Abstract
The neuropeptide pigment-dispersing factor (PDF) is a key transmitter in the circadian clock of Drosophila melanogaster. PDF is necessary for robust activity rhythms and is thought to couple the circadian oscillations of the clock neurons. However, little is known about the action of PDF on individual clock neurons. Here, we combined the period–luciferase reporter system with immunolabeling of clock proteins in wild-type and Pdf01 mutants to dissect the effects of PDF on specific subgroups of clock neurons. Additionally, PDF levels were elevated to higher than normal levels using specific neural mutants, and a correlation analysis of locomotor activity and clock protein staining served to determine the periods of specific clock cells. We found that PDF has multiple effects on the clock neurons: In some groups of clock neurons, PDF was required for maintaining the oscillations of individual cells, and in others, PDF was required for synchronous cycling of the individual members. Other clock neurons cycled with high amplitude in absence of PDF, but PDF affected their intrinsic clock speed. Sometimes PDF shortened and sometimes PDF lengthened period. Our observations indicate that PDF is crucial for adjusting cycling amplitude, period, and phase of the different players in the circadian clock. Under natural conditions PDF may be required for adapting Drosophila's clock to varying photoperiods. Indeed, we show here that Pdf01 mutants are not able to adapt their activity to long photoperiods in a wild-type manner.
Keywords: pigment-dispersing factor, period, timeless, internal desynchronization, circadian rhythms, neuropeptide
Introduction
Animals possess endogenous clocks in the brain that coordinate behavioral and physiological rhythms and help to adapt to the 24 h day/night cycle on earth. These clocks consist of numerous neurons that have the capability to oscillate with periods close to 24 h because of molecular feedback loops generated by the interaction of highly conserved clock genes and proteins (Lowrey and Takahashi, 2004; Hardin, 2005). As signaling molecules, animal clocks use neuropeptides that synchronize the oscillations of individual clock neurons and transfer circadian signals to downstream neurons (Maywood et al., 2006; Nässel and Homberg, 2006). In mammals, the vasoactive intestinal polypeptide (VIP) fulfils this role and is essential for normal behavioral rhythmicity (Aton et al., 2005; Vosko et al., 2007). A similar role has been proposed for the pigment-dispersing factor (PDF) in insects (Renn et al., 1999; Peng et al., 2003; Lin et al., 2004; Schneider and Stengl, 2005; Nitabach et al., 2006). Both, VIP and PDF work via closely related G-protein-coupled receptors of the B1 subfamily (classical hormone receptor) that are present on the clock neurons themselves (Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005; Shafer et al., 2008). Thus, VIP and PDF may couple the oscillations of individual cells as was recently shown for PDF regarding ultradian oscillations in membrane potential within the cockroach clock (Schneider and Stengl, 2005).
In Drosophila, Pdf-null mutants (Pdf01) are arrhythmic or show weak behavioral rhythms with short period (Renn et al., 1999). On the molecular level, individual clock neurons seem to remain rhythmic and to cycle with short period (Wu et al., 2008), and in certain groups of clock cells, the oscillations lose synchrony (Peng et al., 2003; Klarsfeld et al., 2004; Lin et al., 2004). The common hypothesis is that PDF feeds back on the oscillations of individual neurons, lengthens their period, and additionally synchronizes their oscillations.
However, a recent study suggests that the role of PDF is more complex (Wülbeck et al., 2008). When PDF is increased to higher than normal levels, short- and long-period components appeared in the activity pattern. This suggests that PDF not only has period-lengthening effects but also shortens the period of certain clock neurons in a dose-dependent manner (Wülbeck et al., 2008). However, the identity of these neurons remained unknown.
Here, we identify the neurons that respond with period shortening to excessive PDF. Furthermore, we show for the first time that PDF has different effects on the period of the different clock neurons even in wild-type flies. Under natural conditions, this property of PDF might keep the oscillations in the different subsets of clock neurons slightly out of phase. Such a multiphasic clock organization was also found in the mammalian clock (Quintero et al., 2003; Yamaguchi et al., 2003) and proposed to account for many properties of circadian organization as the response to seasonal variations in the photoperiod by altering the coupling of its constituent oscillators (Pittendrigh and Daan, 1976). PDF seems to be critically involved in these processes.
Materials and Methods
Fly rearing and strains.
Flies were reared on cornmeal/agar medium supplemented with yeast and kept in a temperature-constant room at 20°C and in a light/dark cycle with 12 h light and 12 h darkness (LD 12:12). The lab strain CantonS was used as wild-type control. Pdf01 mutants (w +;Pdf01) carry a point mutation in the part of the Pdf-gene that codes for the PAP-precursor of Pdf and lack PDF completely (Renn et al., 1999).
The period–luciferase strain 8.0-luc:9 that expresses the luciferase only in subsets of the pacemaker neurons was used as real-time reporter for oscillations of the clock gene period (Veleri et al., 2003). Furthermore, a stable 8.0-luc:9;Pdf01 line was generated to monitor the molecular oscillations in the absence of PDF. Both strains carried an X chromosome marked with y w, resulting in white eye and yellow body color. To see in which clock neurons the per 8.0-luc:9 transgene is expressed, we brought the 8.0-luc:9 and 8.0-luc:9;Pdf01 lines in the per01 background and performed immunostainings with an antibody against the Period protein (PER) (see below) (Veleri et al., 2003).
To increase PDF levels in the central brain, the following optic lobe affecting mutants were used: sine oculis1 (so1) and sine oculismedusa (somda). The phenotype of these mutants including their rhythmic behavior is described in detail by Wülbeck et al. (2008). Briefly, adult so1 flies have optic lobes that are reduced to 20% of its normal volume (Fischbach, 1983), whereas the optic lobes of somda mutants have <5% of normal size (Wülbeck et al., 2008). As a consequence, both mutations affect the arborizations of a subgroup of the clock neurons, the PDF-positive large ventral lateral neurons (l-LNv) (see Fig. 1). The l-LNv arborize in the most distal layer of the medulla putatively transferring circadian signals to the optic lobes (eyes) (Helfrich-Förster, 1997). In addition, they connect the pacemaker centers, the accessory medullae (aMe), of both brain hemispheres via fibers in the posterior optic tract (see Fig. 1) (Helfrich-Förster et al., 2007a). In the mutants, the size of the medulla is reduced and the l-LNv cells project into the central brain instead of arborizing on the surface of the medulla. This leads to increasing amounts of PDF fibers in the aMe and the dorsal protocerebrum that may release high amounts of PDF into these areas and provoke alterations in the activity rhythms (Wülbeck et al., 2008). so1 mutants show a higher density of PDF fibers in the aMe and exhibit significantly longer periods (24.6 h) than wild-type flies (Table 1). When PDF was absent (so1;Pdf01 double mutants), the period came back to the short value of Pdf01 showing that the period lengthening is caused by PDF (Table 1). somda mutants show additionally many more PDF fibers in the dorsal brain than so1 mutants and in the activity rhythm a short-period component (21.3 h) split off the long (24.7 h) period (Table 1). This short period was significantly shorter than the short period of flies that lack PDF (Pdf01, so1;Pdf01, and somda;Pdf01 mutants), indicating that PDF shortens the period of certain clock neurons in addition to lengthening that of others.
Figure 1.
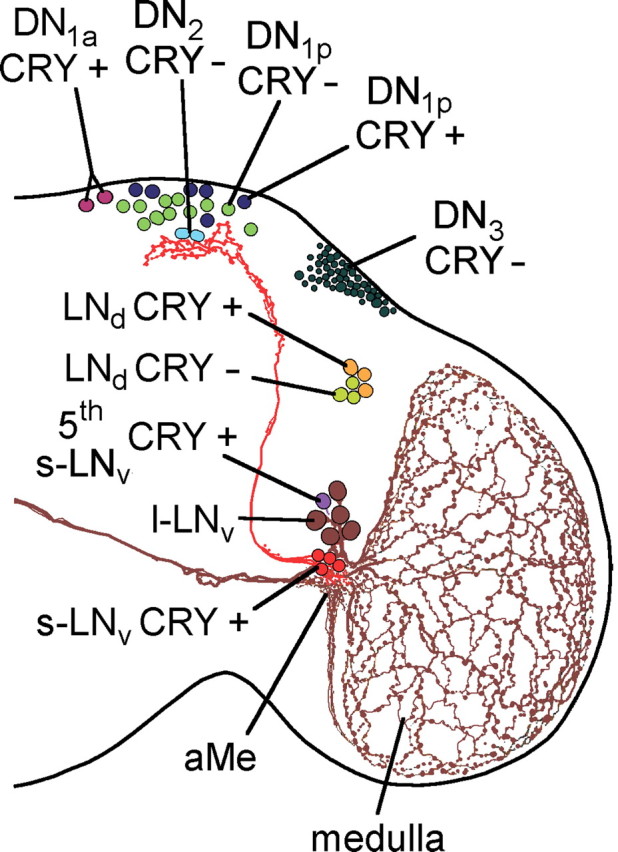
Circadian clock neurons in the right brain hemisphere of the fruit fly. PDF is expressed in two groups of the lateral neurons, the l-LNv and s-LNv. Of these neurons, the arborization patterns are shown; all others are omitted for clarity. The fifth PDF-negative s-LNv is located among the l-LNv, and the LNd are located dorsally of the l-LNv. Whereas all s-LNv and l-LNv cells contain CRY, only three of the six LNd are CRY-positive. The dorsal neurons in the posterior dorsal brain were traditionally divided in three groups (DN1, DN2, and DN3), but they seem to be composed of heterologous cells. Among the DN1, two cells are located in more anterior regions and are therefore called DN1anterior (DN1a), whereas the other up to 15 DN1 are called DN1posterior (DN1p). The DN1a and six of the DN1p cells are CRY-positive. The two DN2 cells are located close to the s-LNv terminals and both are CRY-negative. The DN3 consist of ∼40 neurons of predominantly small size that are all CRY-negative; but there are few larger neurons among them that might have different properties.
