Abstract
Circular RNAs (circRNAs) are a distinct category of single-stranded, covalently closed RNAs formed by backsplicing. The functions of circRNAs are incompletely known and are under active investigation. Here, we report that in addition to traditional linear mRNAs (linRNA), mouse, rat, and human opioid receptor genes generate exonic circRNA isoforms. Using standard molecular biologic methods, Oprm1 circRNAs (circOprm1) were detected in RNAs of rodent and human brains and spinal cords, as well as human neuroblastoma cells, suggesting evolutionary conservation. Sequencing confirmed backsplicing using canonical splice sites. Oprm1 circRNAs were sense-stranded circRNAs resistant to RNase R digestion. The relative abundance of Oprm1 circRNA to linRNA determined by quantitative reverse transcription polymerase chain reaction varied among mouse brain regions, with circRNA isoforms predominating in rostral structures and less abundant in brain stem. Chronic morphine exposure in mice increased brain circOprm1.e2.3 and circOprm1.e2.e3.e4(302) levels by 1.5- to 1.6-fold relative to linRNA. Sequence analysis predicted numerous microRNA binding sites within Oprm1 circRNA sequences, suggesting a potential role in microRNA sequestration through sponging. In addition, we observed that other opioid receptor genes including δ, κ, and nociceptin receptor genes produced similar circRNAs. In conclusion, all members of the opioid receptor gene family express circRNAs, with Oprm1 circRNA levels exceeding those of linear forms in some regions.
Significance Statement
The modulation of Oprm1 circular RNA (circRNA) expression by morphine, coupled with the high abundance and existence of potential miRNA binding sites with circRNA sequences suggests the potential role of Oprm1 circRNAs in chronic opioid effects such as tolerance.
Introduction
Opioids are effective medications, but their prolonged use leads to a number of compensatory changes, including tolerance, which is complex and comprised of many factors (Pasternak and Pan, 2013). The quintessential opioid analgesic, morphine, acts through μ-opioid receptors, encoded by their gene, Oprm1 (Pasternak and Pan, 2013). Although undoubtedly useful for acute pain control, chronic morphine exposure is associated with challenges including tolerance, which is a complex pharmacodynamic phenomenon by which drug action is diminished after serial exposure. In cellulo models have suggested the contribution of μ agonist–induced receptor phosphorylation and internalization to opioid tolerance (Williams et al., 2013). Several systems such as N-methyl-D-aspartate and δ receptors (Pasternak and Pan, 2013) have been involved in morphine tolerance in vivo models. When dosing was extended to 6 weeks in mice, morphine analgesic tolerance increased for 3 weeks, after which it stabilized. This stabilization was associated with profound changes in Oprm1 variant mRNA expression (Xu et al., 2015), suggesting potential involvement of Oprm1 variant mRNA levels in morphine tolerance.
Many factors can impact gene regulation, including mRNA transcription, stability, and translation. The importance of microRNA (miRNA) in modulating gene expression has been extensively validated and recent studies have implicated circular RNA (circRNA) in the control of miRNA function (Hansen et al., 2013; Memczak et al., 2013; Wang et al., 2016; Zheng et al., 2016). While the first descriptions of circular RNAs date back 30 years (Nigro et al., 1991; Cocquerelle et al., 1992; Capel et al., 1993), recent transcriptome surveys vastly expanded the landscape of circRNAs, and 20% of mammalian genes are now estimated to express circRNAs (Salzman et al., 2012; Jeck et al., 2013; Memczak et al., 2013; You et al., 2015). Genome-wide studies revealed circRNAs to be diverse, abundant, and often evolutionarily conserved (Jeck et al., 2013; Rybak-Wolf et al., 2015). Of the three molecularly distinct categories of circular RNAs, the best studied type involves backsplicing of the (downstream) end of an exon to the (upstream) front end of exon, generating covalently closed circular RNAs (Fig. 1A). Two other types include circular intronic RNAs derived from failed debranching of lariat intermediates of mRNA splicing and exon-intron circRNAs, which contain intron and exon sequences. These latter two RNAs are both nuclear enriched, and modulate expression of their parent locus (Zhang et al., 2013; Li et al., 2015b), while exonic circRNAs are predominantly cytoplasmic (You et al., 2015). Although the functions of most circRNAs remain unclear, several competitively antagonize miRNAs, which themselves are noncoding regulatory RNAs that dictate post-transcriptional fates of target mRNAs (Hansen et al., 2013; Memczak et al., 2013; Wang et al., 2016; Zheng et al., 2016). The diversity of circRNAs derives from both the number of genes producing circRNAs as well as the number of variant circles per gene, much like alternative splicing of linear mRNAs (linRNAs) (Salzman et al., 2012; Zhang et al., 2014; Gao et al., 2016). For some genes, circRNA abundance exceeds that of their linRNAs (Salzman et al., 2012), and circRNAs have longer half-lives than linRNAs due to resistance to RNA degradation (Wang and Wang, 2015). circRNAs are enriched in neuronal cells and tissues (Westholm et al., 2014; Rybak-Wolf et al., 2015; You et al., 2015), circRNA encoding genes are enriched for annotations for synaptic function (You et al., 2015), and circRNAs accumulate in synapses (Rybak-Wolf et al., 2015), all suggesting circRNAs may be particularly important in the nervous system. We now test the hypotheses that the Oprm1 gene generates circRNAs and that they are modulated independently of linRNA isoforms. We demonstrate that the μ-opioid receptor gene Oprm1, as well as the δ (Oprd1), κ (Oprk1) and nociceptin (Oprl1) genes, generate circRNA, and that chronic morphine treatment modulates Oprm1 circular RNA (circOprm1) levels.
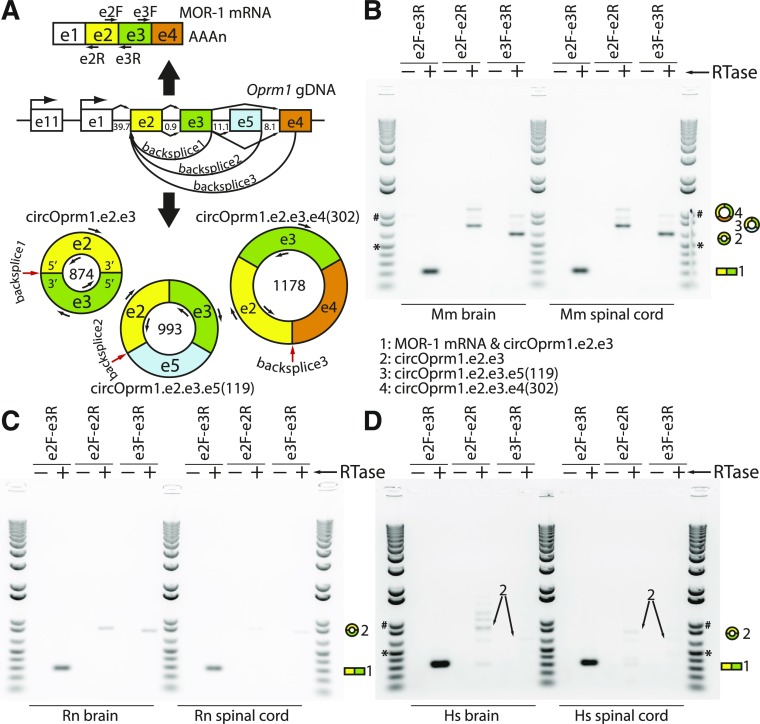
Fig. 1.
Mouse, rat, and human Oprm1/OPRM1 divergent RT-PCRs of circular RNAs. (A) A map of the mouse μ-opioid receptor gene (Oprm1) is shown with select exons in their relative locations (e.g., e1 designates exon 1). The e1 and exon 11 (e11) associated promoters are marked with bent arrows, and intron sizes are labeled in kilobases. The μ-opioid receptor (MOR-1) protein is encoded by the dominant linear mRNA containing e1-e2-e3-e4, shown above the genomic DNA (gDNA) with a polyA tail and locations of forward (F) and reverse (R) primers. The Oprm1 locus also generates multiple circRNA isoforms (shown below the gDNA map, with primer sites; circRNA sizes in nucleotides shown inside circles; red arrows point to backsplice junctions). (B) Divergent RT-PCR products from mouse [Mus musculus (Mm)] brain and spinal cord total RNA were Sanger sequenced and backsplice junctions confirmed (also see Supplemental Figs. 1 and 2). In brain, three variants were sequence confirmed, while only circOprm1.e2.e3 was confirmed in spinal cord. (C) Rat [Rattus norvegicus (Rn)] and (D) human [Homo sapiens (Hs)] divergent RT-PCRs were designed similarly to the mouse. Conventional RT-PCR with convergent primers (e2F-e3R) amplify linear mRNA as well as circRNAs, while divergent primers (e2F-e2R, e3F-e3R) only amplify circRNAs. In all cases, 5 μg total RNA was treated with reverse transcriptase [(RTase), arrow] with 250 ng random hexamers in 20 μl reactions (+), or RTase omitted (−). PCR was done with 1 ng/μl of cDNA input to 35 ampflication cycles. Ten microliters of PCR reaction were loaded against 1 μl of Invitrogen 1 kb Plus ladder; 500 bp (*) and 1 kb (#) are marked as size reference.
