Abstract
Corylin, an phenolic compound from Psoralea corylifolia, has been reported with various pharmacological properties but has poor bioavailability due to massive metabolism. In this study, twelve metabolites of corylin mainly involving in oxidation, hydration, glucuronidation and sulfation were detected in mice. Furthermore, the oxidation and hydration of corylin (M4) in human liver microsomes (HLM) and human intestine microsomes (HIM) were both efficient with high CLint (intrinsic clearance) values of 24.29 and 42.85 μL/min/mg, respectively. CYP1A1, 1B1 and 2C19 contributed most for M4 with the CLint values of 26.63, 33.09 and 132.41 μL/min/mg, respectively. Besides, M4 was strongly correlated with phenacetin-N-deacetylation (r = 0.885, p = 0.0001) and tolbutamide-4-oxidation (r = 0.727, p = 0.001) in twelve individual HLMs, respectively. In addition, corylin was efficiently glucuronidated (M7) in HLM (125.33 μL/min/mg) and in HIM (108.74 μL/min/mg). UGT1A1 contributed the most for M7 with the CLint value of 122.32 μL/min/mg. Meanwhile, M7 was significantly correlated with β-estradiol-3-O-glucuronidation (r = 0.742, p = 0.006) in twelve individual HLMs. Moreover, the metabolism of corylin showed marked species differences. Taken together, corylin was subjected to massive first-pass metabolism in liver and intestine, while CYP1A1, 1B1, 2C19 and UGT1A1 were the main contributors. Finally, the proposed metabolic pathway of corylin involed CYP and UGT isoforms were summarized, which could help to understand the metabolic fate of corylin in vivo.
Keywords: Corylin, Metabolites profile, Cytochromes P450, UDP-glucuronosyltransferase, Species differences
1. Introduction
Corylin is a phenolic compound isolated from the whole plant, fruit and seed of Psoralea corylifolia L., which has been widely used as a kidney tonifying herbal medicine mainly for anti-osteoporosis and also considered as a dietary supplement [1]. Meanwhile, it accounts for 0.06% weight of the dried seeds [2]. Besides, it has attracted increasing interest for significant pharmacological effects in regulating antioxidant activity [3], inhibiting aldose reductase [4], and osteoblastic proliferation-stimulating activity [5]. In addition, corylin could inhibit interleukin-6 (IL-6)-induced signal transducer and activator of transcription 3 (STAT3) activity in hepatocarcinoma Hep3B cells [6]. Recently, corylin was reported to be a novel anti-inflammatory and immunosuppressive drug candidate in the treatment of sepsis and septic shock [7].
To date, numerous researches of corylin-containing herbal medicines have been conducted in the fields of metabolite profiles, pharmacokinetics and drug metabolizing enzyme inhibition. Previous studies indicated that corylin could undergo extensive phase II metabolism (mainly glucuronidation and sulfonation) [8,9]. In addition, it could be quickly absorbed into rat plasma and distributed almost evenly to the cerebral nuclei [10,11]. And also, the reports suggested that the prenylflavonoids could more readily enter the brain than the coumarins in Psoraleae Fructus [11]. Furthermore, it was shown that corylin could exhibit strong inhibition towards human carboxylesterase 1 (IC50 = 0.7 μM) [12] and UDP-glucuronosyltransferase 1A9 (Ki = 3.1 μM) [13,14]. However, in spite of the remarkable biological activities, there are no reports about the metabolism of corylin alone.
For this purpose, the metabolic pathways of corylin in vivo and in vitro would be investigated in the present study. An ultra-performance chromatography coupled with time-of-flight tandem mass spectrometry (UPLC/q-tof-MS) method was applied to (i) elucidate the complete metabolic pathway of corylin, including the identification of all phase I and phase II metabolites, (ii) determine the contribution of human individual CYPs and UGTs enzymes to phase I and phase II metabolism, and (iii) compare the species differences of liver microsomes-mediated metabolism. This study would be helpful for the understanding of the metabolic fate of corylin.
2. Experimental
2.1. Chemicals and reagents
Nicotinamide adenine dinucleotide phosphate (NADPH), uridine diphosphate glucuronic acid (UDPGA), magnesium chloride (MgCl2), alamethicin and D-saccharic-1, 4-lactone were all provided from Sigma-Aldrich (St Louis, MO). Pooled human liver microsomes (HLM), twelve individual pooled human liver micro-somes (iHLM), pooled human intestine microsomes (HIM), rat liver microsomes (RLM), mice liver microsomes (MLM), guinea pig liver microsomes (GpLM), monkey liver microsomes (MkLM), dog liver microsomes (DLM), rabbit liver microsomes (RaLM), expressed human CYPs (CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4 and 3A5) and human UGTs (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 2B4, 2B7, 2B10, 2B15 and 2B17) were all obtained from Corning Biosciences (New York, USA). Corylin (purity > 98%) were purchased from Chengdu Chroma-Biotechnology Co., Ltd. (Chengdu, China). β-estradiol, chenodeoxycholic acid (CDCA), phenacetin and tolbutamide were purchased from Aladdin Chemicals (Shanghai, China). All other chemicals and reagents were analytical grade commercially available.
2.2. In vivo treatment and samples preparation
Male SPF grade KM mice (6- to 8-week-old) were obtained from the Experimental Animal Center of Guangdong Province (Guangzhou, China). The mice were maintained in an animal room with constant temperature (23 ± 2) °C, humidity (60 ± 5) % and a 12 h dark/light cycle for a week before the tests. And all mice had access to water and food ad libitum. The animal protocols were approved and conducted in accordance with the guidelines of Laboratory Animal Ethics Committee of Jinan University. All procedures were in accordance with Guide for the Care and Use of Laboratory Animals (National Institutes of Health).
Eighteen mice were divided into two groups: control group (n = 9) and corylin-treated group (n = 9). Based on our previous study [9], corylin dissolved in 0.3% sodium carboxymethyl cellulose was given by oral administration at a dose of 40 mg/kg body weight, and control mice were treated with 0.3% sodium carboxymethyl cellulose only. After oral gavage, all mice were observed whether abnormal manifestations happened. Blood samples (n = 3) were collected into heparinized tubes by retroorbital bleeding at 0.5 and 6 h after oral administration, and plasma samples were separated by centrifugation at 8000g for 10 min. The mice (n = 3) were implanted with a cannula into the bile duct after an abdominal incision anesthetized with 10% aqueous chloral hydrate to collect bile. Urine and feces samples (n = 3) were collected using metabolic cages. Blank biosamples were obtained in the same way. All biosamples were stored at −80°C until analysis.
Before use, SPE columns (3 cm3, 60 mg, Waters Oasis, Ireland) were conditioned with 3 mL of methanol followed by 3 mL of ionized water which is same as previous study [15]. Plasma (50 μL), urine (1 mL) and bile (50 μL) samples were loaded on pretreated columns, respectively. After being washed off by 3 mL of 5% methanol, the cartridge were eluted using 3 mL of methanol. The methanol filtrate was evaporated to dryness under nitrogen gas at room temperature. The feces (0.2 g) were pulverized and soaked 50 mL methanol in an ultrasonic bath for 30 min. The filtrate was evaporated to dryness at 40°C in vacuum. The residue was reconstituted in 1 mL water. Furthermore, the feces samples (1 mL) were treated by the same way as the plasma samples. Finally, the residue was reconstituted in 100 μL methanol and an aliquot of 4 μL samples were injected into UPLC/q-tof-MS.
2.3. In vitro phase I metabolism assay
As described previously [16], phase I incubation system (200 μL) contained 50 mM Tris-HCl buffer solution (pH = 7.4), 0.5 mg/mL of HLM, 5 mM MgCl2 and 6.25 μM corylin. After 5 min of pre-incubation at 37°C, 1 mM of NADPH solution was added. After 1 h incubation at 37°C, the reactions were terminated using 200 μL cold acetonitrile. After centrifuging at 13800g for 10 min, an aliquot (8 μL) of the supernatant was injected into UPLC/q-tof-MS system. Incubation without NADPH served as negative control to confirm the metabolites produced were NADPH-dependent. Likewise, incubation system for HIM, twelve iHLM, animal liver microsomes and each CYP enzyme was similar to the microsomal incubation system above. All experiments were performed in triplicate. Preliminary experiments were performed to ensure that the rates of phase I metabolism were determined under linear conditions with respect to the incubation time and protein concentration.
2.4. In vitro glucuronidation assay
In brief, a typical incubation mixture (200 μL) contained 50 mM Tris-HCl buffer (pH = 7.4), 0.5 mg/mL of HLM, 0.88 mM MgCl2, 22 μg/mL alamethicin, 4.4 mM saccharolactone and 3.5 mM UDPGA as previously published [17]. After 1 h incubation at 37°C, the reaction was terminated by adding 200 μL ice-cold acetonitrile followed by centrifugation at 13800g for 10 min. An aliquot (8 μL) of supernatant was subjected to UPLC/q-tof-MS analysis. Incubation without UDPGA served as negative control to confirm the metabolites produced were UDPGA-dependent. Similarly, corylin was incubated with HIM, twelve iHLM, animal liver microsomes and recombinant UGT enzymes the above system. All experiments were performed in triplicate. Preliminary experiments were performed to ensure that the rates of glucuronidation were determined under linear conditions with respect to the incubation time and protein concentration.
