodies (shown in red), and observed using a fluorescence microscope. Nuclei were counterstained using Hoechst 33342 (blue). Htt60Q-GFP forms cytoplasmic inclusions in all three cases (shown in green), and colocalization was observed in TIA-1FL-HA and TIA-1C-HA, but not in TIA-1N-HA.
TIA-1 itself is fibrillogenic and integrated into Htt fibrillar aggregates
We next sought to decipher a molecular mechanism of how TIA-1 is recruited into inclusions. Htt exon I with 42 glutamines (Htt42Q) was prepared as a fusion protein with GST (GST-Htt42Q) (Scherzinger et al., 1997). Treatment of GST-Htt42Q with a protease, HRV3C, specifically cleaves the site between Htt42Q and a GST-tag within 10 min (data not shown), and additional incubation at 20°C for 6 h results in the formation of fibrillar aggregates (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). These in vitro Htt fibrils well reproduce the pathological Htt aggregates purified from inclusions in patients and rodent models of HD (Díaz-Hernández et al., 2004).
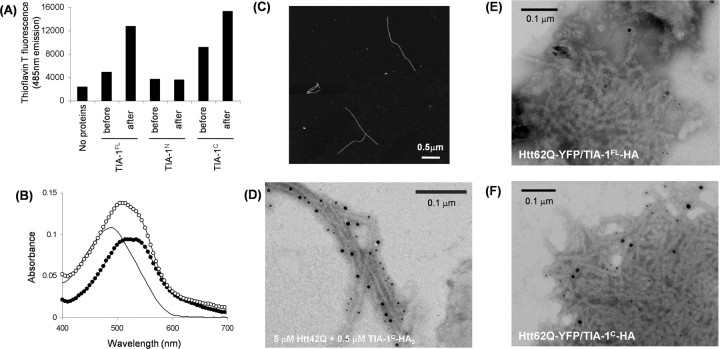
Interestingly, overnight agitation of 10 μm TIA-1FL at 37°C increases the intensity of thioflavin T fluorescence (Fig. 4A). Thioflavin T fluoresces on specific binding to β-sheet rich protein fibrils (Naiki et al., 1989). In addition, the insoluble TIA-1FL aggregates red-shift the absorption peak of Congo red (Fig. 4B). These are typical tinctorial changes on formation of amyloid-like fibrillar aggregates (Harper and Lansbury, 1997). To identify the fibrillogenic region of TIA-1, a soluble form of either TIA-1N or TIA-1C was agitated overnight at 37°C. We found that TIA-1C but not TIA-1N forms the thioflavin T and Congo red-positive aggregates (Fig. 4A,B). Fibrillar morphology of TIA-1C aggregates was further confirmed by an atomic force microscopy (Fig. 4C). Given a high propensity of TIA-1 itself for fibrillation, therefore, we hypothesize that mutant Htt can form a mixed fibril with TIA-1.
Figure 4.
TIA-1 forms amyloid-like fibrils and is integrated in Htt42Q fibril. A, Fluorescence intensity from thioflavin T before and after overnight agitation of 10 μm TIA-1FL, TIA-1N, and TIA-1C at 37°C, 1200 rpm. Excitation and emission wavelengths were 442 and 485 nm, respectively. TIA-1C was prepared by adding an HRV3C protease to GST-TIA-1C. B, Electronic absorption spectra of Congo red in the presence of either TIA-1FL aggregates (curve with open circles) or TIA-1C aggregates (curve with filled circles). A plain curve shows an absorption spectrum of Congo red alone. C, Fibrillar morphologies of TIA-1C aggregates were observed by an atomic force microscopy. D, An immunoelectron microscopic image of Htt42Q aggregated in the presence of TIA-1C-HA3. Htt is decorated with smaller gold particles (5 nm diameter), whereas larger gold particles (10 nm diameter) detect TIA-1C-HA3. E, F, Immunoelectron microscopic images of SDS-insoluble aggregates prepared from Neuro2a cells transfected with Htt62Q-YFP and TIA-1FL-HA (E) or Htt62Q-YFP and TIA-1C-HA (F). Anti-Htt, Smaller gold particles (5 nm diameter); anti-HA, larger gold particles (10 nm diameter).