Materials and Methods
Animals.
Whole brain and spinal cord RNAs were obtained from adult male CD-1 mice (Mus musculus). Chronic morphine treatment involved the subcutaneous implantation of 75 mg morphine free base or placebo pellets (National Institute on Drug Abuse) on the dorsum of 4–6 month old mice under brief isoflurane anesthesia, after which the skin was stapled closed. Animal protocols were approved by the Institutional Animal Care and Use Committee.
RNA Isolation.
Mice anesthetized with isoflurane were sacrificed by cervical dislocation, and brains and spinal cords were quickly dissected while chilled to 4°C as rapidly and consistently as possible. Tissues were homogenized at 4°C in Trizol (ThermoFisher)/chloforom using a Polytron tissue homogenizer per the manufacturer’s recommendation. Total RNA was isopropanol precipitated and quantitated by Nanodrop (ThermoFisher). Rat (Rattus norevegicus) brain and spinal cord total RNAs were purchased from Zyagen (RR-201, RR-230, and Sprague-Dawley), and human (Homo sapiens) male brain and male/female pooled spinal cord total RNAs were purchased from Clontech. For mouse brain region RNA isolations, brains were immediately immersed into ice-cold RNAlater Stabilization Solution (ThermoFisher) for 3–5 minutes, and dissections were performed from coronal slices generated using a brain matrix (Product 15050; Ted Pella) to minimize mRNA degradation. The Brain Explorer 2 software from the Allen Mouse Brain Atlas project was used as an anatomic reference (Lau et al., 2008).
Reverse Transcription Polymerase Chain Reaction.
Reverse transcription (RT) with Superscript3 (18080093; ThermoFisher) was performed per the manufacturer’s recommendation with minor modifications using random hexamer primers (typically, 250 ng per 5 μg total RNA in 20 µl RT reaction) or Oprm1 primers (2 pmol primer per 5 µg total RNA) targeting sense and antisense RNA. Digestion of linear RNA was carried out by treating total RNA with RNase R (Epicentre Biotech), followed by RT with or without Superscript3 enzyme. Primers were obtained from Sigma and Invitrogen. Polymerase chain reactions (PCRs) were performed with Taq polymerase (Sigma) and products were sequenced directly after gel extraction (Zymo) or TOPO cloning (Invitrogen). Sanger sequencing was performed through the MSKCC sequencing core facility.
Mouse Oprm1 quantitative PCRs (qPCRs) were performed using HotStart-IT SYBR Green qPCR Master Mix (Affymetrix) on a Bio-Rad CFX96 Touch Real-Time PCR Detection System. For estimation of linRNA, we employed standard PCR strategies with convergent primer pairs between exon 1 (e1) and exon 3 (e3), after confirming that there are no readily detectable circRNA isoforms by divergent RT-PCR from e1 to exon 2 (e2) (Fig. 2B). The e2.e3 circRNA was specifically amplified with an e3 forward primer, and an e2.e3 backsplice junction-spanning reverse primer, while the e2.e3.e4 circRNA (where e4 denotes exon 4) was amplified from two junction-spanning primers including a backsplice junction-spanning primer (Supplemental Table 1). Analyses of Ct values were performed as described previously (Lu et al., 2014). For the whole brain versus spinal cord comparison, a geometric mean of Ct values for glyceraldehyde-3-phosphate dehydrogenase, succinate dehydrogenase, and β-microglobulin was used to obtain a ΔCt value for each Oprm1 RNA target. Raw ΔCt values are reported in Fig. 4B, and relative ratios of 2−ΔCt values for linear versus circular isoforms are reported in Fig. 4C (qPCR analysis method shown below the graph). For brain region analysis, ΔCt values were obtained against glyceraldehyde-3-phosphate dehydrogenase, and ratios of 2−ΔΔCt values are reported in Fig. 4D. For the morphine treatment study, total RNA was harvested from spinal cord and brain and reverse transcribed with Superscript3, and the geometric means of Ct values for TATA-binding protein, succinate dehydrogenase, and β-microglobulin were used to obtain the ΔCt values reported in Fig. 5. Melt curves and gels indicate single products are generated by these amplification strategies. While it is theoretically possible that divergent RT-PCR strategies could amplify from multiple template sources with internal deletions in the blind spot between divergent primers, screening with multiple divergent primer pairs from e2 and e3 suggests that internal deletions within the Oprm1 e2 and e3 exonic sequence are undetectable under our conditions.
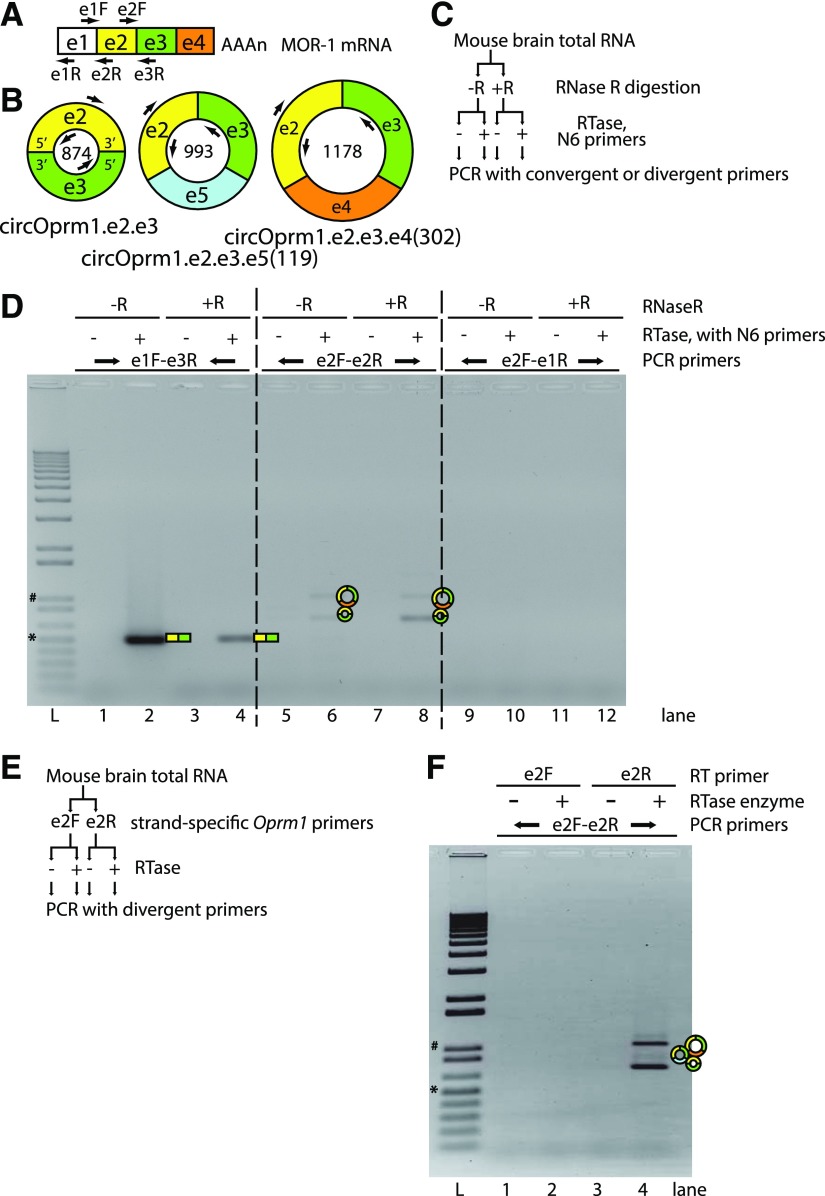
Fig. 2.