Table 1.
Activity rhythms of wild-type strains and Pdf01 mutants in different genetic backgrounds
| Strain | n | Rhythmic flies | t 1 | Power | t 2 | Power |
|---|---|---|---|---|---|---|
| CantonS | 26 | 26 | 24.2 ± 0.04 | 20.4 ± 0.82 | ||
| 8.0-luc:9 | 32 | 32 | 24.1 ± 0.03 | 18.7 ± 0.71 | ||
| Pdf01 | 50 | 25 | 22.8 ± 0.08 | 9.2 ± 0.53 | ||
| 8.0-luc:9;Pdf01 | 32 | 18 | 22.8 ± 0.04 | 9.5 ± 0.64 | ||
| so1 | 50 | 50 | 24.6 ± 0.03 | 27.7 ± 0.75 | ||
| so1;Pdf01 | 47 | 24 | 22.4 ± 0.10 | 8.6 ± 0.67 | ||
| somda | 60 | 58 | 21.1 ± 0.01 | 13.0 ± 0.08 | 24.7 ± 0.01 | 12.9 ± 0.39 |
| somda;Pdf01 | 71 | 29 | 22.2 ± 0.07 | 8.5 ± 0.21 |
The rhythmic values for so1 and somda mutants in the Pdf + and Pdf01 background are taken from the study by Wülbeck et al. (2008).
Luciferase monitoring.
Luciferase expression of individual flies carrying the 8.0-luc transgene was measured as described by Veleri et al. (2003). Before each experiment, flies were entrained for at least 3 d to a LD 12:12 and kept in the same regimen for the first day of the experiment. Subsequently the lights were turned off and the flies were monitored for 6 d in constant darkness (DD). Raw data were plotted and analyzed with Import and Analysis software (Plautz et al., 1997). For details, see Veleri et al. (2003).
To visualize the neurons in which luciferase was expressed, flies were entrained for 5 d to a LD 12:12. Four hours after lights-on, male brains were dissected under M3 Insect Medium (Sigma-Aldrich) and kept in culture medium [M3 Insect Medium with fetal bovine serum, penicillin–streptomycin, insulin, and luciferin (200 μm; Biosynth)]. Four hours after lights-off, the samples were imaged in the Luminoview LV200 (Olympus). Brains were exposed for 40 min at a gain of 255. The bioluminescence intensity was shown in pseudocolors after background subtraction using Rainbow3 (Cell M; Olympus).
Immunohistochemistry for determination of PER-, TIM-, CRY-, and PDF-positive neurons in per0;per-luc flies.
Flies were quickly killed by submersion in 4% freshly prepared paraformaldehyde in phosphate buffer (PB) with 0.5% Triton X, shortly before lights-on, when PER and Timeless (TIM) levels are high. After 2 h fixation, the flies were rinsed three times for 15 min in PB, and their brains were dissected as whole mounts. Then fluorescent immunohistochemistry was applied as described previously (Rieger et al., 2006). The primary antisera were as follows: rat anti-TIM serum (diluted 1:1000) (Sidote et al., 1998), rabbit anti-cryptochrome (CRY) serum (Yoshii et al., 2008), or rabbit anti-PER serum (diluted 1:1000) (Stanewsky et al., 1997), and the monoclonal mouse antibody nb33 (diluted 1:100). The latter is directed against the PDF precursor and thus recognizes the PDF-positive neurons (Veleri et al., 2003). The secondary antisera were as follows: Alexa 488 (goat anti-rabbit), Alexa 568 (goat anti-rat), and Alexa 647 (goat anti-mouse), all diluted 1:200 (Invitrogen). The immunofluorescent brains were embedded in Vectashield mounting medium (Vector Laboratories), directly after rinsing in PB.
Triple labeling (green channel, PER or CRY; red channel converted to magenta, TIM; infrared channel converted to yellow, PDF) was visualized by laser-scanning confocal microscopy (Zeiss LSM 510 META; Carl Zeiss MicroImaging). Confocal stacks of 2 μm thickness were obtained at intervals of 2.4 μm and overlaid (using the LSM Meta software; version 3.2.0.99) to visualize the lateral neurons in the anterior brain and the dorsal neurons in the posterior brain. To exclude bleed-through, we used sequential scans of the three laser lines.
Activity recording.
At the age of 1–3 d, individual male flies were transferred into the recording chambers. Locomotor activity was recorded photoelectrically at 20°C as described previously (Helfrich-Förster, 1998). Halogen photooptic lamps (Osram; XENOPHOTR) served as a light source and intensity was adjusted to 500 μW/cm2 with a dimmer. For judging the free-running locomotor rhythm, activity was recorded for up to 7 d under LD 12:12, and then the flies were transferred to DD. The raw data were displayed as actograms (double plots) to judge the activity pattern, and the periods under DD were determined by the Sokolove–Bushell periodogram analysis (program El Temps, version 1, 226; Antoni Díez-Noguera, Barcelona, Spain).
For judging the activity pattern under different photoperiods, wild-type and Pdf01 flies were recorded for 1 week, respectively, under the following day/night lengths: 8:16 h, 12:12 h, 16:8 h, 20:4 h. Light intensity was adjusted to 100 lux during the day and to 0.03 lux during the night. The dim light during the night prevented the flies from being completely inactive at night. Average activity profiles were calculated for each photoperiod, and the mean phases of morning and evening peaks were calculated as described previously (Rieger et al., 2003).
Immunohistochemistry for determination of PER or TIM staining intensity.
To judge the state of the molecular clock in the different clock neurons during the first 5 d in DD, wild-type flies and Pdf01 mutants were collected at circadian time 11 (CT11) and CT23. The circadian times of each strain were calculated by average free-running periods of each strain in DD (24.2 h for wild-type flies and 22.8 h for Pdf01 mutants) (Table 1). The collected samples of wild-type flies and Pdf01 mutants were subjected to TIM, CRY, and PDF triple immunohistochemistry as described above. Ten brains were stained per time point.
To judge the state of the molecular clock in so1 and somda flies after several days in DD, 10 so1 and 10 somda flies, respectively, were collected at two different time points in DD. For so1 flies, these times were (1) the subjective early morning, just before they became active, and (2) the subjective evening, when they were maximally active on the third to fourth day in DD (see Fig. 5, red and blue points). For somda flies, these times were (1) during the activity peak of the component free-running with short period and (2) during the activity peak of the component free-running with long period on the third to fourth day in DD (see Fig. 5, red and blue points). Furthermore, some somda flies were stained on the sixth day in DD, when both components were again in phase, at the times of low and high activity.
Figure 5.
Actograms of so1 and somda mutants and TIM staining in the clock neurons of both mutants at two times on days 3–4 in DD (red and blue points). Ten flies were stained at each time for each mutant, respectively. As can be seen in the example actograms, so1 mutants show one rhythm free-running with long period as soon as released to DD (A), whereas somda mutants reveal two rhythmic components in DD, which free-run with short and long periods, respectively, and that were 180° out of phase at the times of staining (C). B, In so1 mutants, the CRY-positive clock neurons were strongly TIM-immunoreactive in the early subjective morning, just before the flies became active (A, red point; B, red columns) and less stained in the subjective evening, when the flies were most active (A, blue point; B, blue columns). The CRY-negative neurons did not show this difference. ANOVA revealed a significant influence of the time of staining on labeling intensity (F (1,165) = 30.68; p < 0.001) and that this influence was dissimilar in the different neuronal groups (F (7,165) = 3.96; p < 0.001). The post hoc test showed that only the CRY-positive neurons showed significantly higher TIM staining in the early subjective morning compared with the evening. No significant differences were revealed for the CRY-negative neurons. D, In somda mutants, ANOVA showed again a significant influence of time on staining intensity (F (1,179) = 4.62; p = 0.03), which was strongly dissimilar in the different neurons (F (8,179) = 34.78; p < 0.001). No significant staining differences were only found in the CRY-negative DN1p remain. The other neurons were either strongly TIM-immunoreactive at the activity maximum of the long-period component (C, blue point; D, blue columns) or at the activity maximum of the short-period component (C, red point; D, red columns). For additional explanations, see text. The green point in C indicates the time at which short- and long-period components crossed each other and were consequently in phase again. At this day, the different clock neurons cycled in synchrony with each other (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Error bars indicate SEM.
All brains were subjected to TIM, CRY, and PDF triple immunohistochemistry (see above). One experiment was additionally performed with TIM, PER, and PDF triple immunohistochemistry.
Scoring of staining intensities was performed on single optical sections as described by Rieger et al. (2006), but with the following modifications. Each neuron within each group was assessed separately. If there was no staining in one or more neurons of a given group, these neurons were assigned to have background staining levels. The background staining level was measured in the surrounding field of each neuronal group and subtracted from the pixel intensities measured for the cells. A mean staining intensity was calculated from all neurons within one group for each hemisphere separately. Then for each group an average staining intensity per brain was calculated out of the staining intensities of both hemispheres.
Statistics.
Staining intensities were tested for normal distribution using the Lillifors test. For testing for significant influences of strain and time on PER and TIM stainings in the clock neurons, a two-way ANOVA was applied. In both cases, a subsequent post hoc (Bonferroni adjustment) served for pairwise comparisons of staining intensities. Values were regarded as significantly different at p < 0.05.
