INTRODUCTION
Knowledge of the composition of protein complexes provides key insights into their functions. Immu-noaffinity purification provides an effective means for isolating protein complexes and elucidating their composition. Immunoisolation is achieved with antibodies directed either specifically against the proteins of interest or against tags that are coupled to the proteins of interest. This approach uses immu-noaffinity purification on magnetic beads coated with antibodies for the rapid and efficient purification of protein complexes from cells or tissues. This protocol describes affinity purification of protein complexes using conjugated magnetic beads.
RELATED INFORMATION
A protocol is available for Conjugation of Magnetic Beads for Immunopurification of Protein Complexes (Cristea and Chait 2011).
This method was originally developed using a green fluorescent protein (GFP) tag for the consecutive visualization and isolation of protein complexes in living systems (Cristea et al. 2005). This methodology has been used successfully to look for interactions in both yeast and mammalian systems, such as studies of the dynamic virus–host protein interactions during the course of a viral infection (Cristea et al. 2006).
Three critical concepts are important in performing this protocol. First, work quickly to minimize the dissociation of endogenous complexes and to limit the formation of nonphysiological interactions. Second, work at 4°C and keep solutions on ice to limit unwanted proteolysis and degradation. Third, minimize diluting protein extracts to minimize the dissociation of native complexes and formation of nonspecific interactions. Most protein complexes utilize noncovalent forces to maintain the interactions.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
Reagents
Ammonium hydroxide (0.5 N) (NH4OH), EDTA (0.5 mM) solution
Prepare fresh just before use.
Cell powder, frozen (see Fig. 1)
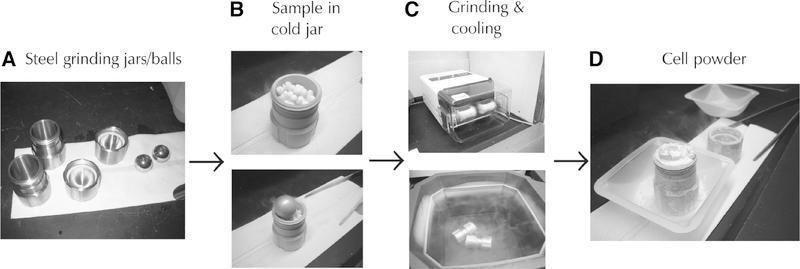
FIGURE 1.
Experimental flow chart for preparing cell extracts by cryogenic grinding of cells or tissues. (A) Stainless steel jars, lids, and balls for cryogenically disrupting frozen cells. The stainless steel jars and balls are used to cryogenically grind frozen cells into a powder for making whole cell protein extracts. Before use, the jars and balls are precooled in liquid nitrogen. (B) Frozen cell pellets added to a frozen steel jar. The pellets are made by dropping a cell slurry into liquid nitrogen and recovering the frozen cell pellets. The precooled steel ball is added to the jar with the cell pellets. The stainless steel cap is then screwed on to seal the chamber. (C) Steel jars inserted into the grinding mill (Retsch MM 301 Mixer Mill). For yeast, the cells are ground into a powder using 10× 3-min cycles at 25 Hz. Between each grinding cycle, the jars are removed from the holders and plunged into liquid nitrogen to keep the cells frozen. (D) Cryogenically disrupted cells. A fine powder is created from the frozen cell pellets and grinding process. The powder is kept frozen and quickly transferred to a 50-mL tube kept cool in liquid nitrogen using a cold spatula. The disrupted cells are stored at −80°C until ready to use.
This protocol uses cells that have been cryogenically disrupted with a ball mill (e.g., Retsch MM301) or a mortar grinder as a starting sample (Fig. 1). However, the same protocol can be used for cells that were disrupted by incubation with lysis buffers, glass beads, or passage through needles of various gauges. We prefer the cryogenic cell disruption because it significantly increases the extraction efficiency and it has proven to be absolutely critical in most of our studies (Cristea et al. 2005, 2006; Wang et al. 2006). For studies that require gentle handling of the cells in order to isolate large intact organelles, the other mentioned techniques might yield better results.
Lysis buffer, optimized (see Step 1)
Magnetic beads conjugated with the antibodies to be used for the affinity isolation (prepared in Conjugation of Magnetic Beads for Immunopurification of Protein Complexes (Cristea and Chait 2011).