2.5. Structural identification of corylin and its metabolites
Metabolite screening was performed using a UPLC-q-tof/MS system (Waters Corporation, Manchester, U.K.). Chromatographic separation was achieved on a BEH C18 column (2.1 mm × 50 mm, 1.7 μm) maintained at 35°C. The mobile phase consisted of water (A) and acetonitrile (B) (both including 0.1% formic acid, v/v), and the flow rate was 0.5 mL/min. The gradient elution program was as follows: 10–50% B from 0 to 2.0 min, 50–100% B from 2.0 to 3.0 min. After holding 100% B for next 0.2 min, the column was returned to its starting condition.
The UPLC system was coupled to a hybrid quadrupole orthogonal time-of-flight (q-tof) tandem mass spectrometer (SYNAPT™ G2 HDMS, Waters, Manchester, U.K.) equipped with electrospray ionization (ESI). The operating parameters were as follow: Capillary voltage of −2.5 kV (ESI-); Sample cone voltage 40 V (ESI-); Ramptrap collision energy of 40–60 V (ESI-); Extraction cone voltage of 4 V, source temperature of 100°C, desolvation temperature 300°C, cone gas flow of 50 L/h and desolvation gas flow of 800 L/h. Argon was used as collision gas for CID in both MSE and MS2 mode. To ensure mass accuracy and reproducibility, the mass spectrometer was calibrated over a range of 50–1500 Da using solution of sodium formate. Leucine-enkephalin (m/z 554.2615 in negative ion mode) was used as external reference of LockSpray™ infused at a constant flow of 5 μL/min and data were centroided during acquisition.
2.6. Quantification corylin and its metabolites
Due to lack of reference standard, quantification of corylin-related metabolites was based on the standard curve of the parent compound (corylin) according to the assumption that parent compound and its phase I metabolites and glucuronide have closely similar UV absorbance maxima [18]. Serial working solutions of corylin were determined on a BEH C18 column (2.1 mm × 50 mm, 1.7 μm, Waters, Ireland, Part NO. 186002350). The detection wavelength was set at 254 nm and the injection volume was 8 μL.
The LOD and LOQ were calculated as 3-fold and 10-fold of the ratio of signal-to-noise (S/N), respectively. The LOD and LOQ for corylin was 0.01 and 0.05 μM, respectively. Calibration curves were constructed by plotting corylin peak area ratios (Y) versus corylin concentrations (X) using a 1/x2 weighting factor. Acceptable linear correlation (Y = 18481X) was confirmed by correlation coefficients (r2) of 0.9995. The linear range was 0.05 ~ 50 μM. The accuracy and precision of the intra-day and inter-day error were both less than 2.6%.
2.7. Enzymes kinetic evaluation
Serial concentrations of corylin (0.39–50 μM) were incubated with pooled HLM, HIM, animal liver microsomes, individual CYP enzyme and expressed UGT enzymes to determine the metabolic rates. The kinetic models Michaelis-Menten equation and substrate inhibition equation were fitted to the data of metabolic rates versus substrate concentrations and displayed in Eqs. (1) and (2), respectively. Appropriate models were selected by visual inspection of the Eadie-Hofstee plot [19]. Model fitting and parameter estimation were performed by Graphpad Prism V5 software (SanDiego, CA).
The parameters are as follows. V is the formation rate of product. Vmax is the maximal velocity. Km is the Michaelis constant and [S] is the substrate. Ksi is the substrate inhibition constant. The intrinsic clearance (CLint) was derived by Vmax/Km for Michaelis-Menten and substrate inhibition models [20].
| (1) |
| (2) |
2.8. Activity correlation analysis
According to the assay protocol reported previously, the metabolic activities of individual HLMs (n = 12) toward corylin, phenacetin (a probe substrate for CYP1A2), tolbutamide (a probe substrate for CYP2C9), β-estradiol (a probe substrate for UGT1A1) and CDCA (a probe substrate for UGT1A3) were determined [20–22]. Corylin (50 μM) was incubated with NADPH-supplemented individual HLM (1.0 mg/mL) for 60 min, whereas phenacetin (200 μM) and tolbutamide (500 μM) were separately treated with NADPH-supplemented individual HLM (1.0 mg/mL) for 120 min. Similarly, corylin (6.25 μM) was treated with UDPGA-supplemented individual HLM (1.0 mg/mL) for 60 min, while β-estradiol (50 μM) and CDCA (250 μM) were separately incubated with UDPGA-supplemented individual HLM (1.0 mg/mL) for 120 min. Correlation analysis were performed between corylin oxidation and hydration (M4), oxidation (M5, M10 and M12), hydration (M6) and phenacetin-N-deacetylation and tolbutamide-4-oxidation, respectively. Similarly, correlation analysis were performed between corylin glucuronidation (M7) and β-estradiol-3-O-glucuronidation and CDCA-24-O-glucuronidation, respectively. Correlation (Pearson) analysis was performed using GraphPad Prism V5 software.
2.9. Contribution of CYP and UGT isoforms
The contribution of individual CYP and UGT enzyme for corylin phase I metabolism and glucuronidation in pooled HLM was evaluated by the relative activity factor (RAF) approach as described in previous study [20]. The RAF value was defined as the activity ratio of a probe substrate in pooled HLM and an expressed CYP or UGT enzyme using Eq. (3). The analytical conditions of phenacetin, tolbutamide, β-estradiol and CDCA were shown in Table S1. The RAFs were derived for CYP1A2, 2C9 and UGT1A1, 1A3 using the well-recognized probe substrates phenacetin, tolbutamide, β-estradiol and CDCA, respectively. The contribution of individual CYP or UGT enzyme were calculated according to Eq.(4).
| (3) |
| (4) |
2.10. Species difference
A series of corylin solutions (0.39–50 μM) were incubated with six animal liver microsomes to determine the phase I metabolism and glucuronidation rates of corylin. Kinetic parameters were derived by appropriate model fitting. The CLint values of corylin in different animal liver microsomes was as evaluation parameter to estimate the species diversity.
2.11. Statistical analysis
Data are expressed as the mean ± SD (standard deviation). Mean differences between treatment and control groups were analyzed by two-tailed Student’s t-test. The level of significance was set at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***).
3. Results
3.1. Structural identification of metabolites in mice
Parent compound (M0)
As reported previously [8], corylin (M0) exhibited a typical [M−H]− ion at m/z 319.0974 (C20H15O4, 1.3 ppm). The MS/MS spectra (Fig. S1a) gave the main fragment ions at m/z 303.0658, 289.0505, 275.0714 and 265.0501. In addition, the fragment ions at m/z 183.0736 ([B1,3−H]−), 167.0522 ([B1,3−CH4−H]−) and 135.0085 ([A1,3−H]−) were originated from an obvious RDA cleavage.
Mono-oxidated corylin (M5, M10 and M12)
M5 (1.83 min, Fig. S1b), M10 (2.27 min, Fig. S1c) and M12 (2.44 min, Fig. S1d) all gave the [M−H]− ions at m/z 335.0923, which were characterized as the oxidated products of M0 with 16 Da (O) more than M0. After MS/MS analysis, M5 exhibited the major fragment ions at m/z 307.0974, 277.0505 and 223.0396 with characteristic ions at m/z 265.0503, 252.0418, 199.0693 and 135.0058 which indicated that the oxidation position was at the D ring. Similarly, the diagnostic fragment ions at m/z 183.0756 and 151.0169 suggested that the oxidation position of M10 and M12 were at the A ring of corylin.
Hydrated corylin (M6) and mono-oxidated hydrated corylin (M4)
M6 exhibited a [M−H]− ion at m/z 337.1079 (C20H17O5, 0.9 ppm) in Fig. S1e, which was 18 Da (H2O) more than M0 and characterized as the hydrated derivate of M0. The MS/MS spectra displayed 319.0994, 265.0498 and 253.0457. The ions at m/z 183.0741, 167.0529 and 135.0086 illustrated that the hydration position was at the double bond of D ring. Besides, M4 gave the [M−H]− ion at m/z 353.1024 (C20H17O6, −0.3 ppm) which is a 34 Da (H2O2) higher than M0. The MS/MS spectra (Fig. S1f) displayed an obvious [M−H-H2O]− ion at m/z 335.0953, which indicated that M1 was the hydrated and oxidated product of M0. The diagnostic ions at m/z 183.0740 and 151.0153 suggested that the oxidation was at the A ring of aglycone.
Mono-glucuronidated corylin (M7) and mono-oxidated and glucuronidated corylin (M1, M3 and M8)
Due to 176 Da (C6H8O6) more than M0, M7 (C26H23O10, 1.4 ppm, Fig. S1 g) were tentatively identified as the glucuronidated conjugate of corylin. The MS/MS spectra gave abundant proof with the diagnostic fragment ions at m/z 175.0242 and 113.0283. Similarly, M1, M3 and M8 were tentatively identified as the mono-oxidated and glucuronidated corylin. Based on the MS/MS spectra, M1 (Fig. S1i) and M3 (Fig. S1j) were characterized as the glucuronidated products of M5, respectively. And M8 (Fig. S1k) was tentatively identified as the glucuronidated conjugate of M10 or M12.