Results
Figure 1 gives an overview of Drosophila's clock neurons. PDF is expressed in the small and large ventral lateral neurons (s-LNv and l-LNv). Both types of neurons arborize in the aMe, which seems to represent the main pacemaker center of the fly. The s-LNv cells project additionally into the dorsal brain, a brain area that comprises additional clock neurons, the dorsal neurons (DN1, DN2, and DN3). The s-LNv cells appear to release PDF rhythmically into this area (Park et al., 2000). There is evidence that the s-LNv cells are the most important clock neurons for rhythmic behavior in DD (Helfrich-Förster, 2005; Stoleru et al., 2005; Picot et al., 2007). There is a fifth PDF-negative s-LNv that is typically located among the l-LNv cells and that seems to have the same arborization patterns as the PDF-positive s-LNv cells and to play a similar crucial role in the circadian clock (Rieger et al., 2006; Helfrich-Förster et al., 2007b). Slightly dorsal of the s-LNv and l-LNv lie the dorsal lateral neurons (LNd) that connect to the aMe and the dorsal brain as do most of the DN cells (Shafer et al., 2006; Helfrich-Förster et al., 2007a). For better clarity, all these connections except those of the PDF neurons are omitted in Figure 1. The described groups of clock cells can be further subdivided in CRY-positive and CRY-negative cells (Fig. 1) (Yoshii et al., 2008). The PDF receptor is most likely expressed in all clock neurons except the l-LNv cells (Shafer et al., 2008).
Our aim was to elucidate how PDF influences the molecular PER/TIM oscillations in the different groups of clock neurons.
Pdf01 mutants show sustained per–luciferase oscillations with short period, whereas these oscillations dampen in wild-type flies
In a first step, we monitored the molecular oscillations in the clock neurons in vivo using the luciferase based real-time reporter assay described previously (Veleri et al., 2003). To specifically reveal the molecular oscillations in certain groups of the clock neurons, we used flies carrying the period–luciferase construct 8.0-luc:9, which is restricted to the dorsal neurons (DN1–DN3) and few dorsal lateral neurons (LNd) (Veleri et al., 2003). The construct was inserted in wild-type and Pdf01 flies (both in the y w background), and the activity rhythm of the transgenic flies was tested to make sure that the construct did not affect behavior. We found that the y w;8.0-luc:9 flies showed wild-type rhythms with endogenous behavioral periods close to 24 h (Fig. 2 A, Table 1), whereas y w;8.0-luc:9;Pdf01 flies behaved like Pdf01 mutants exhibiting, if rhythmic at all, weak rhythmicity with a period of 22.8 h (Fig. 2 B, Table 1).
Figure 2.
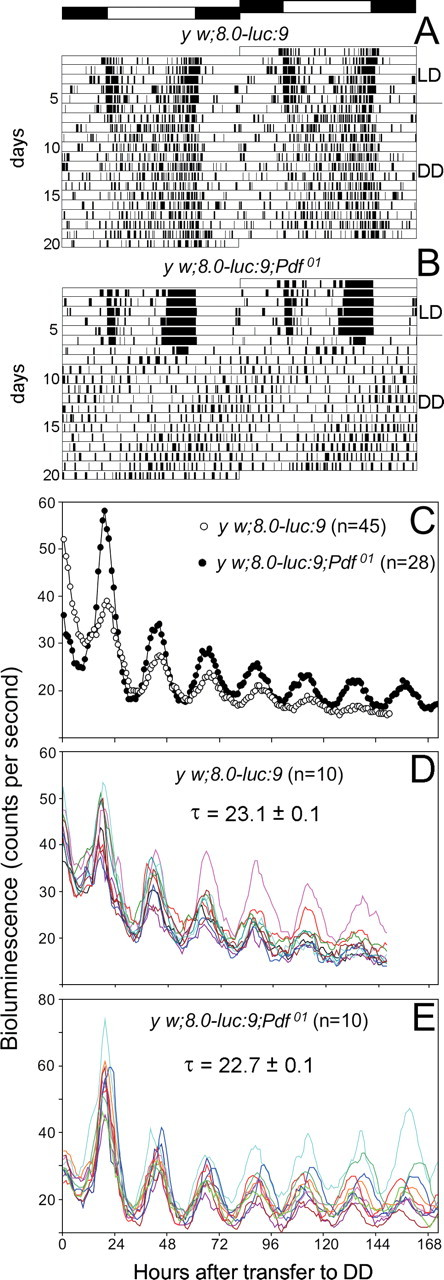
Locomotor activity and bioluminescence rhythm of flies carrying the 8.0-luc:9 transgene in the wild-type and the Pdf01 background. Control flies (y w;8.0-luc:9) show a free-running rhythm with an average period of 24.1 h (A), whereas y w;8.0-luc:9;Pdf01 mutants exhibit weak rhythms with an average period of 22.8 h (B) (compare Table 1). C, Averaged bioluminescence rhythm of 45 control flies and 28 Pdf01 mutants. The flies were recorded for 6 d in DD after initially being entrained to three cycles of 12:12 h LD. The y w;8.0-luc:9;Pdf01 flies showed rather strong oscillations with a mean period of 22.7 ± 0.1 h, whereas y w;8.0-luc:9 flies showed more dampened oscillations with a mean period of 23.1 ± 0.1 h. The weak dampening present in the y w;8.0-luc:9;Pdf01 flies is entirely attributable to slightly different free-running periods of individual flies as can be seen in E, in which the oscillations of individual y w;8.0-luc:9;Pdf01 flies are shown (n = 10). In contrast, a dampening of the oscillations occurs already on the level of individual wild-type flies (n = 10) (D), indicating that the oscillating DN and LNd cells become out of phase in the presence of PDF.
In y w;8.0-luc:9 flies, the luciferase-based real-time reporter assay revealed that PER protein oscillations in the DN and LNd cells continued for 6 d in constant darkness, although with a dampening amplitude (Fig. 2 C,D) (cf. Veleri et al., 2003). Obviously, the per cycling in the luciferase-expressing clock neurons remained more or less in synchrony for several days in DD. This is in ultimate contrast to the per–luciferase cycling in peripheral tissues and organs that became asynchronous shortly after transfer into DD and speaks for a synchronizing factor present in the brain (Veleri et al., 2003). PDF could be this synchronizing agent between the brain clock neurons. Therefore, we expected the per–luciferase oscillations to be more dampened in the Pdf01 background.
To our surprise, these oscillations were even less dampened in y w;8.0-luc:9;Pdf01 mutants. They continued with high amplitude for >6 d in constant darkness (Fig. 2 C,E). This confirms previous results obtained with disco2;8.0-luc:9 flies that lack all LNs and are therefore also PDF-null (Veleri et al., 2003) and indicates that the luciferase-positive cells oscillate rather synchronously in the absence of PDF. Obviously, PDF is not needed as synchronizing agent and its presence even provokes a slight asynchrony among these neurons. Most interestingly, the luciferase rhythm of Pdf01 had a period of 22.7 h that does nicely correspond to the weak 22.8 h period that Pdf01 mutants display in the activity rhythm (Table 1, Fig. 2 B). This suggests that the DN and LNd cells are responsible for the short-period rhythm observed in some Pdf01 mutants. Nevertheless, the behavioral rhythms of Pdf01 mutants were very weak, showing that the short-period oscillations in the DN and LNd cells are not sufficient for strong rhythmicity in DD, a postulation that was already made by previous studies (Blanchardon et al., 2001; Veleri et al., 2003; Stoleru et al., 2005). That the DN and LNd are not the main neurons controlling behavioral rhythmicity in DD becomes also evident from the data gained for wild-type flies (Fig. 2 A,D), which showed rather robust behavioral rhythms with periods around 24 h (Table 1), whereas the per–luciferase oscillations in the DN and LNd had a significantly shorter period (23.1 h) and were dampened (Fig. 2 D). As mentioned, the most important clock neurons for rhythmicity under DD conditions are the s-LNv cells, and these do not express the luciferase in 8.0-luc flies and do therefore not contribute to the per–luciferase rhythm (Veleri et al., 2003).
In the present experiment, we could only see the sum of per oscillations going on in the DN and LNd; in these, PDF did not lengthen the periods to the ∼24 h value observed for the behavioral rhythms; additionally PDF provoked a slight dampening of the oscillations. Two possible reasons for the dampening are as follows: (1) the oscillations in individual neurons stop, and (2) PDF provokes asynchrony among the individual clock neurons (e.g., by affecting the periods of individual neurons differently).
To distinguish between these possibilities, we performed immunohistochemistry with an antibody against TIM over 5 d in DD. TIM shows oscillations that are largely parallel to that of PER (Shafer et al., 2002); and because the anti-TIM antibody was raised in rat, we could additionally stain with anti-CRY (raised in rabbit) to distinguish the TIM cycling in CRY-positive and CRY-negative neurons (Fig. 1). We did not judge the TIM staining intensity in the l-LNv, because previous studies had shown the oscillations to stop in these neurons after transfer into DD (Yang and Sehgal, 2001; Shafer et al., 2002; Veleri et al., 2003).
PDF desynchronizes the molecular oscillations of the CRY-positive and CRY-negative
LNd samples of fly brains were stained for 5 consecutive days in DD at the putative TIM maxima and minima, respectively (Fig. 3, circadian time 23 and 11). Because wild-type flies showed an average free-running period of 24.2 h, and Pdf01 mutant a period of 22.8 h (Table 1), wild-type flies have a circadian day that lasts 24.2 h and Pdf01 mutants one that lasts 22.8 h, and we had to collect wild-type flies every 12.1 h and Pdf01 mutants every 11.4 h over 5 d in DD.
Figure 3.