Mouse Oprm1 divergent RT-PCRs amplify sense-stranded circRNAs. (A) Primer maps with locations matching the MOR-1 mRNA and (B) dominant Oprm1 circRNAs are shown. Exons 1–5 are abbreviated e1–e5, and primer labels designate the target exons and orientation (F/R) with respect to transcription. The numbers inside the circles represent circRNA sizes (nt). (C) To confirm that divergent RT-PCR products are generated from circular RNAs, RNAs were pretreated with RNase R, an exonucleolytic enzyme that preferentially digests linear mRNA templates from the 3′ tails but not circular RNAs. Mouse brain total RNAs were digested with (+R) or without (−R) RNase R and samples were split to RT reactions with random hexamer (N6) primers, with (+) or without (−) reverse transcriptase (RTase) enzyme, and then these cDNAs were input to PCR using convergent or divergent primer pairs. (D) Convergent RT-PCR (primers e1F-e3R, lanes 1–4) amplified the linear MOR-1 mRNA, but RNase R predigestion attenuated this amplification. Divergent RT-PCR products (primers e2F-e2R, lanes 5–8) were stable against RNase R pretreatment. Divergent RT-PCR using primers from e1 to e2 (e2F-e1R primers, lanes 9–12) did not generate products. For labels above the gel image: top, middle, and bottom rows of labels represent RNase R treatment, inclusion/omission of RTase enzyme during RT with N6, and the PCR primer pairs used for each sample, respectively. For lane labels below the gel image, L represents the Invitrogen 1 kb plus ladder, and the 1 kb (#) and 500 bp (*) bands are marked. (E) Some circRNAs are known to derive from antisense transcripts overlapped with annotated genes; therefore, to determine whether the observed Oprm1 divergent RT-PCRs derive from sense or antisense circRNAs, RT reactions using strand-specific Oprm1 primers were performed as shown in the scheme. Mouse brain total RNA was incubated with either the Oprm1 e2 sense (e2F) or antisense (e2R) gene-specific primers, with (+) or without (−) RTase enzyme, and amplified with e2 divergent primers. (F) RT with antisense-stranded primer (e2R) can support divergent RT-PCR from sense-stranded circRNAs e2.e3, e2.e3.e5(119) and e2.e3.e4(302) (lanes 3 to 4), but sense-stranded primer (lanes 1 to 2) did not. Labels above the gel, from top to bottom, are for the RT primers, inclusion (+) or omission (−) of RTase enzyme, and PCR primers, while lane labels and size ladders are the same as for (D).
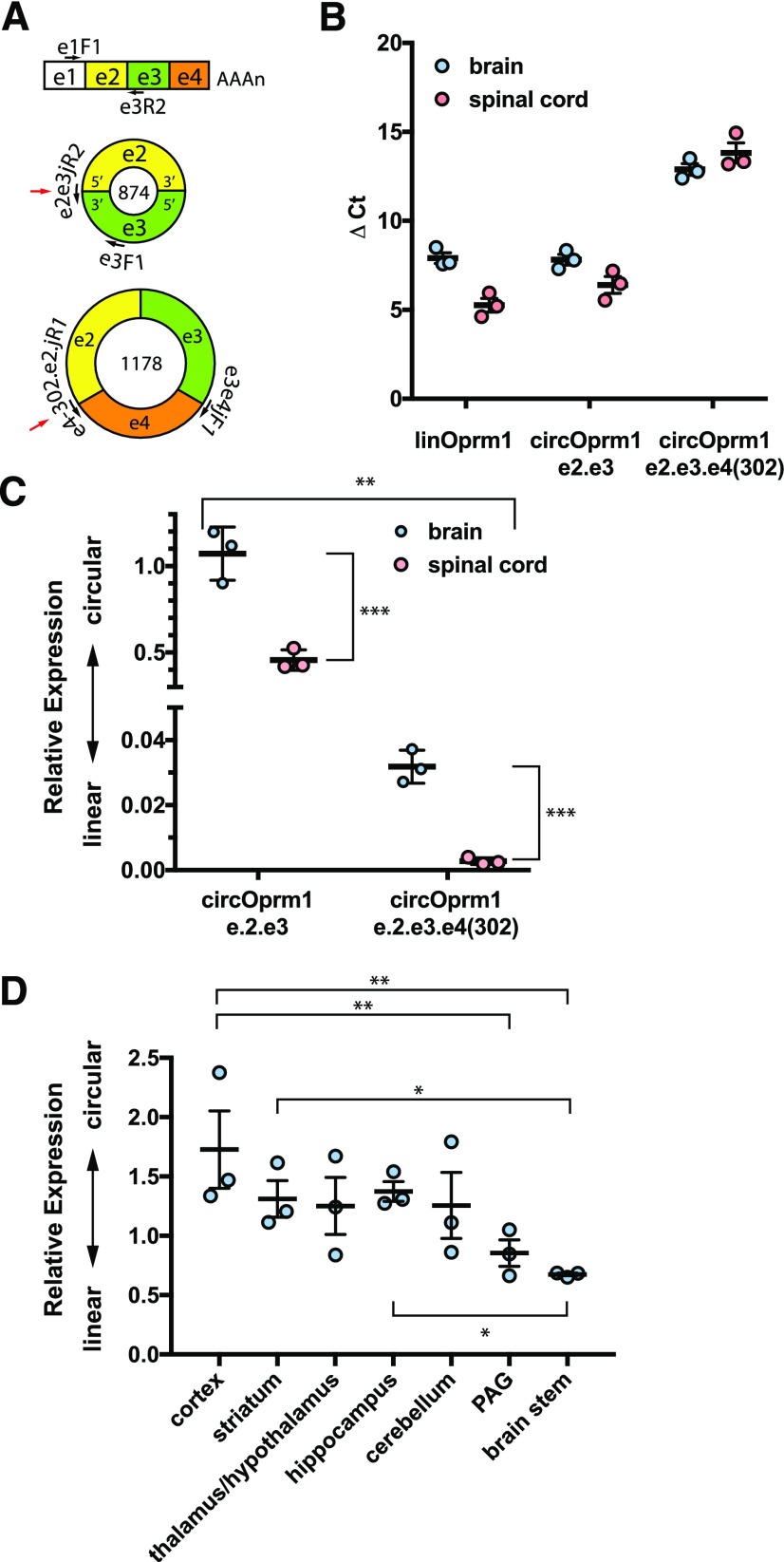
Fig. 4.
Anatomic survey of expression of mouse Oprm1 linear vs. circular RNAs. (A) The target Oprm1 RNAs are shown with exons and primers not drawn to scale. The linear Oprm1 isoforms are detected with primers spanning exon 1 (e1F1) to exon 3 (e3R2). The circOprm1.e2.e3 is detected with a backsplice junction spanning primer (e2e3juncR2) paired with an exon 3 forward primer (e3F1). The circOprm1.e2.e3.e4(302) is selectively amplified with two junction spanning primers. The backsplice junctions for circRNAs are shown with a red arrow. (B) ΔCt values for Oprm1 isoforms in opioid naive mouse whole brain and spinal cord are shown for linear, circOprm1.e2.e3, and circOprm1.e2.e3.e4(302). (C) Relative abundance values for circular isoforms normalized to the linear isoforms, using data shown in (B), are plotted as ΔΔCt values and analyzed by two-way ANOVA (significant variation was attributable to anatomic region, ***P < 0.001; RNA target, **P < 0.01; and interaction, **P < 0.01, see Materials and Methods). (D) Select brain regions were dissected from naive CD-1 male mice, and qPCRs were performed to evaluate relative expression of linear and e2.e3 circRNA levels (overall P < 0.05 by ordinary one-way ANOVA (F6,14 = 2.953; *P = 0.045), followed by post-hoc multiple comparisons analysis with Tukey’s correction significant for cortex vs. brain stem (*P < 0.05).
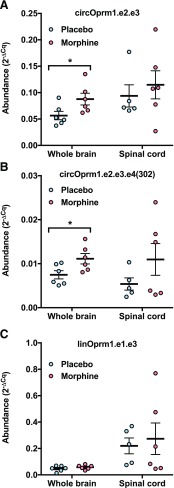
Fig. 5.
circOprm1 RNAs accumulate in mouse whole brain with chronic morphine exposure. CD-1 male mice were subcutaneously implanted with a morphine [(M), 75 mg free base] or placebo (P) pellet every 3 days, for a total of three pellets/animal (n = 5 to 6 mice per group). Tissues were harvested on day 10 for qPCR analysis for Oprm1 RNAs to compare circRNA and linRNA levels in whole brain and spinal cord. Tissues were examined for circOprm1.e2.e3 (A), circOprm1.e2.e3.e4(302) (B), and linear mRNA (linOprm1.e1.e3) (C) levels. Data are shown as 2−ΔCt. For each Oprm1 target RNA for each tissue, morphine vs. placebo comparisons were analyzed by t test, and circOprm1.e2.e3 circOprm1.e2.e3.e4(302) increases in whole brain were significant (*P < 0.05).
Bioinformatics.
