Abstract
Background
Homeobox A10 (HOXA10) has been implicated in the development and progression of various human cancers. However, the precise biological functions of HOXA10 in hepatocellular carcinoma (HCC) have not been defined.
Methods
In this study, we examined mRNA expression by quantitative real-time PCR (qRT-PCR) of HOXA10 as well as histone deacetylase (HDAC) and protein levels by Western blot of HOXA10, HDAC1, Cyclin D1, proliferating cell nuclear antigen (PCNA), Survivin and p53 acetylation in HCC tissues and cell lines. We also assessed cell proliferation using Cell Counting Kit-8 (CCK-8) and analyzed cell cycle by flow cytometry. Furthermore, tumor growth of HCC cells in vivo was monitored using the nude mouse xenograft model. Finally, HDAC1 promoter activity and binding in HCC cell lines were detected by luciferase reporter assay and chromatin immunoprecipitation (ChIP), respectively.
Results
We uncovered the elevated expression of HOXA10 in HCC tissues compared to adjacent normal liver tissues. RNA interference-mediated knockdown of HOXA10 inhibited HCC cell proliferation both in vitro and in vivo. HOXA10 knockdown also induced cell cycle arrest at G0/G1 phase and apoptosis, which were accompanied with the reduced expression of Cyclin D1, PCNA and Survivin. Notably, HOXA10 knockdown enhanced p53 acetylation (Lys382), which is crucial to the activation of p53. Likewise, HOXA10 knockdown suppressed the transcription of HDAC1, a potential deacetylase for p53. In line with these observations, HDAC1 downregulation abrogated the effects of HOXA10 overexpression on proliferation, cell cycle progression, apoptosis and p53 acetylation, indicating the role of HDAC1 in mediating HOXA10 functions.
Conclusion
Our results demonstrate that HOXA10 knockdown inhibits proliferation, induces cell cycle arrest and apoptosis in HCC cells by regulating HDAC1 transcription.
Keywords: hepatocellular carcinoma, HOXA10, HDAC1, p53, acetylation
Introduction
Hepatocellular carcinoma (HCC) is one of the most frequent and lethal malignancies worldwide and represents more than 80% of primary liver tumors.1,2 Viral hepatitis B and C infections, chronic alcohol consumption and exposure to aflatoxin B1 are major risk factors for HCC development.3 Although significant improvements have been made in surgical techniques and perioperative management, the mortality of HCC remains high due to late-stage diagnosis and inefficiency of current therapies.4
Homeobox (HOX) gene family is highly conserved in evolution. HOX genes encode transcription factors that play essential roles in numerous biological processes, such as development, cell differentiation and proliferation.5,6 Among them, Homeobox A10 (HOXA10) has been linked to tumor development and progression. HOXA10 overexpression, for instance, has been observed in acute myeloid leukemia, non-small cell lung cancer, glioblastoma, ovarian cancer, oral squamous cell carcinoma (OSCC) and gastric cancer.7–13 Some studies report that HOXA10 regulates proliferation, migration and invasion of cancer cell lines.13–18 In HCC tissues, the expression of HOXA10, along with other members of HOX gene family, was found to be highly elevated in comparison with normal liver tissues.19–21 The biological functions of HOXA10 in HCC, however, have not been defined.
The p53 tumor suppressor gene responds to a myriad of diverse cellular stress signals to regulate cell cycle progression, senescence, apoptosis and DNA repair.22 The activity and stability of p53 are finely regulated by post-translational modifications, including phosphorylation, ubiquitination, methylation and acetylation. Acetylation is particularly crucial to the activation of p53.23 Histone deacetylase (HDAC) −1, −2 and −3 can directly interact with p53 and deacetylate p53, thus suppressing p53 functions.24
Altered expression and mutation of p53 have been reported in a number of human cancers.25,26 In more advanced stages of HCC, aberrant p53 expression is frequent.27 In other cancer types, such as breast cancer, HOX genes are known to regulate p53. For instance, HOXA5 was shown to promote p53 transcription,28 whereas HOXA10 enhanced p53 expression and invasion in BT20 breast cancer cells.17 However, whether HOXA10 influences p53 functions in HCC has not been investigated.
In this study, we confirmed the elevated expression of HOXA10 in HCC tissues and investigated how HOXA10 knockdown affects proliferation, cell cycle progerssion and apoptosis of HCC cells. We also investigated the mechanism through which HOXA10 exerts its biological functions.
Materials and methods
Human tissue samples
HCC tissue samples from 80 patients and adjacent normal liver tissue samples from 30 patients were obtained from Shanghai Jiao Tong University Affiliated Sixth People’s Hospital. This study was conducted with approval from the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital and in accordance with the Declaration of Helsinki. Prior to study enrollment, written informed consent from each participant was obtained according to the guidelines of the Ethics Committee.