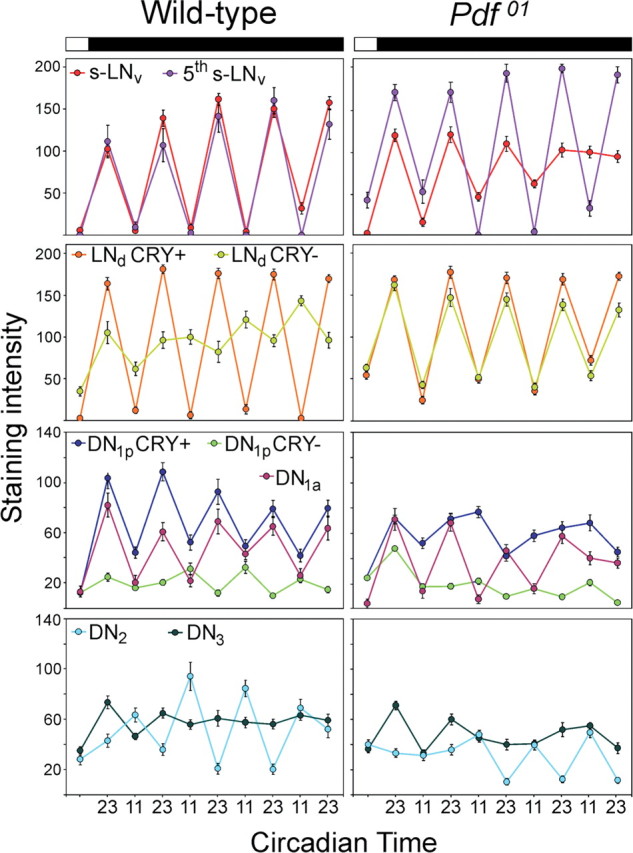
TIM oscillations in the different clock neurons of wild-type flies and Pdf01 mutants for 5 d in constant darkness. The time axis is in circadian time. Wild-type flies and Pdf01 mutants have different periods (Fig. 2); therefore, a circadian day of wild-type flies is 24.2 h long and a circadian day of Pdf01 mutants is 22.8 h long. For both fly strains, circadian time 23 means 1 h before the beginning of the subjective day, and circadian time 11 means 1 h before the beginning of the subjective night. Usually, clock protein levels are high at circadian time 23 and low at circadian time 11. For wild-type flies, this is true for all CRY-positive neurons: the s-LNv, the fifth s-LNv, three LNd, the DN1a, and six DN1p (for significances, see Table 2). However the CRY-negative LNd seem to cycle with different period and are in antiphase with the CRY-positive neurons on the fifth day in DD. The CRY-negative DN1p and DN2 cycled in antiphase to the CRY-positive neurons already earlier, but lost their significant oscillations at the end of the experiment (Table 2). No significant cycling was seen in the CRY-negative DN3 (except for day 1). Note that, in Pdf01 mutants, the CRY-negative and CRY-positive LNd remain in synchrony with each other; the oscillations in the s-LNv and DN1a slowly dampen, and the oscillations in the CRY-positive DN1p disappear quickly after transfer into DD (Table 2 indicates days at which ANOVA revealed significant oscillations); for additional explanations, see text.
In wild-type flies, we found significant oscillations in all CRY-positive clock neurons indicating that they cycle with a 24.2 h period (Fig. 3, Table 2). High-amplitude cycling was present in the 4 s-LNv cells and the fifth s-LNv cell, whereas the DN1a and DN1p cells cycled with slightly lower amplitude. In contrast to the CRY-positive neurons, the CRY-negative cells showed a completely different behavior: In the DN3, no significant oscillations were revealed from the second day in DD onward under the 24.2 h collection regimen (Table 2). Either these cells stop oscillating, move out of phase, or cycle with periods completely different from 24.2 h. The CRY-negative LNd, DN1p, and DN2 cells showed significant cycling only at certain days in DD (Table 2) and changed their phase in respect to the CRY-positive neurons. The CRY-negative LNd cells cycled in phase with the CRY-positive neurons on the first day in DD, showed no visible oscillations on days 2–3, and appeared to oscillate again on days 4–5, but now in antiphase to the CRY-positive LNd cells (Fig. 3). ANOVA followed by a post hoc test revealed significant oscillations only at days 1 and 5 (Table 2). The CRY-negative DN1p cells lost significant oscillations already on the first day in DD, but these came back in antiphase to the CRY-positive DN1p cells on days 3 and 4. On day 5, the oscillations vanished again. The CRY-negative DN2 cells showed significant cycling on days 2–4. On these days, they cycled in antiphase to the CRY-positive neurons (Fig. 3, Table 2). The most likely explanation for this unusual behavior of the CRY-negative neurons is that they cycled with a period different from 24.2 h and therefore switched from in phase cycling to antiphase cycling with times of apparent arrhythmicity in between. We cannot judge the period of these CRY-negative neurons from our immunostainings, but regarding the overall short period of the per–luciferase cycling in wild-type flies (23.1 h) (Fig. 2 D), it is most likely that the CRY-negative neurons cycled with short period. To be in antiphase with the CRY-positive neurons on the fifth day in DD, as was observed for the CRY-negative LNd cells, the neurons must have advanced their phase by 2.5 h per the 24.2 h day. This is equivalent to a free-running period of 21.7 h. For the CRY-negative DN1p and DN2, the free-running periods must be even shorter, because these were in antiphase with the CRY-positive neurons already on days 3–4 in DD. Considering these different periods (24.2 h for the CRY-positive neurons and 21.7 h or shorter for the CRY-negative neurons), dampening oscillations of the per–luciferase with a mean period of ∼23 h are reasonable. But are these different periods and the resulting asynchrony between individual cells caused by PDF?
Table 2.
Significance of TIM cycling revealed by ANOVA and a post hoc test (Bonferroni's) in wild-type flies and Pdf01 mutants during 5 d in DD
| Wild type |
Pdf01
|
|||||
|---|---|---|---|---|---|---|
| F value, F (8,171) | p value | Significant cycling at days | F value, F (8,171) | p value | Significant cycling at days | |
| s-LNv | 99.38 | p < 0.001 | 1–5 | 25.44 | p < 0.001 | 1–4 |
| 5th s-LNv | 24.83 | p < 0.001 | 1–5 | 90.00 | p < 0.001 | 1–5 |
| LNd CRY+ | 258.80 | p < 0.001 | 1–5 | 137.47 | p < 0.001 | 1–5 |
| LNd CRY− | 5.56 | p < 0.001 | 1, 5 | 58.67 | p < 0.001 | 1–5 |
| DN1a | 9.88 | p < 0.001 | 1–5 | 17.64 | p < 0.001 | 1–4 |
| DN1p CRY+ | 14.64 | p < 0.001 | 1–5 | 7.17 | p < 0.001 | 1 a |
| DN1p CRY− | 6.63 | p < 0.001 | 3–4 | 15.19 | p < 0.001 | 3–5 |
| DN2 | 16.81 | p < 0.001 | 2–4 | 19.25 | p < 0.001 | 1, 3–5 |
| DN3 | 2.83 | p = 0.006 | 1 | 10.69 | p < 0.001 | 1 |
aThe post hoc test found an additional significance on days 3–4, but there is no regular circadian cycling visible (Fig. 3).
To answer this question, we have to look at the oscillations in Pdf01 flies. Indeed, we found that the CRY-negative LNd cells cycled with high amplitude and in phase with the CRY-positive LNd cells throughout the 5 d in DD (Fig. 3). This could partly explain why the per–luciferase rhythms were less dampened in Pdf01 mutants compared with wild-type flies. However, the other CRY-negative neurons of Pdf01 mutants behaved principally similar to those of wild-type flies [just differences in amplitude and the days at which significant cycling occurs are evident (Table 3)]. Furthermore, the oscillations in the CRY-positive s-LNv and DN1a continuously lost amplitude and significant oscillations disappeared on day 5 in DD (Fig. 3, Table 2). The oscillations in the CRY-positive DN1p vanished already on the second day in DD (Fig. 3, Table 2). Just the fifth s-LNv cell showed high-amplitude oscillations throughout the recording time. Its cycling amplitude was even higher than in wild-type flies.
Table 3.
Comparison of the effects of PDF on the clock neurons in wild-type flies and somda mutants
| Clock neurons | Total no. | CRY+/− | Action of PDF in |
|
|---|---|---|---|---|
| Wild-type flies | somda mutants | |||
| l-LNv | 4–5 | CRY+ | Not assessed | Not assessed |
| s-LNv | 4 | CRY+ | Period lengthening (+ sustaining cycling) | Period shortening (not assessed) |
| 5th s-LNv | 1 | CRY+ | Period lengthening | Period lengthening |
| LNd | 6 | 3 CRY+ | Period lengthening | Period lengthening |
| 3 CRY− | Period shortening | Period shortening | ||
| DN1a | 2 | CRY+ | Period lengthening | Period shortening |
| DN1p | ∼17 | 6 CRY+ | Period lengthening (+ sustaining cycling) | Period shortening in 2 cells, period lengthening in 4 cells (not assessed) |
| ∼11 CRY− | Period shortening | Period shortening | ||
| DN2 | 2 | CRY− | No effect | Period lengthening in 1 cell |
| DN3 | ∼40 | CRY− | No effect | Not assessed |
The luciferase transgene is expressed in all DN groups and in the LNd, but the expression strength depends on the genetic background (PDF+ or PDF−)
The fact that the CRY-positive and CRY-negative LNd cells of Pdf01 mutants remained coupled and cycled with 22.8 h until the end of the experiment can partly explain why the per–luciferase rhythm of Pdf01 mutants was stronger and not dampened compared with wild-type flies. However, because TIM cycling completely disappears in the six CRY-positive DN1p cells of Pdf01 flies, whereas it persists in those of wild-type flies, it is still puzzling why Pdf01 flies showed a less dampened luciferase rhythm than wild-type flies.
Therefore, we had to make sure that indeed the same cells express the luciferase transgene in the wild-type and Pdf01 background. To do so, we recorded brains with a high-sensitivity camera (Fig. 4 A,B), and we performed PER immunohistochemistry on the brains (Fig. 4 C–M). For the latter, the 8-luc:9 and 8-luc:9;Pdf01 lines were crossed into the per01 background so that only the PER–luciferase fusion protein is revealed.