Mono-sulfated corylin (M11) and mono-oxidated and sulfated corylin (M2 and M9)
M11 was eluted at 2.37 min with [M−H]−ion at m/z 399.0538 (C20H15O7S, 0.1 ppm). In the MS/MS spectra (Fig. S1 h), the mother ion obviously lose a neutral fragment of SO3 (80 Da), which indicated that M11 was the sulfated conjugate of corylin. In addition, M2 (Fig. S1l) was characterized as the sulfated product of M5, whereas M9 (Fig. S1m) was tentatively identified as the sulfated conjugate of M10 or M12.
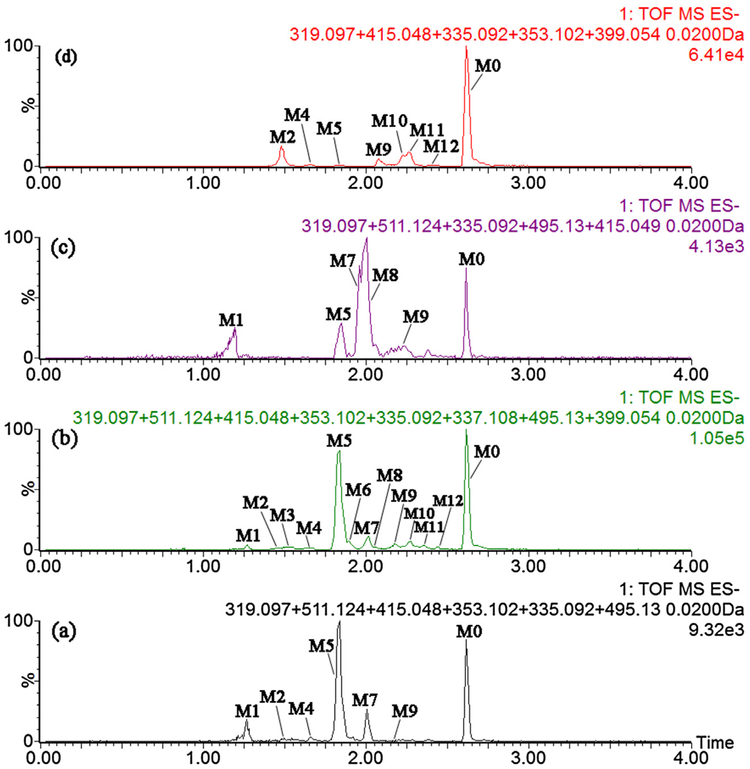
In all, the proposed fragment pathway of M0 ~ M12 were exhibited in Fig. S2. The extracted ion chromatograms and absolute areas of metabolites in mice biosamples were shown in Fig. 1 and Fig. S3, respectively. And the UPLC/q-tof-MS data of M0 ~ M12 was displayed in Table 1.
Fig. 1.
Extracted ion chromatograms of corylin and its metabolites in mice bio-samples. (a), plasma; (b), urine; (c), bile; (d), feces.
Table 1.
UPLC/q-tof-MS data of xenobiotics in mice biosamples after oral administration of corylin.
| NO. | RT(min) | [M-H]− ion | Formula | Error(ppm) | (−) ESI-MS/MS | Identification | Source |
|---|---|---|---|---|---|---|---|
| M0 | 2.62 | 319.0974 | C20H16O4 | 1.3 | 303.0658 289.0505 275.0714 265.0501 183.0736 167.0522 135.0085 | corylin | PUBF |
| Ml | 1.28 | 511.1236 | C26H24O11 | −0.8 | 335.0940307.0997 199.0678 175.0233 135.0065 | mono-oxidated and glucuronidated corylin | PUB |
| M2 | 1.51 | 415.0481 | C20H16O8S | −1.7 | 335.0907 307.0989 135.0083 | mono-oxidated and sulfated corylin | PUF |
| M3 | 1.53 | 511.1246 | C26H24O11 | 1.2 | 335.0914 307.0979 277.0536 252.0449 | mono-oxidated and glucuronidated corylin | U |
| M4 | 1.66 | 353.1024 | C20H18O6 | −0.3 | 335.0953 281.0459 253.0519 183.0740 151.0153 | mono-oxidated and hydrated corylin | PUF |
| M5 | 1.83 | 335.0919 | C20H16O5 | 0 | 307.0974277.0505 265.0503 252.0418 223.0396 199.0693 135.0058 | mono-oxidated corylin | PUBF |
| M6 | l.89 | 337.1079 | C20H18O5 | 0.9 | 319.0994 265.0498 253.0457 183.0741 167.0529 135.0086 | hydrated corylin | U |
| M7 | 2.01 | 495.1298 | C26H24O10 | 1.4 | 319.0974 175.0242 113.0283 | mono-glucuronidated corylin | PUB |
| M8 | 2.05 | 511.1229 | C26H24O11 | −2.2 | 335.0915 167.0526 | mono-oxidated and glucuronidated corylin | UB |
| M9 | 2.18 | 415.0487 | C20H16O8S | −0.2 | 335.0918 291.0770 183.0742 | mono-oxidated and sulfated corylin | PUBF |
| Ml0 | 2.27 | 335.0923 | C20H16O5 | 1.2 | 319.0638 305.0461 291.0788 277.0506 261.0610183.0756 151.0169 | mono-oxidated corylin | UF |
| Mll | 2.37 | 399.0538 | C20H16O7S | 0.1 | 319.0953 | mono-sulfated corylin | UF |
| M12 | 2.44 | 335.0919 | C20H16O5 | 0 | 319.0602305.0480 291.0753 277.0514 183.0759 151.0153 | mono-oxidated corylin | UF |
Note: M: metabolite; RT: retention time; P: plasma; U: urine; B: bile; F: feces.
3.2. Phase I metabolism of corylin in HLM, HIM and expressed CYP enzymes
After incubation of corylin with NADPH-supplemented HLM and HIM, five additional peaks (M4, M5, M6, M10 and M12) were all detected by UPLC/q-tof-MS analysis (Fig. S4). Kinetic profiling revealed that formation of M4 in HLM well modeled by Michaelis-Menten kinetics (Fig. S5a), whereas M5, M10 and M12 in HLM followed the substrate inhibition equation (Fig. S5a). Meanwhile, M4, M5, M10 and M12 in HIM were all well modeled by the substrate inhibition equation (Fig. S5b). Due to the concentration under the limit of quantification, it was unable to determine the kinetic parameters of M6. In HLM, M4 and M5 were the main metabolites with the formation of 38.19 and 38.80 pmol/min/mg, respectively. Similarly, M4 was the main metabolites in HIM with the formation of 57.59 pmol/min/mg, while M5, M10 and M12 were with the formation of 20.20, 19.34 and 28.92 pmol/min/mg, respectively. M4 and M5 in HLM were both efficient (CLint = 24.29 and 11.94 μL/min/mg, respectively), following Km values of 1.57 and 3.25 μM, respectively. The CLint values of M4, M5, M10 and M12 in HIM were 42.85, 9.30, 10.11 and 6.25 μL/min/mg, respectively, whereas the related Km values were 1.34, 2.17, 1.91 and 4.63 μM, respectively. The detailed parameters of the metabolites were listed in Table 2.
Table 2.
Kinetic parameters derived for the phase I metabolites of corylin (M4, M5, M6, M10 and M12) by HLM, HIM, expressed CYP enzymes, MkLM, RLM, MLM, DLM, GpLM and RaLM (Mean ± SD). All experiments were performed in triplicate.
