Abstract
Progranulin (PGRN) is an evolutionarily conserved glycoprotein associated with several disease states, including neurodegeneration, cancer and autoimmune disorders. This protein has recently been implicated in the regulation of lysosome function, whereby PGRN may bind to and promote the maturation and activity of the aspartyl protease cathepsin D (proCTSD – inactive precursor, matCTSD – mature, enzymatically active form). As the full-length PGRN protein can be cleaved into smaller peptides, called granulins, we assessed the function of these granulin peptides in binding to proCTSD and stimulating matCTSD enzyme activity in vitro. Here, we report that full-length PGRN and multi-granulin domain peptides bound to proCTSD with low- to sub-micromolar binding affinities. This binding promoted proCTSD destabilization, which was greater in magnitude for multi-granulin domain peptides than for full-length PGRN. Such destabilization correlated with enhanced matCTSD activity at acidic pH. The presence and function of multi-granulin domain peptides has typically been overlooked in previous studies. This work provides the first in vitro quantification of their binding and activity on proCTSD. Our study highlights the significance of multi-granulin domain peptides in the regulation of proCTSD maturation and enzymatic activity and suggests that attention to PGRN processing will be essential for future understanding of the molecular mechanisms leading to neurodegenerative disease states with loss-of-function mutations in PGRN.
Keywords: Progranulin, cathepsin D, granulin, neurodegeneration
Graphical Abstract
Progranulin (PGRN) is an evolutionarily conserved, multi-functional protein implicated in diverse biological processes, including neurodegeneration, cancer, inflammation and wound healing [1, 2]. Recent studies have highlighted a role for progranulin in regulating lysosome function [3–7]. Specifically, PGRN has been shown to physically interact with the lysosomal aspartyl protease cathepsin D (CTSD), whereby it may stimulate enzymatic maturation and/or activity [8–11]. This implicates a dysregulation of CTSD activity and aberrant lysosomal function in neurodegenerative diseases linked to PGRN loss-of-function mutations [4, 12, 13].
PGRN itself is a pro-protein that can be cleaved into smaller domains called granulins (A through G) [14, 15]. Granulins share a highly disulfide-rich, evolutionarily-conserved beta-sheet fold [16–19]. Such characteristics are often found in highly stable proteins that can withstand heat and pH changes [17, 20]. Indeed, recent studies have highlighted that progranulin cleavage, and therefore granulin production, occurs in the acidic environment of the lysosome [21], through the action of lysosomal cathepsins [22, 23]. Individual granulins may oppose the function of the full-length protein in cell growth and inflammation [17, 24]. Alternatively, some evidence suggests that granulin domains bind to and stimulate CTSD enzymatic activity [8, 10]. Although multi-granulin-sized peptides have been reported in highly degenerative brain regions from AD patients [25], the actual molecular functions of individual granulins, multi-granulin fragments and the full-length protein are still incompletely understood.
We previously demonstrated a role for full-length PGRN in stimulating the maturation and activity of proCTSD [9]. We proposed that this results from a destabilizing effect of PGRN on proCTSD, facilitating CTSD propeptide cleavage and the production of mature, active CTSD. Here, we sought to clarify the role of PGRN and its cleavage products in proCTSD binding and CTSD enzymatic activity. We now report for the first time the binding affinities for full-length PGRN and multi-granulin domain peptides with proCTSD in vitro at neutral pH. These recombinant peptides, particularly the multi-granulin fragments BAC and CDE, induce a significant destabilizing effect on proCTSD, resulting in a negative shift for proCTSD in thermal stability assays. At acidic pH, we demonstrate that multi-granulin domain peptides promote CTSD activity above that of full-length PGRN. We propose a mechanism whereby multi-granulin domain peptides more effectively promote destabilization of the CTSD propeptide from the proCTSD catalytic core.