Cell culture
Human HCC cell lines used in this study were SMMC-7721, Hep3B, MHCC-97L, MHCC-97H and HepG2 (ATCC, Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Gibco, Detroit, MI, USA) and 1% Penicillin-Streptomycin (Solarbio, Beijing, China). All HCC cell lines were maintained in a 37 °C humidified incubator with 5% carbon dioxide (CO2) and 95% air.
Quantitative real-time PCR (qRT-PCR)
Total RNA isolation was conducted using TRIzol reagent following the manufacturer’s protocols (Invitrogen, Carlsbad, CA, USA). This was followed by DNase I treatment to eliminate genomic DNA. Using cDNA Synthesis Kit (Thermo Fisher, St. Louis, MO, USA), total RNA was reverse transcribed into cDNA. qRT-PCR assay was conducted using SYBR Green qPCR Master Mixes (Thermo Fisher) on the ABI 7,300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression was determined using the 2−ΔΔCt method with normalization to the housekeeping gene GAPDH. The primer sequences are listed in Table 1.
Table 1.
Primer pairs used for qRT-PCR
| Genes | Primers (forward/reverse) |
|---|---|
| HOXA10 | 5ʹ- AGAGATTAGCCGCAGCGTCC −3ʹ and 5ʹ- TTCCTGGGCAGAGCCTGAAG −3’ |
| HDAC1 | 5ʹ-CTACTACGACGGGGATGTTGG-3ʹ and 5ʹ- GAGTCATGCGGATTCGGTGAG −3ʹ |
| HDAC2 | 5ʹ- ATGGCGTACAGTCAAGGAGG −3ʹ and 5ʹ- TGCGGATTCTATGAGGCTTCA −3ʹ |
| HDAC3 | 5ʹ- TCTGGCTTCTGCTATGTCAACG-3ʹ and 5ʹ- CCCGGTCAGTGAGGTAGAAAG −3ʹ |
| GAPDH | 5ʹ- CACCCACTCCTCCACCTTTG −3ʹ and 5ʹ-CCACCACCCTGTTGCTGTAG-3ʹ |
| HDAC1 ChIP-primer 1 | 5ʹ-AAAGAAAGGAAACCTGCCCTC-3ʹ and 5ʹ-TGCAGTCACCCAGGATGACTA-3ʹ |
| HDAC1 ChIP-primer 2 | 5ʹ- AGTGCCTCTGGTGCAAAGAA −3ʹ and 5ʹ- TCGGCCAAGGGCTTTTTA −3ʹ |
| HDAC1 ChIP-primer 3 | 5ʹ-CCGCGTGTTGCTTTTCTTTT-3ʹ and 5ʹ- TGAAAAGCGTGCTTGGAGAT −3ʹ |
Abbreviations: qRT-PCR, quantitative real-time PCR; HOXA10, homeobox A10; HDAC, histone deacetylase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ChIP, chromatin immunoprecipitation.
Western blot analysis
HCC cells and liver tissues were lysed in RIPA Buffer (Solarbio) following the manufacturer’s instructions. BCA Protein Assay Kit (Thermo Fisher) was used to measure protein concentration and then equal amounts of proteins were loaded into 10% polyacrylamide gels. Following separation by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were then transferred onto nitrocellulose membranes (Millipore, Bedford, MA, USA), which were subsequently blocked with 5% non-fat milk at room temperature for 1 h and then incubated with primary antibodies overnight at 4 °C. After washing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Beyotime, Nanjing, Jiangsu, China) for 1 h at 37 °C. After final washing, enhanced chemiluminescence (ECL) detection Kit (Pierce Biotechnology, Rockford, IL, USA) was used to detect protein bands. Antibodies against HOXA10 (catalog: Ab23392), histone deacetylase 1 (HDAC1, catalog: Ab53091) and acetylated p53 (Ace-p53, catalog: Ab75754) were obtained from Abcam (Cambridge, MA, USA). Antibodies against p53 (catalog: #9282), Cyclin D1 (catalog: #2922), proliferating cell nuclear antigen (PCNA, catalog: #2586), Survivin (catalog: #2803) and GAPDH (catalog: #5174) were obtained from Cell Signaling Technology (Danvers, MA, USA). The band intensity of detected proteins was quantified with Image J software (http://rsb.info.nih.gov/ij/, Bethesda, MD, USA).