| Meta. | Vmax (pmol/min/mg) | Km (μM) | Ki (μM) | CLint (μL/min/mg) | Model | |
|---|---|---|---|---|---|---|
| HLM | M4 | 38.19 ±0.24 | 1.57 ±0.04 | N.A. | 24.29 ± 0.67 | MM |
| M5 | 38.80 ±1.12 | 3.25 ± 0.20 | 68.03 ±6.18 | 11.94 ±0.80 | SI | |
| M6 | ++ | ++ | ++ | ++ | ||
| M10 | 5.46 ±0.26 | 1.85 ±0.21 | 74.05 ±12.89 | 2.95 ± 0.37 | SI | |
| M12 | 5.37 ± 0.12 | 2.64 ±0.14 | 78.25 ±6.37 | 2.03 ±0.11 | SI | |
| HIM | M4 | 57.59 ±1.23 | 1.34 ± 0.08 | 219.70± 37.61 | 42.85 ±2.79 | SI |
| M5 | 20.20 ±0.93 | 2.17 ± 0.23 | 68.74 ±10.79 | 9.30 ±1.08 | SI | |
| M6 | + | + | + | + | ||
| M10 | 19.34 ±0.96 | 1.91 ± 0.24 | 96.05 ± 20.27 | 10.11 ± 1.34 | SI | |
| M12 | 28.92 ±3.55 | 4.63 ±1.00 | 29.86 ±7.58 | 6.25 ± 1.55 | SI | |
| CYP1A1 | M4 | 165.90 ±13.49 | 6.47 ± 0.85 | 26.17 ± 3.99 | 26.63 ±3.96 | SI |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | 189.90 ±9.61 | 12.11 ± 0.85 | 16.05 ± 1.22 | 15.68 ± 1.35 | SI | |
| M12 | 48.97 ± 4.30 | 7.41 ± 1.01 | 22.00 ±3.37 | 6.61 ± 1.07 | SI | |
| CYP1A2 | M4 | 60.51 ± 1.75 | 8.79 ±0.73 | N.A. | 6.89 ± 0.60 | MM |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | 72.97 ±3.22 | 10.50 ±1.26 | N.A. | 6.94 ± 0.89 | MM | |
| M12 | 36.19 ±1.51 | 10.03 ± 1.15 | N.A. | 3.61 ± 0.44 | MM | |
| CYP1B1 | M4 | 36.10 ±0.41 | 1.09 ± 0.06 | N.A. | 33.09 ± 1.78 | MM |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | 4.12 ± 0.49 | 5.48 ± 1.13 | 39.29 ± 10.28 | 0.75 ±0.18 | SI | |
| M12 | + | + | + | + | ||
| CYP2C8 | M4 | 18.36 ± 0.40 | 1.31 ± 0.13 | N.A. | 14.05 ± 1.39 | MM |
| M5 | 15.1 ± 0.77 | 2.19 ± 0.23 | 30.29 ± 3.66 | 6.90 ±0.81 | SI | |
| M6 | − | − | − | − | ||
| M10 | − | − | − | − | ||
| M12 | − | − | − | − | ||
| CYP2C9 | M4 | 10.03 ±0.50 | 1.70± 0.21 | 85.55 ±17.23 | 5.92 ± 0.80 | SI |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | + | + | + | + | ||
| M12 | 6.62 ± 0.08 | 1.38 ± 0.08 | N.A. | 4.79 ±0.27 | MM | |
| CYP2C19 | M4 | 587.50 ±11.09 | 4.44 ± 0.28 | N.A. | 132.41 ± 8.85 | MM |
| M5 | − | − | − | − | ||
| M6 | + | + | + | + | ||
| M10 | 25.14 ±0.78 | 1.75 ±0.22 | N.A. | 14.39 ± 1.90 | MM | |
| M12 | + | + | + | + | ||
| CYP2D6 | M4 | + | + | + | + | |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | 6.35 ±0.18 | 1.31 ± 0.10 | 55.11 ± 5.27 | 4.86 ±0.38 | SI | |
| M12 | + | + | + | + | ||
| CYP2E1 | M4 | + | + | + | + | |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | − | − | − | − | ||
| M12 | + | + | + | + | ||
| CYP3A4 | M4 | + | + | + | + | |
| M5 | − | − | − | − | ||
| M6 | − | − | − | − | ||
| M10 | + | + | + | + | ||
| M12 | + | + | + | + | ||
| MkLM | M4 | 45.49 ±1.85 | 1.32 ± 0.14 | 70.04 ±10.79 | 34.49 ± 3.97 | SI |
| M5 | 159.60 ±8.71 | 2.16 ± 0.27 | 63.32 ± 11.32 | 73.79 ±10.08 | SI | |
| M6 | + | + | + | + | ||
| M10 | 27.71 ± 1.79 | 2.27 ± 0.31 | 37.25 ±6.16 | 12.18 ± 1.85 | SI | |
| M12 | 17.73 ±1.71 | 2.54 ±0.52 | 48.02 ±12.88 | 6.97 ± 1.58 | SI | |
| RLM | M4 | 74.12 ±7.05 | 8.10± 1.32 | 76.43 ± 20.64 | 9.15 ± 1.73 | SI |
| M5 | 6.0 ±0.1 | 0.78 ±0.07 | N.A. | 7.65 ± 0.65 | MM | |
| M6 | + | + | + | + | ||
| M10 | 11.88 ±1.47 | 5.86 ±1.32 | 62.56 ± 20.98 | 2.03 ±0.52 | SI | |
| M12 | 14.31 ± 3.07 | 13.70 ±4.16 | 25.52 ± 9.15 | 1.04 ± 0.39 | SI | |
| MLM | M4 | 34.25 ±0.46 | 1.22 ±0.07 | N.A. | 28.20 ±1.75 | MM |
| M5 | + | + | + | + | ||
| M6 | + | + | + | + | ||
| M10 | 18.5 ± 0.33 | 2.11 ± 0.15 | N.A. | 8.75 ±0.64 | MM | |
| M12 | 12.12 ± 0.19 | 1.64 ±0.11 | N.A. | 7.38 ±0.49 | MM | |
| DLM | M4 | 30.90 ±1.21 | 1.05 ± 0.12 | 130.50 ±29.32 | 29.37 ± 3.63 | SI |
| M5 | − | − | − | − | ||
| M6 | 13.14± 0.20 | 1.19 ± 0.08 | N.A. | 11.03 ± 0.79 | MM | |
| M10 | 72.38 ±9.75 | 9.21 ± 1.84 | 21.92 ± 4.98 | 7.86 ±1.90 | SI | |
| M12 | 13.11 ± 0.90 | 5.31 ± 0.68 | 67.90 ± 13.3 | 2.47 ± 0.36 | SI | |
| GpLM | M4 | 153.70 ±7.00 | 1.61 ± 0.19 | 121.50 ±28.12 | 95.58 ± 12.3 | SI |
| M5 | 19.37 ± 1.28 | 0.88 ±0.17 | 71.94 ±19.59 | 22.02 ± 4.59 | SI | |
| M6 | 16.37 ± 0.24 | 0.70 ± 0.05 | N.A. | 23.36 ± 1.79 | MM | |
| M10 | 67.1 ± 3.82 | 2.82 ±0.35 | 67.49 ±12.39 | 23.81 ± 3.24 | SI | |
| M12 | 90.55 ±6.11 | 3.16 ± 0.45 | 64.83 ± 13.52 | 28.68 ±4.50 | SI | |
| RaLM | M4 | 223.4 ±10.19 | 1.66 ± 0.20 | 128.3 ± 30.62 | 134.98 ± 17.32 | SI |
| M5 | + | + | + | + | ||
| M6 | ++ | ++ | ++ | ++ | ||
| M10 | 150.9 ±16.27 | 9.47 ± 1.67 | 65.01 ± 17.81 | 15.94 ±3.29 | SI | |
| M12 | 19.9 ± 0.14 | 4.10± 0.10 | N.A. | 4.86 ±0.12 | MM |
Note: Meta., metabolites; N.A., not available; −, not detected; +, under the limit of quantification; ++, unable to determine the kinetic parameters in the absence of a full kinetic profile; HLM, human liver microsomes; HIM, human intestine microsomes; MkLM, monkey liver microsomes; RLM, rat liver microsomes; DLM, dog liver microsomes; GpLM, guinea pig liver microsomes; RaLM, rabbit liver microsomes; SI, substrate inhibition model; MM, Michaelis-Menten model.
To identify the enzymes involving in the phase I metabolism of corylin, twelve expressed CYP enzymes were analyzed for their catalysis activities (expressed as pmol/min/mg protein). As shown in Fig. S4, CYP1A1, 1A2, 1B1, 2C8, 2C9, 2C19, 2D6, 2E1 and 3A4 were responsible for the formation of phase I metabolites of corylin. Other three CYP enzymes were not capable of the production toward phase I metabolites of corylin.