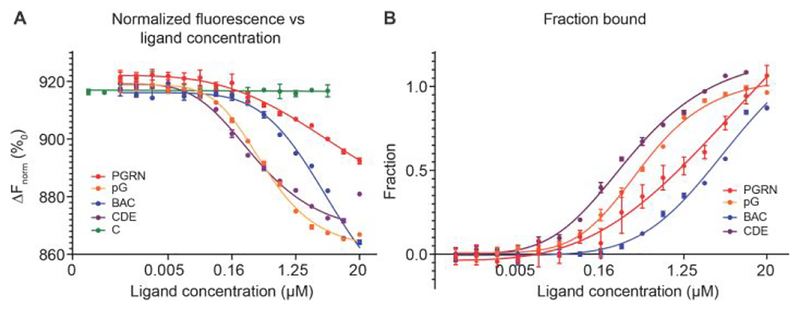
In recent studies, our lab and others have demonstrated that full-length PGRN can bind to proCTSD [8–11]. To quantitatively analyze this binding interaction, we utilized microscale thermophoresis (MST) [26]. MST measures the directed movement of a fluorescently-labeled protein and/or protein-ligand complex within a temperature gradient. Interaction with a ligand changes the diffusion of the protein–ligand complex with respect to the unbound protein, and this can be used to derive equilibrium binding constants. We performed MST assays at neutral pH to prevent the auto-activation of proCTSD, which occurs at acidic pH [27–29]. After confirming the binding of fluorescently-labeled proCTSD with full-length PGRN, we measured the binding affinity between proCTSD and the recombinant multi-granulin (pG, BAC, CDE) or granulin (C) peptides that are commercially available (Figure 1, Table 1 and Supplementary Figure 1). PGRN and multi-granulin domain peptides bound to proCTSD at low- or sub-micromolar affinity. Interestingly, peptides pG and CDE bound to proCTSD with a higher affinity than that of full-length PGRN. Furthermore, ligand addition induced a change in the initial fluorescence of these protein complexes during MST capillary scans (Supplementary Figure 2), which was eliminated after protein denaturation (Supplementary Figure 3), suggesting that a physical interaction between proCTSD and ligand results in a conformational change in proCTSD.
Figure 1.
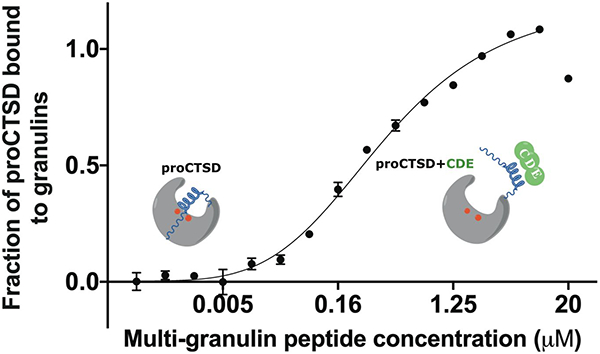
Multi-granulin domain peptides bind to proCTSD at low- to sub-micromolar affinity. (A) Normalized MST fluorescence (ΔFnorm) of labeled proCTSD (12.5 nM) is plotted against ligand concentration (0.6 nM to 20 μM) for PGRN (red), pG (orange), BAC (blue), CDE (purple) and C (green). (B) MST binding response plotted as fraction bound (ΔFnorm values divided by curve amplitude). Curves were fitted to derive dissociation constants (Kd). Assays were run in triplicate at pH 7.4 (except granulin C which was run in duplicate) and data plotted are mean ± SD.
Table 1.
Summary table showing binding and activity of multi-granulin domain peptides on proCTSD.a
| Neutral pH (7.4) | Acidic pH (3.5) | |||
|---|---|---|---|---|
| proCTSD | - | - | 1.00 ± 0.00 | 1712 ± 31.42 |
| + PGRN | 0.95 ± 0.34 | −0.41 ± 0.05 | 1.29 ± 0.06 | 1585 ± 34.65 |
| + pG | 0.33 ± 0.03 | −0.23 ± 0.02 | 1.27 ± 0.11 | 1611 ± 30.01 |
| + BAC | 2.80 ± 0.89 | −3.38 ± 0.08 | 1.89 ± 0.09*** | 1343 ± 51.57** |
| + CDE | 0.13 ± 0.01 | −2.78 ± 0.36 | 1.84 ± 0.09*** | 1376 ± 48.23* |
| + C | - | 0.05 ± 0.07 | 1.13 ± 0.17 | 1488 ± 117.20 |
Values shown are mean ± SEM. Normalized Vmax and lag time were analyzed for significant difference to proCTSD alone using one-way ANOVA with Tukey’s multiple comparisons test,
= P<0.001,
= P<0.01,
= P<0.05.