RNA interference
Three shRNAs targeting HOXA10 (shHOXA10#1, #2 and #3), one shRNA targeting HDAC1 (shHDAC1) and scramble shRNA (shNC) were cloned into linearized pLKO.1-puro (Addgene, Cambridge, MA, USA), respectively. shRNAs and lentiviral packaging vectors were co-transfected into HEK293T cells using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocols. Lentivirus supernatant was collected at 48–72 h after transfection to infect the targeting cells. Western blot confirmed the knockdown efficiency after 48 h. The shRNA sequences were shown below:
shHOXA10#1: 5ʹ-CTTTCGCGCAGAACATCAA-3ʹ;
shHOXA10#2: 5ʹ-TATGTACCTTACTCGAGAG-3ʹ;
shHOXA10#3: 5ʹ-TGAATCGAGAAAACCGGAT-3ʹ;
shHDAC1: 5ʹ-GGCAAGTATTATGCTGTTA-3ʹ.29
Ectopic expression of HOXA10
The coding sequence of HOXA10 was amplified from human cDNA using primers as follows: forward primer: 5ʹ-CCCAAGCTTATGTCAGCCAGAAAGGGCTATCTG-3ʹ; reverse primer: 5ʹ-CCGGAATTCTCAGGAAAAATTAAAGTTGGCTGTGAGC-3ʹ, digested with HindIII/EcoRI (Thermo Fisher), and inserted into HindIII/EcoRI-digested pcDNA3.1 (+) (Addgene). HOXA10-pcDNA3.1 was transfected into MHCC-97H cells and the overexpression of HOXA10 was confirmed by Western blot at 48 h after transfection.
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was performed to examine cell proliferation. Briefly, SMMC-7721 and HepG2 cells were seeded in 96-well plates (3×103 cells/well) and transfected with shHOXA10#3 or shNC after incubation overnight. Each group had three replicate wells. Then, CCK-8 solution (Signalway Antibody, College Park, MD, USA) was added to each well after 0, 24, 48 and 72 h, and incubated for 1 h. The optical density (OD) was detected at 450 nm on a microplate spectrophotometer (Thermo Fisher).
Flow cytometry analyses of cell cycle and apoptosis
SMMC-7721 and HepG2 cells were transfected with shHOXA10#3 or shNC. MHCC-97H cells were divided into four groups: Group 1, transfected with pcDNA3.1 and shNC; Group 2, transfected with HOXA10-pcDNA3.1 and shNC; Group 3, transfected with pcDNA3.1 and shHDAC1; and Group 4, transfected with HOXA10-pcDNA3.1 and shHDAC1. After 48 h, cells were harvested. For cell cycle analysis, cells were fixed in 70% ethanol overnight at −20 °C and washed once in ice-cold PBS. Cells were then stained with propidium iodide (PI)-RNase solution (Sigma-Aldrich, St. Louis, MO, USA). For cell apoptosis analysis, cells were stained with Annexin V-FITC Kit (Beyotime) following the manufacturer’s instructions. Cells in early apoptosis were Annexin V-FITC+ and PI-. DNA content and apoptosis were analyzed by FACScan flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Luciferase assay
The human HDAC1 promoter30 was inserted into linearized pGL3-basic vector (Promega, Madison, WI, USA). SMMC-7721 and HepG2 cells expressing pRL-TK (Promega) and pGL3-HDAC1 promoter were transfected with shHOXA10#3 or shNC. After 48 h, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was normalized to that of Renilla luciferase. The activity control, which was co-transduced with pRL-RK and pGL3-HDAC1 promoter only, was set at 1.0.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were conducted in SMMC-7721 and HepG2 cells following the manufacturer’s protocols (Abcam). Cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Following lysis and sonication, the supernatant was obtained by centrifugation. Samples were incubated overnight with anti-HOXA10 (Abcam) or control IgG (Abcam) in the presence of protein A/G beads. After washing, the enrichment of specific DNA sequences was measured by qRT-PCR. The qRT-PCR primers for the HDAC1 promoter are listed in Table 1.
Xenografts in nude mice
Animal care and experimentation were carried out according to the animal ethics guidelines and approved protocols of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (DWSY2018-028). Twelve 4–5 week-old BALB/c nude mice (Shanghai SLAC laboratory animal Co., Ltd., Shanghai, China) were maintained with free access to sterile water and chow. SMMC-7721 cells transfected with shHOXA10#3 or shNC were injected subcutaneously into the nude mice (2×106 cells per mouse; n=6 per group). Tumor volume was calculated using the formula: volume =0.5× (the longest diameter) × (the shortest diameter)2. After 33 days, the mice were sacrificed and the tumors were weighted.
Statistical analysis
Statistical analysis was conducted using the GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA). All assays had three independent replicates and the data were presented as mean ± standard deviation (SD). Statistical evaluation of two or more groups was conducted using Student’s t-test and one-way ANOVA with Bonferroni’s post hoc test, respectively. Pearson’s correlation analysis between HOXA10 and HDAC1 mRNA expression in HCC tissues was conducted. P-values of less than 0.05 were regarded as statistically significant.
Results
HCC tissues displayed elevated HOXA10 expression
From our analysis of 80 HCC tissues and 30 adjacent normal liver tissues, we observed that HCC tissues displayed elevated HOXA10 mRNA expression (Figure 1A). We then analyzed TCGA (the cancer genome atlas) LIHC (liver hepatocellular carcinoma) dataset, and likewise observed upregulation of HOXA10 mRNA in HCC tissues (n=191) compared to normal liver tissues (n=50) (Figure 1B). Western blot detected HOXA10 protein in four pairs of HCC and adjacent normal liver tissues, and the results similarly showed elevated HOXA10 expression in HCC tissues at the protein level (Figure 1C). These results suggest the possible involvement of HOXA10 in liver carcinogenesis.