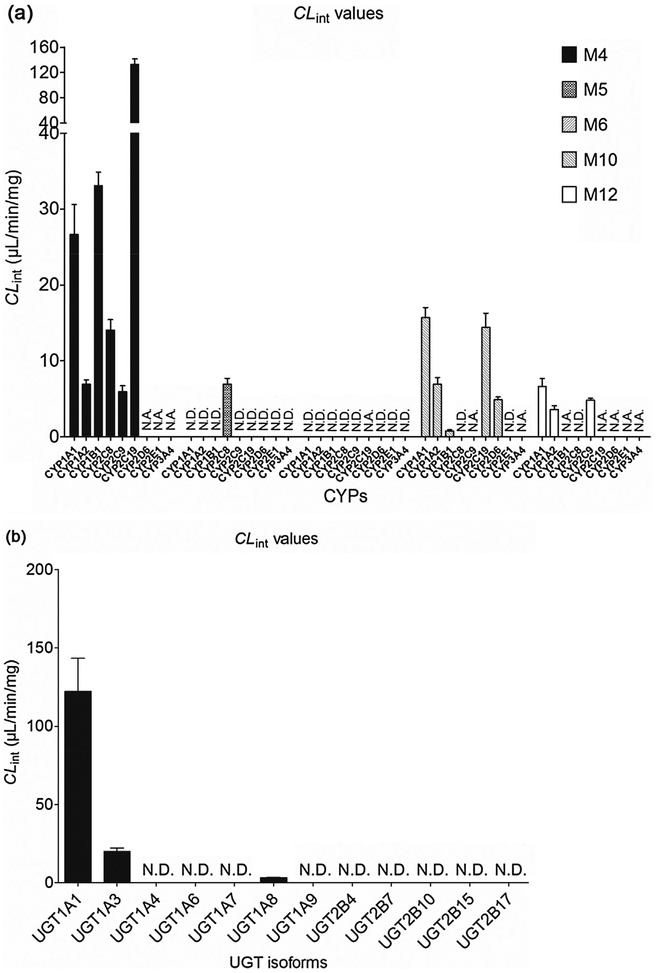
Based on reaction phenotyping results, kinetic profiles of active recombinant CYP enzymes were analyzed using a series of substrate concentrations. Obviously, M4 was the main metabolite of corylin in all these active expressed CYP enzymes. The kinetic profile of M4 by CYP1A1 (Fig. S5c) and 2C9 (Fig. S5d) were both well modeled by substrate inhibition equation, which did not always followed the same kinetics as the formation of M4 in HLM. In contrast, the kinetic profile of M4 by CYP1A2 (Fig. S5e), 1B1 (Fig. S5f), 2C8 (Fig. S5g) and 2C19 (Fig. S5h) were well modeled by Michaelis-Menten kinetics, which was in line with its kinetic profiles in HLM. In addition, the CLint values (Fig. 2a) of M4 by CYP1A1, 1A2, 1B1, 2C8, 2C9 and 2C19 were 26.63, 6.89, 33.09, 14.05, 5.92 and 132.41 μL/min/mg, respectively, which indicated that CYP2C19 exhibited the highest activity toward the formation of M4. Similarly, only CYP2C8 could catalyze the formation of M5, which was with a CLint value of 6.90 μL/min/mg. For the formation of M6, it was unable to obtain the kinetic parameters. The catalyze efficiencies (reflected by CLint values, Fig. 2a) for M10 by CYP enzymes followed the order of CYP1A1 (15.68 μL/min/mg) > CYP2C19(14.39 μL/min/mg) > CYP1A2 (6.94 μL/min/mg) > CYP2D6(4.86 μL/min/mg) > CYP1B1 (0.75 μL/min/mg). For M12, CYP1A1 showed the highest activity with a CLint value of 6.61 μL/min/mg, followed with CYP2C9 (CLint = 4.79 μL/min/mg) and CYP1A2 (CLint = 3.61 μL/min/mg). Hence, CYP1A1, 1B1 and 2C19 were the main CYP enzymes for the formation of M4, the main phase I metabolite.
Fig. 2.
The CLint values of expressed CYPs (a) and UGTs (b). All experiments were performed in triplicate. N.D. mean not detected. N.A. means that it was unable to determine the kinetic parameters due to the concentration under the limit of quantification.
3.3. In vitro glucuronidation of corylin in HLM, HIM and expressed UGT enzymes
Unlike the phase I metabolism, only one glucuronic acid conjugate of corylin (M7) was detected in glucuronidation assay. Glucuronidation of corylin (M7) in pooled HLM followed the substrate inhibition kinetics with a small Km value of 5.27 μM (Table 3), suggesting that corylin was a high affinity substrate for HLM (Fig. S6a). The derived intrinsic clearance value (CLint) was as high as 125.33 μL/min/mg, suggesting that corylin was a good substrate for UGT metabolism. On the other hand, kinetic profiling revealed that glucuronidation of corylin in HIM followed the classical Michaelis-Menten kinetics (Fig. S6b). The derived Km value was 3.25 μM and the CLint value was 108.74 μL/min/mg (Table 3), which indicated that glucuronidation may be a significant metabolic pathway for corylin in intestine.
Table 3.
Kinetic parameters derived for the corylin glucuronides (M7) of corylin by HLM, HIM, expressed UGT enzymes, MkLM, RLM, MLM, DLM, GpLM and RaLM (Mean ± SD). All experiments were performed in triplicate.
| Enzyme | Meta. | Vmax (pmol/min/mg) | Km (mM) | Ki(μM) | CLint (μL/min/mg) | Model |
|---|---|---|---|---|---|---|
| HLM | M7 | 660.1 ± 31.75 | 5.27 ±0.46 | 54.97 ± 6.85 | 125.33 ± 12.58 | SI |
| HIM | M7 | 353.2 ± 9.71 | 3.25 ±0.33 | N.A. | 108.74± 11.30 | MM |
| UGT1A1 | M7 | 390.8 ± 29.15 | 3.20 ±0.50 | 62.12 ±13.98 | 122.32 ± 21.07 | SI |
| UGT1A3 | M7 | 76.99 ±3.80 | 3.83 ±0.37 | 41.63 ± 4.95 | 20.11 ± 2.16 | SI |
| UGT1A8 | M7 | 83.76 ±3.06 | 25.59 ±1.94 | N.A. | 3.27 ± 0.28 | MM |
| MkLM | M7 | 1477.0± 94.32 | 9.80 ±0.90 | 17.32 ± 1.7 | 150.78 ±16.83 | SI |
| RLM | M7 | 211.1 ± 17.75 | 4.20 ±0.57 | 13.71 ± 1.95 | 50.24 ± 8.00 | SI |
| MLM | M7 | 929.9 ± 124.3 | 8.22 ±1.65 | 20.07 ± 4.50 | 113.07 ± 27.24 | SI |
| DLM | M7 | 534.2 ± 8.68 | 5.84 ±0.30 | N.A. | 91.52 ± 4.00 | MM |
| GpLM | M7 | 1424.0 ±27.57 | 7.73 ±0.44 | N.A. | 184.24 ±11.13 | MM |
| RaLM | M7 | 654.7 ± 40.13 | 7.34 ±0.73 | 33.93 ±4.17 | 89.26 ±10.43 | SI |
Note: Meta., metabolites; N.A., not available; HLM, human liver microsomes; HIM, human intestine microsomes; MkLM, monkey liver microsomes; RLM, rat liver microsomes; DLM, dog liver microsomes; GpLM, guinea pig liver microsomes; RaLM, rabbit liver microsomes; SI, substrate inhibition model; MM, Michaelis-Menten model.
Of tested UGT enzymes, UGT1A1, 1A3 and 1A8 were three main isoforms responsible for generating glucuronide from corylin. Glucuronidation mediated by UGT1A1 (Fig. S6c) and 1A3 (Fig. S6d) followed the substrate inhibition kinetics, which were in line with its glucuronidation profiles in HLM (Fig. S6a). In addition, the glucuronidation of corylin mediated by UGT1A8 (Fig. S6e) followed the Michaelis-Menten profile, which also followed the same kinetics as HIM (Fig. S6b). Of note, a comparison of the CLint values revealed that UGT1A1 was the UGT enzyme with highest activities toward corylin (Fig. 2b). The CLint value for UGT1A1 was 122.32 μL/min/mg. By contrast, glucuronidation of corylin by UGT1A3 (20.11 μL/min/mg) and 1A8 (3.27 μL/min/mg) was less efficient. In general, UGT1A1 showed the highest glucuronidation activities for the formation of M7.
3.4. Activity correlation analysis by CYPs and UGTs
Involvement of CYP1A2 and 2C9 in corylin phase I metabolism was further confirmed by activity correlation analysis using a bank of individual HLMs (n = 12). It was shown that M4 and M5 were strongly correlated with phenacetin-N-deacetylation with correlation factors (r = 0.885, p = 0.0001; Fig. S7a) and (r = 0.777, p = 0.003; Fig. S7b), respectively. Besides, M4 and M5 were also significantly correlated with tolbutamide-4-oxidation (r = 0.727, p = 0.008; Fig. S8a) and (r = 0.743, p = 0.006; Fig. S8b), respectively. In addition, activity correlation analysis between corylin oxidation (M10 and M12), and phenacetin-N-deacetylation and tolbutamide-4-oxidation were all evaluated and exhibited in (Figs. S7 & S8), respectively. Moreover, the detailed results were shown in Table 4.
Table 4.
Metabolic activities correlation analysis of individual HLM (n = 12) toward corylin and corresponding substrates.
| Enzyme | Substrate | Metabolite | r value | p value |
|---|---|---|---|---|
| CYP1A2 | phenacetin | M4 (oxidation and hydration) | 0.885 | 0.0001,*** |
| CYP1A2 | phenacetin | M5 (oxidation) | 0.777 | 0.003, ** |
| CYP1A2 | phenacetin | M10 (oxidation) | 0.579 | 0.049, * |
| CYP1A2 | phenacetin | M12 (oxidation) | 0.652 | 0.022, * |
| CYP2C9 | tolbutamide | M4 (oxidation and hydration) | 0.727 | 0.008, ** |
| CYP2C9 | tolbutamide | M5 (oxidation) | 0.743 | 0.006, ** |
| CYP2C9 | tolbutamide | M10 (oxidation) | 0.697 | 0.012, * |
| CYP2C9 | tolbutamide | M12 (oxidation) | 0.624 | 0.030, * |
| UGT1A1 | β-estradiol | M7 (glucuronidation) | 0.742 | 0.006, ** |
| UGT1A3 | CDCA | M7 (glucuronidation) | 0.626 | 0.029, * |
Note: CDCA, chenodeoxycholic acid. All experiments were performed in triplicate.
p < 0.05,
p < 0.01,
p < 0.001.