To test this hypothesis in in vitro conditions, 5 μm GST-Htt42Q was incubated with 0.5 μm of a GST-TIA-1C protein that is further C-terminally fused with a triple HA tag (GST-TIA-1C-HA3). A GST tag in both proteins was liberated by adding a specific protease, HRV3C, during incubation for aggregation. Htt42Q/TIA-1C mixed aggregates were then observed by an electron microscopy after immunodecoration by anti-Htt and anti-HA antibodies. As shown in Figure 4D, we found that TIA-1C-HA3 is integrated in Htt42Q fibrils. Because TIA-1 remains SDS-soluble in the absence of Htt inclusion formation (Figs. 1E, 2), mutant Htt/TIA-1 mixed fibrils would form in the pathological inclusions found in HD.
Indeed, recruitment of TIA-1 in in vivo Htt fibrils was supported by immunoelectron micrograms of SDS-insoluble pellets that were prepared from Neuro2a cells doubly transfected with Htt60Q-GFP and TIA-1FL or C-HA. Htt60Q-GFP forms fibrillar aggregates, which were reacted with an anti-Htt (EM48) antibody, and an anti-HA antibody detects TIA-1FL-HA or TIA-1C-HA in SDS-insoluble Htt fibrils (Fig. 4E,F; supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Such fibrillar structures were not found in the SDS-insoluble fraction of cells transfected with TIA-1FL or C-HA alone (data not shown). These results hence suggest that Htt aggregates induce the conformational transition of TIA-1 from a soluble state into insoluble aggregates in vitro and in vivo.
Htt fibril functions as an efficient seed for in vitro fibrillation of TIA-1C
In in vitro mutant Htt fibrillation, a nucleation step precedes a fibril elongation step, which was monitored by fluorescence increase of thioflavin T after a lag time (Fig. 5A, blue curve). Interestingly, addition of soluble 0.5 μm TIA-1C to the solution containing 5 μm Htt42Q extended the lag time of Htt fibrillation (from ∼100 to ∼400 min) (Fig. 5A, blue and black curves), implying that soluble TIA-1 decelerates fibrillation of mutant Htt by protein–protein interactions. Once mutant Htt starts to fibrillate, however, TIA-1 becomes intertwined in a fibrillar structure of mutant Htt aggregates as observed in Figure 4D.
Figure 5.
Htt aggregates function as an efficient seed for accelerating TIA-1C fibrillation. A, B, A cross-seeding activity of TIA-1C and Htt42Q aggregates toward Htt42Q and TIA-1C fibrillation, respectively. A, GST-Htt42Q (5 μm) was incubated with 0.5 μm of either soluble GST-TIA-1C, Htt42Q aggregates, or TIA-1C aggregates at 20°C in 100 mm Na-Pi/100 mm NaCl/1 mm EDTA/16.7 μm thioflavin T/2 U of HRV3C, pH 8.0. B, GST-TIA-1C (5 μm) was incubated with 0.5 μm of either soluble GST-Htt42Q, Htt42Q aggregates, or TIA-1C aggregates in the same experimental conditions. Fibrillation was monitored by increase of thioflavin T fluorescence (442 nm excitation and 485 nm emission wavelength). C–E, Direct observation of TIA-1C fibrils (shown in green) seeded with Htt42Q aggregates (shown in red) by a fluorescence microscope (Olympus IX70, UPLAPO100x013). Soluble TIA-1C modified with Alexa 488 was incubated with seeds of Alexa 555-modified Htt42Q. A cross-seeding reaction proceeds on a glass slide covered with a cover glass at 37°C overnight. Magnified images are shown in D and E.