Figure 1.
HCC tissues displayed elevated HOXA10 expression.
Notes: (A) HOXA10 mRNA levels were detected by qRT-PCR in HCC tissues (n=80) and adjacent normal liver tissues (n=30). (B) HOXA10 expression in HCC tissues (n=191) and normal liver tissues (n=50) in TCGA LIHC dataset. (C) HOXA10 protein levels were detected in four pairs of HCC tissues (T1-T4) and adjacent normal liver tissues (N1-N4) by Western blot. GAPDH was used as the loading control.
Abbreviations: HCC, hepatocellular carcinoma; HOXA10, homeobox A10; mRNA, messenger RNA; qRT-PCR, quantitative real-time PCR; TCGA, the cancer genome atlas; LIHC, liver hepatocellular carcinoma; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
HOXA10 knockdown inhibited HCC cell proliferation both in vitro and in vivo
HOXA10 protein was examined by Western blot in five HCC cell lines (Figure 2A). SMMC-7721 and HepG2 cells displayed the highest protein levels of HOXA10, while MHCC-97H cells showed the lowest protein level. To probe the functions of HOXA10, we generated the lentiviral constructs for HOXA10 shRNAs (shHOXA10#1, #2 and #3). SMMC-7721 and HepG2 cells were transfected with HOXA10 shRNAs or scramble shRNA (shNC). As indicated by Western blot, shNC did not affect HOXA10 expression compared to untreated cells (WT), whereas HOXA10 shRNAs repressed HOXA10 expression compared to shNC (Figure 2B). Of the three shRNAs, shHOXA10#3 showed the best knockdown efficiency and was used in subsequent assays.
Figure 2.
HOXA10 knockdown inhibited HCC cell proliferation both in vitro and in vivo.
Notes: (A) HOXA10 protein levels were detected by Western blot in five HCC cell lines. (B) SMMC-7721 and HepG2 cells were transfected with HOXA10 shRNA (shHOXA10#1, #2 and #3) or scramble shRNA (shNC). HOXA10 protein was examined by Western blot. (C) CCK-8 assays of SMMC-7721 and HepG2 cells after transfected with shHOXA10#3 or shNC for 24, 48 and 72 h. (D) SMMC-7721 cells stably expressing shHOXA10#3 or shNC were injected subcutaneously into the nude mice. Tumor volume was measured every 3 days for 33 days. (E) On Day 33, tumors were harvested and weighed. **P<0.01 and ***P<0.001 compared to shNC.
Abbreviations: HOXA10, homeobox A10; HCC, hepatocellular carcinoma; shRNA, short hairpin RNA; shNC, negative control shRNA; CCK-8, cell counting kit-8.
Next, we conducted CCK-8 assays to determine how HOXA10 affects HCC cell proliferation in vitro. After transfected with HOXA10 shRNA#3 for 24, 48 and 72 h, the OD values of SMMC-7721 and HepG2 cells at 450 nm were significantly attenuated, suggesting the inhibitory effects of HOXA10 knockdown on HCC cell proliferation (Figure 2C). Furthermore, we examined whether reduction of HOXA10 expression in HCC cells would suppress tumor growth in vivo. SMMC-7721 cells transfected with shHOXA10#3 or shNC were injected subcutaneously into the nude mice. Interestingly, the growth rates of xenografts formed from HOXA10 knockdown cells were slower compared to those formed from control cells (shNC) (Figure 2D). At 33 days after inoculation, the size and weight of xenografts formed from HOXA10 knockdown cells were remarkably reduced compared to those formed from control cells (shNC) (Figure 2E). Thus, these results suggest HOXA10 knockdown inhibits HCC cell proliferation both in vitro and in vivo.
