Abstract
Neurofilaments assemble from three intermediate-filament proteins, contribute to the radial growth of axons, and are exceptionally stable. Microtubules are dynamic structures that assemble from tubulin dimers to support intracellular transport of molecules and organelles. We show here that neurofilaments, and other intermediate-filament proteins, contain motifs in their N-terminal domains that bind unassembled tubulin. Peptides containing such motifs inhibit the in vitro polymerization of microtubules and can be taken up by cultured cells in which they disrupt microtubules leading to altered cell shapes and an arrest of division. In transgenic mice in which neurofilaments are withheld from the axonal compartment, axonal tubulin accumulation is normal but microtubules assemble in excessive numbers. These observations suggest a model in which axonal neurofilaments modulate local microtubule assembly. This capacity also suggests novel mechanisms through which inherited or acquired disruptions in intermediate filaments might contribute to pathogenesis in multiple conditions.
Introduction
Neurofilaments (NFs) are prominent components of the cytoskeleton in mature neurons (Perrot et al., 2007, 2008). They are composed of three independently encoded subunits, light (NFL), medium (NFM), and heavy (NFH), of 68, 150, and 200 kDa, respectively. The NF core consists of NFL on which NFM and NFH anchor through coil–coil interactions. The C-terminal domains of NFM and NFH project from the filament and interact with other NFs and organelles (Hirokawa, 1982). The phosphorylation state of KSP motifs repeated in these lateral projections (Julien and Mushynski, 1982; Lee et al., 1988) is thought to regulate interfilament affinity, interactions with microtubules (MTs), the rate of NF transport, and the radial growth of axons (Carden et al., 1987; Eyer and Leterrier, 1988; Hisanaga and Hirokawa, 1990; de Waegh et al., 1992; Ohara et al., 1993; Eyer and Peterson, 1994; Zhu et al., 1997).
MTs play an essential role in axonal transport, functioning as rails along which organelles are transported in anterograde and retrograde directions by specific motor proteins. MTs are in dynamic equilibrium with αβ-tubulin dimers. In neurons, the assembly and stability of these noncentrosomal MTs are regulated through tubulin modifications and association with several additional proteins [e.g., microtubule-associated proteins (MAPs), STOP, and OP18] (Mandelkow and Mandelkow, 1995; Guillaud et al.., 1998; Cassimeris, 2002; Baas and Qiang, 2005; Baas et al., 2006; Bartolini and Gundersen, 2006; Conde and Caceres, 2009; Fukushima et al., 2009). NFs also are associated with tubulin polymerizing activity (Minami et al., 1982; Aamodt and Williams, 1984; Hisanaga and Hirokawa, 1990). Notably, dephosphorylation of NFH modifies the proportion of cold-soluble and cold-insoluble MTs, suggesting a role for the phosphorylation state of the local NF cytoskeleton in MT stability (Tashiro et al., 1994). Here, we demonstrate that tubulin and NFs also are binding partners, and in mature neurons, this interaction appears to play an important role in sculpting the axonal MT cytoskeleton.
In classical biochemical assays, unpolymerized tubulin copurifies with assembled NFs, and we show here that conserved sites present in the N-terminal domain of all three NF subunits are capable of mediating this interaction. Although peptides corresponding to these conserved sequences have no effect on the stability of assembled MTs, they inhibit MT polymerization in vitro. Furthermore, such peptides can enter cells in culture leading to disruption of the MT cytoskeleton and altered cell proliferation. In contrast, peptides in which the normal amino acid order has been reversed or scrambled lose such activities.
To characterize the in vivo significance of NF–tubulin binding, we investigated transgenic mice in which NFs aggregate in cell bodies preventing their export to axons. Tubulin expression, accumulation, and distribution are not affected by this alteration in NF distribution. However, MTs are present in abnormal abundance within the NF-deficient axons. These observations lead us to propose a model in which axonal NFs modulate the local assembly of the MT cytoskeleton. Motifs with similar tubulin-binding capacity are located on multiple intermediate-filament proteins suggesting that MT assembly might be similarly modulated in diverse cell types.
Materials and Methods
Neurofilament, microtubule, and tubulin isolation.
Brains, spinal cords, and sciatic nerves were dissected from adult mice, weighed, and homogenized in RB buffer (100 mm MES, pH 6.8, 1 mm EGTA, and 1 mm MgCl2). The homogenates were centrifuged at 105 × g at 4°C for 1 h in Beckman LE-80k or TLX100 ultracentrifuges. For MT isolation, the first supernatant (S1) was incubated with 4 m glycerol and 0.1 mm GTP at 37°C for 1 h, and then centrifuged at 105 × g for 1 h at 37°C to generate the second pellet (P2). MTs and NFs present in the pellet (P2) were resuspended in RB buffer, incubated at 4°C for 1 h to depolymerize MTs, and centrifuged at 105 × g for 1 h at 4°C. The unpolymerized tubulin fraction was recovered in the third supernatant (S3), whereas NFs were present in the pellet (P3). Tubulin was further purified from the third supernatant (S3) by passage on a phosphocellulose column as described previously (Weingarten et al., 1975). For NF purification, a similar protocol was used, but no GTP was added, and all steps were realized at 4°C to prevent MT polymerization, as previously described (Leterrier and Eyer, 1987).
Protein quantification and Western blot analysis.
Protein concentration was determined by the BCA assay kit (Pierce) and separation was performed on a 7.5% SDS-PAGE and revealed by Coomassie blue (Laemmli, 1970). Alternatively, proteins were transferred onto nitrocellulose membranes (Immobilon; Millipore) for blot-overlay experiments or immunoblotting analysis (Towbin et al., 1979). NF subunits were identified using monoclonal anti-NFH, anti-NFM, and anti-NFL antibodies (Sigma-Aldrich N5389, N5264, N5139), and an anti-NFH polyclonal antibody (Sigma-Aldrich N4142). Tubulin isotypes were identified using antibodies recognizing α-tubulin, β-tubulin, the neuronal specific βIII-tubulin, γ-tubulin (Sigma-Aldrich T9026, T4026, T8660), and polyglutamylated tubulin epitopes (GT335; generously provided by Dr. P. Denoulet, Paris, France). Experiments were performed in triplicate and membranes were incubated sequentially with each antibody (Fasani et al., 2004). Each reaction was revealed using a chemiluminescent protocol (ECL; GE Healthcare). To evaluate the intensity of signals, the exposed films were scanned and quantified by measuring the area and density of the spot with ImageQuant software (GE Healthcare) or NIH Image. For each experiment, the control sample signal was used as the reference.
RNA analysis by Northern blots.
Brains and spinal cords were obtained from NFHLacZ transgenic and control mice at 2 d, 4 weeks, 6 months, and 18 months of age, frozen in liquid nitrogen, and stored at −80°C. RNA extraction and Northern blots were performed as previously described (Robert et al., 2001). Northern blots were quantified using a PhosphorImager (GE Healthcare) and exposed to autoradiographic films (Biomax MR; Kodak) at −80°C.
To analyze the mRNA encoding each tubulin isotype (Wang et al., 1986), specific probes were generated from 3′-untranslated sequences in the pUC vector by the PCR using pUC primers (5′–3′, CTGCAAGGCGATTAAGTTGG and GTTGTGTGGAATTGTGAGCG).
mRNA isolated from the brains of 2-d-old control animals was used as reference, and band intensity was defined arbitrarily as 1. Exceptions were for α3/7 and β1 tubulin mRNA isolated from the testis and spleen of adult control mice because of their absence of expression in other tissues. As glyceraldehyde-3-phosphate dehydrogenase (GAPDH) values did not vary between control and transgenic samples, correction factors that equalized the GAPDH value were calculated to normalize mRNA levels for each Northern blot as previously described (Robert et al., 2001). Mean values from three different experiments at each postnatal stage were calculated and expressed as mean ± SE.
Peptide array membranes and synthetic peptides.
Immobilized peptide arrays corresponding to the sequence of mouse intermediate filaments were produced with an ABIMED ASP 222 automated spot robot (Frank, 1992; Frank and Overwin, 1996). Each sequence was arrayed as 15-mer peptide fragments, with each peptide overlapping the next one by 12 aa. Peptides were synthesized spotwise as an array (spot diameter of 2 mm; distance of 4 mm; 25 spots per row) on a specially manufactured cellulose membrane (AC01; AIMS Scientific Products). The peptides are linked to the membrane via a polyethylene glycol spacer through their C-terminal amino acid and are N-terminally acetylated. The membrane was saturated overnight at 4°C with 10% milk powder diluted in TBS, washed three times 10 min with TBS, and incubated 1 h at 4°C with freshly purified phosphocellulose tubulin (3 mg/ml), and the excess of tubulin was removed by three washes with TBS at 4°C. Tubulin binding was revealed using classical Western blot protocol by anti-βIII-tubulin (Sigma-Aldrich) and a secondary anti-mouse peroxidase-labeled antibody (Dako) as previously described (Fasani et al., 2004). Soluble peptides corresponding to tubulin-binding sites (TBSs) and their reverse and scrambled sequences were synthesized by Millegen and added to MTs (S3 fraction from a MT preparation) to test their effect on MT polymerization. Data were obtained with peptides (alone or biotinylated) synthesized by Millegen (>90% purity) and were reproduced using peptides synthesized by the laboratory of R. Frank, by Sequentia, or by Genepep.
Computational analysis.