As a possible mechanism of the recruitment of TIA-1 into Htt fibrils, furthermore, we have noted a seeding phenomenon of protein fibrils, in which existing fibrils induce subsequent fibrillation of a soluble protein (Harper and Lansbury, 1997). Effective concentrations of fibrillar species can be increased by shearing fibrils with ultrasonication, and sheared fibrils are used as “seeds” (Harper and Lansbury, 1997). When Htt42Q seeds are added to a soluble form of Htt42Q, thioflavin T fluorescence increases without a lag time, supporting a seeding reaction (Fig. 5A, blue and red curves). TIA-1C also exhibits a seeding reaction, in which addition of TIA-1C seeds diminishes the lag time in TIA-1C fibrillation (Fig. 5B, blue and red curves). Interestingly, albeit with less efficiency compared with the self-seeding of TIA-1C, Htt42Q seeds also accelerate fibrillation of TIA-1C by shortening the lag time (Fig. 5B, blue and green curves), whereas addition of soluble Htt42Q does not alter the fibrillation kinetics of TIA-1C (Fig. 5B, blue and black curves). Such a “cross-seeding” reaction between heterologous proteins was not, however, observed in the fibrillation of Htt42Q with TIA-1C seeds (Fig. 5A, blue and green curves). Htt aggregate can thus act as a nucleus (or seed) to accelerate fibrillation of TIA-1C.
To further confirm the cross-seeding reaction of TIA-1C by Htt42Q aggregates, we performed direct observation of protein fibrils using a fluorescence microscope. Alexa 555-modified Htt42Q and Alexa 488-modified TIA-1C were first prepared and then incubated for aggregation. When Alexa 555-Htt42Q seeds were incubated with soluble Alexa 488-TIA-1C on a glass slide, it is clearly observed that a green TIA-1C fibril grows from a red Alexa 555-Htt42Q seed (Fig. 5C–E). A cross-seeding activity of Htt aggregates in TIA-1 fibrillation is, therefore, proposed to be one of molecular origins to describe the recruitment of TIA-1 into Htt fibrillar aggregates.
Htt inclusions induce recruitment of TIA-1 in a cultured cell, but not vice versa
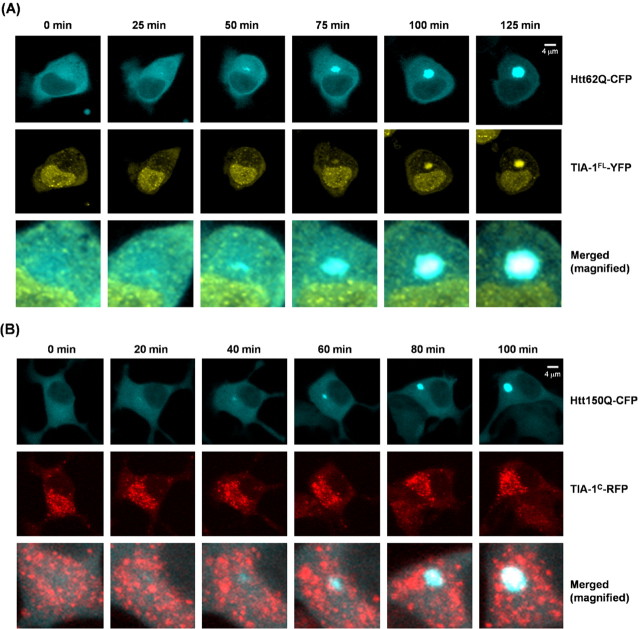
A seeding role of Htt aggregates in the in vitro recruitment of TIA-1 is tested by the time-lapse live-cell imaging experiments. First, Neuro2a cells are cotransfected with Htt62Q-CFP and TIA-1FL-YFP. Htt62Q-CFP inclusions first appear, but no colocalization of TIA-1FL-YFP occurs in an early phase (Fig. 6A, 0–50 min). TIA-1FL-YFP becomes sequestered into Htt62Q-CFP inclusions in the later phase of observation (Fig. 6A, 75–125 min). These results appear to reproduce the observations that Htt inclusion formation precedes insolubilization of TIA-1 in a mouse brain (Fig. 2).