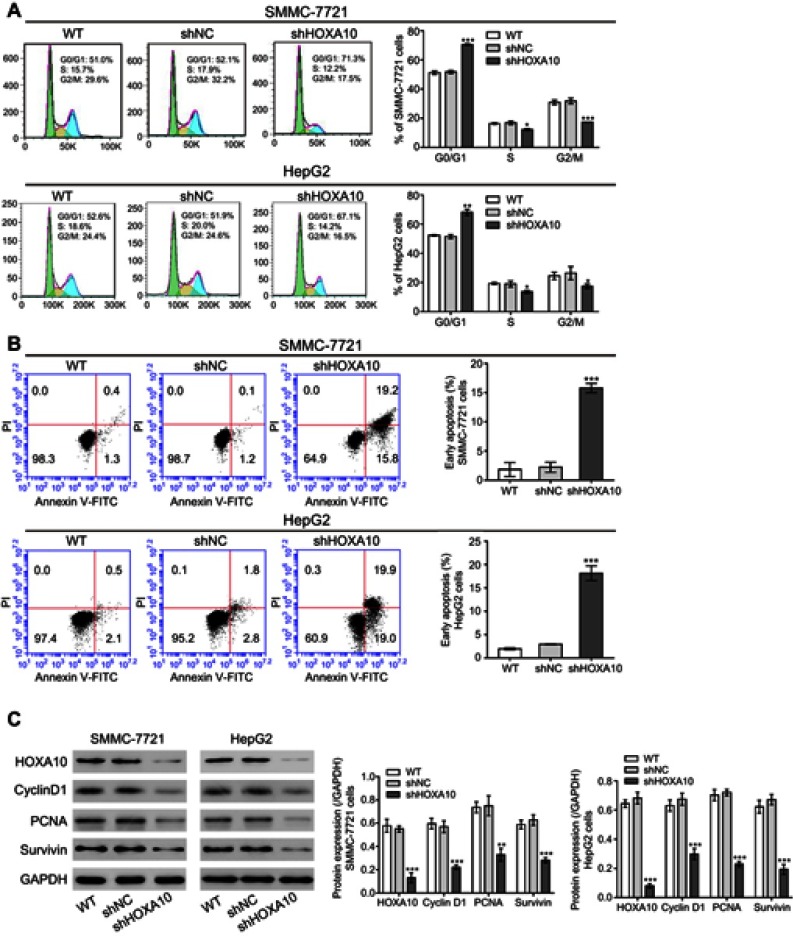
HOXA10 knockdown induced cell cycle arrest and apoptosis in HCC cells
To assess the percentages of cells at different cell cycle stages and cells undergoing apoptosis, we conducted flow cytometry analysis. The results showed that HOXA10 inhibition in both SMMC-7721 and HepG2 cells blocked cell cycle at G0/G1 phase (Figure 3A). Furthermore, compared to the control group (shNC), shHOXA10#3 treatment increased the apoptotic rates in both SMMC-7721 and HepG2 cells (Figure 3B). HOXA10 knockdown cells (SMMC-7721 and HepG2 cells) also displayed reduced levels of Cyclin D1, PCNA and Survivin (Figure 3C). Based on the above findings, we propose that HOXA10 knockdown induces cell cycle arrest and apoptosis in HCC cells, thus suppressing tumor cell growth.
Figure 3.
HOXA10 knockdown induced cell cycle arrest and apoptosis in HCC cells.
Notes: Flow cytometry analyses of cell cycle (A) and apoptosis (B) in SMMC-7721 and HepG2 cells at 48 h after shHOXA10#3 or shNC transfection. (C) Western blot showed that HOXA10 knockdown resulted in altered expression of cell cycle-related (Cyclin D1 and PCNA) and apoptosis-related (Survivin) proteins. *P<0.05, **P<0.01 and ***P<0.001 compared to shNC.
Abbreviations: HOXA10, homeobox A10; HCC, hepatocellular carcinoma; shNC, negative control shRNA; PCNA, proliferating cell nuclear antigen.
Effects of HOXA10 knockdown on p53 acetylation and HDAC1 transcription
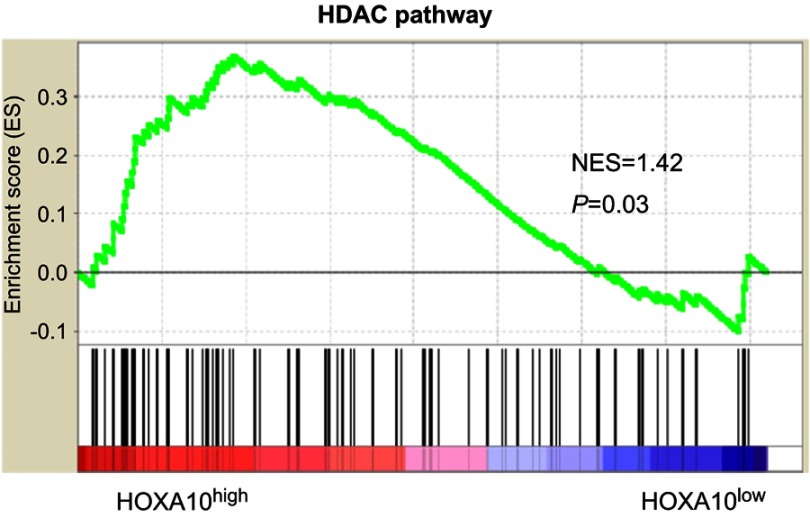
HOX genes are known to regulate p53 in breast cancer cell lines.17,28 We found that downregulation of HOXA10 enhanced p53 acetylation (Lys382) in both SMMC-7721 and HepG2 cells, without affecting total p53 protein (Figure 4A). Gene set enrichment analysis (GSEA) based on TCGA LIHC dataset showed a significant association between the HDAC pathway and HOXA10 expression in HCC tissues (Figure 5). HDAC1, HDAC2 and HDAC3 mRNA expression was analyzed in both SMMC-7721 and HepG2 cells. Interestingly, HDAC1 expression was markedly decreased by HOXA10 knockdown (Figure 4B). Inhibition of HDAC1 protein by HOXA10 knockdown was confirmed by Western blot (Figure 4C).