Multiple sequence alignment was performed with ClustalW and manually refined. The rod domain of each intermediate filament was used to anchor the alignment. The alignment was drawn with genedoc software (http://www.psc.edu/biomed/genedoc).
Evaluation of microtubule polymerization.
Polymerization of tubulin was followed by turbidimetry (OD, 350 nm) using a Thermospectronic Helios spectrophotometer. MT formation, which produced an increase of the OD at 350 nm, was monitored at 1 min intervals.
Proliferation assay.
T98G cells were cultured in RPMI medium with 10% fetal calf serum. Cells were platted in 96-well plates at a density of 500 cells per well and treated with 10 μm peptide during 12, 24, and 48 h, or with vehicle. Bioreduction of 3-carboxymethoxyphenyl-2-(4-sulfophrenyl)-2H-tetrazolium (MTS) (Promega) yields a color change. The intensity of coloration measured by the amount of 490 nm light absorbance is directly proportional to the number of living cells in culture. To enhance the cellular reduction of MTS, phenazine methosulfate (PMS) was added to MTS immediately before use (MTS-to-PMS ratio, 1:20). The mixture (25 μl) was added to each well. After a 2 h incubation at 37°C in a humidified atmosphere with 5% CO2, the optical density was measured by spectrophotometry (490 nm). For DNA content analysis, 2 × 105 cells were washed twice with PBS and fixed in ethanol 70%. Cells were then treated with 100 U/ml ribonuclease A for 20 min at 37°C. Cells were resuspended in PBS containing 50 μg/ml propidium iodide and immediately analyzed by flow cytometry.
Immunohistochemical analysis.
Cells were plated on coverslips and cultured in media containing biotinylated peptides (10 μm) for 6 h. Cells were then washed in PBS one time, fixed in 4% PFA for 15 min, washed three times in PBS, and permeabilized for 15 min in 70% ethanol. After extensive washes, cells were incubated with anti-β-tubulin antibody at 1:500 overnight. Tubulin and biotinylated peptides were localized using Alexa 568 nm anti-mouse antibody and streptavidin Alexa 488 nm (Invitrogen) at 1:200 for 1.5 h, respectively. Tissues were processed as previously described (Fasani et al., 2004). Primary antibodies and dilutions used were as follows: monoclonal anti-α-tubulin, anti-β-tubulin, and anti-βIII-tubulin were diluted 1:100; polyclonal anti-NFH and anti-peripherin were diluted 1:1000 and 1:100, respectively. Fluorescent secondary antibodies (Alexa 468 nm anti-mouse antibody, and Alexa 568 nm anti-rabbit antibodies from Invitrogen) were diluted 1:200. Immunolabeled slides were analyzed with an Olympus confocal microscope (BX50) using Fluoview 3.1 software.
Electron microscopy and quantification.
Electron microscopy (50,000×) followed previously described protocols using a JEOL JEM2010 electron microscope (Perrot et al., 2007). Axonal MT density was evaluated in randomly selected axons as previously described (Perrot et al., 2007).
NFH-LacZ transgenic mice.
The production and general characteristics of the NFHlacZ transgenic mice is described by Eyer and Peterson (1994). The mice used in this investigation, as well as control littermates, had an inbred FVB genetic background obtained through an extensive backcross program.
Results
Unpolymerized tubulin coisolates with NFs
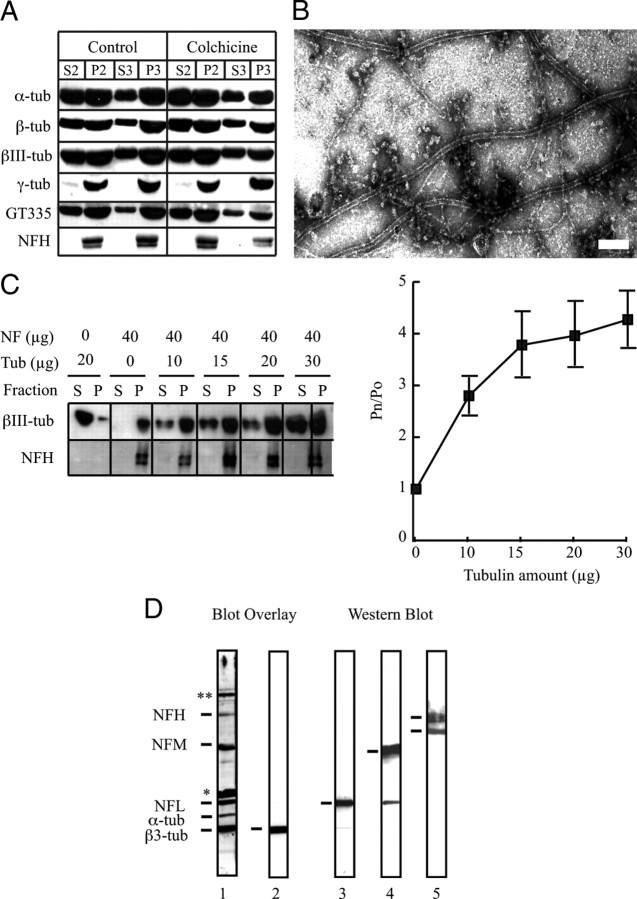
NFs were isolated by centrifugation from glycerol-treated brain and spinal cord homogenates (Leterrier and Eyer, 1987). When glycerol is added to the first supernatant (S1) inter-NF cross-bridges form leading to a reticulated NF network that sediments on centrifugation [referred to as pellet 2 (P2)]. If glycerol incubation is repeated with resuspended P2, additional NFs are recovered on centrifugation in P3. Western blot analysis reveals that such NF-enriched fractions contain α-, β-, and γ-tubulin. As assembled MTs also pellet using this technique, care was taken to perform all procedures under conditions that prevent MT formation (4°C or with 3 μm colchicine). As similar amounts of copurifying tubulin were observed under all conditions (Fig. 1A), the tubulin copurifying with pelleted NFs is unlikely to represent MT contamination.
Figure 1.
Tubulin coisolates with NFs and is a binding partner of the three NF subunits. A, Tubulin was detected in each fraction of a classical preparation of brain NFs using specific antibodies against α-, β-, βIII-, and γ-tubulin or polyglutamylated epitopes of tubulin (GT335). To prevent MT assembly, all procedures were conducted at 4°C or in the presence of colchicine. B, When the third pellet (P3) from such a NF purification, performed at 4°C but without colchicine, was examined at the electron microscope, no MTs were observed (data not shown), but they were abundant when such preparations are incubated at 37°C in the presence of 1 mm GTP. Scale bar, 100 nm. C, NFs (40 μg) were incubated for 30 min alone (Po) or with increasing quantities of purified tubulin (Pn) and centrifuged, and each pellet and supernatant were analyzed for tubulin content. Tubulin present in the NF pellet increases as an increased amount of tubulin is added. Error bars indicate SEM. D, NFs present in the third pellet (P3) were separated on a 7.5% SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with phosphocellulose-purified tubulin (3 mg/ml). The membrane was incubated with anti-βIII-tubulin and processed like a normal Western blot. The blot overlay (lane 1) revealed that tubulin interacted with bands migrating at the same position as NFL, NFM, and the phosphorylated isoform of NFH, together with α- and β-tubulin, as well as *synapsin and **MAPs. Without the addition of purified tubulin, only the band corresponding to βIII-tubulin was revealed (lane 2). The same membrane was stripped to remove tubulin and anti-βIII-tubulin antibodies, and reprobed sequentially with anti-NFL (lane 3), anti-NFM (lane 4), and anti-NFH (lane 5) antibodies. The anti-NFH antibody recognizes both phosphorylated and dephosphorylated isoforms of NFH. P, Pellet; S, supernatant; tub, tubulin.
We next show that tubulin copurifying with the NF fraction retains polymerization potential. When 10 μg of the P3 fraction, prepared under conditions preventing MT formation (4°C), was loaded onto 300 mesh grids, abundant NF profiles heavily decorated with amorphous material were observed by electron microscopy. In such preparations, short MT profiles were observed but only on rare grid squares. In contrast, under conditions favoring tubulin polymerization (37°C and 1 mm GTP), long MT profiles were encountered in high number (up to 50) in all grid squares examined and such MTs were frequently associated with NF profiles (Fig. 1B).
To evaluate the tubulin-binding capacity of NFs, 40 μg of the P3 NF-enriched fraction was incubated (4°C for 1 h) with increasing amounts of pure tubulin [isolated by phosphocellulose chromatography (Weingarten et al., 1975)]. The mixture was then sedimented at 100,000 × g for 30 min, and the amounts of supernatant versus pelleted tubulin were estimated by Western blot analysis. In the absence of NFs, tubulin remained soluble. In the presence of NFs, the amount of tubulin sedimenting with NFs increased with the quantity of added tubulin until saturation was reached at 20 μg of tubulin (Fig. 1C).
We next determined by blot overlay which of the NF proteins interacts with tubulin (Fig. 1D). Proteins present in the NF-enriched fraction (P3) were resolved by 7.5% SDS-PAGE and transferred to nitrocellulose. Using classical Western blot methods and anti-tubulin antibody, only the copurifying endogenous tubulin was detected (lane 2). However, when the blot was incubated with phosphocellulose purified tubulin (3 mg/ml; 1 h at 4°C), additional tubulin-binding bands colabeling with the three NF subunits were observed [NFL (lane 3), NFM (lane 4), and NFH (lane 5)]. Additional tubulin-binding bands were detected and identified by mass spectroscopy as synapsin I and II and various MAPs. However, these molecules did not colocalize with NF aggregates in NFHLacZ transgenic mice (see below) and did not copurify with NFs.