Figure 4.
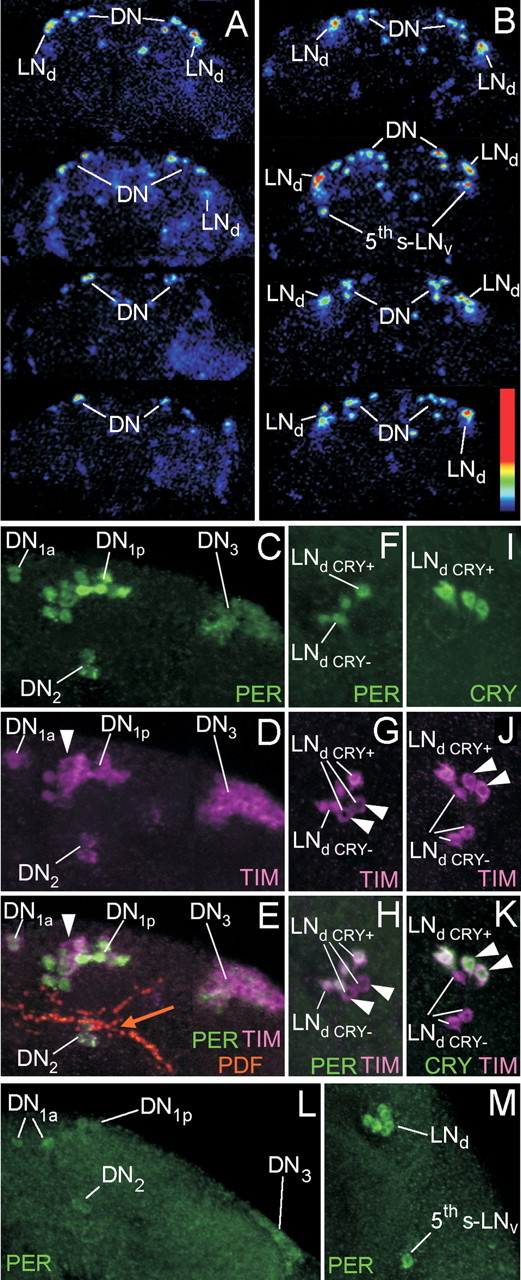
Spatial expression pattern of the PER–LUC (luciferase) fusion protein in brains of 8.0-luc:9 transgenics. A–B, Monitoring of the luciferase expression with a high-sensitive camera show luminescent spots in the dorsal and dorsolateral brain of control (A) and Pdf01 (B) brains. For each genotype, four brains are shown. Note that, in the Pdf01 background, the LNd are stronger luminescent than the DN. C–K, To reveal transgene expression at higher resolution per01;8.0-luc:9 brains are triple labeled with anti-PER, anti-TIM, and anti-PDF or with anti-CRY, anti-TIM, and anti-PDF at the peak point of staining (1 h before lights-on). Overlays of all labelings are found in E, H, and K. (The orange arrow in E points to the fiber tract originating from the s-LNv and terminating in the distal dorsal protocerebrum close to the DN1 and DN2). PER labeling is only present in neurons that express the transgene (C, F), whereas endogenous TIM is found in all clock neurons (D, G, J. However, nuclear TIM occurs only in the PER-positive cells (arrowheads point to cells that had cytoplasmic TIM and consequently were PER-negative). C, F, PER labeling shows that the 8.0-luc:9 transgene is expressed in the DN1a, most DN1p, the DN2, most DN3, and in four LNd cells. E, H, K, CRY and TIM labeling show that three of the four PER-positive LNd cells are CRY-negative (the arrowheads in D–K point to the CRY-negative cells that have cytoplasmic TIM). L, M, Transgene expression in the Pdf01 background. Note that the DN cells (L) are much weaker labeled as in the wild-type background, but that all six LNd are strongly labeled (M). Additionally, PER is always present in the fifth s-LNv cell (M).
The high-sensitivity camera could not reveal single cells but showed luminescent cell clusters in the dorsal and dorsolateral brain that are most likely identical with the DN and the LNd. These cell clusters were present in wild-type (Fig. 4 A) and Pdf01 (Fig. 4 B) brains. The main difference between both genotypes was the luminescence intensity. The latter was higher in the Pdf01 brains, and this was especially true for the dorsolateral cluster that probably corresponds to the LNd (Fig. 4 B). In addition, some Pdf01 brains showed a luminescent spot in the region of the l-LNv that was not present in the wild-type brains (Fig. 4 B).
PER immunostaining gave clearer results that pointed into the same direction. In the wild-type background, all DN clusters clearly expressed the transgene, whereas staining in the LNd was notably weaker (Fig. 4 C–K). We counted four to five stained LNd cells and CRY double labeling revealed that the transgene was always expressed in the three CRY-negative LNd cells and additionally in one to two CRY-positive cells (Fig. 4 H,K). In rare cases (∼10% of the brains), we found the transgene additionally in the CRY-positive fifth s-LNv (data not shown). In the Pdf01 background, all six LNd cells plus the fifth s-LNv expressed the transgene, and the staining intensity was much higher than in the wild-type background (Fig. 4 M). In contrast, the staining intensity appeared considerable weaker in the DN clusters (Fig. 4 L).
Our results indicate that the transgene expression level was influenced by the genetic background. This could fully explain the different per–luciferase oscillations we got for wild-type and Pdf01 flies (Fig. 2 C). In wild-type flies, mainly the DN clusters and the CRY-negative LNd cells contribute to the luciferase cycling. Because the CRY-negative LNd and DN cells most probably cycle with shorter period than the CRY-positive DN cells, it is understandable that the overall oscillation is dampening. In Pdf01 mutants, the situation is different. Here, mainly the LNd cluster and the fifth s-LNv seem to contribute to the per–luciferase cycling. These cells continue to cycle in synchrony with 22.8 h period under DD conditions (Fig. 3), and this may explain the high-amplitude not-dampened oscillation of the per–luciferase (Fig. 2 C,E).
In summary, we can say that PDF affects the clock neurons in a complex manner: (1) PDF mediates synchrony within certain groups of clock neurons as the s-LNv, and the DN1a. Simultaneously, PDF seems to lengthen the period of these neurons from 22.8 to 24.2 h. (2) PDF similarly lengthens the periods of some clock neurons as the CRY-positive LNd and the fifth s-LNv; but within these groups, PDF is not necessary for synchronous high-amplitude cycling. (3) PDF shortens the periods of other clock neurons as the CRY-negative LNd; again PDF is not necessary for synchronous high-amplitude cycling in this group. (4) PDF appears necessary for rhythmicity in the CRY-positive DN1p, because cycling in this group stops immediately after transfer into DD. (5) PDF has no obvious effect on the CRY-negative DN1p, DN2, and DN3 cells, because these behaved similarly in wild-type flies and Pdf01 mutants.
Enhanced PDF levels in the dorsal brain lead to period shortening in the CRY-positive DN1a, in two DN1p, and in the four PDF-positive s-LNv cells
Despite the period-shortening effect of PDF on the CRY-negative LNd cells, no short period component is visible in the free-running rhythm of wild-type flies. Obviously, these neurons do not visibly contribute to the behavioral rhythm under DD conditions in wild-type flies. The same seems to apply to so1 mutants that have elevated PDF levels in the aMe and show behavioral rhythms with 24.6 h periods (Fig. 5 A) (Wülbeck et al., 2008). Again, only the period-lengthening effects of PDF are visible in the behavior. This is obviously different when PDF is elevated to very high levels in the dorsal brain as true in somda mutants (Wülbeck et al., 2008). Under these circumstances, a PDF-dependent short period (21.3 h) component appeared in the activity rhythm (Fig. 5 C). Most interestingly, this short period is similar to the short period we calculated for the CRY-negative neurons in wild-type flies. We wondered why in somda mutants this short period appeared in the behavioral rhythm. A possible explanation would be that, in somda mutants, more clock neurons were accelerated by PDF and consequently contributed to the activity rhythm. We therefore asked whether there are subsets of pacemakers that are accelerated by increased PDF levels in the brain.
To find out which neurons run with short and which with long period in somda mutants, we applied a method that has been previously proven to work successfully for internally desynchronized rhythms (de la Iglesia et al., 2004; Rieger et al., 2006). We immunostained somda brains at two time points in DD at which the two activity components that free-ran with short and long periods in somda mutants were 180° out of phase (Fig. 5 C). Fortunately, somda individual flies show all a rather stereotyped activity pattern desynchronizing into two components with short and long periods, respectively. The two activity components became completely out of phase with each other on days 3–4 in DD, crossed each other, and remained together for a short while on days 6–8, and then separated again and were once more out of phase on days 10–11. The intersection of the two components continued until the end of the experiment (Fig. 5 C).
so1 mutants that showed just a long period, but no internal desynchronization in the activity patterns served as controls. For these, we chose the times of highest and lowest activity on days 3–4, respectively (Fig. 5 A). We expected from data of previous studies (Yoshii et al., 2004; Rieger et al., 2006) that high TIM immunoreactivity is present in the early subjective morning (slightly before the morning peak of activity) (Fig. 5 A, red point) and low TIM immunoreactivity is present in the subjective evening (during the evening activity peak) (Fig. 5 A, blue point) in all the cells that contribute to the activity (the CRY-positive neurons). This was indeed the case (Fig. 5 B). ANOVA revealed significant differences in staining intensity between both time points for all CRY-positive neurons. Additionally, we found TIM to be mainly nuclear at the times of strong staining, whereas TIM was mainly cytoplasmic at the time of low staining.