SDS-PAGE sample buffer
Stain, mass spectrometry-compatible (for staining proteins in SDS-PAGE gels; see Step 15)
Equipment
Centrifuge (3000 rpm) at 4°C
Liquid nitrogen
Magnetic particle concentrator, 1.5-mL tube type (Dynal/Invitrogen 12020D)
Microcentrifuge tubes
Micropipettor and tips
Neodymium magnets (Dynal/Invitrogen or National Imports MAGCRAFT)
Rare-earth neodymium magnets are used to capture the Dynabeads. Dynal/Invitrogen offers different size magnetic particle concentrators. In addition, National Imports offers a large variety of economical MAGCRAFT neodymium magnets in different shapes and sizes that can be used to capture Dynabeads. The magnets can be temporarily attached to the sides of tubes using rubber bands.
Polytron
Rotator
Rotor chilled to 4°C
Round-bottomed tubes for affinity isolations (e.g., culture tubes for small volumes or 50-mL Falcon tubes for larger volumes)
SDS-PAGE equipment for 1D gels
Shaking incubator set at 70°C
Tube shaker (Tomy)
Vacuum evaporator (e.g., SpeedVac)
METHOD
-
Prepare the lysis buffer. The buffer should be optimized to give efficient extraction of the tagged protein while being mild enough to maintain the sought after protein–protein interactions.
During the affinity purification step, the yield of the isolated tagged protein (and any associated macromolecules) is highly dependent on the conditions used for extraction and purification. Optimize the extraction and purification conditions individually for each different protein complex of interest. Examples of detergents to test are Triton, Tween, deoxycholate, octylglucoside, and digitonin. Various concentrations of salt should also be tested. Examples of buffers are described in Cristea et al. (2005). Begin by testing several buffers of various stringencies. This provides a good indication for what it takes to isolate the tagged protein without losing interacting partners and without accumulating nonspecific interactions. While trying only one buffer is not recommended, a possible starting lysis buffer is 20 mM HEPES-KOH (pH 7.4), 110 mM potassium phosphate, 2 mM MgCl2, 0.1% Tween-20, 0.1% Triton, 150 mM NaCl, and1/100 (v/v) protease inhibitor cocktail. There is no guarantee that this would work on a specific protein complex.
-
Resuspend the frozen cell powder in the optimized lysis buffer (Fig. 1). Use 5 mL of buffer per gram of cells and gently mix to ensure resuspension. Do not let the cell pellet thaw before adding the buffer containing the protease inhibitors.
If the sample derives from cells that were cryogenically ground inside round-bottomed microcentrifuge tubes, then open the tubes in a fume hood, add the buffer in small aliquots (e.g., 200μL), and shake the tube. In this way, the stainless steel grinding ball is used to wash the tube several times.
-
To improve the extraction, homogenize the cells for 10–15 sec using a Polytron.
Keep the cell lysate on ice. This step will generate foam. It is advisable to use a container in which the cell lysate will occupy less than one-third of the volume during the Polytron step. This will ensure that sample will not be lost during the procedure.
If necessary, place the cell lysate on a rotator (gentle rotation) for 5–10 min at 4°C to reduce the amount of foam.
-
Centrifuge the cell lysate at 3000 rpm for 10 min at 4°C.
Any remaining foam will disappear during the centrifugation.
While the cell lysate is being centrifuged, measure the amount of conjugated beads needed to perform the immunoprecipitation. Place the beads on a magnet, remove the PBS, 0.02% NaN3 solution, and wash the beads three times each with 1 mL of lysis buffer. After the third wash, resuspend the beads in a small volume of lysis buffer (e.g., 50–100 μL).
Transfer cleared cell lysate to a clean container. Preferably, use a round-bottomed or a wide-bottomed tube (e.g., culture tubes for smaller volumes, or 50-mL Falcon tubes for larger volumes).
-
Check the cleared cell lysate carefully for any particles that might not have pelleted.
The supernatant should be clear. If any particles are present, remove them, because these can bind/block the beads and interfere with the isolation efficiency or cause a large unwanted background. Additional centrifugation may not pellet the particles (e.g., lipids), depending on the cells or tissue. In these cases, the particles can be removed using a micropipettor.