Identification of NF-TBSs
To identify the amino acid sequences in NF proteins that bind tubulin, the complete sequence of each NF protein was displayed on cellulose membranes as spots of 15 aa peptides, overlapping by 12 aa. The peptide-binding capacity of the cellulose membrane is from 0.1 to 1 μmol/cm2 depending on the size of the peptide, with the highest concentration (1 μmol/cm2) realized for peptides <10 aa. Here, all the peptides were produced by the SPOT synthesis technique and are precisely 15 aa long. Consequently, their concentration is identical at each spot, contributing to the reproducibility, accuracy, and reliability of the method (Frank, 1992; Frank and Overwin, 1996; Weiser et al., 2005). The membranes were incubated with phosphocellulose-purified tubulin and those peptides binding tubulin were revealed as above by a classical Western blot protocol. As shown in Figure 2A, TBSs were located in NFL between amino acids 4 and 27, and 40 and 63; in NFM between amino acids 13 and 51; and in NFH between amino acids 37 and 75. A tubulin-binding signal was detected with 8 of the 177 peptides derived from NFL, with 6 of the 291 peptides derived from NFM, and with 5 of the 377 peptides derived from NFH.
Figure 2.
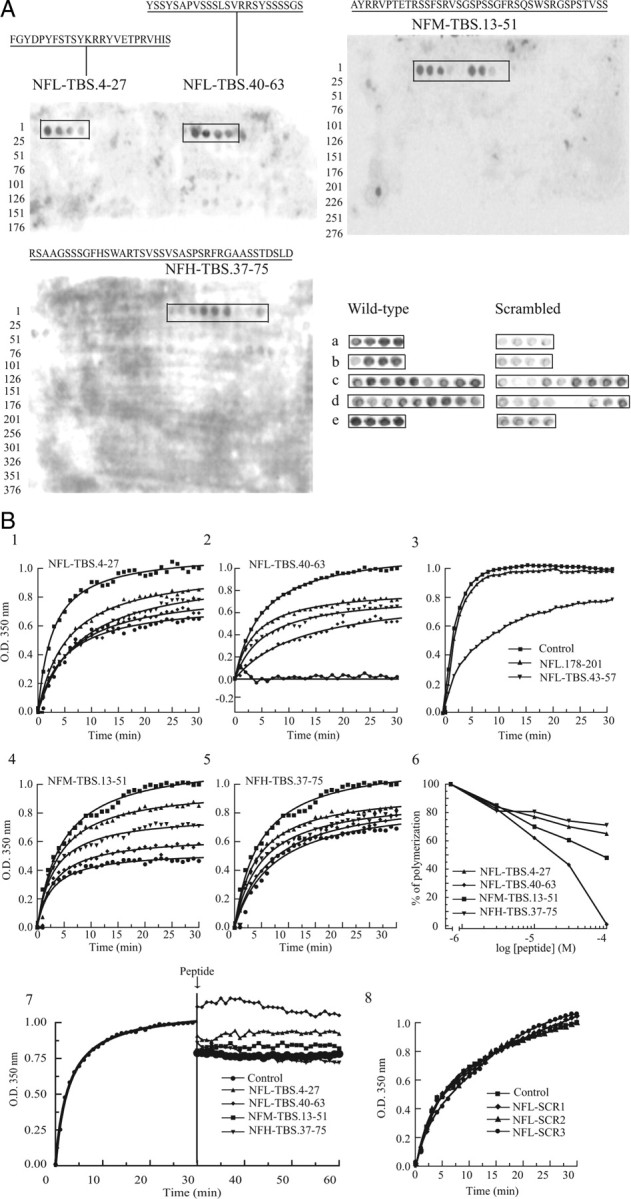
Tubulin binds to the N-terminal domain of each NF subunit, and peptides corresponding to these TBSs affect MT polymerization. A, Peptide array membranes were incubated with phosphocellulose-purified tubulin (3 mg/ml) overnight at 4°C, and bound tubulin was detected with an anti-βIII-tubulin antibody and a peroxidase-labeled secondary antibody. The NFL subunit contains two 24-aa-long TBSs (NFL-TBS.4-27 and NFL-TBS.40-63), whereas NFM and NFH contain one 39-aa-long TBS (NFM-TBS.13-51 and NFH-TBS.37-75). Bottom, When the sequence of peptides corresponding to the TBS was scrambled (right), their capacity to bind tubulin was typically abolished or greatly diminished. The sequences of peptides are as follows: (a) NFL-TBS (4-27), FGYDPYFSTSYKRRYVETPRVHIS; and NFL-TBS (4-27) scrambled, SDETRHFGVPISKYSYRYPFVYRT; (b) NFL-TBS (40-63), YSSYSAPVSSSLSVRRSYSSSSGS; and NFL-TBS2 (40-63) scrambled, SSASLSYSPSRSVSRSYSGSYSVS; (c) NFM-TBS (13-51), AYRRVPTETRSSFSRVSGSPSSGFRSQSWSRGSPSTVSS; and NFM-TBS (13-51) scrambled, SVWRASYGSRSVSPGTSERTSFSRPSFSRSPRSGVSTQS; (d) NFH-TBS (37-75), RSAAGSSSGFHSWARTSVSSVSASPSRFRGAASSTDSLD; and NFH-TBS (37-75) scrambled, SHSPSTSASRSASWSGSDGRFSADRVATSFSRLGSASAV; (e) Ker-TBS (1-24), MSIRVTQKSYKMSTSGPRAFSSRS; and Ker-TBS1 (1-24) scrambled, QTGAKSISFMRPSVSYRSKTMSRS. B1–B5, Peptides corresponding to the TBS of each NF subunit were added at different concentrations to unpolymerized MTs (S3 fraction from an MT preparation), and their effect on MT assembly was evaluated. ●, 100 μm; ♦, 30 μm; ▼, 10 μm; ▲, 3 μm; or ■, without peptide. B6, The percentage of the MT assembly inhibition after 30 min was plotted against peptide concentration. B7, When peptides were added to fully polymerized MTs (arrow), no depolymerization occurred within 30 min. B8, When the sequence of NFL-TBS.40-63 peptide was scrambled, it lost its capacity to affect MT assembly. The sequences of NFL.SCR1, NFL.SCR2, and NFL.SCR3 are presented in Table 2.
To test the specificity of these peptide–tubulin interactions, we first varied experimental conditions using BSA or dry milk as blocking agents and MES or PIPES as buffer. Although the absolute intensity of the binding signal differed between experimental conditions, the overall pattern of tubulin binding was similar under all conditions (data not shown). Finally, when their amino acid sequence was scrambled or reversed, a strong decrease or complete elimination of tubulin-binding capacity was observed for most TBS peptides (Fig. 2A).
To evaluate the functional relevance of this binding relationship, peptides corresponding to TBS sequences were added to purified MTs (S3 fraction from a MT preparation) under conditions favoring polymerization (37°C and 1 mm GTP). The rate of MT formation was measured by turbidimetry (OD, 350 nm). All NF-TBS peptides exerted a concentration-dependent inhibitory effect (Fig. 2B). At 100 μm, the NFH-TBS.37-75 peptide reduced polymerization by 30% (Fig. 2B5). The NFL-TBS.40-63 caused a robust interference causing a 40% reduction at 3 μm, and it abolished all polymerization at 100 μm (Fig. 2B2). However, when NFL-TBS.40-63 was truncated to 43-57, such inhibitory activity was reduced (Fig. 2B3). Peptides demonstrating no affinity for tubulin on peptide arrays [e.g., NFL.178-201, truncated (NFL.43-57), or scrambled peptides (Fig. 2B3,B8)] had no effect on MT polymerization at concentrations up to 100 μm. Finally, none of the tested peptides had a detectable effect on preformed MTs (Fig. 2B7).
Biochemical properties of tubulin-binding sites
Isoelectric point (pI) values for each peptide present on the peptide-arrays were calculated and plotted. Whereas the pI values of all TBSs were relatively high, peptides with no binding activity had similar or even higher values (Fig. 3A–C). Additional analysis of the TBS sequences did not reveal an exceptional content of positively or negatively charged amino acids (Fig. 3D). One control NFL.178-201 peptide is rich for both negative (37%) and positive amino acids (21%) but has no tubulin-binding activity (Fig. 2A) and does not affect MT polymerization (Fig. 2B3). Interestingly, the pI value of the head domain of each NF subunit was markedly higher when compared with the rod and tail domains (Fig. 3D). Together, these data indicate that tubulin is binding to a limited number of specific sites along the NF sequence, and this interaction is mediated in large measure by amino acid sequence rather than peptide charge characteristics.
Figure 3.