We did not see any significant differences in staining intensity between the two time points in the CRY-negative neurons. This is in agreement with our present results for wild-type flies, because these cells seem to free-run with short period and might neither be in phase nor in complete antiphase to the CRY-positive neurons on days 3–4 in DD (note that the CRY-positive neurons of so1 mutants cycle with a period of 24.6 h and not with a 24.2 h period; therefore, we could not deduce the days with in phase and antiphase cycling between CRY-positive and CRY-negative neurons from the wild type shown in Fig. 3).
For somda, we chose the times of high activity of the short component and that of high activity of the long component on days 3–4 in DD, respectively (Fig. 5 C). According to our previous results (Rieger et al., 2006), we expected the neurons that control the short component to be stained the strongest at the activity maximum of the long component (Fig. 5 C, blue point) and those that control the long component to be stained the strongest at the activity maximum of the short component (Fig. 5 C, red point).
As found for so1 mutants and wild-type flies, the majority of CRY-negative neurons appeared to cycle with short period in somda mutants (most evident for the CRY-negative LNd cells), whereas mainly CRY-positive neurons (the three LNd cells, the fifth s-LNv, and two CRY-positive DN1p cells) ran with long period (Fig. 5 D). However, most interestingly, several CRY-positive neurons did now join the CRY-negative neurons and did also cycle with short period. These were the four s-LNv cells, the two DN1a cells, and two cells of the DN1p that were strongly CRY-positive (Fig. 5 D). However, one DN2 cell appeared to cycle with long period (Figs. 5 D, 6).
Figure 6.

TIM labeling showing the internal desynchronization among the dorsal and lateral neurons in somda mutants stained at the two time points marked by red and blue in Figure 5 (red time point, left panel; blue time point, right panel). Both brains were triple stained with anti-TIM (green), anti-CRY (magenta), and anti-PDF precursor (orange). The large pictures in the center of both panels depict the overlay of all three labelings (combination of 10 confocal stacks). Depending on their amount of TIM and CRY, the PDF-positive LNv cells appear in yellow to orange, neurons that have no PDF (e.g., some LNd and DN1) but equal amounts of TIM and CRY appear in white, those that contain only TIM are in green, and those that contain CRY but no or only tiny amounts of TIM are in magenta. To reveal TIM and CRY staining more clearly, single labeling of TIM and CRY of several clock neurons were enlarged and shown separately above and below the central pictures (DN1–2 and LNd on top, LNv on bottom; see insets in the large pictures). At the “red” time point (left panels), TIM was high in one pair of CRY-positive DN1p (the DN1p2) (Fig. 6) and in one cell of the DN2 (note that the DN2 cells are CRY-negative). No TIM labeling was found in the CRY-positive DN1a, and only weak TIM labeling in the CRY-positive DN1p1 and some CRY-negative DN1p. Among the LNd, TIM was high in the three CRY-positive LNd, and no TIM-staining at all was present in the three CRY-negative LNd. Among the LNv, TIM was high in the fifth s-LNv and in the l-LNv that were not evaluated. Almost no TIM was found in the PDF-positive s-LNv. At the “blue” time point (right panels), TIM was high in the DN1a and the DN1p1. Only low amounts of TIM were found in the remaining DN1p and in the DN2. Among the LNd, the CRY-negative LNd were strongly stained by TIM, whereas the CRY-positive LNd contained only little TIM. Of the LNv, again the l-LNv were strongly stained (not evaluated), but now the PDF-positive s-LNv were highly TIM-immunoreactive and little TIM was present in the fifth s-LNv.
The asynchrony within the different groups of clock neurons can be nicely seen in Figure 6. This was especially evident for the LNd cluster: At the time marked with the red point, the three CRY-positive LNd cells were strongly immunoreactive against TIM and the three CRY-negative LNd cells were not stained at all, whereas at the blue marked time point, the three CRY-negative LNd cells were strongly TIM-positive and the three CRY-positive cells were only weakly stained. The same asynchrony occurred between the PDF-positive s-LNv cells and the PDF-negative fifth s-LNv (Fig. 6): At the time marked with the red point, the fifth s-LNv was strongly immunoreactive against TIM and the PDF-positive s-LNv cells were not, whereas at the blue marked time point, the PDF-positive s-LNv cells were strongly TIM-positive and the fifth s-LNv was not stained.
To make sure that the internal desynchronization of the molecular oscillations we found on days 3–4 in DD is not a permanent state in somda mutants, we also stained few flies on day 6 in DD. On day 6, the long and short period component crossed each other, and as a consequence both were in phase for a short time interval (Fig. 5 C, green point). Accordingly, no asynchrony among the CRY-positive and CRY-negative LNd cells should be visible. This was indeed the case. We found prominent TIM staining in all LNd cells during the activity trough and weaker staining during the activity peak (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Similarly, the PDF-positive s-LNv cells and the PDF-negative fifth s-LNv cell seemed to be in phase with each other. As was expected, both were more prominently stained during the activity trough and weaker stained during the activity peak (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
In summary, we found that high PDF levels in the dorsal brain correlated with a permanently shortened period of the CRY-negative LNd and DN1p cells plus that of the CRY-positive DN1a, two CRY-positive DN1p, and the PDF-positive s-LNv cells. The short-period rhythm of the s-LNv is probably the reason why the short-period component was visible in the activity pattern of somda mutants. However, PDF appeared to lengthen the period of the PDF-negative fifth-sLNv, the CRY-positive LNd cells, and one DN2 cell, and these may control the long-period component in the activity rhythm. Figure 7 shows the neurons that seemed to shorten their period on PDF in red and those that seemed to lengthen their period in blue for somda mutants. Table 3 compares the effects of PDF on the different neurons in wild-type and somda flies.
Figure 7.
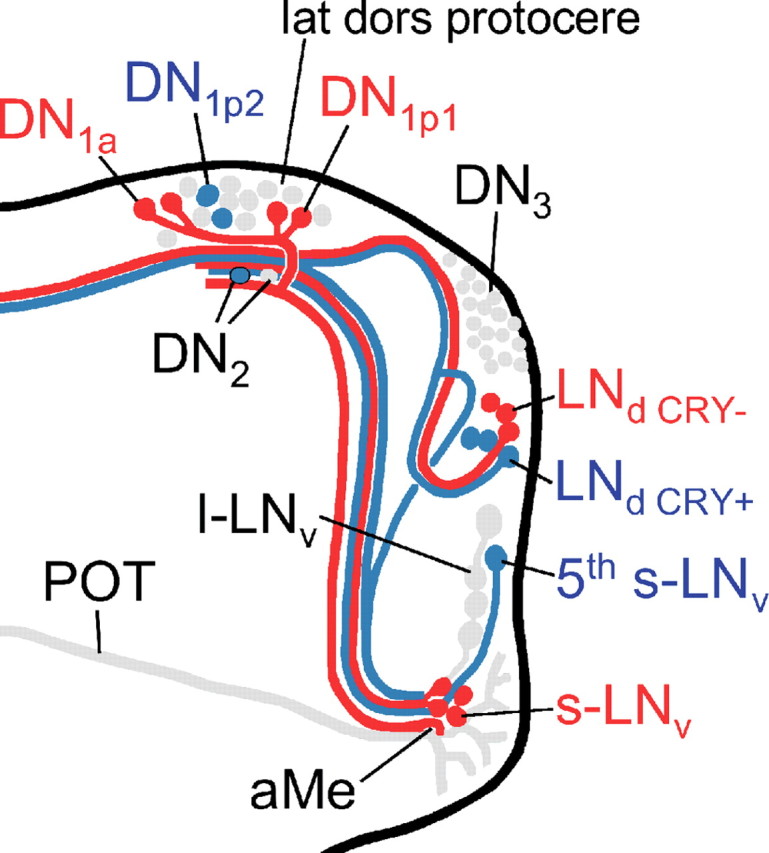
Diagram summarizing our present view on the differential action of PDF on the clock neurons in somda mutants (only the right brain hemisphere is shown). Neurons that lengthen their period on excessive amounts of PDF in the dorsal brain are shown in blue. Neurons that shorten their period under the same conditions are shown in red. Neurons not assessed in the present study or neurons with unclear response to PDF are shown in gray. Note that the aberrant arborizations of the l-LNv are omitted for better clarity. The s-LNv, fifth s-LNv, l-LNv, DN1a, and the three CRY-positive LNd neurons arborize in the aMe, in which PDF secreted by the l-LNv cells might serve as coupling factor in wild-type flies (Wülbeck et al., 2008, their Discussion). Furthermore, fibers from the s-LNv, the DN1a, DN1p1, and the CRY-positive and -negative LNd cells may contact each other in the lateral dorsal protocerebrum (lat dors protocere), in which PDF seems to be rhythmically released from the s-LNv cells (Park et al., 2000). The fifth s-LNv cell was also found to project toward the dorsolateral protocerebrum, but it is not clear whether it reached this area or whether it terminated before (Helfrich-Förster et al., 2007). The l-LNv cells do not project into the dorsolateral brain in wild-type flies, but do so extensively in somda mutants (Wülbeck et al., 2008). We suppose that extensive amounts of PDF secreted into this brain region shorten the periods of the CRY-positive DN1a and DN1p1 cells. The latter two groups may feedback on the s-LNv forcing them also to free-run with short period (for details, see supplemental Discussion, available at www.jneurosci.org as supplemental material).
PDF is necessary for adaptation of the activity rhythm to different photoperiods
Most interestingly, the same scenario that the PDF-positive s-LNv and the fifth PDF-negative s-LNv (together with one-half of the LNd cells) showed short and long periods respectively was observed previously for wild-type flies and cryb mutants under constant-light conditions (Rieger et al., 2006; Helfrich-Förster et al., 2007b). In this case, light was the factor that speeded up and slowed down the relevant neurons.