The +PGRN condition is significantly different for Vmax when compared with a Student’s t-test (P = 0.0076).
Several studies have reported the binding of individual granulins to CTSD in immunoprecipitation assays from cells [8, 10, 11]. Thus we were surprised that granulin C did not bind proCTSD in this assay (Figure 1 and Supplementary Figure 1). It is possible that different conditions are required for single granulin binding to proCTSD (i.e. pH, buffers, cofactors) or that the earlier IP interactions were indirect. In addition, all of the individual granulin domains were not tested, therefore, we cannot definitively conclude that individual granulin domains do not bind to proCTSD in vivo.
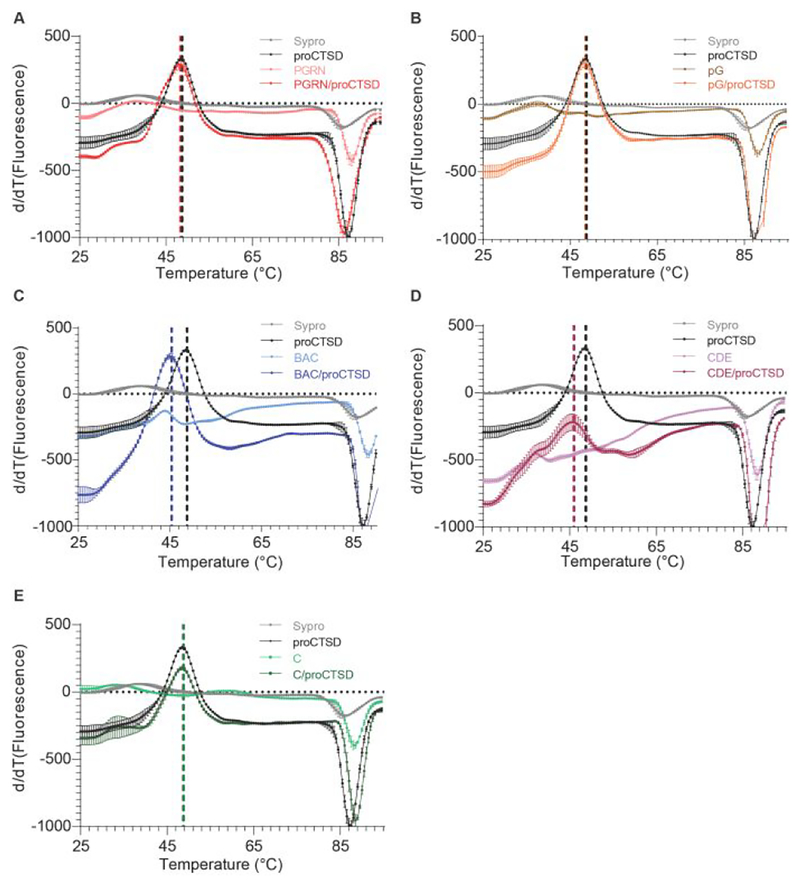
We next sought to evaluate how granulin peptide binding might impact the conformational stability of proCTSD. Previously, we showed that full-length PGRN can induce a destabilizing effect on proCTSD utilizing differential scanning fluorimetry (DSF) [9]. This is a technique that can measure the melting temperature (Tm), or stability, of proteins alone or in complexes. As we observed that multi-granulin domains bind to proCTSD with higher affinity that full-length PGRN, we hypothesized that these smaller peptides might also induce a destabilizing effect on the protein. We confirmed this result, finding that peptides BAC and CDE both induced a significant destabilizing effect on proCTSD and reduced its Tm by over 2.5 °C when present at a 2:1 molar ratio (Table 1, Figure 2 and Supplementary Figure 4). DSF with peptide CDE required optimization due to the very high initial fluorescence of this protein. A Tm reduction of 2.75 °C for proCTSD was observed in the presence of CDE at a 1:1 molar ratio (Supplementary Figure 5).
Figure 2.