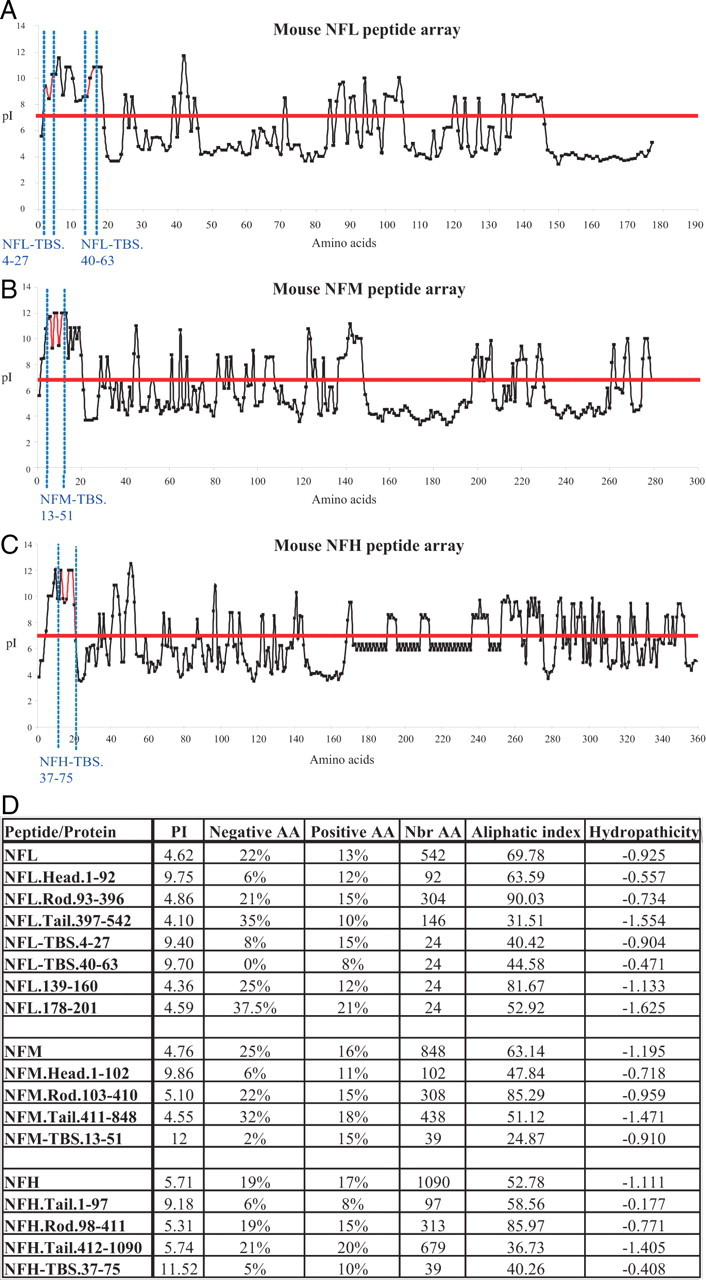
Calculation of the pI (isoelectric point) for each peptide spotted on the peptide arrays. A–C, The pI value was calculated for each 15-aa-long peptide present on the peptide array corresponding to NFL (A), NFM (B), and NFH (C). The graphs show that the pI of the TBS is slightly higher when compared with the rest of the sequence. However, some peptides have high pI values, but they do not bind tubulin. D, Several biochemical characteristics of each NF subunit and their intramolecular domains are summarized. Interestingly, the pI value of the “head” domain of each NF subunit is higher compared with the rest of the sequence. Calculations were performed online using “ProtParam tool” available at www.expasy.org.
Diverse intermediate-filament proteins also contain tubulin-binding sites
Online basic local alignment search tool (BLAST) analysis (www.ncbi.nlm.nih.gov:80/blast) using mouse TBS sequences revealed amino acid sequences with 90% or greater identities in the corresponding NF proteins of several mammals including human, rat, bovine, and dog (Table 1). Furthermore, for some peptides, obvious motif conservation extends to species with more divergent NF proteins (e.g., Xenopus and Danio).
Table 1.
BLAST analysis of the NF-TBS homology

Online BLAST analysis reveals a high conservation of the TBS sequences among different species.
ClustalW multiple alignment of the NF tubulin-binding sites also revealed conservation between the TBS sequences on the three NF subunits of mice (Fig. 4A). Like NFs, the TBS motifs identified in additional intermediate-filament proteins were all located in N-terminal domains (Fig. 4A). To evaluate the potential functional significance of the motifs found in desmin (Des), vimentin (Vim), cytokeratin (Ker), and glial fibrillary acid protein (GFAP), similar peptide arrays were prepared from each protein and evaluated for tubulin-binding capacity. All motifs identified by amino acid sequence conservation demonstrated tubulin-binding capacity (Fig. 4B). In addition, when tested for their ability to interfere with in vitro MT assembly, some of these TBS peptides (e.g., Ker-TBS.1-24) were as effective as NF-derived TBSs (Fig. 4C). Although the comparative amino acid content of these TBSs revealed no exceptional charge characteristics or amino acid composition, like their conserved NF derived TBS counterparts, most had rather high pI values (Fig. 4D).
Figure 4.
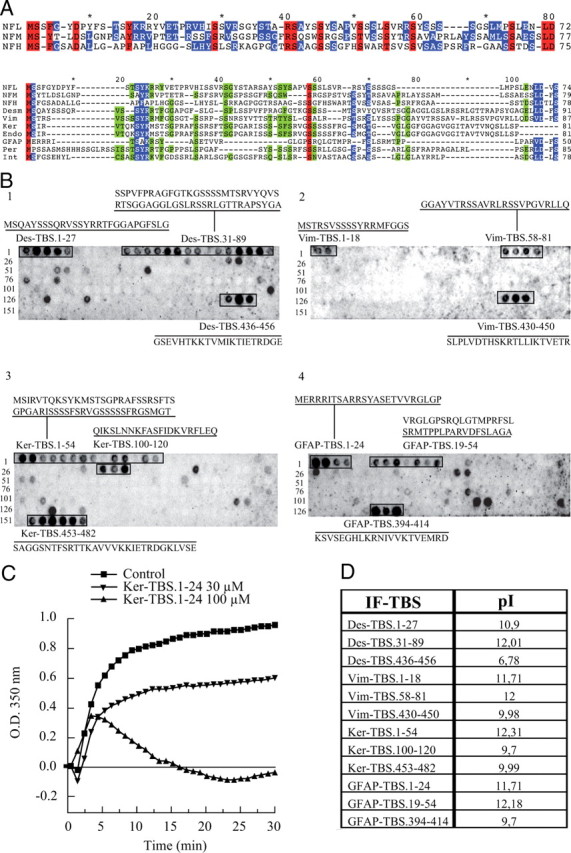
Sequence analysis of the tubulin-binding sites and their presence in other intermediate-filament proteins. A, Sequences of the three mouse NF subunits were aligned together, or with several intermediate-filament sequences, according to their homology by the ClustalW program. Alignment of NF subunits with the other N-terminal domains of intermediate filaments shows several conserved amino acids (red, 100%; blue, 80%; and green, 60% homology). B, Using the same experimental approach as described for NF subunits (compare Fig. 2A), the peptide arrays corresponding to the sequence of several intermediate filaments (desmin, vimentin, keratin, and GFAP) were tested for the possible presence of tubulin-binding sites. For all the intermediate filaments tested, a tubulin-binding site (alignment of 2 or more spots) was located in the N-terminal domain. C, The effect of Ker.TBS.1-24 was tested on MT polymerization, as previously described in Figure 2B, and demonstrates a typical inhibition of MT polymerization. D, Summary of the pI values for each intermediate-filament TBS as calculated online.
TBS peptides enter cells, disrupt the MT cytoskeleton, and affect proliferation
As TBS peptides alter the in vitro polymerization of tubulin, we investigated their capacity to affect the assembly of MTs in living cells. When glioblastoma-derived T98G cells were incubated with 10 μm biotinylated NFL-TBS.40-63 for 6 h, >75% of the cells contained detectable peptide and this subpopulation demonstrated an atypical spherical shape (Fig. 5A1–A3). Cells affected by the peptide also displayed a disrupted MT network with tubulin and peptide colocalized in amorphous masses. In contrast, those cells in which peptide was undetectable maintained a typical MT cytoskeleton and cell shape. The same effect was observed with the Ker-TBS.1-24 peptide (Fig. 5A4–A6). The mechanism through which these peptides penetrate cells remains to be characterized.
Figure 5.