Light is the most important Zeitgeber for synchronizing circadian clocks to the 24 h day. Prolonged light exposure in the summer makes the morning activity of flies to advance (speed up) to the beginning of the day and the evening activity to delay (slow down) to the end of the day to avoid the midday heat (Majercak et al., 1999; Rieger et al., 2003, 2006). This adaptation is known for many animals including humans, and it is important for survival (Foster and Roenneberg, 2008). Pittendrigh and Daan (1976) proposed a model in which this adaptation to long summer days is achieved by two circadian oscillators that respond differently to light. Drosophila was the first animal in which the anatomical substrates of the two oscillators could be traced to certain clock neurons (Grima et al., 2004; Stoleru et al., 2004, 2005, 2007), and in which it was shown that certain clock neurons indeed speeded up or slowed down in response to light (Rieger et al., 2006).
According to our present results, PDF may also be crucially involved in the speed control of the clock neurons, and thus also in the adaptation of the activity patterns to long days. If true, Pdf01 mutants should not be able to adapt to long days in the normal manner. To test this, we subjected wild-type flies and Pdf01 mutants to different photoperiods.
We found that wild-type flies phase-advanced their morning activity and phase-delayed their evening activity with increasing photoperiod as reported previously (Fig. 8 A–E) (Rieger et al., 2003). However, Pdf01 mutants were not able to behave in this manner (Fig. 8 F–J). As reported previously, they showed little activity in the morning (Renn et al., 1999). Therefore, we did concentrate our analysis on the evening activity. The latter was very pronounced in Pdf01 mutants and occurred earlier in the day than in wild-type flies fitting to their short period under DD conditions (Renn et al., 1999). However, in clear contrast to wild-type flies, the evening activity did not show any phase adaptations in respect to the photoperiod (Fig. 8 F–J). Instead of delaying with increasing day length, the maximum of the evening peak did even slightly advance. We conclude that PDF is necessary for the flies to adapt normally to different photoperiods. Thus, the major role of PDF under natural conditions might be to adjust the phasing of the different players in the clock and perhaps mediate the effects of light on the clock neurons under long days. Future studies have to reveal the oscillations of the clock neurons in Pdf01 mutants under long and short days.
Figure 8.
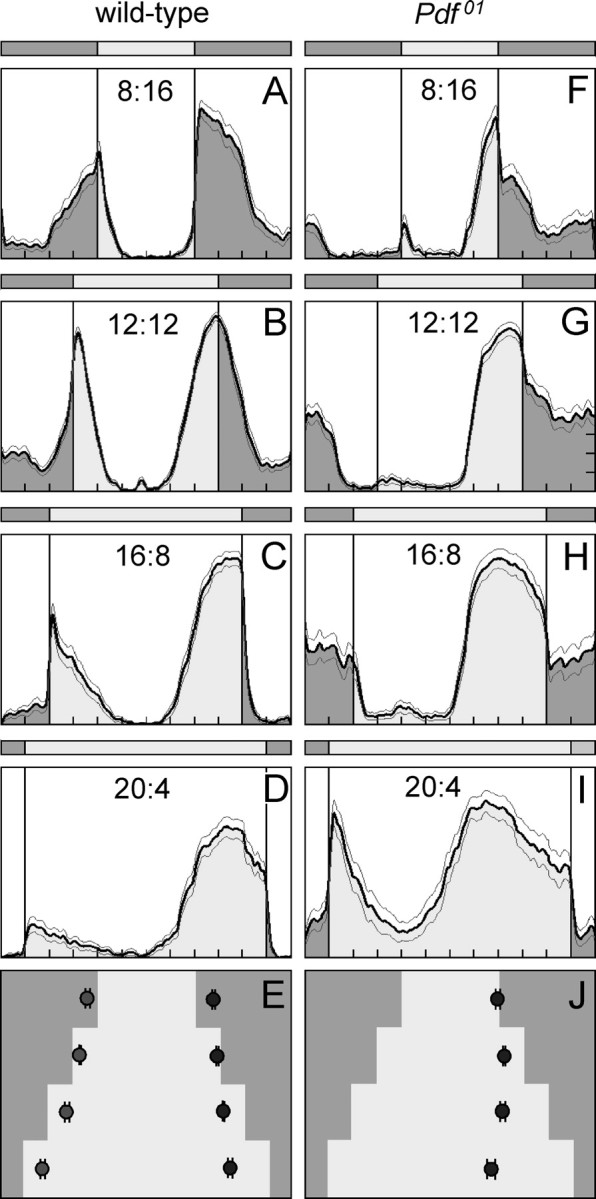
Average activity profiles of wild-type flies and Pdf01 mutants under different photoperiods. Photoperiods are indicated in the diagrams, whereby 8:16 means 8 h day and 16 h night; 12:12, 12 h day and 12 h night; 16:8, 16 h day and 8 h night; and 20:4, 20 h day and 4 h night. Each activity profile represents the average of 30 flies. Wild-type flies showed prominent morning and evening activity peaks under all photoperiods. The morning peaks advanced with increasing day length, whereas the evening peaks delayed. As a consequence, the midday trough became larger the longer the day length. This can nicely be seen in the phase plot of morning and evening peaks (±SEM) (E). Pdf01 mutants did not show a clear morning peak. Under LD 8:16 (F) and 20:4 (I), the flies seem to respond to lights-on with an increase of activity, and in LD 16:8 (H), a small peak was present ∼4 h after lights-on; furthermore, an activity bout was present in the middle of the night under all photoperiods except 20:4 (I). None of these peaks could be unequivocally regarded as morning peak, and therefore no average phases of the morning peak was calculated (J). The evening peak was well pronounced in Pdf01 mutants. It occurred earlier than in wild-type flies, and it did not delay with increasing day length (J). According to these results, Pdf01 mutants are unable to avoid the midday heat in long summer days.
Discussion
Our data underline previous findings that PDF has multiple roles in the fruit fly clock. First, PDF is necessary for the maintenance of robust rhythms within certain groups of clock neurons; simultaneously, PDF lengthens the period of most of these clock cells. Second, in other groups of clock neurons, PDF has no influence on the maintenance of rhythmicity, but it clearly influences clock speed; either PDF lengthens or it shortens period.
The first effect of PDF is in agreement with the hypothesis that PDF lengthens the periods of the clock neurons and simultaneously couples their oscillations so that a coherent rhythm is produced (Peng et al., 2003). This hypothesis is supported by the observations that Pdf01 and PDF receptor mutants are arrhythmic or show weak short-period behavioral rhythms (Renn et al., 1999; Hyun et al., 2005) fitting to an asynchronous cycling of several clock neurons and short-period rhythms in other neurons in absence of PDF (Klarsfeld et al., 2004; Lin et al., 2004; Wu et al., 2008). Furthermore, PDF fulfils the prerequisites of a synchronizing clock factor: PDF is expressed in ∼10% of the clock neurons that are highly connected to the other clock cells (Fig. 7), PDF seems to be released rhythmically (Park et al., 2000), and its receptor is expressed by most clock neurons (Shafer et al., 2008).
Synchronizing and period-lengthening effects of PDF on the clock have also been reported for the cockroach (Schneider and Stengl, 2005). Here, PDF injections into the pacemaker center phase-delayed the activity rhythm (Petri and Stengl, 1997). Permanent phase delays are equivalent to a period lengthening, and Petri and Stengl (2001) showed that phase delays can couple the oscillators. Thus, the decelerating and coupling effects of PDF on the clock may be more generally valid.
Furthermore, the just-described effects of PDF strongly resemble those described for VIP in the circadian clock of mammals, the suprachiasmatic nucleus (SCN) (Aton and Herzog, 2005). VIP is expressed in 15% and the VIP receptor is expressed in 60% of the SCN neurons (Abrahamson and Moore, 2001); VIP is rhythmically released from the rat SCN in vitro (Shinohara et al., 1995) and shifts both behavioral and SCN firing rhythms (Piggins et al., 1995; Reed et al., 2001). VIP has two distinct roles in the SCN: maintaining circadian rhythmicity in a subset of clock neurons and maintaining synchrony between intrinsically rhythmic neurons (Aton et al., 2005). Approximately 50% of the rhythmically firing neurons in the SCN require VIP to do so; the other one-half do not depend on VIP concerning rhythmic firing, but VIP is necessary to synchronize their firing rhythms.
In Drosophila, we cannot easily distinguish between clock neurons that need PDF to be rhythmic and clock neurons that remain rhythmic without PDF, but lose internal synchrony, because we could not record individual neurons. Nevertheless, it is very likely that these two populations do also exist in the fly. The CRY-positive DN1p may belong to the first population, because their cycling is lost immediately after transfer into DD (Fig. 3). However, the oscillations in the s-LNv and the CRY-positive DN1a dampen slowly, indicating that here PDF is needed for mutually synchronizing individual oscillations within the group. Without PDF, these became slowly asynchronous.
Nevertheless, in Drosophila, a third group of neurons seems to exist that are independent of PDF concerning rhythmicity and internal synchronization, but that are strongly influenced by PDF regarding their period. These are the LNd and the fifth s-LNv. Most interestingly, PDF lengthened the period of the fifth s-LNv and the CRY-positive LNd, whereas it shortened that of the CRY-negative LNd. The period-shortening effect of PDF is completely unexpected, especially because we never observed a short-period component in the activity rhythm of wild-type flies. We have to conclude that the CRY-negative LNd (and other CRY-negative neurons as the DN1p that run with short period) do not obviously contribute to the locomotor rhythms in DD.