-
Add the washed beads (from Step 6) to the cell lysate. Incubate at 4°C with gentle rotation for 5 min–1 h.
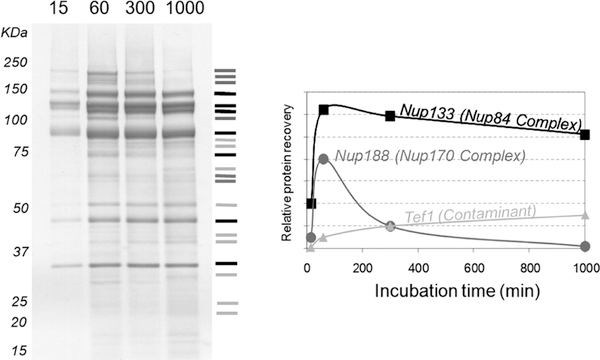
Ensure that beads are in contact with the cell lysate and do not get trapped on the walls or cap of the container. Do not use longer incubation times, because this will promote the accumulation of nonspecific binding and the loss of weak interacting partners (Fig. 2). The optimal incubation time must be determined empirically.
After incubation, place the tube on a magnet. Transfer the flowthrough to another tube to use for western blot analyses to measure the efficiency of protein recovery.
Wash the beads six times with 1 mL of lysis buffer. During the first wash, transfer the beads to a clean microcentrifuge tube. After the fourth wash, transfer the beads to another clean microcentrifuge tube. This will ensure that any components of the cell lysate that might have bound to the walls of the tube will not contaminate the eluate.
-
After the sixth wash, elute the isolated proteins from the beads.
Add 500 μL of freshly made aqueous 0.5 N NH4OH, 0.5 mM EDTA solution to the beads.
Shake the tube (e.g., using a Tomy shaker) for 20 min at room temperature.
-
Place the tube on a magnet and transfer the eluate to a clean microcentrifuge tube. Keep the beads to test for the efficiency of elution.
Use a larger volume when the amount of beads used for the isolation exceeds 10 mg. The elution can also be performed with other solutions. For example, acid elutions can be achieved with 0.1 M citrate (pH 3.1; as recommended by Dynal) or 0.1% trifluoroacetic acid (TFA; pH 1.5). In our hands, the elution with citrate is not very efficient.
Snap-freeze the eluate in liquid nitrogen and dry it in a vacuum evaporator overnight (a minimum of 4 h). Ensure that the sample is fully dried before proceeding.
Add SDS-PAGE sample buffer (e.g., 20 μL) to the dried sample. Shake on a Tomy shaker for 10 min, and place the sample for 10 min at 70°C (if possible with shaking). Keep 10% (e.g., 2 μL) of the sample for western blot analysis to test for immunoprecipitation efficiency.
-
Run the sample on a one-dimensional SDS-PAGE gel. Stain the gel with a stain that is compatible with mass spectrometric analysis (e.g., colloidal Coomassie Blue or zinc staining).
To identify the purified proteins, individual bands can be trypsin digested in the gel and the recovered peptides analyzed by tandem mass spectrometry. Alternatively, the entire gel lane of the sample can be cut into equal pieces, each gel piece in-gel trypsin digested, and recovered peptides analyzed by tandem mass spectrometry. Because of the high sensitivity of mass spectrometry, it is common to detect proteins in unstained regions of the gel that appear to be void of proteins.
FIGURE 2.
Effects of incubation times on protein recovery and nonspecific interactions. Coomassie-stained SDS-PAGE showing isolation of GFP-tagged Nup84 and its associated proteins using various incubation times (left). Recoveries of the indicated proteins as a function of time. Proteins were identified by mass spectrometry (right). (Reprinted, with permission, from the American Society for Biochemistry and Molecular Biology, Inc.)
REFERENCES
- Cristea IM, Chait BT. 2011. Conjugation of magnetic beads for immu-nopurification of protein complexes. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP. 2005. Fluorescent proteins as proteomic probes. Mol Cell Proteomics 4: 19331941. [DOI] [PubMed] [Google Scholar]
- Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem 281: 30269–30278. [DOI] [PubMed] [Google Scholar]
- Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, Chait BT, Zhong Y, Heintz N, Yue Z. 2006. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci 26: 8057–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]