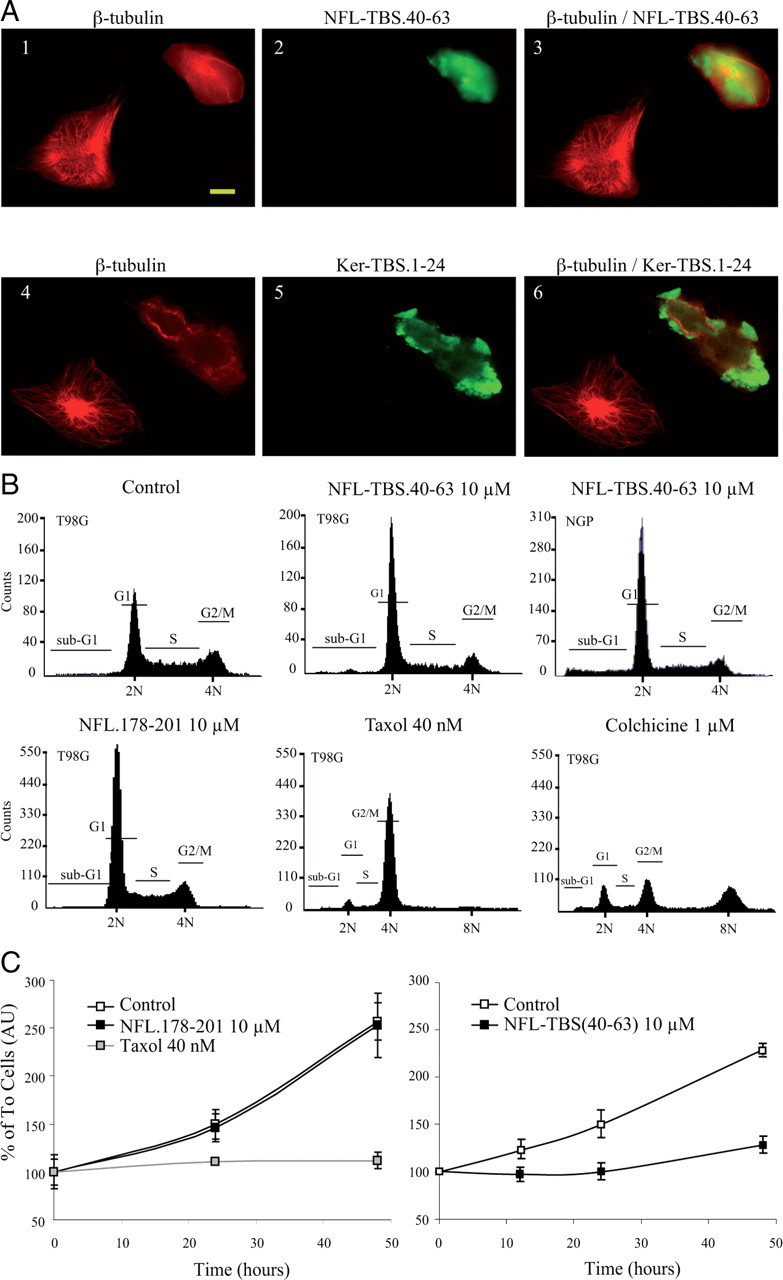
Effect of NFL-TBS.40-63 and Ker-TBS.1-24 peptides on the MT cytoskeleton and cell division. A, T98G cells were grown in the presence of biotinylated NFL-TBS.40-63 peptide (10 μm) for 6 h (A1–A3). MTs were detected using an anti-tubulin antibody (red), and NFL-TBS.40-63 was detected using Alexa-labeled avidin (green). Cells containing the peptide lack a typical MT network and have a round shape. Similar results were observed using biotinylated Ker-TBS.1-24 (A4–A6). B, Flow cytometric analysis of untreated and NFL-TBS.40-63-treated cells. T98G or NGP cells (third panel) treated with NFL-TBS.40-63 peptides (10 μm) during 48 h showed a typical G1 phase arrest. In the presence of NFL.178-201, no major effect was observed. Taxol and colchicine were used as controls. C, T98G cells were treated with NFL-TBS.40-63, NFL.178-201, or Taxol. Proliferation of NFL-TBS.40-63 peptide-treated cells (10 μm for 48 h) is reduced compared with cells grown in normal media. No effect was observed for NFL.178-201. Error bars indicate SEM.
To determine whether exposure to the NFL-TBS.40-63 peptide (10 μm) affects cell division, we evaluated the cell cycle distribution of control and peptide-treated T98G cultures using FACS analysis (after 50 μg/ml propidium iodide incubation to label DNA). In cultures treated with Taxol and colchicine, the typical perturbation of the G2/M transition was observed, whereas in untreated cultures, 39% of the cells were at G1 and this proportion was unchanged after exposure to the control NFL.178-201 peptide. In contrast, in cultures treated with the NFL-TBS.40-63 peptide, 61% of the cells were arrested at the G1 phase (Fig. 5B), and the G2/M and S phase populations were decreased proportionally. Consistent with this cell cycle perturbation, the MTS assay revealed a lower cell proliferation (Fig. 5C). Multiple cell lines demonstrated a similarly marked response to the NFL-TBS.40-63 peptide including NGP neuroblastoma (Fig. 5B3), MCF7 breast cancer, LS187 colorectal cancer, and 3T3 cells (data not shown). Additionally, we show that the NFL-TBS.40-63 peptide is taken up ex vivo by primary postmitotic hippocampal neurons from rat and mouse in which it also leads to disruption of the perikaryial microtubule network (supplemental material 1, available at www.jneurosci.org).
Peptides with composition identical with NFL-TBS.40-63, but with amino acid order reversed or scrambled (SCR), were tested for their activity in cultured cells. The reversed version of peptide (NFL-SBT.63-40) lost all capacity to enter cells, whereas the scrambled peptides NFL-SCR1, NFL-SCR2, and NFL-SCR3 demonstrated a diminished capacity to enter cells. Ker-SCR1 and Ker-SCR2 failed to enter cells, whereas Ker-SBT.24-1 entered cells but demonstrated a diminished capacity to disrupt MTs (Table 2).
Table 2.
Reversing or scrambling the amino acid order in TBS peptides abolishes their capacity to enter in cells and/or perturb the MT network
| Sequence | Cells with peptide (%) | Cells with intact MT (%) | |
|---|---|---|---|
| Buffer alone | 0.00 ± 0.00 | 85.26 ± 4.68 | |
| NFL-TBS0.40-63 | YSSYSAPVSSSLSVRRSYSSSSGS | 53.41 ± 9.18 | 39.29 ± 1.65 |
| NFL-SBT0.63-40 | SGSSSSYSRRVSLSSSVPASYSSY | 0.00 ± 0.00 | 67.65 ± 2.95 |
| NFL-SCR1 | SLGSPSSSVRASYSSSRSYVYSSS | 42.24 ± 0.28 | 81.92 ± 4.44 |
| NFL-SCR2 | VRPLSASRSSSGSYSSSYSSSVYS | 27.34 ± 13.82 | 62.81 ± 15.58 |
| NFL-SCR3 | SPSVSYSRSGSYSASRSLSYSVSS | 18.63 ± 9.78 | 63.04 ± 8.78 |
| Ker-TBS0.1-24 | MSIRVTQKSYKMSTSGPRAFSSRS | 31.44 ± 19.15 | 22.70 ± 6.62 |
| Ker-SBT0.24-1 | SRSSFARPGSTSMKYSKQTVRISM | 25.67 ± 14.47 | 67.72 ± 5.67 |
| Ker-SCR1 | KVTASMFYPIGRSRSTMSSRQSSK | 0.00 ± 0.00 | 74.59 ± 7.94 |
| Ker-SCR2 | RYSVSMSKSRGSTMRIFSPKSQAT | 0.00 ± 0.00 | 78.74 ± 4.91 |
The capacity of normal, reverse (NFL-SBT0.63-40 and Ker-SBT0.24-1), or scrambled peptides (SCR) to enter in cells and/or to affect the MT network was tested by incubating T98G cells for 6 h with 10 μm peptide. For each peptide treatment, at least 200 cells were examined for the presence of internalized peptide and disrupted MTs. The experiment was reproduced three times with cell phenotypes evaluated blindly by three independent examiners. The results are expressed as the mean ± SD. Approximately 15% of the total cell population demonstrates an abnormal MT network characterized by prominent tubulin labeling around the nucleus but with few or no MTs extending in the cytoplasm. NFL-SBT0.63-40, Ker-SCR1, and Ker-SCR2 lost all capacity to enter cells. NFL-SCR1, NFL-SCR2, NFL-SCR3, and Ker-SBT0.24-1 retained some capacity to enter cells, but their capacity to disrupt MT was significantly less than that displayed by the parental NFL-TBS0.40-63 and Ker-TBS0.1-24 sequences.
An enhanced microtubule cytoskeleton in axons of NFHLacZ transgenic mice
To investigate the potential in vivo significance of NF–tubulin interactions, we investigated the neuronal cell bodies and axons in NFHLacZ transgenic mice. Mice bearing the NFHLacZ transgene express an NFH–β-galactosidase fusion protein, and, through interactions mediated by β-galactosidase dimerization domains, NFs cross-link in the cell body, form aggregates, and are not transported to the axonal compartment. Despite the maldistribution of their entire NF cytoskeleton, such neurons are viable until advanced age (Eyer and Peterson, 1994). Although α-, β-, and γ-tubulin epitopes were readily detected within the perikaryal NF aggregates (Fig. 6B), no MT profiles were observable by electron microscopy (50 aggregates examined at 50,000×) (Fig. 6C). In contrast, in the NF-deficient axons, MTs were exceptionally prominent (Fig. 6A) and to measure their density, profiles within a predetermined axon cross-sectional area were counted in 500 sciatic nerve axons from 3-month-old animals. Control axons contained an average of 9.44 ± 1.4 MTs/μm2, whereas in transgenic samples, 99.06 ± 5.1 MTs/μm2 were observed. Note that the 10-fold higher density of MTs in the transgenic samples is not accommodated by the 50% reduction observed in the calibers of transgenic axons (see below).
Figure 6.