Multi-granulin domain peptides reduce the melting temperature of proCTSD. Derivatives of the raw DSF fluorescent intensity versus temperature are plotted for 1 μM proCTSD in the absence and presence of: (A) 2 μM PGRN, (B) 2 μM peptide pG, (C) 2 μM peptide BAC, (D) 2 μM peptide CDE, and (E) granulin C. DSF was performed at neutral pH 7.4 with 1× Sypro Orange. Assays were run in triplicate and data plotted are mean + SEM.
To further understanding the surface accessibility of full-length PGRN and granulin peptides for binding to proCTSD, we analyzed the initial fluorescent signals resulting from the interaction of Sypro Orange dye with the hydrophobic residues of each recombinant protein at 25 °C. More dye binding leads to a higher fluorescence, which correlates with more exposed hydrophobic residues. PGRN displayed a higher fluorescence than the individual granulin C domain (~1.5-fold), but a similar fluorescence to its N-terminal peptide pG (Table S1). However, the multi-granulin domain peptides BAC and CDE showed approximately 2-fold more fluorescence than PGRN. This suggests that compared to the multi-granulin peptides, PGRN may exist in a compact, folded conformation, with fewer exposed hydrophobic residues. Alternatively, the recombinant full-length PGRN used in this assay may exist as higher-order structures, such as dimers, which PGRN is known to form [30]. While the NMR-based structures of several individual granulin domains (PDB IDs: 2JYV, 2JYE, 2JYT, 2JYU) have been reported [17], it is important to highlight that, to date, there is no detailed structural study of full-length PGRN or multi-granulin domain peptides.
We have previously shown that full-length PGRN stimulates the maturation and activity of proCTSD at acidic pH in vitro [9]. We therefore investigated whether multi-granulin domain peptides might behave similarly. Maximal CTSD activity (Vmax) and time to reach Vmax (lag time) were measured in the absence or presence of PGRN and multi-granulin domain peptides at a ~3:1 molar ratio in acidic pH (Figure 3A). We observed that CTSD activity was increased approximately 30% in the presence of PGRN or peptide pG, and approximately 80% in the presence of peptides BAC or CDE (Table 1 and Figure 3B). This was accompanied by a decrease in lag time to reach Vmax (Table 1 and Figure 3B) suggesting that maturation of proCTSD to its active form is stimulated in the presence of these multi-granulin domain peptides. Granulin C had no significant effect on the enzymatic activity of proCTSD. Overall, these data suggest that multi-granulin domain peptides may stimulate the in vitro maturation and activity of proCTSD more effectively than full-length PGRN.
Figure 3.
The in vitro activity of proCTSD activity is increased by recombinant multi-granulin domain peptides. (A) Representative kinetic curves for the measurement of enzymatic activity of 20 nM proCTSD in vitro at pH 3.5 in the absence (black) or presence of 65 nM recombinant protein: PGRN (red), peptide pG (orange), peptide BAC (blue), peptide CDE (purple) and peptide C (green). 5 μM of a fluorescent substrate was used in these assays. (B) Lag time to reach Vmax is plotted against normalized maximal velocity (Vmax) for CTSD activity (n = 3). (C) pH-dependent charge predictions (propKa3.0) for the individual granulin domains A-G. (D) 3-color charge scale for granulin domains, colored from red (negative) to blue (positive) using a percentile scale.
The seven granulin domains of PGRN (A-G) share approximately 40% sequence similarity, including the presence of a network of cysteine residues that are predicted to form up to six disulfide bonds per domain [17, 19]. Electrostatic charge differences are observed between individual granulins at both neutral and acidic pH (Figure 3C). The most positively charged regions (pG and DE) are in the N- and C-terminal domains of the protein, respectively, while the central region (FBAC) contains the most negatively charged domains (Figure 3D). We predict that both termini may interact with the central core, keeping the full-length protein in a conformation that is less accessible to solvent. In addition, such electrostatic differences may facilitate interaction between PGRN molecules. Upon cleavage of either (or both) terminal domains, the multi-granulin domain peptides may adopt a more open conformation than the holoprotein, increasing their accessibility for protein-protein interactions. While more studies are required for a deeper understanding of PGRN structure, our preliminary in silico and DSF results suggest that PGRN conformation may be driven by electrostatic interactions among its domains. This raises an interesting possibility that PGRN function may be regulated by protein cleavage and/or pH changes that occur during its trafficking through the endolysosomal pathway.