Custom miRNA target predictions were performed using the online mirDB interface (Wong and Wang, 2015). Exon sequences were permuted in all possible orderings to ensure capture of predictions near the backsplice junction since the software interface assumes linear sequence inputs. By default, predictions scoring below 50 were not reported by mirDB, but any prediction scoring above 50 for at least one permutation is presented here. Evolutionary conservation of sequences at predicted miRNA binding sites was computed using bootstrapping (discussed subsequently).
Experimental Design and Statistical Analysis.
The divergent RT-PCRs shown in Fig. 1B were replicated from n > 6 animals, with data shown deriving from young (<3 months) adult CD-1 male mice. The rat data shown in Fig. 1C were derived from a commercial RNA source (gender unspecified), with similar results from total RNA isolated from brain and spinal cords of male Sprague-Dawley rats (data not shown). The human divergent RT-PCRs were performed on one brain and one spinal cord sample each, purchased commercially as described previously. Statistical size was based on previous experience.
Statistical analysis of the qPCRs was assessed using Prism 7 (GraphPad), with statistical significance set at P < 0.05. Analysis for Fig. 4C was carried out using two-way ANOVA of 2−ΔΔCt values (circRNA Ct − linRNA Ct) with matching for each animal (n = 3) and no multiple comparisons testing. Two-way ANOVA P values for Fig. 4C are reported in Results. Statistical analysis of the qPCR 2−ΔΔCt values of Fig. 4D used one-way ANOVA to analyze the brain region–specific differences in linear/circular Oprm1 RNA relative ratios followed by post-hoc multiple comparisons testing between brain regions with Tukey’s test (see Results). For analyses of morphine versus placebo treatment qPCRs in Fig. 5, 2−ΔCt values were computed (where ΔCt = Oprm1 RNA Ct − control RNAs’ geometric mean Ct). The t tests were performed for morphine versus placebo for each Oprm1 primer target at each anatomic site tested (brain and spinal cord).
Calculation of P values for the evolutionary conservation of sequences at predicted miRNA binding sites was performed with bootstrapping using Python3.6 with NumPy version 1.11.3 as follows. Mouse and human e2.e3 circRNA sequences were aligned with BLAST2seq, and the match/mismatch information from the alignment was concatenated into a binary string of length 874 [mouse, rat, and human e2.e3 circRNA lengths are all exactly 874 nucleotides (nt)]. This string was extended at the terminus by a copy of the first six positions and then resampled using numpy.random.choice without replacement for a total of 25 samplings (corresponding to the number of miRNA sites predicted). For each of these position identities, the 7mer beginning at that position was inspected and categorized as 100% conserved or not conserved. This process was repeated for 100,000 trials, and the fraction of those trials with greater than eight out of 25 samplings having 100% identity is reported as the P value. Scripts are available upon request.
Results
Discovery of the Mouse Oprm1 Circular RNAs.
In standard RT-PCR design, pairs of primers are chosen with one sense stranded primer directed downstream and an antisense primer directed upstream of a downstream location (i.e., e2F-e3R) (Fig. 1A). Using an RT-PCR strategy with primer pairs oriented divergently (wherein the antisense primer is anchored upstream of the sense primer) from exon 2 of mouse Oprm1, and a second independent pair of primers directed divergently from exon 3 (Fig. 1, A and B), we detected circular RNA variants of Oprm1 from mouse whole brain total RNA. Sanger sequencing of the PCR products confirmed a backsplice junction connecting the 3′ end of e3 with the 5′ end of e2, employing the typical splice-donor and splice-acceptor sites of e2 and e3 as with canonical linear splicing by the spliceosome (Fig. 1; Supplemental Fig. 1). This dominant circular RNA, which we refer to as circOprm1.e2.e3, was amplified from Oprm1 e2 and e3, and is an 874-nt circle (Fig. 1, A and B). In addition, we amplified larger products from both the e2- and e3-based primer pairs, albeit less efficiently.
The second strongest signal in these assays corresponded to circRNAs containing e2, e3, and the first 302 nt of e4. This product derives from a 1178-nt circRNA [circOprm1.e2.e3.e4(302)] (Fig. 1, B and C; Supplemental Fig. 2). The 3′ end of this backsplice junction coincides perfectly with the terminus of several RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/) and UCSC genome database (https://genome.ucsd.edu) entries (Supplemental Fig. 2C) (Kent et al., 2002), suggesting these annotations do not mark true ends of mRNAs and in fact may be sequence entries derived from circular templates. The third most readily detected circRNA was sequence verified through TOPO cloning and contained e2, e3, and 119 nt of exon 5 (e5) derived from a 993-nt circRNA [circOprm1.e2.e3.e5(119)] (Fig. 1, B and C). By employing divergent primers at the 5′ end of e2 against primers at the 3′ end of e3, we observed numerous other amplicons, suggesting an unexpected diversity of circRNA isoforms from this locus (Supplemental Fig. 3). We determined the sequences of two more isoforms of lesser abundance as circOprm1.e2.e3.e5(94).e4(302) and circOprm1.e2.e3.e5(119).e4(302) (Supplemental Fig. 3). All of these circRNAs adhere the GT-AG rule of consensus splice site recognition (Scotti and Swanson, 2016), suggesting that these circRNAs are generated through mechanisms of canonical splicing or alternative splicing. Other divergent RT-PCR products remain to be sequence verified, and likely represent unknown Oprm1 circRNAs. Thus, the mouse Oprm1 gene generates a dominant circRNA isoform containing e2 and e3, while other variant circRNAs are also generated at lower levels. Although not quantified, circRNA isoforms were also observed in female mice (Supplemental Fig. 3).
Evolutionary Conservation of Oprm1 e2-e3 Circular RNA Isoforms.
Evolutionary conservation of many circRNA has been noticed in large-scale transcriptomic projects (Jeck et al., 2013). To evaluate the evolutionary conservation of the circOprm1.e2.e3 isoform, we performed divergent RT-PCR using primers oriented outward from e2 and separately from e3 of the rat and human Oprm1/OPRM1 genes. Using total RNA from whole brain, we confirmed the existence of homologous e2.e3 circRNA isoforms in rat and human brains by divergent RT-PCR and Sanger sequencing (Fig. 1, C and D). Furthermore, total RNA from spinal cord of mouse, rat, and human also contained Oprm1/OPRM1 e2.e3 circRNA (Fig. 1, B–D). Of note, the e2.e3 circRNAs are exactly 874 nt in all three species due to conservation of e2 and e3 lengths among these species. The mouse e2.e3 circRNA has an open reading frame that theoretically could encode a six-transmembrane variant of 365 amino acids, from a start codon in e3 reading through e2 and e3 until reaching a stop codon in the 5′ half of e2 during a second pass through the circRNA sequence (Supplemental Fig. 1). The rat and human e2.e3 circRNAs have longer theoretical open reading frames of 583 and 396 amino acids, respectively, with both retaining six-transmembrane spans predicted by TMHMM2 (Sonnhammer et al., 1998), albeit with a long C-terminal sequence in the rat open reading frame (Supplemental Fig. 4). The rat brain and spinal cord also weakly express circRNA isoforms roughly 100 and 200 nt larger than the e2.e3 isoform, while the human RNAs reveal more bands, suggesting a more diverse repertoire of circRNA isoforms (Fig. 1D). Overall, these studies indicate the Oprm1/OPRM1 genes of all three vertebrates express a conserved e2.e3 circRNA.
Confirmation of Circularity and Strand of Origin.
As further evidence supporting the existence of a circRNA variant of Oprm1, we used the RNase R/RT-PCR assay (Salzman et al., 2012) to selectively degrade linear RNA from our total RNA pool prior to RT-PCR. The RNase R enzyme is 10-fold more active against linear RNA than against circular RNA (Suzuki et al., 2006). We observed a reduction of the e1-e2 convergent RT-PCR product with RNase R pretreatment, while the e2- and e3-based divergent RT-PCRs were resistant to RNase R degradation (Fig. 2, C and D), confirming the circular nature of these RNA templates.
Exonic circRNAs predominantly originate from the sense strand of genes, with only approximately 10% of circRNAs detected by RNA-sequencing studies antisense to the primary annotations at their gene of origin (Memczak et al., 2013). To confirm the strand of origin of circOprm1.e2.e3, we used strand-specific primers to the Oprm1 exonic sequence for reverse transcription, and then evaluated the e2 divergent RT-PCR amplicons. Using this strand-specific RT-PCR strategy, we confirmed that circOprm1.e2.e3 derived from the sense strand Oprm1 transcript (Fig. 2, E and F).
Analyses of Candidate cis Regulatory Sequences within Oprm1 Introns that May Facilitate Circularization.