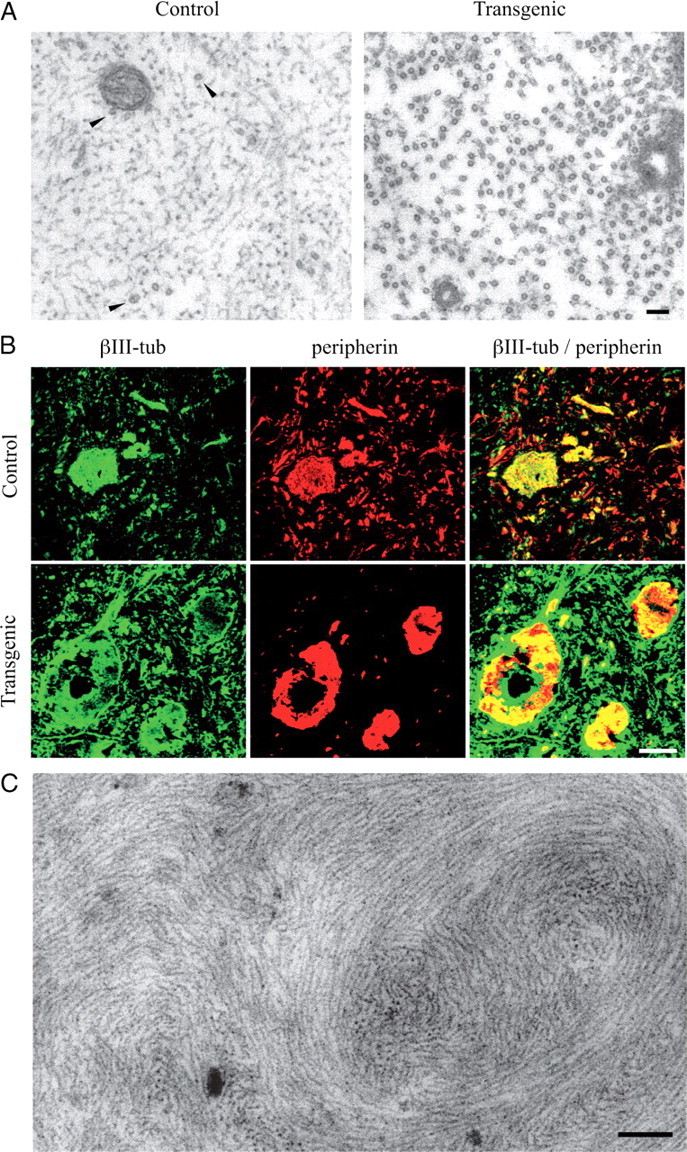
Analysis of the cytoskeleton in neurons of control and NFHLacZ transgenic mice. A, Typical axons in control samples are filled with a densely packed NF cytoskeleton and sparse MTs, whereas in transgenic samples they lack NFs and are filled with MTs. Scale bar, 100 nm. B, Confocal immunofluorescence analysis of spinal cords from control and NFHLacZ transgenic mice with antibodies recognizing βIII-tubulin and peripherin. In NFHLacZ transgenic samples, intermediate-filament labeling is restricted to cell body aggregates, and these aggregates also label with anti-tubulin antibodies. Scale bar, 12.5 μm. C, Electron micrograph of a typical NF aggregate present in a motor neuron cell body in NFHLacZ mice. Despite the presence of tubulin epitopes in such aggregates, no MT profile is detectable. Scale bar, 200 nm.
Accumulation of tubulin transcripts and proteins
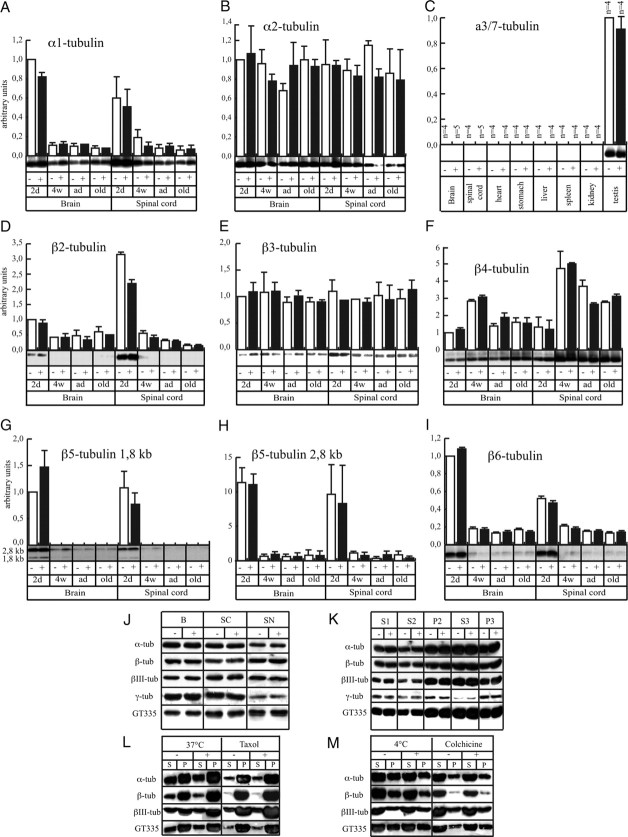
Neuronal development and axonal regeneration are associated with complex programs of temporally and spatially regulated tubulin isotype expression and such changes are reflected in the density of the axonal MT network (Ginzburg et al., 1985; Faundez and Alvarez, 1986; Hoffman and Cleveland, 1988; Tashiro and Komiya, 1992). Previous investigation of NFHLacZ transgenic mice showed that the amount of accumulated tubulin and MAPs was not dramatically changed (Tu et al., 1997; Riederer et al., 2003). To further evaluate the level at which the increased density and total number of axonal MTs in NFHLacZ mice is controlled, we compared accumulated tubulin mRNA in the nervous system of NFHLacZ transgenic and control mice at different postnatal ages. In contrast to the marked differences in axonal MT content observed by electron microscopy, brain and spinal cord samples from transgenic and control mice revealed no significant differences in tubulin mRNA levels (Fig. 7A–I).
Figure 7.
The amount of α- and β-tubulin mRNA and protein is similar between transgenic (+) and control (−) samples. A, During aging, a similar 10-fold decrease of mouse α-tubulin isoform 1 (Mα1) tubulin mRNA occurred both in brain and spinal cord of control and transgenic mice (2d, 2 d; 4w, 4 weeks; ad, adult; old, >24-month-old animals). Mean ± SE is shown. B, Mα2 mRNA remained constant during postnatal development of NFHLacZ mice. C, Mα3/7 tubulin mRNA was detected only in testis. D, Mouse β-tubulin isoform 2 (Mβ2) tubulin mRNA decreased 3-fold in brain and 10-fold in spinal cord in both control and transgenic samples. E, Mβ3 tubulin accumulated to similar levels in both normal and transgenic mice. F, Mβ4 tubulin mRNA showed a two-phase profile in developing brain and spinal cord samples. First, Mβ4 mRNA levels increased in brain threefold between 2 d and 4 weeks, and fivefold in spinal cord. Thereafter, Mβ4 tubulin mRNA decreased by a factor of 2 both in brain and spinal cord. G, H, Two Mβ5 transcripts exhibited a similar pattern of expression in control and transgenic spinal cords. The 2.8 kb transcript was expressed 10-fold higher than the 1.8 kb transcript, and during aging, both decreased by 10-fold. I, Mβ6 tubulin mRNA decreased by fivefold in brain and threefold in spinal cord in early postnatal development. J, Specific antibodies against different tubulin epitopes revealed no major difference in tubulin quantity present in total homogenates of brain (B), spinal cord (SC), or sciatic nerves (SN) between control and transgenic samples. K, After a classical preparation of MTs from the brains of normal and transgenic mice, each fraction was analyzed for tubulin content. No obvious difference was observed between the two genotypes. L, M, The first supernatants (S1) from brain of control and transgenic mice were exposed 1 h at 37°C with 1 mm GTP alone (37°C), or with Taxol to induce MT polymerization. Alternatively, the same S1 fraction was incubated at 4°C to block polymerization, or with colchicine. These mixtures were centrifuged at 100,000 × g for 30 min in the same incubation conditions, and supernatants and pellets were analyzed for tubulin epitopes by Western blotting. Control and transgenic samples yielded similar results.
We next estimated the total amount of tubulin recovered from samples of transgenic and control animals during a typical MT preparation. The same amount of protein from the crude extract was analyzed by Western blot and similar tubulin isotype ratios were observed in both transgenic and control samples (Fig. 7J). MT proteins then were isolated by one cycle of polymerization at 37°C and one cycle of depolymerization at 4°C (Weingarten et al., 1975). After each step, an aliquot was removed and analyzed by Western blotting using the panel of anti-tubulin antibodies described above. No quantitative differences were observed between transgenic and control samples at any of these steps (Fig. 7K). For sciatic nerves, limited sample size precluded accurate measurement of S3/P3 ratios but the S2/P2 ratios in control and transgenic samples were indistinguishable. Finally, to compare the assembly properties of the tubulin present in transgenic and control brains, S1 crude extracts were incubated under conditions that promote MT assembly (37°C or Taxol) or disassembly (4°C or colchicine) and centrifuged. No difference in the amount of tubulin present in the supernatant and pellet was apparent between transgenic and control samples (Fig. 7L,M). Thus, the 10-fold increase in axonal MT density in axons of NFHLacZ transgenic mice is not accompanied by detectable quantitative or qualitative changes in tubulin mRNA or protein accumulation.