We report here for the first time that PGRN and multi-granulin peptides bound to proCTSD in vitro at neutral pH with low- to sub-micromolar binding affinities. This binding promoted proCTSD destabilization, which was greater in magnitude for multi-granulin domain peptides than for full-length PGRN. This destabilization correlated with enhanced CTSD maturation and activity at acidic pH. Such low micromolar binding affinities are within the range commonly reported for regulatory proteins [31, 32]. Based on our DSF and in silico results, we suggest the surface of multi-granulin domain peptides may be more accessible to interact with the propeptide region of proCTSD than PGRN. We propose that this interaction may facilitate propeptide cleavage and promote proCTSD conversion to its mature, fully enzymatically active form. Indeed, there are several examples from biology where smaller protein domains exert greater efficacy than the holoprotein from which they are derived. Such examples include fibrinogen, which undergoes multiple cleavages during its activation in blood clotting [33], caspase 9 [34] and even procathepsins themselves [29]. The in vitro assays reported here are influenced by the folding, structural arrangement and potential aggregation states of recombinant proteins [35]. However, our MST results do not indicate aggregate formation for PGRN or granulin peptides at the concentrations used in DSF and CTSD activity assays (< 4.5 μM). A comparison between recombinant proteins from different sources would be desirable, but the availability of commercially purified human PGRN peptides is limited. Further studies will be necessary to assess the impact of other individual granulin domains and multi-granulin domain peptides in proCTSD maturation and activity.
This study reveals the complex functional interactions that can occur between proCTSD and either intact PGRN or specific multi-granulin peptides. As these interactions are potentially regulated by pH and sequential PGRN cleavage, the biological context of these interactions within the endolysosomal compartment would be critical and could serve as a means to titrate CTSD activity with high precision. With age, or with the lowered PGRN levels that are seen in genetic haploinsufficiency, perturbations in this fine balance could transpire. Thus, relative levels of PGRN and its cleavage peptides should be considered in the etiology of neurodegenerative diseases due to PGRN mutations.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institutes of Health R21NS082709, R01NS095257, R01AG059052, the Alzheimer’s Disease Research Center and the Paul G. Allen Family Foundation (A.W.K.). We also thank The James and Barbara Knuppe Family Foundation for research support. We thank Charles Craik and Sam Ivry for technical advice with enzymatic assays, Joseph Lobel for technical advice with DSF assays, Brett Thurlow for technical advice with MST assays and members of the Kao lab for helpful discussions.
ABBREVIATIONS
- PGRN
progranulin
- CTSD
cathepsin D
- MST
microscale thermophoresis
- DSF
differential scanning fluorimetry
- Tm
thermal shift
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
Conflict of Interest
M.P.J. is a consultant to and shareholder of Schrodinger LLC, which licenses software used in this work.