Figure 6.
Htt inclusions sequester TIA-1 in a Neuro2a cell, but not vice versa. A, Representative time-lapsed images of Neuro2a cells cotransfected with Htt62Q-CFP and TIA-1FL-YFP. After overnight transfection, the confocal live-cell imaging was started and performed at intervals of 5 min. A time point when the cell contains no aggregates was arbitrarily set as 0 min. Scale bar, 4 μm. B, Time-lapse confocal imaging of a live Neuro2a cell cotransfected with Htt150Q-CFP and TIA-1C-RFP. Most of the cells expressing TIA-1C-RFP form inclusions after overnight transfection, but those TIA-1C-RFP inclusions do not induce Htt150Q-CFP aggregation. Scale bar, 4 μm.
To examine whether preexisting TIA-1 aggregates affect the formation of Htt inclusions in the cellular environment, we also performed time-lapse imaging of Neuro2a cells transfected with Htt150Q-CFP and TIA-1C-RFP. Unlike TIA-1FL-YFP, TIA-1C-RFP forms cytoplasmic inclusions before Htt150Q-CFP starts to aggregate. Htt inclusion formation occurs at the site at which TIA-1C-RFP inclusions are not evident (Fig. 6B, 0–60 min). At a later stage of the observation (Fig. 6B, 80–100 min), however, TIA-1C-RFP is eventually colocalized with Htt inclusions.
Inclusion formation observed in cultured cells is supposed to be a distinct process from protein fibrillation in a molecular level and has been known as a complex cellular process involving ubiquitination, active protein transport, and interactions with molecular chaperones (Kopito, 2000). For example, a proteasomal inhibition caused by insoluble Htt aggregates (Jana et al., 2001) may also be a possible mechanism of Htt/TIA-1 colocalization in the pathological inclusions. In Neuro2a cells, Htt62Q-YFP and TIA-1FL-HA form coaggregates that are trapped on a 0.2 μm cellulose acetate membrane even after a wash with 2% SDS (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material). When Neuro2a cells were transiently transfected with TIA-1FL-HA alone and treated with a specific proteasomal inhibitor, epoxomycin, we could not find any SDS-insoluble TIA-1 aggregates trapped on the membrane (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Although other cellular processes might regulate colocalization of Htt and TIA-1 in pathological inclusions, it is unlikely that proteasomal inhibition by Htt aggregates plays major roles in decreasing the solubility of a TIA-1 protein. As shown in in vitro/in vivo experiments, instead, we suppose that a cross-seeding reaction can be appended as one of molecular mechanisms for the recruitment of TIA-1 into SDS-insoluble Htt aggregates. Based on these results, our next question is whether the recruitment of TIA-1 into Htt inclusions is associated with dysregulation of its physiological function.