The biogenesis of circRNAs involves interactions between introns flanking the circRNA exons that may be due to base pairing of inverted repeat elements such as Alu (human) and B1 (mouse) repetitive elements or through the presence of binding sites for sequence-specific RNA binding proteins such as Quaking (Qki) and Muscleblind (Ashwal-Fluss et al., 2014; Conn et al., 2015). Since the Oprm1 intron between e1 and e2 is ∼40 kb long and the intron between e3 and e4 is ∼19 kb long (with e5 interjected 11 kb downstream from e3) (Fig. 1A), respectively, many B1 elements and other repeat sequences were readily visible via the UCSC genome browser, consistent with their possible role in the biogenesis of these circRNAs. On the other hand, the high number of repeat elements in these introns makes the evaluation of their individual contributions to circRNA biogenesis difficult.
We also investigated the flanking intron sequences for Qki and Mbnl2 binding sites using the MEME Suite of motif analyses tools; Mbnl2 is reported to be the brain-dominant mammalian homolog of Drosophila Muscleblind dominant in the brain. Four instances of potential Qki binding sites were detected within the 20 kb of sequence upstream of Oprm1 e2 (14.1, 11.9, 7.7, and 3.3 kb upstream of e2), and again in the 20 kb downstream from e3 (1.8, 6.1, 13.9, and 15.2 kb downstream), suggesting that the mouse e2.e3 circRNA may employ Qki-dependent mechanisms for circularization (Supplemental Fig. 5 and Supplemental Material for Supplemental Figs. 5 and 6). Similar analyses for Mbnl2 binding sites within the same Oprm1 intronic sequences also revealed consensus Mbnl2 binding sites within both the upstream and downstream introns (one site 7.6 kb upstream of e2 and two sites downstream from e3 at 13.6 and 16.4 kb downstream of e3) (Supplemental Fig. 6 and Supplemental Material for Supplemental Figs. 5 and 6). Experimental verification of these predicted RNA binding protein interactions is warranted.
δ (Oprd1) and κ (Oprk1) Opioid and Nociceptin (Oprl1) Receptor circRNAs.
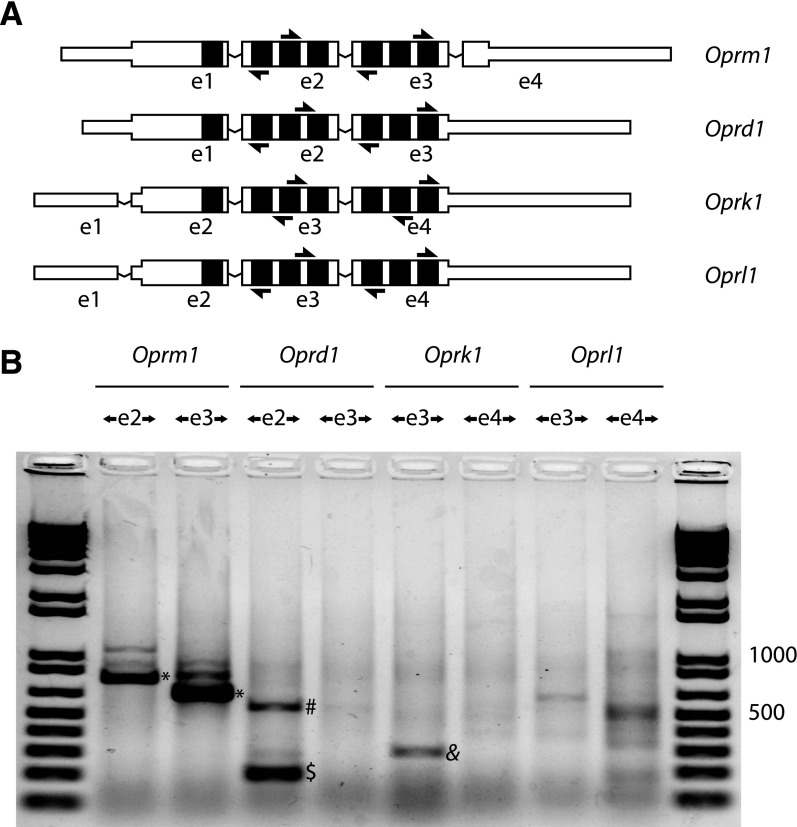
We also observed circRNAs for the δ (Oprd1), κ (Oprk1), and nociceptin (Oprl1) receptor genes as well (Fig. 3). These studies used divergent RT-PCR from primers within the exons homologous to Oprm1 e2 and e3 (see Fig. 3A). Oprd1 e2 and Oprk1 e3 (which are homologous to Oprm1 e2) generated single exon circRNAs, although we failed to observe any single exon circRNAs from Oprm1 (Fig. 3B). Oprl1 also generated a low yield divergent RT-PCR product containing e3 and two discontinuous portions of e4, with splice junctions adhering to the GT-AG rule, but this amplicon was detected less reproducibly (data not shown).
Fig. 3.
Mouse non-μ-opioid receptor divergent RT-PCRs. (A) A schematic of mouse μ (Oprm1), δ (Oprd1), κ (Oprk1), and nociceptin (Oprl1) receptor mRNAs for the dominant splice isoforms is shown, with exons labeled below (exon 1 is designated as e1). The mRNAs are organized by alignment of homologous exons, with transmembrane span coding regions colored black. Divergent RT-PCR primers for each target sequence are shown as arrows. Exons are not drawn to scale; narrow portions of exons represent untranslated regions. (B) Divergent RT-PCR was performed on mouse whole brain total RNA with PCR primer pairs directed outwardly from exons as labeled. For each gene, two primer pairs were used, targeting two separate exons. The Oprm1 locus generates several circRNAs, of which the e2.e3 circRNA appears dominant (*). Oprd1 e2 generates a single exon circRNA (dollar), and this template supported detection of a dimeric RT-PCR product consistent with processive reverse transcription of a single exon circRNA (#). Oprk1 also generates a single exon e3 circRNA (&). These divergent RT-PCRs were confirmed to derive from circRNAs by examination of backsplice sequences from direct Sanger sequencing, or after TOPO cloning, to confirm the existence of a backsplice junction and adherence to canonical GT-AG splice junctions. Five micrograms of total RNA were treated with reverse transcriptase using 250 ng random hexamers in 20 μl reactions, and 35 cycles of PCR amplification were done with 25 ng/μl input cDNA. Five microliters of PCR reaction were loaded against 1 μl of Invitrogen 1 kb Plus ladder; 500 and 1000 bp are labeled as size reference.
Anatomic Specificity of circOprm1 Abundance.
Oprm1 alternative splice variants are differentially expressed in various brain regions (Xu et al., 2015). To determine if the regulation of circRNA expression also was region dependent, we evaluated mouse whole brain and spinal cord total RNAs for linear versus circRNA variants of Oprm1 by quantitative RT-PCR. SYBR green qPCRs targeting three Oprm1 amplicons were performed. Since divergent RT-PCR from e1 to e2 failed to generate amplicons (Fig. 2C), we interpreted products from e1 to e3 as deriving from linRNA isoforms, while circOprm1.e2.e3 and circOprm1.e2.e3.e4(302) were detected using backsplice junction spanning primers (Fig. 2A). The ΔCt values are shown in Fig. 4B, and these values are shown again as relative RNA abundance (2−ΔΔCt values) in Fig. 4C. The latter values were analyzed by two-way ANOVA, with significant variation attributable to anatomic region [whole brain vs. spinal cord; F(1,4) = 92.68, P = 0.0007] and RNA target [circOprm1.e2.e3 vs. circOprm1.e2.e3.e4(302); F(1,4) = 163.5, P = 0.002] as well as interaction [F(1,4) = 76.73, P = 0.009]. Subjects (matching) were not a significant source of variation [F(4,4) = 3.045, P = 0.1531]. The abundances of both circRNA isoforms were greater in brain than spinal cord, with circOprm1.e2.e3 being more abundant of the two. The observation that the abundance of circOprm1.e.2.e3 in brain was similar to linear mRNA levels was unanticipated, as was the marked differences between circRNA expression in brain and spinal cord. Together, these results imply that linear and circRNA isoforms are differentially regulated post-transcriptionally.
We also examined mouse brain regions for circRNA expression, including cortex, striatum, diencephalon (thalamus/hypothalamus), periaqueductal gray, and brain stem, and found significant differences in the ratios of Oprm1 linRNA versus e2.e3 circRNA in brain regions (Fig. 4D) [one-way ANOVA with no matching, F(6,14) = 2.953, overall P = 0.0446], with the highest ratio of circOprm1.e2.e3 isoforms in the cortex and the lowest in the brain stem (post-hoc multiple comparisons testing with Tukey correction, P = 0.0306 for cortex vs. brain stem) (Fig. 4D). In the cortex, circOprm1.e2.e3 levels exceeded linRNA levels, whereas in the brainstem the opposite was true. The regional distributions in brain revealed a trend, with higher fractions of circOprm1.e2.e3 in rostral regions that declined in more caudal regions, consistent with the comparison of whole brain and spinal cord (Fig. 4C). The high level of circOprm1.e2.e3 observed in whole brain may be in part due to high cortical contribution to whole brain mass, although other technical factors may have contributed as well.