Figure 4.
Effects of HOXA10 knockdown on p53 acetylation and HDAC1 transcription.
Notes: Experiments were conducted in both HCC cell lines (SMMC-7721 and HepG2). (A) Western blot showed that HOXA10 knockdown enhanced p53 acetylation (Lys382). (B) qRT-PCR showed the effects of HOXA10 knockdown on HDAC1-3 mRNA expression. (C) Western blot showed the effects of HOXA10 knockdown on HDAC1 protein expression. (D) Luciferase assays showed that HOXA10 knockdown remarkably reduced the activity of the HDAC1 promoter. (E) ChIP analysis of HOXA10 binding to the HDAC1 promoter. (F) Pearson’s correlation analysis between HOXA10 and HDAC1 mRNA expression in HCC tissues (n=80). *P<0.05, **P<0.01 and ***P<0.001 compared to shNC. ^^P<0.01 and ^^^P<0.001 compared to IgG.
Abbreviations: HOXA10, homeobox A10; HDAC1, histone deacetylase 1; HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time PCR; mRNA, messenger RNA; ChIP, chromatin immunoprecipitation; shNC, negative control shRNA.
Figure 5.
The HDAC pathway was significantly associated with HOXA10 expression.
Notes: GSEA was carried out in TCGA LIHC dataset to analyze pathways associated with HOXA10 expression. NES: normalized enrichment score.
Abbreviations: HDAC, histone deacetylase; HOXA10, homeobox A10; GSEA, gene set enrichment analysis; TCGA, the cancer genome atlas; LIHC, liver hepatocellular carcinoma.
Considering that HOX genes encode transcription factors, we speculated that HOXA10 directly bound to the HDAC1 promoter and regulated the transcription of HDAC1. To test this, we cloned the human HDAC1 promoter into a luciferase reporter vector and luciferase assays were conducted. As shown in Figure 4D, HOXA10 knockdown suppressed the activity of the HDAC1 promoter. The consensus binding site for HOXA10, containing a TTAT core sequence, has been identified.31 Four potential sites were found in the HDAC1 promoter sequence (Figure 4E), and ChIP assays showed that HOXA10 bound to the regions at −463~-275 and −729~-586 of the HDAC1 promoter, but not to the regions at −874~-785. Furthermore, Pearson’s correlation analysis showed a positive correlation between HOXA10 and HDAC1 mRNA expression in 80 HCC tissues (Figure 4F).
HDAC1 mediated the effects of HOXA10 on proliferation, cell cycle progression, apoptosis and p53 acetylation
To test whether HDAC1 mediated the effects of HOXA10 on proliferation, cell cycle progression and apoptosis, we knocked down HDAC1 in MHCC-97H cells with HOXA10 overexpression. CCK-8 assays showed that HOXA10 overexpression increased the OD values of MHCC-97H cells after 24, 48 and 72 h, however the effects were abrogated by HDAC1 knockdown (Figure 6A). Flow cytometry analysis showed that HOXA10 overexpression increased the percentage of MHCC-97H cells at S phase, whereas decreased the percentage of early apoptotic cells. Nevertheless, the effects were abrogated by HDAC1 knockdown (Figure 6B and C). HOXA10 overexpression also increased the levels of Cyclin D1, PCNA and Survivin, but the effects were similarly abolished by HDAC1 knockdown (Figure 6D). Overexpression of HOXA10 and downregulation of HDAC1 were verified by Western blot (Figure 6E). Importantly, HDAC1 knockdown blocked the inhibitory effects of HOXA10 on p53 acetylation (Figure 6E). Together, these results indicate that HOXA10 affects proliferation, cell cycle progression, apoptosis and p53 acetylation through regulating HDAC1.
Figure 6.
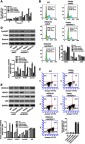
HDAC1 mediated the effects of HOXA10 on proliferation, cell cycle progression, apoptosis and p53 acetylation.
Notes: MHCC-97H cells were treated with Vector + shNC, HOXA10+ shNC, Vector + shHDAC1 and HOXA10+ shHDAC1. (A) CCK-8 assays were conducted at 0, 24, 48 and 72 h after transfection. (B) Flow cytometry analysis of cell cycle and (C) apoptosis at 48 h after transfection. (D) Western blot showed the expression of cell cycle-related (Cyclin D1 and PCNA) and apoptosis-related (Survivin) proteins at 48 h after transfection. (E) Western blot showed the expression of HOXA10, HDAC1, Ace-p53 and total p53 at 48 h after transfection. *P<0.05, **P<0.01 and ***P<0.001 compared to Vector+shNC. ##P<0.01 and ###P<0.001 compared to HOXA10+shNC.