Functional inactivation of TIA-1 on recruitment into Htt inclusions
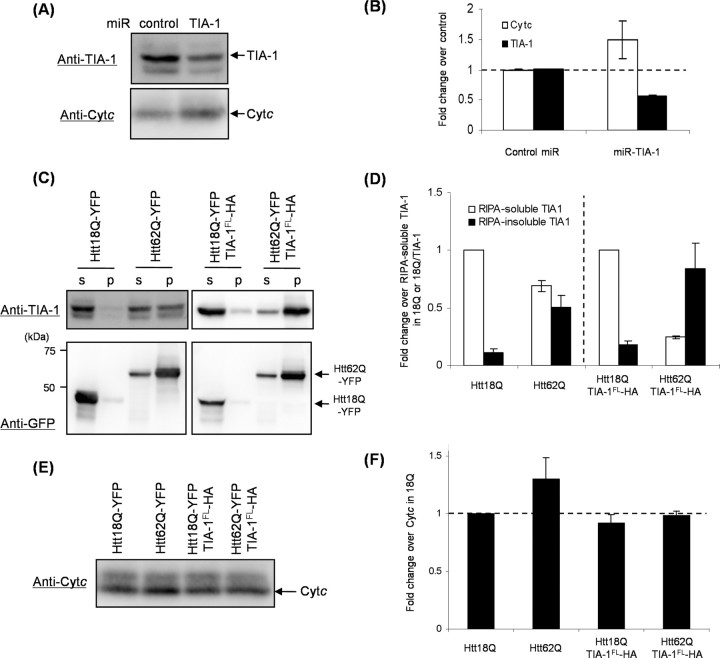
One of the physiological functions of TIA-1 is to suppress a translation of a target mRNA by binding its 3′-untranslated region (3′-UTR) (López de Silanes et al., 2005). It has been shown that translation of Cytc is suppressed by binding of TIA-1 to 3′-UTR of Cytc mRNA (Kawai et al., 2006). To test whether intracellular amounts of TIA-1 modulate expression of a Cytc protein, endogenous TIA-1 was decreased in human embryonic kidney cells, HEK293T, by using miRNA. Our miRNA targeting a human TIA-1 gene reduced amounts of an endogenous TIA-1 protein (∼50% reduction), resulting in the increase in expression level of Cytc (∼50% increase) (Fig. 7A,B). These results confirm the suppressive role of TIA-1 on expression of a Cytc protein (Kawai et al., 2006).
Figure 7.
Functional repression of TIA-1 as a translational silencer on coaggregation with mutant Htt. A, B, HEK293T cells transfected with negative control or miR-TIA-1 were lysed in a RIPA buffer. After ultracentrifugation, a supernatant containing 3 μg (for TIA-1) or 10 μg (for Cytc) of total proteins was analyzed by Western blotting (A) and densitometric quantification of band intensity (B). C–F, HEK293T cells transfected with Htt18/62Q-YFP and pPUR were lysed in a RIPA buffer. For coexpression of TIA-1FL-HA, Htt18Q-YFP or Htt62Q-YFP in pIRES-TIA-1 was used. After ultracentrifugation, supernatant (s, RIPA-soluble fraction) and pellet (p, RIPA-insoluble fraction) were analyzed by Western blotting: TIA-1 and GFP (C), Cytc (E). D, Amounts of TIA-1 in each sample were quantified and normalized by those of RIPA-soluble TIA-1 in either the sample transfected with Htt18Q-YFP or the sample with Htt18Q-YFP plus TIA-1FL-HA. F, Amounts of Cytc were quantified and normalized by those of the sample transfected with Htt18Q-YFP. Three independent experiments were performed to estimate errors (B, D, F).
We next attempt to reveal functional dysregulation of TIA-1 on its recruitment into Htt inclusions. Either Htt18Q-YFP or Htt62Q-YFP was transiently transfected with HEK293T cells together with a pPUR plasmid that contains a puromycin-resistant gene; thereby, contributions from untransfected cells can be reduced by treatment with puromycin. After 3 d of incubation, Htt62Q-YFP but not Htt18Q-YFP has accumulated in a RIPA-insoluble fraction (Fig. 7C, bottom), which agrees with the fact that abnormal length of a poly(Q) tract (62Q) causes Htt aggregation. Aggregation of Htt62Q-YFP also associates with insolubilization of endogenous TIA-1 and concomitantly reduces amounts of RIPA-soluble TIA-1 compared with those in the cells with an aggregation-free Htt18Q-YFP (∼30% decrease) (Fig. 7C, top, Fig. 7D). Furthermore, significant increase (∼30%) in the amount of Cytc was observed in Htt62Q-YFP compared with Htt18Q-YFP, but its increase was suppressed when TIA-1FL-HA was coexpressed (Fig. 7E,F). Even though significant fractions (∼70%) of TIA-1FL-HA becomes RIPA-insoluble on coexpression of TIA-1FL-HA with Htt62Q-YFP (Fig. 7C,D), remaining amounts of soluble TIA-1FL-HA are ∼2.5-fold higher than those of endogenous TIA-1 in the cells transfected with Htt18Q-YFP alone (data not shown) and appear to be enough to complement the function of recruited endogenous TIA-1. These results are thus consistent with the idea that coaggregation with mutant Htt reduces intracellular soluble amounts of TIA-1 and represses a physiological function of TIA-1.