Discussion
Observations made here with in vitro and in vivo preparations indicate that intermediate-filament proteins bind unpolymerized tubulin. The in vivo relevance of this interaction is suggested by the balance between tubulin and MTs in NF-deficient axons in which MT number exceeds that predicted by tubulin concentration alone. Consistent with these observations is a model in which intermediate filaments act as a tubulin reservoir modulating the local availability of tubulin (supplemental material 1, available at www.jneurosci.org). Although numerous MAPs alter the stability of assembled MTs, our observations suggest that axonal NFs exert a novel effect on MT formation by binding unpolymerized tubulin. By association with the dense and stable NF axoskeleton, a significant source of tubulin could be maintained throughout the axon, potentially limiting the need to transport tubulin proteins to regions of MT assembly.
Our finding that TBSs are present in all NF proteins is consistent with the axonal phenotypes reported in numerous mutant and experimental animal models. In quiver quail, a null NFL allele leads to a failure of NF assembly and, similar to NFHLacZ transgenic mice, axons fail to undergo normal radial growth and demonstrate a prominent MT cytoskeleton (Zhao et al., 1995). In mice, inactivation of each NF gene, either alone or in combination, also results in an enriched axonal MT cytoskeleton (Zhu et al., 1997, 1998; Elder et al., 1998; Rao et al., 1998; Jacomy et al., 1999). Finally, deletion of the C-terminal tail of NFH, a mutation that does not remove the NFH-TBS, has no consequence on the axonal MT cytoskeleton (Rao et al., 2002). In one conflicting report, the number of MTs in axons without NFs was considered to be normal (Elder et al., 1999). However, in that study, normalized axon sizes were predicted by reference to the number of myelin lamellae on a small number of axons. Thus, we consider it likely that an enriched axonal MT phenotype develops in most, if not all, circumstances in which axonal NFs are lacking.
In mature NFHLacZ transgenic mice, the average reduction of axon diameter is 50% (Eyer and Peterson, 1994). However, this reduction is not sufficient to account for the observed increase in axonal MT density. Assuming a circular profile, a normal axon with a diameter of 10 μm has a cross-sectional area of 78.5 μm2, and with 9.44 MTs/μm2 it would contain ∼741 microtubule profiles (9.44 × 78.5 = 741). If the same axon was rendered NF deficient, its diameter would reduce to 5 μm, yielding a cross-sectional area of 19.6 μm2. If the number of MTs remained unaltered, their density would increase to (741 MTs/19.6 μm2) 37.8 MTs/μm2, which represents a density increase of fourfold (37.8/9.44 = 4.0). In contrast, in the NF-deficient axons, the MT density was increased 10-fold and the absolute number of MTs was 2.5 times normal.
Theoretically, the increased number of MTs observed in NF-deficient axons could result from multiple perturbations secondary to the absence of axonal NFs. However, the absolute amount of tubulin in the axonal compartment is unaffected by the presence or absence of NFs requiring that normal axons maintain a pool of unpolymerized tubulin. Consistent with the view that NFs exert a direct effect on the pool of available tubulin to modulate the local assembly of MTs, a dynamic relationship between NFs and MTs is observed in both developing and regenerating axons (supplemental material, available at www.jneurosci.org). Developing axons are mostly devoid of NFs and, as predicted by the above model, demonstrate a high density of MTs. As peripheral fibers begin to mature, demonstrated by NF investment, radial growth, and myelination, MT density declines. This inverse relationship is realized in axons throughout the vertebrates including the large caliber (500 μm) CNS Mauthner axons of fish (Faundez and Alvarez, 1986). Although there are multiple changes in the synthesis of cytoskeletal elements in axotomized neurons, the same inverse relationship between MT density and NF content is observed as fibers regenerate and mature (Ginzburg et al., 1985; Hoffman and Cleveland, 1988; Tashiro and Komiya, 1992).
MT profiles were not observed within the dense NF aggregates seen in the neuronal perikarya of NFHLacZ transgenic mice. The local tubulin-binding capacity of the dense NF network may be sufficient to prevent the assembly of MTs. Alternatively, the highly phosphorylated status of NFs within the axonal compartment is not displayed by NFs residing in the cell body (de Waegh et al., 1992) nor in the aggregates present in these mice (Eyer and Peterson, 1994), and therefore the tubulin-binding properties of NFs in axons and perikaryal aggregates may differ. It is also possible that the density or configuration of the NFs within the aggregates prevents stable MT assembly by physical constraints.
Conservation of TBS motifs across the intermediate-filament family suggests that tubulin–intermediate filament binding relationships may exist in many cell types. Consistent with the prediction that such a partnership is physiologically relevant, diverse human diseases are associated with mutations affecting TBS motifs in multiple intermediate-filament proteins (www.interfil.org). Of particular interest, mutations changing the amino acid sequence of NFL-TBS.4-27 are associated with Charcot–Marie–Tooth type II neuropathy (Georgiou et al., 2002; Yoshihara et al., 2002). Similarly, mutation of prolines 8 and 22 in NFL-TBS.4-27 were shown to affect axonal transport (Pérez-Ollé et al., 2005). Thus, multiple levels of axon biology in both health and disease may depend on tubulin–NF interactions. Additionally, of the TBS motifs identified so far, several harbor residues subject to cell cycle-dependent phosphorylation (Chou et al., 1996; Goto et al., 1998; Ku et al., 1998; Toivola et al., 2002). Thus, the tubulin-binding capacity of intermediate filaments, shown here for mature neurons, may be a key regulator contributing to the assembly dynamics of MTs in diverse cell types.
Footnotes
This work was supported by grants from the Association Francaise contre les Myopathies, Association pour la Recherche sur le Cancer, Fonds Europeen de Developement Regional, and Institut National du Cancer (J.E.), the Canadian Institutes of Health Research (A.C.P.), and the German National Genome Research Net (R.F.). A.B. and R.B. were supported by Angers Agglomeration and Association pour la Recherche sur le Cancer. We thank Pr. N. Hirokawa and S. Terada (University of Tokyo, Tokyo, Japan) for helpful comments on this manuscript. We gratefully acknowledge Dr. D. Job, R. Margolis, and E. Denarier (Inserm and Centre National de la Recherche Scientifique, Grenoble, France) and H. Friedman (Laboratory of Developmental Biology, McGill University, Montreal, Quebec, Canada) for helpful comments. We thank Dr. J. Garin (Commissariat à l'Energie Atomique, Grenoble, France) for mass spectroscopy, and S. Daenicke (Helmholtz Centre for Infection Research, Braunschweig, Germany) for peptide array synthesis. We are grateful to Pr. O. Coqueret, B. Barré, and A. Vigneron (University of Angers, Angers, France) for their expertise in cell culture, to the Service Commun d'Imageries et d'Analyses Microscopiques de l'Universite d'Angers for assistance in electron and confocal microscopy, and to P. Chiron (animal facility).
References
- Aamodt EJ, Williams RC., Jr Microtubule-associated proteins connect microtubules and neurofilaments in vitro. Biochemistry. 1984;23:6023–6031. doi: 10.1021/bi00320a019. [DOI] [PubMed] [Google Scholar]
- Baas PW, Qiang L. Neuronal microtubules: when the MAP is the roadblock. Trends Cell Biol. 2005;15:183–187. doi: 10.1016/j.tcb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baas PW, Vidya Nadar C, Myers KA. Axonal transport of microtubules: the long and short of it. Traffic. 2006;7:490–498. doi: 10.1111/j.1600-0854.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Carden MJ, Trojanowski JQ, Schlaepfer WW, Lee VMY. Two stage expression of neurofilament polypeptides during rat neurogenesis with early establishment of adult phosphorylation patterns. J Neurosci. 1987;7:3489–3504. doi: 10.1523/JNEUROSCI.07-11-03489.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Chou YH, Opal P, Quinlan RA, Goldman RD. The relative roles of specific N- and C-terminal phosphorylation sites in the disassembly of intermediate filament in mitotic BHK-21 cells. J Cell Sci. 1996;109:817–826. doi: 10.1242/jcs.109.4.817. [DOI] [PubMed] [Google Scholar]
- Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- de Waegh SM, Lee VM, Brady ST. Local modulation of neurofilament phosphorylation, axonal caliber, and slow axonal transport by myelinating Schwann cells. Cell. 1992;68:451–463. doi: 10.1016/0092-8674(92)90183-d. [DOI] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL, Jr, Bosco P, Kang C, Gourov A, Tu PH, Lee VM, Lazzarini RA. Absence of the mid-sized neurofilament subunit decreases axonal calibers, levels of light neurofilament (NF-L), and neurofilament content. J Cell Biol. 1998;141:727–739. doi: 10.1083/jcb.141.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL, Jr, Pereira D, Tu PH, Zhang B, Lee VM, Lazzarini RA. Mice with disrupted midsized and heavy neurofilament genes lack axonal neurofilaments but have unaltered numbers of axonal microtubules. J Neurosci Res. 1999;57:23–32. doi: 10.1002/(SICI)1097-4547(19990701)57:1<23::AID-JNR3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Eyer J, Leterrier JF. Influence of the phosphorylation state of neurofilament proteins on the in vitro interactions between purified filaments. Biochem J. 1988;252:655–660. doi: 10.1042/bj2520655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer J, Peterson AC. Neurofilament-deficient axons and perikaryal aggregates in viable transgenic mice expressing a NF-beta-galactosidase fusion protein. Neuron. 1994;12:389–405. doi: 10.1016/0896-6273(94)90280-1. [DOI] [PubMed] [Google Scholar]