In a similar manner, glucuronidation activities between corylin glucuronidation (M7) and β-estradiol-3-O-glucuronidation, CDCA-24-O-glucuronidation were performed using twelve individual HLM. It was shown that M7 were strongly correlated with β-estradiol-3-O-glucuronidation with correlation factors (r = 0.742, p = 0.006) (Fig. S9a). Similarly, M7 were correlated with CDCA-24-O-glucuronidation (r = 0.626, p = 0.029) (Fig. S9b). The results indicated that contribution of UGT1A1 to corylin glucuronidation in the liver was appreciable.
3.5. Contribution of CYP and UGT isoforms
The RAF approach was used to estimate the exact contribution of CYP1A2 and 2C9 to corylin phase I metabolism in HLM. The RAF values of CYP1A2 and 2C9 were calculated by the CLint values of phenacetin-N-deacetylation and tolbutamide-4-oxidation in HLM and corresponding individual CYP enzyme, respectively. As a result, kinetic profiles of phenacetin-N-deacetylation (Fig. S10a) and tolbutamide-4-oxidation (Fig. S10b) were modeled by the classical Michaelis-Menten kinetics. The derived RAFs for CYP1A2 and 2C9 were 0.121 and 0.070, respectively (Table S2). The scaled CLint values of M4, M10 and M12 were0.83 (=6.89 × 0.121), 0.84 (=6.94 × 0.121) and 0.44 (=3.61 0.121) μL/min/mg for CYP1A2, that was 3.2%, 28.5%, and 21.7%×of total phase I metabolism activity in pooled HLM. The scaled CLint values of M4 and M12 were 0.41 (=5.92 × 0.070) and 0.34 (=4.79 0.070) μL/min/mg for CYP2C9, that was 1.5% and 16.7% of total× phase I metabolism activity in pooled HLM. The remaining phase I metabolism activity for M4, M10 and M12 was probably contributed by other CYP enzymes, such as CYP2C8, 2C19, 2D6, 2E1 and 3A4.
Similarly, the RAF approach was calculated by CLint values of β-estradiol-3-O-glucuronidation and CDCA-24-O-glucuronidation in HLM and expressed UGT1A1 and 1A3 enzyme, respectively. The derived RAFs for UGT1A1 and 1A3 were 0.554 and 0.478 based on previous study [17]. The scaled CLint value of M7 was 67.77 (=122.32 × 0.554) μL/min/mg for UGT1A1 that represented 54.1% of the CLint values (125.33 μL/min/mg) in HLM. The scaled CLint value of M7 was 9.61 (=20.11 × 0.478) ml/min/mg for UGT1A3 that represented 7.7% of the total glucuronidation activity in HLM. Obviously, UGT1A1 contributed most for the glucuronidation of corylin in HLM.
3.6. Species difference
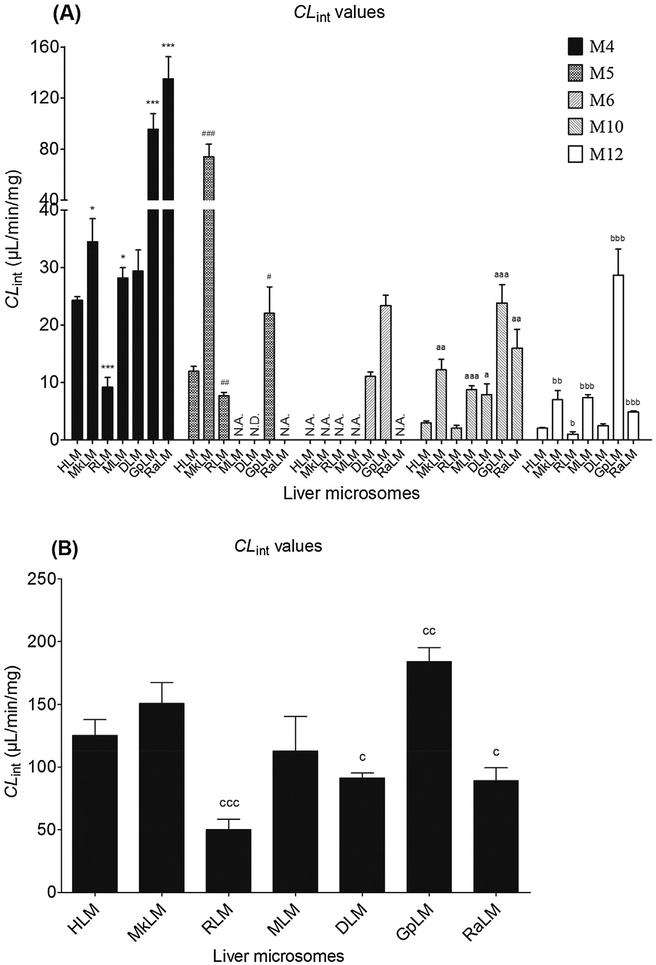
Phase I metabolism kinetics of corylin were determined using liver microsomes from various animal species including monkey, rat, mouse, dog, guinea pig and rabbit (Table 2). On the whole, M4 and M5 were still the main metabolites by these six animal liver microsomes. Except MLM (Michaelis–Menten equation, Fig. S11a), kinetic profiles of other five liver microsomes were all well followed the substrate inhibition kinetics (Figs. S11b–S11f). The apparent Vmax and Km values were determined for the formation of M4, M5, M6, M10 and M12 by animal microsomes (Table 2). Corylin underwent phase I metabolism in human and five types of animal microsomes with the CLint values of 9.15–134.98, 2.03–23.81 and 1.04–28.68 μL/min/mg for M4, M10 and M12, respectively. The catalyze efficiencies (reflected by CLint values, Fig. 3A) for M4 of human and animal microsomes followed the order of RaLM (134.98 μL/min/mg) > GpLM (95.58 μL/min/mg) > MkLM (34.49 μL/min/mg) > DLM (29.37 μL/min/mg) > MLM (28.20 μL/min/mg) > HLM (24.29 μL/min/mg) > RLM (9.15 μL/min/mg). Similarly, the order of CLint values for M10 were GpLM (23.81 μL/min/mg) > RaLM (15.94 μL/min/mg) > MkLM (12.18 μL/min/mg) > MLM (8.75 μL/min/mg) > DLM (7.86 μL/min/mg) > HLM(2.95 μL/min/mg) > RLM (2.03 μL/min/mg). In addition, the CLint values for M12 were 28.68, 7.38, 6.97, 4.86, 2.47, 2.03 and 1.04 by GpLM, MLM, MkLM, RaLM, DLM, HLM and RLM, respectively.
Fig. 3.
Comparison of the CLint values of human liver microsomes and five animal microsomes for phase I metabolism (A) and glucuronidation of corylin (B). (*,#,a,b and c compared with the CLint values of M4, M5, M10, M12 and M7 in HLM, respectively. (*,#,a,b,c p < 0.05, **,##,aa,bb,cc p < 0.01, ***,###,aaa,bbb,ccc p < 0.001).
Glucuronidation of corylin in DLM (Fig. S12a) and GpLM (Fig. S12b) followed the classical Michaelis-Menten kinetics, whereas corylin glucuronidation by MkLM (Fig. S12c), RLM (Fig. S12d), MLM (Fig. S12e) and RaLM (Fig. S12f) well modeled by the substrate inhibition kinetics. Marked species differences (reflected by CLint values, Fig. 3B) were noted for the derived kinetic parameters (Table 3). The Km values ranged from 4.20 to 9.80 μM; the Vmax values ranged from 211.1 to 1477.0 pmol/min/mg and the CLint values ranged from 50.24 to 184.24 μL/min/mg.
Clearly, there were marked species differences in phase I metabolism of corylin (Fig. 3A). Up to 14.75, 11.73 and 27.58 fold of CLint values for M4, M10 and M12 were obtained. Only 3.67-fold of CLint values for M7 were between human and other five types of animal liver microsomes (Fig. 3B). For the main metabolites M4 and M7, monkey, mice and dog showed the calculated parameters that were closest to those of humans (Tables 2 and 3). The kinetic parameters differed by 0.73–1.42-fold between humans and the three animal species. Hence, monkey, mice and dog were probably the best models for the main phase I metabolism and glucuronidation studies of corylin in humans.
4. Discussion
As a bioactive phenolic compound isolated in Fructus Psoraleae, corylin has drawn increasing attentions in the fields of pharmacological activities including antioxidant, anti-proliferation, anti-inflammatory properties [3–7]. However, its metabolic pathways and pharmacokinetics properties have not been fully characterized. To the best of our knowledge, only four literatures reported the metabolism and pharmacokinetics behaviors of corylin in rats after oral administration of corylin-containing herbal preparations [8–11]. The pharmacokinetics parameters of corylin in rats have been characterizeds as rapid absorption and poor bioavailability [10,11], whereas extensive phase II metabolites could be detected in rat bio-samples [8,9]. These findings inspired us to further investigate the metabolic pathways of corylin in vivo and in vitro.