Discussion
Our studies show that an RNA-binding protein, TIA-1, is a member of the proteins recruited into insoluble Htt inclusions. In vivo and in vitro experimental results suggest that Htt aggregates function as an efficient seed for the conversion of a soluble TIA-1 protein into an insoluble structure. On recruitment into Htt inclusions, furthermore, we have shown in a cultured cell model that intracellular amounts of soluble TIA-1 are decreased and that the physiological function of TIA-1 is repressed. A cross-seeding activity of Htt aggregates could hence be one of molecular mechanisms to describe how mutant Htt causes functional dysregulation of coaggregated proteins. Based on the results presented here, we propose that cross-seeded fibrillation of Q/N-rich proteins leads to a pathological diversity in a poly(Q) disease (Fig. 8).
Figure 8.
Our concept of pathological diversity based on a cross-seeding mechanism. An abnormally expanded poly(Q) tract forms fibrillar aggregates, which can induce fibrillation of Q/N-rich proteins by acting as a seed (cross-seeding). Various kinds of Q/N-rich proteins (designated as different shapes and/or colors) can be potentially recruited into the fibrillar aggregates through a cross-seeding reaction. Cross-seeded fibrils of Q/N-rich proteins may further facilitate fibrillation of the other proteins that cannot be directly seeded by poly(Q) aggregates (cross-cross-seeding). Given that physiological functions of recruited proteins are repressed, cross-seeding reactions will describe diverse pathologies observed in the poly(Q) diseases.
Precedents of a coaggregation mechanism: proteins with a benign length of poly(Q) in Htt fibrils
So far, coaggregation with a mutant Htt protein has been reported in several proteins containing a nonpathogenic length of a poly(Q) tract. Important examples include the TATA box binding protein (TBP) and the transcriptional coactivator CBP (CREB binding protein), which possesses 38 and 19 consecutive glutamine residues, respectively (Huang et al., 1998; Nucifora et al., 2001; Schaffar et al., 2004). In addition, Htt with a normal length of poly(Q) (Q20, Q32, and Q37) is coaggregated with mutant Htt in vitro (Busch et al., 2003). These benign poly(Q)-containing proteins themselves are not fibrillogenic but become an integral component of fibrils on coaggregation with mutant Htt (Busch et al., 2003; Schaffar et al., 2004). Although Htt aggregates do not trigger fibrillation of these benign poly(Q) proteins in vitro (Busch et al., 2003), poly(Q)–poly(Q) interactions in the soluble state would be enough to intertwine benign poly(Q) proteins into Htt fibrils. In a human proteome, 64 proteins have more than five consecutive glutamines in their amino acid sequences (Butland et al., 2007), although not all of those poly(Q) proteins are necessarily sequestered in HD inclusions (Busch et al., 2003). Compared with poly(Q) proteins, the total number of Q/N-rich domains in a human proteome has been estimated to be ∼200 (Harrison and Gerstein, 2003). As evidenced in TIA-1 as a model of Q/N-rich proteins, we suppose that more numbers of Q/N-rich proteins are potentially involved in HD inclusions by adopting a fibrillar structure (Fig. 8).