Effect of Chronic Morphine Exposure on Relative Abundance of Oprm1 circRNAs.
Chronic morphine produces a range of compensatory responses, including physical dependence and tolerance. These physiologic changes are associated with changes in the regulation of Oprm1 mRNA, with levels of specific isoforms in select brain regions increasing as much as 300-fold (Xu et al., 2015). In view of the potential role of circRNA in regulation of μ-opioid receptor expression, we examined the effects of chronic morphine on Oprm1 circRNA and linRNA isoforms. We used a 10-day chronic morphine treatment paradigm consisting of subcutaneously implanted 75 mg morphine or placebo pellets (Fig. 5). Morphine treatment significantly increased circOprm1.e.2.e.3 (1.6-fold, P < 0.05 by t test) and circOprm1.e2.d3.e4(302) levels (1.5-fold, P < 0.05 by t test) in brain, but not spinal cord (Fig. 5, A and B). Meanwhile, no change was observed in whole brain linRNA in either region (Fig. 5C). The lack of change in whole brain linRNA is consistent with our earlier study employing 6-week treatments (Xu et al., 2015). While that study did observe changes for Oprm1 mRNA expression, they were only seen when assessing specific individual regions. Here, the ratio of circular/linear forms in whole brain increased also [circOprm1.e.2.e.3/linRNA, 1.6-fold, circOprm1.e2.d3.e4(302)/linRNA, 1.5-fold], indicating circRNA regulation was independent of linear Oprm1 mRNA expression. This relative increase in circular isoforms raises questions about their potential involvement in μ receptor gene regulation with chronic opioid treatment.
miRNA Binding Site Predictions.
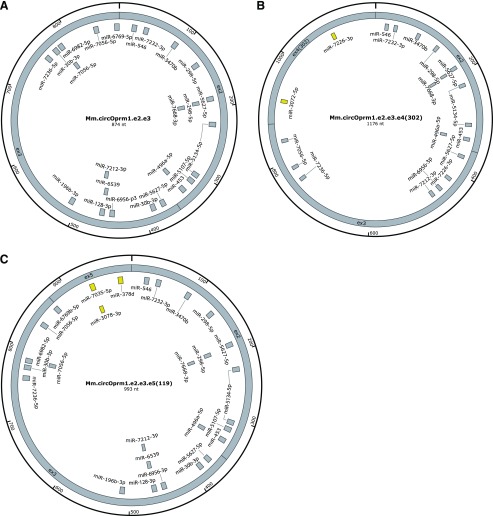
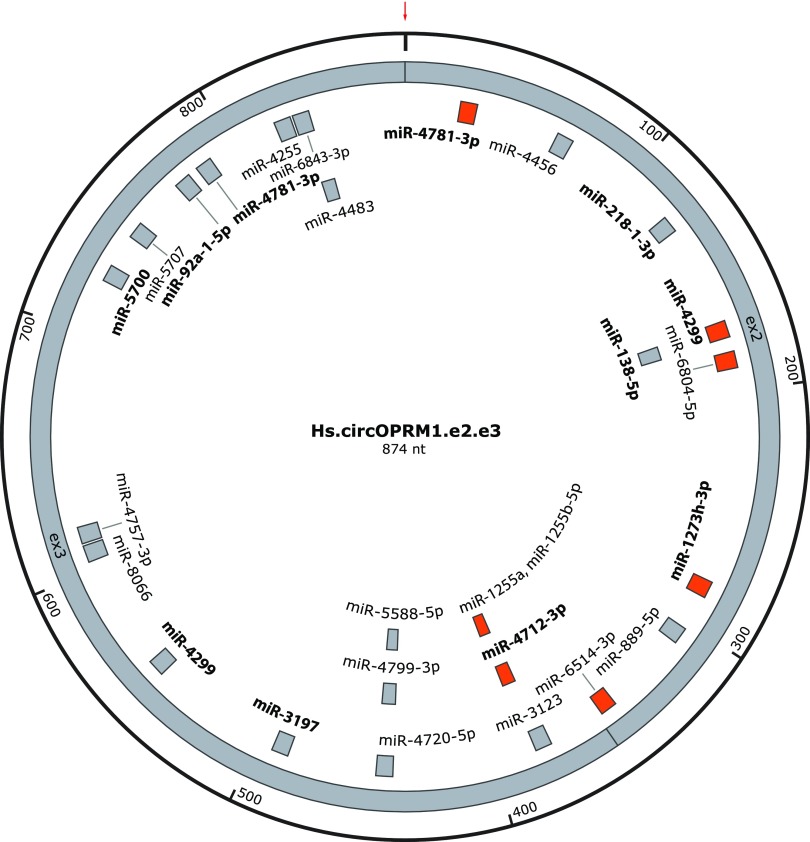
miRNAs can modulate gene expression by selectively binding to specific mRNA sequences and thereby influencing mRNA stability and/or translatability. Thus, altering miRNA levels provides a mechanism to influence protein expression. Typically, miRNAs are 22 bases long and hybridize with their target RNAs through binding mediated by a core 6 to 7 bp interaction through the seed sequence (Gorski et al., 2017). Some circRNAs operate as decoys for miRNA binding, titrating the availability of important regulatory miRNAs and achieving an indirect regulatory effect. Bioinformatic miRNA binding site analysis of the Oprm1 circRNAs predicted numerous putative miRNA binding sites (Fig. 6). The miRNA binding sites predicted within e2 and e3 are shared by all three circRNAs, with the sole exception of miR-6769b-5p, which was predicted only for the e2.e3 and e2.e3.e5(119) circRNAs (Fig. 6). We also predicted miRNAs specific for the e2.e3.e4(302) and e2.e3.e5(119) circRNAs, and absent in the e2.e3 circRNA. For example, miR-3072-5p and miR-7226-3p are specific for the exon 4 portion of e2.e3.e4(302) (Fig. 6B), while miR-7035-5p, miR-3078-3p, and miR-378d are predicted to target the exon 5 portion included within e2.e3.e5(119) (Fig. 6C). Predictions for miRNA binding sites within the human OPRM1 e2.e3 circRNAs (Fig. 7) revealed several predicted miRNA binding sites with 100% nucleotide level conservation between the mouse and human e2.e3 circRNAs. For miRNAs predicted in mouse, eight out of 25 sites have 100% nucleotide conservation of the miRNA seed sequences in the human e2.e3 circRNA, while for miRNAs predicted in the human e2.e3 circRNA, seven out of 27 are 100% conserved in mouse at the seed sequence positions. The contribution of high nucleotide level conservation of the 874 nucleotides in the e2.e3 circRNA (770 of 874 between mouse and human) to the fraction of predicted miRNA binding sites with 100% interspecies conservation could not be excluded (P = 0.68 for the eight out of 25 sites of mouse versus human sequence, P = 0.88 for human to mouse). Collectively, these analyses suggest several candidate miRNA binding sites exist within the evolutionarily conserved Oprm1/OPRM1 e2.e3 circRNA, as well as others specific for the lesser variant circRNAs.
Fig. 6.
Predicted miRNA binding sites in mouse Oprm1 circRNAs. Potential miRNAs targeted by circOprm1.e2.e3 (A), circOprm1.e2.e3.e4(302) (B), and circOprm1.e2.e3.e5(119) (C) are shown. Shared miRNA binding sites are colored in gray, while sites unique for the circOprm1.e2.e3.e4(302) and circOprm1.e2.e3.e5(119) circRNAs are yellow. Circle sizes not to scale.
Fig. 7.
Predicted miRNA binding sites within human circOPRM1.e2.e3. A map of predicted miRNA binding sites within the human [Homo sapiens (Hs)] Hs.circOPRM1.e2.e3 is shown. Those miRNAs with experimental validation (in other contexts) are labeled in bold. Shown in orange are the predicted miRNA binding sites that are 100% conserved for the full heptamer miRNA seed sequence vs. the mouse homolog (Mm.circOPRM1.e2.e3). The red arrow depicts the location of the backsplice junction.