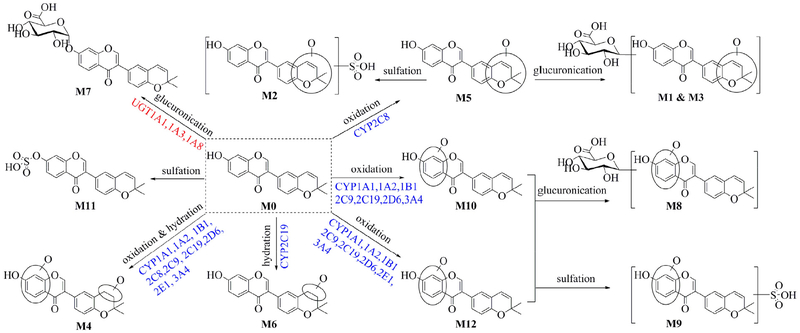
In this study, we have elucidated the phase I metabolism and glucuronidation of corylin in vivo and in vitro (Fig. 4). It was revealed that corylin was subjected to efficient oxidation and glucuronidation, producing the main metabolites M1, M5 and M7 in mice (Fig. 1). Notably, these three metabolites could be detected in mouse plasma and urine, wihch could be considered as the potential effective components for pharmacological benefits based on the theory of serum pharmacochemistry [23]. Besides, in vitro assays also demonstrated that corylin could be rapidly oxidated and glucuronidated in both HLM and HIM in the presence of NADPH or UDPGA (Fig. 3). All of these findings significantly suggested that oxidation and glucuronidation were the major metabolic pathways of corylin in both mice and humans.
Fig. 4.
Complete metabolic pathway of corylin involving in CYPs and UGTs after oral administration.
Taking into consideration that corylin-containing herbal preparations are usually administrated orally, it is necessary to investigate the metabolic clearance of corylin in human intestine and liver in depth. Hence, the identification of individual CYPs and UGTs involving in corylin metabolism is another important task for deciphering the metabolic pathway of corylin (Fig. 2). It was clearly shown that CYP1A1, 1B1 and 2C19 were the main contributors to hydration and oxidation of corylin (Fig. 2a), while UGT1A1 contributed most to the formation of glucuronide (Fig. 2b). It has been reported that CYP1A1 and 1B1 cannot be detected in HLM and mainly expressed in human extrahepatic tissues (intestine, lung, skin, and kidney, etc), while CYP2C19 was mainly expressed in human liver and intestine [24]. In addition, UGT1A1 and UGT1A3 were mainly expressed in the human liver but the content of the former is about 10-fold of that of UGT1A3 [25], whereas UGT1A8 was absent in human liver and mainly detected in human intestine [26]. These results from reaction phenotyping (Fig. 2) combined with kinetics assays (Tables 2 and 3), activity correlation analysis assays (Table 4) and RAF approach suggested that massive metabolism of corylin in extrahepatic tissues, especially human intestine, cannot be ignored.
As mentioned above, CYP1A1, 1B1 and UGT1A1 all participated in the phase I metabolism and glucuronidation of corylin in human intestine, which would assume a great role in the understanding of oral bioavailability. From the view of the intrinsic clearance (CLint value), the CLint values of M4 and M7 in HLM were 24.29 and 125.33 μL/min/mg, respectively, whereas the CLint values of M4 and M7 in HIM were 42.85 and 108.74 μL/min/mg, respectively (Tables 2 and 3). Furthermore, CYP2C19 is more efficient than CYP1A1 in the oxidation of corylin, its inherent clearance (132.41 μL/min/mg protein) is about 5-fold of that in recombinant CYP1A1. Similarly, the CLint values of M7 in UGT1A1 (122.32 μL/min/mg protein) is about 6-fold of that in UGT1A3. These findings implied that massive first-pass metabolism in human liver and intestine would be a significant limiting factor to oral absorption of corylin. In addition, intestine also abundantly expresses the catalytically active CYPs (e.g., 1A1, 2C9, 2C19, 2D6 and 3A4) [24] and UGT enzymes (e.g., UGT1A1 and 1A8) [25]. It is certain that corylin was subjected to massive intestinal phase I metabolism and glucuronidation.
In this study, to explore further the metabolic mechanism, metabolic activities of the CYPs and UGTs enzymes toward corylin were determined based on the CLint values (Fig. 2) derived from kinetic modeling. Use of CLint values was more advantageous in characterization of enzyme activities as follows. (1) CLint represents the catalytic efficiency of the enzyme and is independent of the substrate concentration; (2) compared with other kinetic parameters such as Km and Vmax, CLint is more relevant in an attempt to predict clearance in vivo [27]. Besides, it was noteworthy that substrate concentrations in microsomal incubations (and kinetic parameters) were not corrected due to the presence of protein binding. This was because binding of corylin (log P = 3.15) to microsomal proteins was negligible according to the Hallifax and Houston model [28,29]. The model consisting of log P and microsomal protein concentration has been shown to provide accurate predictions on fu values for the compounds with intermediate lipophilicity (log P = 2.5–5.0) [30]. Also, comparative assessment of several predictive models of fu has revealed that the Halifax and Houston model is one of the best performing prediction methods [31].
It is well-known that common genetic polymorphisms among different people, often lead to different levels and activities of CYPs and UGTs enzyme, which could influence the occurrence of diseases and efficacy of clinical drugs in vivo. In this study, the main contributors for the metabolism of corylin, CYP1A1, 1B1, 2C19 and UGT1A1 are all polymorphic enzymes. For example, light smokers with the susceptible genotype CYP1A1 have a 7-fold higher risk of developing lung cancer compared to light smokers with the normal CYP1A1 genotype [32]. Similarly, CYP2C19 polymorphism (CYP2C19*2, CYP2C19*3 and CYP2C19*17) exists in approximately 3–5% of Caucasian and 15–20% of Asian populations being poor metabolizers with no CYP2C19 function [33,34], and this could reduce the efficacy of clopidogrel (an antiplatelet agent). Similarly, the UGT1A1 polymorphism (UGT1A1*28) has been associated with the dose-limiting toxicities in irinotecan (also known as CPT-11) chemotherapy [35,36]. Therefore, it is readily conceivable that individuals with different polymorphisms most likely exhibited different metabolic activities of corylin, leading to different bioavailability in vivo.
5. Conclusion
In conclusion, a total of twelve metabolites involving in oxidation, hydration, glucuronidation and sulfation were characterized in mice based on retention times and characteristic fragment ions. Furthermore, CYP1A1, 1B1 and 2C19 were the main contributors for corylin phase I metabolism, while UGT1A1 was also the main UGTs for corylin glucuronidation. In addition, M4, M5, M10 and M12 were all correlated with phenacetin-N-deacetylation and tolbutamide-4-oxidation, respectively. Similarly, M7 was also correlated with β-estradiol-3-O-glucuronidation and CDCA-24-O-glucuronidation in twelve individual HLMs. Based on RAF approach, the contributions of CYP1A2, 2C9 toward phase I metabolites and UGT1A1, 1A3 toward M7 were all calculated, respectively. Moreover, phase I metabolism and glucuronidation of corylin by liver microsomes showed marked species differences. Taken altogether, this study proved that corylin was subjected to efficient metabolism in liver and intestine, where in CYP1A1, 1B1, 2C19 and UGT1A1 were the main contributing enzymes, following the complete metabolic pathway of corylin. And these findings provide further justification for the metabolic fate of corylin.
Acknowledgements
The authors would like to thank Guangzhou Research and Creativity Biotechnology Co. Ltd (Guangzhou, China) for the samples tests. This work was supported by Major Project for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 81220108028), State Key Program of National Natural Science Foundation of China (81630097), Program of Introducing Talents of Discipline to Universities (B13038), Guangdong Provincial Science and Technology Project (2016B090921005) and National Natural Science Foundation of Guangdong (2017A03031387).
Footnotes
Declaration of interest
All the authors report no declarations of interest.