Abbreviations: HDAC1, histone deacetylase 1; HOXA10, homeobox A10; shNC, negative control shRNA; PCNA, proliferating cell nuclear antigen; Ace-p53, acetylated p53.
Discussion
HOXA10 overexpression has been observed in diverse cancer types and shown to regulate proliferation of cancer cell lines. In this study, we demonstrated that HCC tissues displayed elevated HOXA10 expression, consistent with results from the previous study.21 For the first time, we investigated how HOXA10 functioned in hepatocellular carcinogenesis in vitro as well as in vivo using lentiviral-mediated RNA interference. Our study showed that two human HCC cell lines (SMMC-7721 and HepG2 cells) with HOXA10 knockdown had low cell proliferation rates, compared to cells with scramble shRNA (shNC). In the nude mouse xenograft model, the volume and weight of tumors formed in HOXA10 knockdown group were markedly reduced, compared to shNC group. Moreover, HOXA10 knockdown increased the percentages of cells at G0/G1 phase, as well as the percentages of apoptotic cells. In summary, our data provide evidence for a critical role of HOXA10 in HCC carcinogenesis.
p53, a widely recognized tumor suppressor, executes several important functions including regulation of cell cycle, apoptosis and DNA repair.22 Aberrant expression and mutations of p53 have been found in HCC tissues.25–27 HOX genes, such as HOXA5 and HOXA10, are known to regulate p53 expression in breast cancer cell lines.17,28 Similar to the above observations, our present study showed that HOXA10 knockdown in two HCC cell lines enhanced the acetylation of p53, which was crucial to p53 activity.23 In addition, downregulation of HOXA10 in HCC cells did not affect p53 protein, which was rather inconsistent with data from breast cancer BT20 cells.17 The discrepancy may be due to tissue differences. Cyclin D1 and Survivin which regulate G0/G1 to S phase transition and apoptosis respectively have previously been found as direct or indirect targets of p53.32–34 Here, HOXA10 knockdown led to reduced expression of Cyclin D1 and Survivin, which was consistent with the results of flow cytometry analysis. Our data suggest that HOXA10 influences cell cycle progression and apoptosis in HCC cells through regulating p53 functions.
We then explored the molecular mechanism through which HOXA10 knockdown enhanced p53 acetylation. Previously, HDAC1 has been demonstrated to directly bind to p53, which deacetylates p53 and downregulates p53 functions.24,35 Our results showed that HOXA10 downregulation dramatically suppressed HDAC1 expression and promoter activity. Furthermore, ChIP assays revealed that HOXA10 protein bound to the regions of HDAC1 promoter. The binding was stronger in the region containing three TTAT motifs than that in the region with only one TTAT motif.31 A positive correlation was observed between HOXA10 and HDAC1 mRNA expression in 80 HCC tissues. Moreover, HDAC1 knockdown partially abrogated the effects of HOXA10 overexpression on proliferation, cell cycle progression, apoptosis and p53 acetylation. Thus, we propose that HOXA10 might induce the deacetylation of p53 by promoting HDAC1 transcription, thus downregulating the functions of p53 on cell cycle progression and apoptosis.
Conclusion
In summary, our study provided evidence for the strong association between HOXA10 expression and HCC progression via in vitro and in vivo experiments. Furthermore, HOXA10 knockdown downregulated HDAC1 transcription, thus enhancing p53 acetylation. HDAC1 mediated the effects of HOXA10 on proliferation, cell cycle progression, apoptosis and p53 acetylation. Our findings provide potential insights into the mechanisms of HCC pathogenesis and the development of novel therapeutics for HCC.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (No. 81770589 and 81702050), the Natural Science Foundation of Shanghai (No. 17ZR1421500) and the Medical Education Research Project of Shanghai Jiao Tong University School of Medicine (No. YB150712).