Discussion
Oprm1 circRNA is abundant and differentially modulated by chronic morphine, independently of Oprm1 linear RNAs. The dominant mouse circOprm1 backsplices exon 3 to exon 2 (circOprm1.e2.e3), and a secondary isoform backsplices the first 302 nucleotides of exon 4 to exon 2 [circOprm1.e2.e3.e4(302)]. Exon 5 inclusion into the third most abundant isoform confirms Oprm1 circRNAs can incorporate alternative exons; other incompletely characterized forms were observed. Human OPRM1 e2.e3 circRNAs were also detected in brain and spinal cord albeit at levels lower than rodents, possibly reflecting postmortem changes in RNA, differences in anatomic sampling, evolutionary divergence, or individual variation. Regardless of quantitative differences, e2.e3 circRNAs from mouse, rat, and human are evolutionarily conserved. They join a list of circRNAs with potential involvement in physiologic and pathologic states, including pain (Cao et al., 2017), ischemic stroke (Mehta et al., 2017), cancer (Li et al., 2015a; Xia et al., 2018), neurodevelopment (Szabo et al., 2015), and cardiac injury (Gupta et al., 2018).
Oprm1 generates circRNAs at levels equal to or exceeding linRNAs in rostral brains, raising questions about previous analyses of Oprm1 mRNA that did not distinguish linRNAs versus circRNAs (Meuser et al., 2003). Our observation that circOprm1 levels increased in brain by ∼50% after 10-day morphine exposure highlights the importance of disambiguating Oprm1 linRNA from circRNAs when studying Oprm1 gene expression. Previously, 6 weeks of morphine treatment robustly increased specific Oprm1 variant mRNAs in selected brain regions (Xu et al., 2015). However, circOprm1 contributions to those changes are not known, since the probes employed cannot differentiate circOprm1 versus linRNAs. It will be interesting to revisit this long-term morphine treatment paradigm to quantitate Oprm1 circRNAs versus linRNAs. circRNAs are more stable than linear RNAs (Wang and Wang, 2015), thus are particularly well poised to affect long-term pharmacological outcomes such as opioid tolerance (Meuser et al., 2003; Xu et al., 2015).
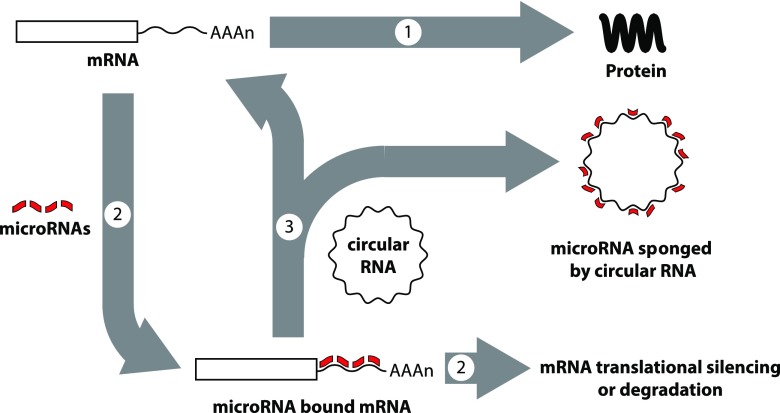
Overall, several functions of circRNAs are reported, including as miRNA sponges (Fig. 8) (Hansen et al., 2013; Memczak et al., 2013; Wang et al., 2016; Zheng et al., 2016), RNA binding protein depots, splicing modulators (Ashwal-Fluss et al., 2014), and a template for protein translation (Abe et al., 2015; Wang and Wang, 2015). The model circRNA CDR1as has 60–70 miR-7 binding sites, and transgenic overexpression of CDR1as circRNA in zebrafish impaired midbrain development, phenocopying miR-7 morpholino knockdown (Memczak et al., 2013). High copy numbers of miRNA sites may be atypical of circRNAs (Guo et al., 2014; Jeck and Sharpless, 2014) but low copy numbers of miRNA sites can be compensated by high levels of circRNA expression and circRNA stability. The heart-related circRNA only encodes six miR-223 binding sites, but its overexpression blocked cardiac hypertrophy in a heart failure model (Wang et al., 2016). CircHIPK3 is abundantly expressed in liver cancer and sponges nine different miRNAs, some of which exist as single copy miRNA sites (Zheng et al., 2016).
Fig. 8.
Post-transcriptional gene regulation by microRNAs and circular RNAs. Gene expression involves translation of mRNAs to proteins (arrow 1), but the process can be blocked when microRNAs bind to mRNAs through sequence complementarity (arrow 2). Depending on the microRNA sequences and the presence or absence of bulges in the binding due to the presence or absence of perfect matches in complementarity, mRNAs may become translationally silenced or targeted for degradation (arrow 2). However, circular RNAs also can bind and sequester microRNAs, lowering their ability to target mRNAs, a process termed sponging, which can block the inhibitory effects of microRNAs (arrow 3).
Many predicted miRNA binding sites within Oprm1/OPRM1 e2.e3 circRNA are conserved between mouse and human at the nucleotide level, suggesting evolutionary significance. Of the predicted miRNA binding sites conserved between mouse and human e2-e3 sequences, several are experimentally verified miRNAs (although not yet verified to interact with circOprm1), including mouse miR-546 and miR-3470b as well as human miR-4781-3p and miR-4299. Hippocampal neuronal plasticity in cellulo is regulated by miR-546 (van Spronsen et al., 2013), lentiviral transgenesis of miR-3470b in peripheral tissues led to increased inflammatory pain behaviors in mice (Segall et al., 2015), dysregulation of miR-4781-3p is present in serum from Alzheimer’s patients (Satoh et al., 2015), and miR-4299 levels are associated with thyroid cancer pathogenesis and colon cancer chemoresistance (Hu et al., 2015; Miao et al., 2016). The high abundance of circOprm1.e2.e3 could empower it to sponge important miRNAs despite low copy numbers of each miRNA binding site. It will be important to test these predicted circRNA-miRNA interactions directly.
Rarely, circRNAs can encode proteins (Legnini et al., 2017; Pamudurti et al., 2017). Typical eukaryotic protein translation requires ribosome recruitment to 5′ ends of mRNAs via interactions between translation initiation factors and the 7-methylguanosine cap at the 5′ termini (Sonenberg and Hinnebusch, 2009), and is aided by de facto circularization by bridging protein-protein interactions between translation initiation factors and poly-A binding protein (Wells et al., 1998). circRNAs lack 5′ caps and are largely excluded from polysomes (Nigro et al., 1991; Jeck et al., 2013), but some are polysomal and their encoded proteins are detectable by mass spectrometry (Legnini et al., 2017; Pamudurti et al., 2017). Translation of circRNAs may be mediated by internal ribosome binding sites or 6-methyladenine RNA modifications, which both bypass cap-dependent translation (Meyer et al., 2015; Legnini et al., 2017; Yang et al., 2017). Alternatively, some circRNAs might be translated after linearization (Otsuka et al., 2009; Mukherjee et al., 2012), cytoplasmic capping, and polyadenylation (for a review, see Elkon et al., 2013). Such a mechanism could allow for long-term RNA storage and delayed protein translation. The circRNA CDR1as can be linearized by binding of miR-671 at a single copy binding site (Hansen et al., 2011). circOprm1.e2.e3 has a predicted miRNA binding site at the end of e3. RNA linearized at this position could encode six-transmembrane opioid receptor isoforms similar to the MOR-1K isoform implicated in opioid-induced hyperalgesia and tolerance (Oladosu et al., 2015; Marrone et al., 2017). Although speculative, translation of Oprm1 circRNAs is an intriguing possibility.