References
- [1].Chopra B, Dhingra AK, Dhar KL, Psoralea corylifolia L. (Buguchi) – folklore to modern evidence: review, Fitoterapia 90 (2013) 44–56. [DOI] [PubMed] [Google Scholar]
- [2].Zhang Y, Chen Z, Xu X, Zhou Q, Liu X, Liao L, Zhang Z, Wang Z, Rapid separation and simultaneous quantitative determination of 13 constituents in Psoraleae Fructus by a single marker using high-performance liquid chromatography with diode array detection, J. Sep. Sci 40 (2017) 4191–4202. [DOI] [PubMed] [Google Scholar]
- [3].Guo J, Weng X, Wu H, Li Q, Bi K, Antioxidants from a Chinese medicinal herb-Psoralea corylifolia L, Food Chem 91 (2005) 287–292. [Google Scholar]
- [4].Shim SH, Aldose reductase inhibitory activity of the compounds from the seed of Psoralea corylifolia, J. Korean Soc. Appl. Biol. Chem 52 (2009) 568–572. [Google Scholar]
- [5].Li WD, Yan CP, Wu Y, Weng ZB, Yin FZ, Yang GM, Cai BC, Chen ZP, Osteoblasts proliferation and differentiation stimulating activities of the main components of Fructus Psoraleae corylifoliae, Phytomedicine 21 (2014) 400–405. [DOI] [PubMed] [Google Scholar]
- [6].Lee SW, Yun BR, Kim MH, Park CS, Lee WS, Oh HM, Rho MC, Phenolic compounds isolated from Psoralea corylifolia inhibit IL-6-induced STAT3 activation, Planta Med 78 (2012) 903–906. [DOI] [PubMed] [Google Scholar]
- [7].Hung YL, Fang SH, Wang SC, Cheng WC, Liu PL, Su CC, Chen CS, Huang MY, Hua KF, Shen KH, Wang YT, Suzuki K, Li CY, Corylin protects LPS-induced sepsis and attenuates LPS-induced inflammatory response, Sci. Rep 7 (2017) 46299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Geng JL, Dai Y, Yao ZH, Qin ZF, Wang XL, Qin L, Yao XS, Metabolites profile of Xian-Ling-Gu-Bao capsule, a traditional Chinese medicine prescription, in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry analysis, J. Pharm. Biomed. Anal 96 (2014) 90–103. [DOI] [PubMed] [Google Scholar]
- [9].Wang PL, Yao ZH, Zhang FX, Shen XY, Dai Y, Qin L, Yao XS, Identification of metabolites of PSORALEAE FRUCTUS in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry analysis, J. Pharm. Biomed. Anal 112 (2015) 23–35. [DOI] [PubMed] [Google Scholar]
- [10].Yao ZH, Qin ZF, He LL, Wang XL, Dai Y, Qin L, Gonzalez FJ, Ye WC, Yao XS, Identification bioactivity evaluation and pharmacokinetics of multiple components in rat serum after oral administration of Xian-Ling-Gu-Bao capsule by ultra performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry, J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 1041–1042 (2017) 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang YF, Zhang YB, Chen ZJ, Zhang YT, Yang XW, Plasma pharmacokinetics and cerebral nuclei distribution of major constituents of Psoraleae Fructus in rats after oral administration, Phytomedicine 38 (2018) 166–174. [DOI] [PubMed] [Google Scholar]
- [12].Sun DX, Ge GB, Dong PP, Cao YF, Fu ZW, Ran RX, Wu X, Zhang YY, Hua HM, Zhao Z, Fang ZZ, Inhibition behavior of fructus psoraleae’s ingredients towards human carboxylesterase 1 (hCES1), Xenobiotica 46 (2016) 503–510. [DOI] [PubMed] [Google Scholar]
- [13].Yu ML, Wang CM, Guo YL, Tu YY, Zhang XM, Bai MC, Jiang LM, Liu C, Corylin exhibits strong inhibition towards In vitro glucuronidation metabolism of propofol, Lat. Am. J. Pharm 33 (2014) 1051–1053. [Google Scholar]
- [14].Ding YB, Yu ZY, Chen LJ, Pan WZ, Jin J, Chi YY, Li Z, Li KZ, Ma JH, Yu JH, Shi CX, Optimized incubation system for Psoralea corylifolia linn.-propofol interaction, Lat. Am. J. Pharm 34 (2015) 1886–1890. [Google Scholar]
- [15].Qin ZF, Dai Y, Yao ZH, He LL, Wang QY, Geng JL, Chen HF, Yao XS, Study on chemical profiles and metabolites of Allii Macrostemonis Bulbus as well as its representative steroidal saponins in rats by ultra-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry, Food Chem 192 (2016) 499–515. [DOI] [PubMed] [Google Scholar]
- [16].Wang H, Fang ZZ, Zheng Y, Zhou K, Hu C, Krausz KW, Sun D, Idle JR, Gonzalez FJ, Metabolic profiling of praziquantel enantiomers, Biochem. Pharmacol 90 (2014) 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang L, Hong X, Yao Z, Dai Y, Zhao G, Qin Z, Wu B, Gonzalez FJ, Yao X, Glucuronidation of icaritin by human liver microsomes, human intestine microsomes and expressed UDP-glucuronosyltransferase enzymes: identification of UGT1A3 1A9 and 2B7 as the main contributing enzymes, Xenobiotica 48 (2018) 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Troberg J, Jarvinen E, Ge GB, Yang L, Finel M, UGT1A10 is a high activity and important extrahepatic enzyme: why has its role in intestinal glucuronidation been frequently underestimated? Mol. Pharm 14 (2017) 2875–2883. [DOI] [PubMed] [Google Scholar]
- [19].Hutzler J, Tracy T, Atypical kinetic profiles in drug metabolism reactions, Drug Metab. Dispos 30 (202) (2016) 355–362. [DOI] [PubMed] [Google Scholar]
- [20].Sun H, Wang H, Liu H, Zhang X, Wu B, Glucuronidation of capsaicin by liver microsomes and expressed UGT enzymes: reaction kinetics, contribution of individual enzymes and marked species differences, Expert Opin. Drug Metab. Toxicol 10 (2014) 1325–1336. [DOI] [PubMed] [Google Scholar]
- [21].Romand S, Spaggiari D, Marsousi N, Samer C, Desmeules J, Daali Y, Rudaz S, Characterization of oxycodone in vitro metabolism by human cytochromes P450 and UDP-glucuronosyltransferases, J. Pharm. Biomed. Anal 144 (2017) 129–137. [DOI] [PubMed] [Google Scholar]
- [22].Yao HT, Chang YW, Uramaru N, Watanabe Y, Kitamura S, Kuo YH, Lii CK, Yeh TK, Effects of Bu-Zhong-Yi-Qi-Tang on hepatic drug-metabolizing enzymes and plasma tolbutamide concentration in rats, J. Ethnopharmacol 142 (2012) 121–128. [DOI] [PubMed] [Google Scholar]
- [23].Wang XJ, Studies on serum pharmacochemistry of traditional Chinese medicine, World Sci. Technol./Modern. Tradit. Chin. Med 4 (2002) 1–4. [Google Scholar]
- [24].Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC, The human intestinal cytochrome P450 pie, Drug Metab. Dispos 34 (2006) 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Harbourt DE, Fallon JK, Ito S, Baba T, Ritter JK, Glish GL, Smith PC, Quantification of human uridine-diphosphate glucuronosyl transferase 1A isoforms in liver intestine, and kidney using nanobore liquid chromatography-tandem mass spectrometry, Anal. Chem 84 (2012) 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ohno S, Nakajin S, Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction, Drug Metab. Dispos 37 (2009) 32–40. [DOI] [PubMed] [Google Scholar]
- [27].Wu B, Dong D, Hu M, Zhang S, Quantitative prediction of glucuronidation in humans using the in vitro-in vivo extrapolation approach, Curr. Top Med. Chem 13 (2013) 1343. [DOI] [PubMed] [Google Scholar]
- [28].Austin RP, Response to binding of drugs to hepatic microsomes: comment and assessment of current prediction methodology with recommendation for improvement, Drug Metab. Dispos 34 (2006) (727–727). [DOI] [PubMed] [Google Scholar]
- [29].Zhou J, Tracy TS, Remmel RP, Glucuronidation of dihydrotestosterone and trans-androsterone by recombinant UDP-glucuronosyltransferase (UGT) 1A4: evidence for multiple UGT1A4 aglycone binding sites, Drug Metab. Dispos 38 (2010) 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gao H, Steyn SJ, Chang G, Lin J, Assessment of in silico models for fraction of unbound drug in human liver microsomes, Expert Opin. Drug Metab. Toxicol 6 (2010) 533–542. [DOI] [PubMed] [Google Scholar]
- [31].Poulin P, Haddad S, Microsome composition-based model as a mechanistic tool to predict nonspecific binding of drugs in liver microsomes, J. Pharm. Sci 100 (2011) 4501–4517. [DOI] [PubMed] [Google Scholar]
- [32].Nie Q, Yang XN, An SJ, Zhang XC, Yang JJ, Zhong WZ, Liao RQ, Chen ZH,Su J, Xie Z, Wu YL, CYP1A1*2A polymorphism as a predictor of clinical outcome in advanced lung cancer patients treated with EGFR-TKI and its combined effects with EGFR intron 1 (CA)n polymorphism, Eur. J. Cancer 47 (2011) 1962–1970. [DOI] [PubMed] [Google Scholar]
- [33].Bertilsson L, Geographical/interracial differences in polymorphic drug oxidation. Current state of knowledge of cytochromes P450 (CYP) 2D6 and 2C19, Clin. Pharmacokinet 29 (1995) 192–209. [DOI] [PubMed] [Google Scholar]
- [34].Desta Z, Zhao X, Shin JG, Flockhart DA, Clinical significance of the cytochrome P450 2C19 genetic polymorphism, Clin. Pharmacokinet 41 (2002) 913–958. [DOI] [PubMed] [Google Scholar]
- [35].Fujita KI, Sparreboom A, Pharmacogenetics of irinotecan disposition and toxicity: a review, Curr. Clin. Pharmacol 5 (2010) 209–217. [DOI] [PubMed] [Google Scholar]
- [36].Nagar S, Blanchard RL, Pharmacogenetics of uridine diphosphoglucuronosyltransferase (UGT) 1A family members and its role in patient response to irinotecan, Drug Metab. Rev 38 (2006) 393–409. [DOI] [PubMed] [Google Scholar]