- Fasani F, Bocquet A, Robert P, Peterson A, Eyer J. The amount of neurofilament aggregated in the cell body is controlled by their increased sensitivity to trypsin-like proteases. J Cell Sci. 2004;117:861–869. doi: 10.1242/jcs.00940. [DOI] [PubMed] [Google Scholar]
- Faundez V, Alvarez J. Microtubules and calibers in developing axons. J Comp Neurol. 1986;250:73–80. doi: 10.1002/cne.902500107. [DOI] [PubMed] [Google Scholar]
- Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- Fukushima N, Furuta D, Hidaka Y, Moriyama R, Tsujiuchi T. Post-translational modifications of tubulin in the nervous system. J Neurochem. 2009;109:683–693. doi: 10.1111/j.1471-4159.2009.06013.x. [DOI] [PubMed] [Google Scholar]
- Georgiou DM, Zidar J, Korosec M, Middleton LT, Kyriakides T, Christodoulou K. A novel NF-L mutation Pro22Ser is associated with CMT2 in a large Slovenian family. Neurogenetics. 2002;4:93–96. doi: 10.1007/s10048-002-0138-4. [DOI] [PubMed] [Google Scholar]
- Ginzburg I, Teichman A, Dodemont HJ, Behar L, Littauer UZ. Regulation of three beta-tubulin mRNAs during rat brain development. EMBO J. 1985;30:3667–3673. doi: 10.1002/j.1460-2075.1985.tb04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kosako H, Tanabe K, Yanagida M, Sakurai M, Amano M, Kaibuchi K, Inagaki M. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J Biol Chem. 1998;273:11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- Guillaud L, Bosc C, Fourest-Lieuvin A, Denarier E, Pirollet F, Lafanechere L, Job D. STOP proteins are responsible for the high degree of microtubule stabilization observed in neuronal cells. J Cell Biol. 1998;142:167–179. doi: 10.1083/jcb.142.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Cross-linker system between neurofilament, microtubule, and membranous organelles in frog axons revealed by the quick-freeze, deep-etching method. J Cell Biol. 1982;94:129–142. doi: 10.1083/jcb.94.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga S, Hirokawa N. Dephosphorylation-induced interactions of neurofilaments with microtubules. J Biol Chem. 1990;265:21852–21858. [PubMed] [Google Scholar]
- Hoffman PN, Cleveland DW. Neurofilament and tubulin expression recapitulates the developmental program during axonal regeneration: induction of a specific beta-tubulin isotype. Proc Natl Acad Sci U S A. 1988;85:4530–4533. doi: 10.1073/pnas.85.12.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H, Zhu Q, Couillard-Després S, Beaulieu JM, Julien JP. Disruption of type IV intermediate filament network in mice lacking the neurofilament medium and heavy subunits. J Neurochem. 1999;73:972–984. doi: 10.1046/j.1471-4159.1999.0730972.x. [DOI] [PubMed] [Google Scholar]
- Julien JP, Mushynski WE. Multiple phosphorylation sites in mammalian neurofilament polypeptides. J Biol Chem. 1982;257:10467–10470. [PubMed] [Google Scholar]
- Ku NO, Liao J, Omary MB. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 1998;17:1892–1906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee VMY, Carden MJ, Schlaepfer WW, Trojanowski JQ, Otvos L, Hollosi M, Dietzchold B, Lazzarini RA. Identification of the major multiphosphorylation sites in mammalian neurofilaments. Proc Natl Acad Sci U S A. 1988;85:1998–2002. doi: 10.1073/pnas.85.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier JF, Eyer J. Properties of highly viscous gels formed by neurofilaments in vitro. A possible consequence of a specific inter-filament cross-bridging. Biochem J. 1987;245:93–101. doi: 10.1042/bj2450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Mandelkow EM. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Minami Y, Murofushi H, Sakai H. Interaction of tubulin with neurofilaments: formation of networks by NF-dependent tubulin polymerization. J Biochem. 1982;92:889–898. doi: 10.1093/oxfordjournals.jbchem.a134003. [DOI] [PubMed] [Google Scholar]
- Ohara O, Gahara Y, Miyake T, Teraoka H, Kitamura T. Neurofilament deficiency in quail caused by nonsense mutation in neurofilament-L gene. J Cell Biol. 1993;121:387–395. doi: 10.1083/jcb.121.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ollé R, López-Toledano MA, Goryunov D, Cabrera-Poch N, Stefanis L, Brown K, Liem RK. Mutations in the neurofilament light gene linked to Charcot-Marie-Tooth disease cause defects in transport. J Neurochem. 2005;93:861–874. doi: 10.1111/j.1471-4159.2005.03095.x. [DOI] [PubMed] [Google Scholar]
- Perrot R, Lonchampt P, Peterson AC, Eyer J. Axonal neurofilaments control multiple fiber properties but do not influence structure or spacing of nodes of Ranvier. J Neurosci. 2007;27:9573–9584. doi: 10.1523/JNEUROSCI.1224-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot R, Berges R, Bocquet A, Eyer J. Review of the multiple aspects of neurofilaments functions, and their possible contribution to neurodegeneration. Mol Neurobiol. 2008;38:27–65. doi: 10.1007/s12035-008-8033-0. [DOI] [PubMed] [Google Scholar]
- Rao MV, Houseweart MK, Williamson TL, Crawford TO, Folmer J, Cleveland DW. Neurofilament-dependant radial growth of motor axons and axonal organization of neurofilaments does not require the neurofilament heavy subunit (NF-H) or its phosphorylation. J Cell Biol. 1998;143:171–181. doi: 10.1083/jcb.143.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Garcia ML, Miyazaki Y, Gotow T, Yuan A, Mattina S, Ward CM, Calcutt NA, Uchiyama Y, Nixon RA, Cleveland DW. Gene replacement in mice reveals that the heavily phosphorylated tail of neurofilament heavy subunit (NF-H) does not affect axonal caliber or the transit of cargoes in slow axonal transport. J Cell Biol. 2002;158:681–693. doi: 10.1083/jcb.200202037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer IM, Robert P, Porchet R, Eyer J, Riederer BM. Selective changes in the neurofilament and microtubule cytoskeleton of NFHLacZ mice. J Neurosci Res. 2003;71:196–207. doi: 10.1002/jnr.10485. [DOI] [PubMed] [Google Scholar]
- Robert P, Peterson AC, Eyer J. Neurofilament cytoskeleton does not modify accumulation of trophic factors mRNA. J Neurosci Res. 2001;64:487–492. doi: 10.1002/jnr.1100. [DOI] [PubMed] [Google Scholar]
- Tashiro T, Komiya Y. Changes in organization and axonal transport of cytoskeletal proteins under normal and regenerating conditions. Mol Neurobiol. 1992;6:301–311. doi: 10.1007/BF02780559. [DOI] [PubMed] [Google Scholar]
- Tashiro T, Imai R, Komiya Y. Early effects of beta, beta′-iminodipropionitrile on tubulin solubility and neurofilament phosphorylation in the axon. J Neurochem. 1994;63:291–300. doi: 10.1046/j.1471-4159.1994.63010291.x. [DOI] [PubMed] [Google Scholar]
- Toivola DM, Zhou Q, English LS, Omary MB. Type II keratins are phosphorylated on a unique motif during stress and mitosis in tissues and cultured cells. Mol Biol Cell. 2002;13:1857–1870. doi: 10.1091/mbc.01-12-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu PH, Robinson KA, de Snoo F, Eyer J, Peterson A, Lee VM, Trojanowski JQ. Selective degeneration fo Purkinje cells with Lewy body-like inclusions in aged NFHLACZ transgenic mice. J Neurosci. 1997;17:1064–1074. doi: 10.1523/JNEUROSCI.17-03-01064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Villasante A, Lewis SA, Cowan NJ. The mammalian beta-tubulin repertoire: hematopoietic expression of a novel, heterologous beta-tubulin isotype. J Cell Biol. 1986;103:1903–1910. doi: 10.1083/jcb.103.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser AA, Or-Guil M, Tapia V, Leichsenring A, Schuchhardt J, Frömmel C, Volkmer-Engert R. SPOT synthesis: reliability of array-based measurement of peptide binding affinity. Anal Biochem. 2005;342:300–311. doi: 10.1016/j.ab.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Yamamoto M, Hattori N, Misu K, Mori K, Koike H, Sobue G. Identification of novel sequence variants in the neurofilament-light gene in a Japanese population: analysis of Charcot-Marie-Tooth disease patients and normal individuals. J Peripher Nerv Syst. 2002;7:221–224. doi: 10.1046/j.1529-8027.2002.02028.x. [DOI] [PubMed] [Google Scholar]
- Zhao JX, Ohnishi A, Itakura C, Mizutani M, Yamamoto T, Hojo T, Murai Y. Smaller axon and unaltered numbers of microtubules per axon in relation to number of myelin lamellae of myelinated fibers in the mutant quail deficient in neurofilaments. Acta Neuropathol. 1995;89:305–312. doi: 10.1007/BF00309623. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Couillard-Després S, Julien JP. Delayed maturation of regenerating myelinated axons in mice lacking neurofilaments. Exp Neurol. 1997;148:299–316. doi: 10.1006/exnr.1997.6654. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Lindenbaum M, Levavasseur F, Jacomy H, Julien JP. Disruption of the NFH gene increases axonal microtubule content and velocity of neurofilament transport: relief of axonopathy resulting from the toxin beta,beta′-iminodipropionitrile. J Cell Biol. 1998;143:183–193. doi: 10.1083/jcb.143.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]