Our hypothesis that PDF shortens the period of some clock neurons and lengthened that of others is strongly supported by the appearance of short- and long-period components in the activity rhythm of somda mutants. somda mutants have elevated PDF levels in the dorsal brain and in the aMe that are caused by misrouted l-LNv fibers (Wülbeck et al., 2008). Most interestingly, a correlation analysis with neural mutants showing variable numbers of PDF fibers in the dorsal brain and the aMe showed that the period of the short component became shorter the more PDF fibers were present in the dorsal brain, and that of the long period became longer with increasing number of PDF fibers in the aMe, suggesting that PDF has different effects in different brain areas (Wülbeck et al., 2008). Here, we identified the neuronal groups that shortened and lengthened periods in response to PDF, respectively (Fig. 7, Table 3).
But how can we explain that the same peptide influences the periods differently in various clock neurons? Shafer et al. (2008) found that all clock neurons (except the l-LNv that do not seem to express the PDF receptor) respond to PDF with an increase in cAMP levels. Certainly, increased cAMP levels do not necessarily lead to the same cellular responses. The clock cells may express different sets of kinases and phosphatases that may change the phosphorylation of the clock proteins in opposite manners. But how can we explain that certain neurons like the DN1a, DN1p1, and the s-LNv cells seem to be able to respond with period lengthening at moderate PDF levels (so1) and with period shortening at high PDF levels (somda)?
Notably, the extra-PDF in the dorsal protocerebrum of somda flies stems from misrouted l-LNv and not from the s-LNv cells that normally arborize in this brain area. In contrast to the s-LNv, the l-LNv cells express the dimmed gene that leads to amidated PDF (Park et al., 2008). Amidated PDF is more active and has a longer half-life than nonamidated PDF. Little is known about the properties of the PDF receptor, but it is possible that the PDF receptor in the DN1a or DN1p1 gets saturated at certain PDF levels, and consequently other signaling pathways to the molecular clockwork overtake. For example, the DN1a and DN1p1 may get input from clock neurons that use other messengers. Good candidates are two of the three CRY-negative LNd that express Neuropeptide F (NPF), the homolog for the mammalian Neuropeptide Y (NPY) (Lee et al., 2006; Yoshii et al., 2008). NPY receptors mediate NPY action through coupling to G-proteins that downregulate the intracellular level of cAMP (Wen et al., 2005) and would thus counteract the PDF effect. The DN1a or DN1p1 that run now with short period may consecutively feedback on the s-LNv and make them run also with shorter period (see supplemental Discussion, available at www.jneurosci.org as supplemental material). As a consequence, the short-period component appeared in the activity pattern.
Internal desynchronization into short- and long-period components was also found after electrical hyperexcitation of the LNv cells (Nitabach et al., 2006; Sheeba et al., 2008). The CRY-positive and CRY-negative LNd cells were not assessed separately in these studies; but the treatment provoked an internal desynchronization among the DN and between the fifth s-LNv and the s-LNv. Whereas the s-LNv cells were associated with the short-period component in the present study, they were associated with the long-period component after hyperexcitation of the LNv cells (Sheeba et al., 2008). Obviously, the association of clock neurons with short- and long-period components is variable and depends on environmental or intercellular signals. Together with the above-outlined putative interactions between different groups of clock neurons, this points to a flexible network of the clock. To understand the properties of the entire clock network, it will be necessary to reveal the physiological properties of single clock neurons as well as the interactions of all clock neurons (for review, see Nitabach and Taghert, 2008). Here, we propose a critical role of PDF in the network that can be tested in the future.
Under natural conditions, the role of PDF may be to keep the oscillations in the different subsets of clock neurons slightly out of phase. Different phases of clock neurons were also observed in the SCN (Quintero et al., 2003; Yamaguchi et al., 2003). This multiphasic SCN organization accounts for many properties of circadian organization, including the ability of the SCN to produce multiple-phase peak rhythms in vitro and in vivo (Jagota et al., 2000; Mrugala et al., 2000; Schwartz and Meijer, 2004) as well as to trigger various physiological output rhythms, and to respond to seasonal variations in the photoperiod by altering the coupling of its constituent oscillators (Pittendrigh and Daan, 1976). The same seems to apply for Drosophila (Grima et al., 2004; Shafer et al., 2004; Stoleru et al., 2004, 2005; Bachleitner et al., 2007; Fernández et al., 2007; Murad et al., 2007). We show here that Pdf01 mutants are unable to adapt their activity pattern to long photoperiods, indicating that PDF plays a major role in the seasonal adaptation of the clock.
Footnotes
This work was supported by Deutsche Forschungs Gemeinschaft Grants Fo207/9-1,2 (C.H.-F.) and Sta421/4 and Sta421/5 (R.S.); the Graduate College 640; and EUCLOCK. We thank A. Hofbauer for the nb33 monoclonal antibody, I. Edery for the TIM antiserum, T. Todo for the CRY antiserum, K.-F. Fischbach for the so1 mutants, J. C. Hall for the somda mutants, P. Taghert for the Pdf01 mutants, A. Hofbauer, and O. Shafer for comments on this manuscript, as well as members of our laboratories for fruitful discussions. We are very grateful to Olympus UK Ltd., namely to Alan Kidger and Werner Kammerloher for generously providing Lumnoview LV200.
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse; retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together, right… now: synchronization of rhythms in a mammalian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachleitner W, Kempinger L, Wülbeck C, Rieger D, Helfrich-Förster C. Moonlight shifts the endogenous clock of Drosophila melanogaster . Proc Natl Acad Sci U S A. 2007;104:3538–3543. doi: 10.1073/pnas.0606870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chélot E, Hardin PE, Préat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Díez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Fernández MP, Chu J, Villella A, Atkinson N, Kay SA, Ceriani MF. Impaired clock output by altered connectivity in the circadian network. Proc Natl Acad Sci U S A. 2007;104:5650–5655. doi: 10.1073/pnas.0608260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KF. Neural cell types surviving congenital sensory deprivation in the optic lobes of Drosophila melanogaster . Dev Biol. 1983;95:1–18. doi: 10.1016/0012-1606(83)90002-7. [DOI] [PubMed] [Google Scholar]
- Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18:R784–R794. doi: 10.1016/j.cub.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila . Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster . J Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Neurobiology of the fruit fly's circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Shafer OT, Wülbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster . J Comp Neurol. 2007a;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Yoshii T, Wülbeck C, Grieshaber E, Rieger D, Bachleitner W, Cusamano P, Rouyer F. The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb Symp Quant Biol. 2007b;72:517–525. doi: 10.1101/sqb.2007.72.063. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Jagota A, de la Iglesia HO, Schwartz WJ. Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro . Nat Neurosci. 2000;3:372–376. doi: 10.1038/73943. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhée C, Picot M, Chélot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila . J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH. Sex and clock-controlled expression of the neuropeptide F gene in Drosophila . Proc Natl Acad Sci U S A. 2006;103:12580–12585. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mrugala M, Zlomanczuk P, Jagota A, Schwartz WJ. Rhythmic multiunit neural activity in slices of hamster suprachiasmatic nucleus reflect prior photoperiod. Am J Physiol Regul Integr Comp Physiol. 2000;278:R987–R994. doi: 10.1152/ajpregu.2000.278.4.R987. [DOI] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila . Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Homberg U. Neuropeptides in interneurons of the insect brain. Cell Tissue Res. 2006;326:1–24. doi: 10.1007/s00441-006-0210-8. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Shafer OT, Shepherd SP, Suh H, Trigg JS, Taghert PH. The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol Cell Biol. 2008;28:410–421. doi: 10.1128/MCB.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila . Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri B, Stengl M. Pigment-dispersing hormone shifts the phase of the circadian pacemaker of the cockroach Leucophaea maderae . J Neurosci. 1997;17:4087–4093. doi: 10.1523/JNEUROSCI.17-11-04087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri B, Stengl M. Phase response curves of a molecular model oscillator: implications for mutual coupling of paired oscillators. J Biol Rhythms. 2001;16:125–141. doi: 10.1177/074873001129001836. [DOI] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: a clock for all seasons. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1976;106:333–355. [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed HE, Meyer-Spasche A, Cutler DJ, Coen CW, Piggins HD. Vasoactive intestinal polypeptide (VIP) phase shifts the rat suprachiasmatic nucleus in vitro . Eur J Neurosci. 2001;13:839–843. doi: 10.1046/j.0953-816x.2000.01437.x. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A Pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila . Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Förster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster . J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Förster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster . J Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider NL, Stengl M. Pigment-dispersing factor and GABA synchronize cells of the isolated circadian clock of the cockroach Leucophaea maderae . J Neurosci. 2005;25:5138–5147. doi: 10.1523/JNEUROSCI.5138-A-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz WJ, Meijer JH. Real-time imaging reveals spatiotemporal dynamics of cellular circadian clocks. Trends Neurosci. 2004;27:513–516. doi: 10.1016/j.tins.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster . J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Levine JD, Truman JW, Hall JC. Flies by night: effects of changing day length on Drosophila's circadian clock. Curr Biol. 2004;14:424–432. doi: 10.1016/j.cub.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Förster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster's neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by realtime cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Sharma VK, Gu H, Chou YT, O'Dowd DK, Holmes TC. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J Neurosci. 2008;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Honma S, Katsuno Y, Abe H, Honma K-I. Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc Natl Acad Sci U S A. 1995;92:7396–7400. doi: 10.1073/pnas.92.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila . Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernández MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Förster C, Hall JC, Stanewsky R. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol. 2007;152:165–175. doi: 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2142–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cao G, Nitabach MN. Accelerated free-running cellular oscillation of non-LNv clock neurons in the absence of LNv electrical activity. J Biol Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- Wülbeck C, Grieshaber E, Helfrich-Förster C. Pigment-dispersing factor (PDF) has different effects on Drosophila's circadian clocks in the accessory medulla and in the dorsal brain. J Biol Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila . Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Funada Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveness to light. J Insect Physiol. 2004;50:479–488. doi: 10.1016/j.jinsphys.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila's clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]