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 3.Thun MJ, DeLancey JO, Center MM, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31(1):100–110. doi: 10.1093/carcin/bgp263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motola-Kuba D, Zamora-Valdés D, Uribe M, Méndez-Sánchez N. Hepatocellular carcinoma. An overview. Ann Hepatol. 2006;5(1):16–24. [PubMed] [Google Scholar]
- 5.Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–278. doi: 10.1016/S0070-2153(09)88009-5 [DOI] [PubMed] [Google Scholar]
- 6.Shah N, Sukumar S. The hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826 [DOI] [PubMed] [Google Scholar]
- 7.Thorsteinsdottir U, Sauvageau G, Hough MR, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17(1):495–505. doi: 10.1128/mcb.17.1.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe M, Hamada J, Takahashi O, et al. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006;15(4):797–802. doi: 10.3892/or.15.4.797 [DOI] [PubMed] [Google Scholar]
- 9.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164 [DOI] [PubMed] [Google Scholar]
- 10.Cheng W, Jiang Y, Liu C, Shen O, Tang W, Wang X. Identification of aberrant promoter hypomethylation of HOXA10 in ovarian cancer. J Cancer Res Clin Oncol. 2010;136(8):1221–1227. doi: 10.1007/s00432-010-0772-4 [DOI] [PubMed] [Google Scholar]
- 11.Yamatoji M, Kasamatsu A, Yamano Y, et al. State of homeobox A10 expression as a putative prognostic marker for oral squamous cell carcinoma. Oncol Rep. 2010;23(1):61–67. doi: 10.3892/or_00000606 [DOI] [PubMed] [Google Scholar]
- 12.Lim JY, Yoon SO, Seol SY, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19(41):7078–7088. doi: 10.3748/wjg.v19.i41.7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Y, Lu S, Wen YG, et al. Overexpression of HOXA10 promotes gastric cancer cells proliferation and HOXA10(+)/CD44(+) is potential prognostic biomarker for gastric cancer. Eur J Cell Biol. 2015;94(12):642–652. doi: 10.1016/j.ejcb.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 14.Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21(5):945–952. [PubMed] [Google Scholar]
- 15.Li B, Cao X, Weng C, et al. HoxA10 induces proliferation in human prostate carcinoma PC-3 cell line. Cell Biochem Biophys. 2014;70(2):1363–1368. doi: 10.1007/s12013-014-0065-7 [DOI] [PubMed] [Google Scholar]
- 16.Carrera M, Bitu CC, de Oliveira CE, et al. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8(4):3613–3623. [PMC free article] [PubMed] [Google Scholar]
- 17.Chu MC, Selam FB, Taylor HS. HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol Ther. 2004;3(6):568–572. doi: 10.4161/cbt.3.6.848 [DOI] [PubMed] [Google Scholar]
- 18.Cui XP, Qin CK, Zhang ZH, et al. HOXA10 promotes cell invasion and MMP-3 expression via TGFβ2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci. 2014;59(7):1442–1451. doi: 10.1007/s10620-014-3033-6 [DOI] [PubMed] [Google Scholar]
- 19.Quagliata L, Matter MS, Piscuoglio S, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi: 10.1002/hep.26740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zha TZ, Hu BS, Yu HF, Tan YF, Zhang Y, Zhang K. Overexpression of HOXA1 correlates with poor prognosis in patients with hepatocellular carcinoma. Tumor Biol. 2012;33(6):2125–2134. doi: 10.1007/s13277-012-0472-6 [DOI] [PubMed] [Google Scholar]
- 21.Cillo C, Schiavo G, Cantile M, et al. The HOX gene network in hepatocellular carcinoma. Int J Cancer. 2011;129(11):2577–2587. doi: 10.1002/ijc.25941 [DOI] [PubMed] [Google Scholar]
- 22.Farnebo M, Bykov VJ, Wiman KG. The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun. 2010;396(1):85–89. doi: 10.1016/j.bbrc.2010.02.152 [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juan LJ, Shia WJ, Chen MH, et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275(27):20436–20443. doi: 10.1074/jbc.M000202200 [DOI] [PubMed] [Google Scholar]
- 25.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641 [DOI] [PubMed] [Google Scholar]
- 26.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naka T, Toyota N, Kaneko T, Kaibara N. Protein expression of p53, p21WAF1, and Rb as prognostic indicators in patients with surgically treated hepatocellular carcinoma. Anticancer Res. 1998;18(1B):555–564. [PubMed] [Google Scholar]
- 28.Raman V, Martensen SA, Reisman D, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405(6789):974–978. doi: 10.1038/35016125 [DOI] [PubMed] [Google Scholar]
- 29.Xie HJ, Noh JH, Kim JK, et al. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7(4):e34265. doi: 10.1371/journal.pone.0034265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Salz T, Zajac-Kaye M, Liao D, Huang S, Qiu Y. Overexpression of histone deacetylases in cancer cells is controlled by interplay of transcription factors and epigenetic modulators. Faseb J. 2014;28(10):4265–4279. doi: 10.1096/fj.14-250654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benson GV, Nguyen TH, Maas RL. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol. 1995;15(3):1591–1601. doi: 10.1128/mcb.15.3.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-κB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23(13):4713–4727. doi: 10.1128/mcb.23.13.4713-4727.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guardavaccaro D, Corrente G, Covone F, et al. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol. 2000;20(5):1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21(17):2613–2622. doi: 10.1038/sj.onc.1205353 [DOI] [PubMed] [Google Scholar]
- 35.Ito A, Kawaguchi Y, Lai CH, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. Embo J. 2002;21(22):6236–6245. doi: 10.1093/emboj/cdf616 [DOI] [PMC free article] [PubMed] [Google Scholar]