REFERENCES
- 1.Bateman A and Bennett HP, The granulin gene family: from cancer to dementia. Bioessays, 2009. 31(11): p. 1245–54. [DOI] [PubMed] [Google Scholar]
- 2.Kao AW, et al. , Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka Y, et al. , Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet, 2017. [DOI] [PubMed] [Google Scholar]
- 4.Smith KR, et al. , Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet, 2012. 90(6): p. 1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evers BM, et al. , Lipidomic and Transcriptomic Basis of Lysosomal Dysfunction in Progranulin Deficiency. Cell Rep, 2017. 20(11): p. 2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paushter DH, et al. , The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol, 2018. 136(1): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrant AE, et al. , Progranulin Gene Therapy Improves Lysosomal Dysfunction and Microglial Pathology Associated with Frontotemporal Dementia and Neuronal Ceroid Lipofuscinosis. J Neurosci, 2018. 38(9): p. 2341–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beel S, et al. , Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet, 2017. 26(15): p. 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler VJ, et al. , Progranulin Stimulates the In Vitro Maturation of Pro-Cathepsin D at Acidic pH. J Mol Biol, 2019. 431(5): p. 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valdez C, et al. , Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, et al. , Regulation of cathepsin D activity by the FTLD protein progranulin. Acta Neuropathol, 2017. 134(1): p. 151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker M, et al. , Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature, 2006. 442(7105): p. 916–9. [DOI] [PubMed] [Google Scholar]
- 13.Cruts M, et al. , Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature, 2006. 442(7501): p. 920–4. [DOI] [PubMed] [Google Scholar]
- 14.Plowman GD, et al. , The epithelin precursor encodes two proteins with opposing activities on epithelial cell growth. J Biol Chem, 1992. 267(18): p. 13073–8. [PubMed] [Google Scholar]
- 15.Cenik B, et al. , Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem, 2012. 287(39): p. 32298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman A and Bennett HP, Granulins: the structure and function of an emerging family of growth factors. J Endocrinol, 1998. 158(2): p. 145–51. [DOI] [PubMed] [Google Scholar]
- 17.Tolkatchev D, et al. , Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci, 2008. 17(4): p. 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolkatchev D, et al. , Design and solution structure of a well-folded stack of two beta-hairpins based on the amino-terminal fragment of human granulin A. Biochemistry, 2000. 39(11): p. 2878–86. [DOI] [PubMed] [Google Scholar]
- 19.Palfree RG, Bennett HP, and Bateman A, The Evolution of the Secreted Regulatory Protein Progranulin. PLoS One, 2015. 10(8): p. e0133749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavergne V, Taft RJ, and Alewood PF, Cysteine-rich mini-proteins in human biology. Curr Top Med Chem, 2012. 12(14): p. 1514–33. [DOI] [PubMed] [Google Scholar]
- 21.Holler CJ, et al. , Intracellular Proteolysis of Progranulin Generates Stable, Lysosomal Granulins that Are Haploinsufficient in Patients with Frontotemporal Dementia Caused by GRN Mutations. eNeuro, 2017. 4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee CW, et al. , The lysosomal protein cathepsin L is a progranulin protease. Mol Neurodegener, 2017. 12(1): p. 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, et al. , Lysosomal processing of progranulin. Mol Neurodegener, 2017. 12(1): p. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, et al. , Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell, 2002. 111(6): p. 867–78. [DOI] [PubMed] [Google Scholar]
- 25.Salazar N, et al. , The progranulin cleavage products, granulins, exacerbate TDP-43 toxicity and increase TDP-43 levels. J. Neurosci, 2015. 35(25): p. 9315–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wienken CJ, et al. , Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun, 2010. 1: p. 100. [DOI] [PubMed] [Google Scholar]
- 27.Hasilik A, et al. , Lysosomal enzyme precursors in human fibroblasts. Activation of cathepsin D precursor in vitro and activity of beta-hexosaminidase A precursor towards ganglioside GM2. Eur J Biochem, 1982. 125(2): p. 317–21. [DOI] [PubMed] [Google Scholar]
- 28.James MN and Sielecki AR, Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 A resolution. Nature, 1986. 319(6048): p. 33–8. [DOI] [PubMed] [Google Scholar]
- 29.Wittlin S, et al. , Mechanisms and kinetics of procathepsin D activation. Eur J Biochem, 1999. 265(1): p. 384–93. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen AD, et al. , Secreted progranulin is a homodimer and is not a component of high density lipoproteins (HDL). J Biol Chem, 2013. 288(12): p. 8627–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinke AW, et al. , Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science, 2013. 340(6133): p. 730–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, et al. , Protein interaction affinity determination by quantitative FRET technology. Biotechnol Bioeng, 2012. 109(11): p. 2875–83. [DOI] [PubMed] [Google Scholar]
- 33.Mosesson MW, Fibrinogen and fibrin structure and functions. J Thromb Haemost, 2005. 3(8): p. 1894–904. [DOI] [PubMed] [Google Scholar]
- 34.Boatright KM and Salvesen GS, Mechanisms of caspase activation. Curr Opin Cell Biol, 2003. 15(6): p. 725–31. [DOI] [PubMed] [Google Scholar]
- 35.Jian J, et al. , Progranulin directly binds to the CRD2 and CRD3 of TNFR extracellular domains. FEBS Lett, 2013. 587(21): p. 3428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.