The cis-regulatory sequences and trans-acting factors required for biogenesis of some circRNAs are known, although not for circOprm1. Long intronic inverted repeat (IR) sequences flanking the circularized exon in the mouse sex-determining gene Sry are necessary for circular Syr RNA formation (Capel et al., 1993; Dubin et al., 1995). Genome-wide RNA-sequencing extended this observation, and exons included in human circRNAs are flanked by longer introns enriched for IRs (Jeck et al., 2013; Salzman et al., 2013; Zhang et al., 2014) and these IRs can be Alu elements (Tsirigos and Rigoutsos, 2009) or nonrepeat inverted complementary sequences (Zhang et al., 2014). circRNAs typically do not include first and last exons (Zhang et al., 2014). RNA binding proteins also contribute to circRNA formation, including sequence-specific RNA binding proteins Quaking and Muscleblind that bind to introns bracketing the circRNA exons (Ashwal-Fluss et al., 2014; Conn et al., 2015), as well as serine-arginine proteins and heterogeneous nuclear ribonucleoproteins in fruit fly circRNA formation (Kramer et al., 2015). The structure of mouse circOprm1.e2.e3 is consistent with known general rules of circRNA formation, such as exclusion of the first exon, presence of long introns (introns flanking e2 and e3 of Oprm1 are 40 and 19 kb, and the intron between e2 and e3 is 0.9kb) (Supplemental Fig. 1A), and existence of numerous repetitive elements in these spans that could contribute to the circularization of these exons through an IR-dependent mechanism. Sequences for Quaking and Mbnl2 binding sites are present in mouse Oprm1 introns flanking the e2-e3 exon region (Supplemental Fig. 3), suggesting their possible involvement in e2.e3 circRNA formation, but these observations warrant validation. The intron spans of non-μ-opioid receptor genes are smaller than those of Oprm1, possibly contributing to lower circRNA levels of non-μ genes. Our divergent RT-PCR primers for non-μ genes targeted only two exons, chosen based on protein level homology, leaving open the possibility of other circRNAs outside the scope of our designs.
circRNAs may be eliminated by dilution, enzymatic degradation, and cellular extrusion. Eukaryotic mRNA stability depends largely on 3′ exonucleolytic degradation by the exosome (Kilchert et al., 2016). circRNAs are long lived due to their lack of free 3′ termini for exonucleolytic attack (Wang and Wang, 2015). Accumulation in aged fly and mouse brains may result from minimal dilution by cell division (Westholm et al., 2014; Gruner et al., 2016). The lack of dilution in postmitotic neurons may be more important than in other cell types since accumulation of circRNAs can be cytotoxic (Errichelli et al., 2017). Conversion to linear forms via endonucleolytic cleavage, seen with CDR1as circRNA via a miRNA/Ago2-mediated mechanism (Hansen et al., 2011), may restore access to standard RNA outcomes. Finally, vesicular extrusion of circRNA in exosomes (not the mRNA degrading enzyme complex of the same name) is also documented (Li et al., 2015a). Exosomal circRNAs enriched in plasma of colon cancer patients may be useful biomarkers (Li et al., 2015a). The potential utility of exosomal circOprm1 as a tolerance biomarker remains unexplored.
In conclusion, the high abundance of μ-opioid receptor circRNA isoforms, their conservation from rodent to human, their differential expression in various brain regions, and their modulation by morphine imply the functional significance of Oprm1 circRNAs, and open questions regarding their potential role in regulation of miRNAs and linear Oprm1 mRNAs, as well as morphine tolerance. The detectability of circRNA isoforms of the other opioid receptor genes as well as the abundance of circRNAs of Drosophila homologs of the serotonin receptor gene (5-HT2A) and the metabotropic glutamate receptor gene (mGluRA) (Salzman et al., 2013; Westholm et al., 2014) argue for the general importance of circRNAs in G protein-coupled receptor pharmacology.
Abbreviations
- circOprm1
Oprm1 circular RNA
- circRNA
circular RNA
- e1
exon 1
- e2
exon 2
- e3
exon 3
- e4
exon 4
- e5
exon 5
- IR
inverted repeat
- linRNA
linear mRNA
- miRNA
microRNA
- nt
nucleotide
- PCR
polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- RT
reverse transcription
Authorship Contributions
Participated in research design: Irie, Wilson, Fischer, Pasternak, Pan.
Conducted experiments: Irie, Shum, Deni, Hunkele, Le Rouzic, Xu.
Performed data analysis: Irie, Pasternak, Pan.
Wrote or contributed to the writing of the manuscript: Irie, Wilson, Fischer, Pasternak, Pan.
Footnotes
This work was supported, in part, by funds from the Department of Anesthesiology (to T.I.), the National Institutes of Health National Institute on Drug Abuse [Grants DA06241, DA07242, DA042888, DA040858 and DA046714], the Mayday Foundation, the Peter F. McManus Charitable Trust, and a core grant from the National Institutes of Health National Cancer Institute [Grant CA008748] to the Memorial Sloan-Kettering Cancer Center.
This article is dedicated to the memory of our dear mentor, colleague, and friend, Dr. Gavril W. Pasternak (deceased February 22, 2019), who contributed immensely to our understanding of opioid pharmacology.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al. (2015) Rolling circle translation of circular RNA in living human cells. Sci Rep 5:16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. [DOI] [PubMed] [Google Scholar]
- Cao S, Deng W, Li Y, Qin B, Zhang L, Yu S, Xie P, Xiao Z, Yu T. (2017) Chronic constriction injury of sciatic nerve changes circular RNA expression in rat spinal dorsal horn. J Pain Res 10:1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–1030. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B. (1992) Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. (1995) Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167:245–248. [DOI] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14:496–506. [DOI] [PubMed] [Google Scholar]
- Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R, Peruzzi G, et al. (2017) FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun 8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. (2016) Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 7:12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Vogel J, Doudna JA. (2017) RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol 18:215–228. [DOI] [PubMed] [Google Scholar]
- Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P. (2016) CircRNA accumulation in the aging mouse brain. Sci Rep 6:38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J, Thum T. (2018) Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res 122:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. (2011) miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30:4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Xu Y, Cai S. (2015) Specific microRNAs as novel biomarkers for combination chemotherapy resistance detection of colon adenocarcinoma. Eur J Med Res 20:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. (2002) The human genome browser at UCSC. Genome Res 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L. (2016) The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol 17:227–239. [DOI] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C, Ng L, Thompson C, Pathak S, Kuan L, Jones A, Hawrylycz M. (2008) Exploration and visualization of gene expression with neuroanatomy in the adult mouse brain. BMC Bioinformatics 9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. (2017) Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol Cell 66:22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. (2015a) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. (2015b) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu J, Xu M, Pasternak GW, Pan YX. (2014) Morphine regulates expression of μ-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the μ-opioid receptor (OPRM1) gene via miR-103/miR-107. Mol Pharmacol 85:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone GF, Le Rouzic V, Varadi A, Xu J, Rajadhyaksha AM, Majumdar S, Pan YX, Pasternak GW. (2017) Genetic dissociation of morphine analgesia from hyperalgesia in mice. Psychopharmacology (Berl) 234:1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SL, Pandi G, Vemuganti R. (2017) Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke 48:2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. [DOI] [PubMed] [Google Scholar]
- Meuser T, Giesecke T, Gabriel A, Horsch M, Sabatowski R, Hescheler J, Grond S, Palmer PP. (2003) Mu-opioid receptor mRNA regulation during morphine tolerance in the rat peripheral nervous system. Anesth Analg 97:1458–1463. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. (2015) 5′ UTR m6A promotes cap-independent translation. Cell 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X, Jia L, Zhou H, Song X, Zhou M, Xu J, Zhao L, Feng X, Zhao Y. (2016) miR-4299 mediates the invasive properties and tumorigenicity of human follicular thyroid carcinoma by targeting ST6GALNAC4. IUBMB Life 68:136–144. [DOI] [PubMed] [Google Scholar]
- Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR. (2012) Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep 2:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. (1991) Scrambled exons. Cell 64:607–613. [DOI] [PubMed] [Google Scholar]
- Oladosu FA, Conrad MS, O’Buckley SC, Rashid NU, Slade GD, Nackley AG. (2015) Mu opioid splice variant MOR-1K contributes to the development of opioid-induced hyperalgesia. PLoS One 10:e0135711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR. (2009) Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29:2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. (2017) Translation of circRNAs. Mol Cell 66:9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Pan YX. (2013) Mu opioids and their receptors: evolution of a concept. Pharmacol Rev 65:1257–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J, Kino Y, Niida S. (2015) MicroRNA-seq data analysis pipeline to identify blood biomarkers for Alzheimer’s disease from public data. Biomark Insights 10:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. (2016) RNA mis-splicing in disease. Nat Rev Genet 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall SK, Shabalina SA, Meloto CB, Wen X, Cunningham D, Tarantino LM, Wiltshire T, Gauthier J, Tohyama S, Martin LJ, et al. (2015) Molecular genetic mechanisms of allelic specific regulation of murine Comt expression. Pain 156:1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos A, Rigoutsos I. (2009) Alu and B1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLOS Comput Biol 5:e1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, van Battum EY, Kuijpers M, Vangoor VR, Rietman ML, Pothof J, Gumy LF, van Ijcken WF, Akhmanova A, Pasterkamp RJ, et al. (2013) Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS One 8:e74907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Z. (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2:135–140. [DOI] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S, Koch T, Evans CJ, Christie MJ. (2013) Regulation of μ-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol Rev 65:223–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Wang X. (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, et al. (2018) CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res 46:D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Faskowitz AJ, Rossi GC, Xu M, Lu Z, Pan YX, Pasternak GW. (2015) Stabilization of morphine tolerance with long-term dosing: association with selective upregulation of mu-opioid receptor splice variant mRNAs. Proc Natl Acad Sci USA 112:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]