Cross-seeding activity of Htt aggregates leads to recruitment of Q/N-rich proteins in HD inclusions
TIA-1 has no poly(Q) tract (at most three consecutive glutamines), but its C-terminal domain is rich in Q/N residues (31% abundance) (Fig. 3A), given that the average abundance of Q and N for the eukaryotic proteomes is ∼5% each per open reading frame (Michelitsch and Weissman, 2000). Fibrillation of a Q/N-rich protein sequence has been shown in several yeast prion proteins (Ross et al., 2005) such as Sup35 and New1. Q/N richness of the prion domain of Sup35 and New1 is higher than (48.8%) or comparable with (26.1%) that of TIA-1 C-terminal domain (31.0%), respectively. Sup35 exhibits weak sequence homology with TIA-1 at the Q/N-rich domain; TIA-1C (97 aa) and Sup35 N-terminal NM domain (124 aa) share 26.1% identity and 53.4% similarity in the overlapped 88 aa. TIA-1C also exhibits a limited homology with exon I of Htt with 42Q (112 aa): 25.0% identity and 36.0% similarity in the overlapped 72 aa. As implied in the species barrier of the prion transmission, a seeding specificity does not always correlate with the primary sequence similarities; rather, conformational roles of fibrils are more evident in a seeding reaction (Vanik et al., 2004). A structure of poly(Q) fibrils has been described by a “polar zipper” model, in which hydrogen bonding interactions among both side and main chains of glutamine residues constitute an ordered β-sheet structure (Perutz et al., 1994). Although it remains unclear how many glutamines can sustain a polar zipper interaction in the poly(Q)-Q/N mixed fibrils, our results imply that ∼30% abundance of Q/N residues as found in TIA-1 is sufficient for being zipped with a poly(Q) sequence. Although a Q/N-rich sequence with low complexity would play a role in interacting with another protein molecule including a hydrogen-bonding interaction (Wright and Dyson, 1999), there is a caveat that presence of Q/N-rich region(s) in a certain protein molecule does not necessarily mean the ability to interact with poly(Q) aggregates. Nonetheless, it is worth noting that the coaggregated proteins our group has recently reported contain Q/N-rich sequences; an N-terminal part (1–162) of an RNA-binding protein, TLS, exhibits 26% in the Q/N abundance, and 33 and 34% of an N-terminal part of a transcription factor, NF-YA (1–153), and a C-terminal part of NF-YC (150–186) is composed of Q/N residues, respectively (Doi et al., 2008; Yamanaka et al., 2008). In contrast, it has been reported that fibrillation of Aβ(1–40), which contains only one each of glutamine and asparagine residues, are not efficiently seeded with poly(Q) aggregates (O'Nuallain et al., 2004). A poly(Q) fibrillar structure could, therefore, provide a seeding scaffold to induce fibrillation of some Q/N-rich proteins.
Pathological outcome of cross-seeding reactions in neurodegenerative diseases
Compared with a transient protein–protein association reaction, one of the kinetic features of a cross-seeding reaction is to explosively promote protein aggregation once the seeding starts. Pathological inclusion bodies in HD contain many kinds of protein molecules such as chaperones and proteasome components (Jana et al., 2001), which are possibly recruited through a cellular process distinct from cross-seeding fibrillation. A molecular chaperone, Hsp70, for example, is one of the proteins found in the HD inclusions, but its interaction with inclusions is transient and highly dynamic (Kim et al., 2002). Unlike TIA-1 and the other Q/N-rich proteins such as TBP, NF-Y, and TLS, Hsc70 as well as Hsp84 do not contain a Q/N-rich region in its primary sequence and are easily dissociated from Htt inclusions in the presence of SDS (Mitsui et al., 2002). Although it remains unknown how functional repression of TIA-1 is related to HD pathologies, a cross-seeding behavior of mutant Htt bears some analogy with phenotypic changes in HD; diseases rapidly progress after the onset, and neurological symptoms often follow appearance of inclusions in the affected tissues (Davies et al., 1997).
In addition to a translational silencing of specific targeted genes including Cytc, TIA-1 has been known to play an essential role in forming stress granules (SGs) under stressed conditions (Kedersha et al., 1999) and is also a constituent of intracytoplasmic basophilic inclusions found in the patients with adult-onset motor neuron disease (Fujita et al., 2008). TIA-1 recruits untranslated mRNAs into SGs without apparent specificity and self-aggregates within SGs (Gilks et al., 2004). Formation of SGs promotes disassembly of a functional polysome and arrests the translation of housekeeping genes. A cross-seeded aggregation of TIA-1 by Htt fibrils may lead to global suppression of translation by converging SGs and protein aggregates. As is the case with TIA-1, many DNA/RNA-binding proteins possess a Q/N-rich domain, which has been proposed to support protein–protein interactions (Michelitsch and Weissman, 2000). We thus suppose that cross-seeded fibrillation of a Q/N-rich domain with mutant Htt disrupts a canonical interaction in the transcriptional/translational complexes in particular and contributes to produce pathological phenotypes (Fig. 8).
Moreover, pathological phenotypes could become more diverse when a cross-seeded product further induces fibrillation of proteins that are not directly seeded with Htt aggregates (Fig. 8). Such a “cross-cross-seeding” initiated by Htt aggregates may affect physiological functions of more numbers of proteins (Fig. 8). Given a role of Htt aggregates as a structural template toward fibrillation of certain Q/N-rich sequences, therefore, we propose that a cross-seeding reaction by mutant Htt is one of molecular origins of diverse pathological phenotypes in HD patients.
Footnotes
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant-in-Aid 17025044 for Scientific Research on Priority Areas (Research on Pathomechanisms of Brain Disorders) (N.N.) and Grant-in-Aid 18031044 (Y.F.), and Ministry of Health, Welfare, and Labor (Japan) Grant-in-Aid for the Research on Measures for Intractable Diseases (N.N.). We thank Dr. Shoji Watanabe for technical assistance and fruitful discussion.
References
- Busch A, Engemann S, Lurz R, Okazawa H, Lehrach H, Wanker EE. Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J Biol Chem. 2003;278:41452–41461. doi: 10.1074/jbc.M303354200. [DOI] [PubMed] [Google Scholar]
- Butland SL, Devon RS, Huang Y, Mead CL, Meynert AM, Neal SJ, Lee SS, Wilkinson A, Yang GS, Yuen MM, Hayden MR, Holt RA, Leavitt BR, Ouellette BF. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Díaz-Hernández M, Moreno-Herrero F, Gómez-Ramos P, Morán MA, Ferrer I, Baró AM, Avila J, Hernández F, Lucas JJ. Biochemical, ultrastructural, and reversibility studies on huntingtin filaments isolated from mouse and human brain. J Neurosci. 2004;24:9361–9371. doi: 10.1523/JNEUROSCI.2365-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, Machida Y, Miyazaki H, Mitsui K, Kuroiwa Y, Nukina N. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem. 2008;283:6489–6500. doi: 10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- Fujita K, Ito H, Nakano S, Kinoshita Y, Wate R, Kusaka H. Immunohistochemical identification of messenger RNA-related proteins in basophilic inclusions of adult-onset atypical motor neuron disease. Acta Neuropathol. 2008;116:439–445. doi: 10.1007/s00401-008-0415-x. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Yamanaka K, O'Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in familial form of ALS. J Biol Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JD, Lansbury PT., Jr Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeki N, Tukamoto T, Goto J, Kanazawa I. Formic acid dissolves aggregates of an N-terminal huntingtin fragment containing an expanded polyglutamine tract: applying to quantification of protein components of the aggregates. Biochem Biophys Res Commun. 2000;277:386–393. doi: 10.1006/bbrc.2000.3682. [DOI] [PubMed] [Google Scholar]
- Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel JP, MacDonald ME, Gusella JF. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- Jana NR, Zemskov EA, Wang Gh, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10:1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Nakayama H, Akagi T, Nekooki M, Ohtawa K, Takio K, Hashikawa T, Nukina N. Purification of polyglutamine aggregates and identification of elongation factor-1α and heat shock protein 84 as aggregate-interacting proteins. J Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Naiki H, Higuchi K, Hosokawa M, Takeda T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- O'Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J Biol Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell. 2004;14:139–145. doi: 10.1016/s1097-2765(04)00155-8. [DOI] [PubMed] [Google Scholar]
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, Doi H, Nukina N. Mutant huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]