Abstract
Understanding early predictors of sow fertility has the potential to improve genomic predictions. A custom SNP array (SowPro90 produced by Affymetrix) was developed to include genetic variants overlapping quantitative trait loci for age at puberty, one of the earliest indicators of sow fertility, as well as variants related to innate and adaptive immunity. The polymorphisms included in the custom genotyping array were identified using multiple genomic approaches including deep genomic and transcriptomic sequencing and genome-wide associations. Animals from research and commercial populations (n = 2,586) were genotyped for 103,476 SNPs included in SowPro90. To assess the quality of data generated, genotype concordance was evaluated between the SowPro90 and Porcine SNP60 BeadArray using a subset of common SNP (n = 44,708) and animals (n = 277). The mean genotype concordance rate per SNP was 98.4%. Differences in distribution of data quality were observed between the platforms indicating the need for platform specific thresholds for quality parameters. The optimal thresholds for SowPro90 (≥97% SNP and ≥93% sample call rate) were obtained by analyzing the data quality distribution and genotype concordance per SNP across platforms. At ≥97% SNP call rate, there were 42,151 SNPs (94.3%) retained with a mean genotype concordance of 98.6% across platforms. Similarly, ≥94% SNPs and ≥85% sample call rates were established as thresholds for Porcine SNP60 BeadArray. At ≥94% SNPs call rate, there were 41,043 SNPs (91.8%) retained with a mean genotype concordance of 98.6% across platforms. Final evaluation of SowPro90 array content (n = 103,476) at ≥97% SNPs and ≥93% sample call rates allowed retention of 89,040 SNPs (86%) for downstream analysis. The findings and strategy for quality control could be helpful in identifying consistent, high-quality genotypes for genomic evaluations, especially when integrating genotype data from different platforms.
Keywords: custom, pig, puberty, quality, reproductive longevity, SNP genotype, SowPro90
INTRODUCTION
Sow fertility, innate and adaptive immunity are critical factors that could significantly impact productivity of swine operations, especially following exposure to environmental stressors (Serenius and Stalder, 2006; Rowland et al., 2012; Engle et al., 2014). Sow reproductive traits are generally lowly heritable (Trenhaile et al., 2016) and expressed late in life making early selection for these traits difficult. Age at puberty is the earliest indicator of reproductive longevity (Tart et al., 2013). Late onset of puberty was associated with a decrease in service rate (Graves, 2015) and a decreased probability to generate multiple parities (Tart et al., 2013). As a result, identification of pleiotropic sources that influence phenotypic variation of age at puberty and other fertility traits have the potential to improve phenotypic prediction of these traits expressed late in life.
Using multiple approaches including transcriptomic and genomic sequencing and genome-wide association study (GWAS), we have identified potential genetic variants influencing fertility (Tart et al., 2013; Nonneman et al., 2016b; Trenhaile et al., 2016; Wijesena et al., 2017), immune response (Engle et al., 2014; Kreikemeier et al., 2015; Walker et al., 2018), and SNP with predicted loss of function (Keel et al., 2017). These variants were integrated into “SowPro90,” a custom Affymetrix Axiom myDesign SNP array.
Diverse genotyping platforms with varying SNP densities are often used across various subsets of animals for genomic evaluation. Previous reports showed that quality metrics and distribution of quality data differ across genotyping platforms in human (Hong et al., 2012) and livestock (Berry et al., 2016). This study evaluated data obtained from two genotyping platforms, SowPro90 and Porcine SNP60 BeadArray, and established the optimal quality control parameters across platforms to identify high-confidence genotypes for downstream analysis.
MATERIALS AND METHODS
This study was approved by the University of Nebraska-Lincoln (UNL) Institutional Animal Care and Use Committee (Project ID: 1677).
Animal Populations
Tissue and DNA samples were available from 1,644 experimental sows from the UNL resource population. The experimental sows were developed to investigate the effect of genetics and diet on age at puberty and their reproductive potential. The genetic makeup of dams of the experimental sows was comprised of Nebraska Index Line and commercial maternal Landrace × Large White crossbred lines while the sires were from two unrelated commercial Landrace lines. A detailed description of the resource population and the phenotypic data collected was previously reported (Miller et al., 2011; Wijesena et al., 2017). In addition, tissue and DNA samples were available from 2,309 animals from two commercial populations with different genetics including Landrace and Yorkshire pigs as well as maternal Landrace × Large White crossbred sows.
Genotypic Data Collection
The DNA was isolated from tail tissue samples collected from the 1,644 sows in the UNL population generated in 14 batches as described in Wijesena et al. (2017). Genotyping was completed with Porcine SNP60 BeadArray versions 1 and 2 (Illumina Inc., San Diego, CA), and SNP with a GenCall score ≥0.4, and SNP and samples with a call rate ≥ 80% were retained for downstream analysis (n = 53,529; Wijesena et al., 2017). In addition, 277 sows in the UNL population representing both extremes of the distribution for their genomic prediction values for age at puberty (~10% of the gilts with genomic prediction values representing early age at puberty and ~8% of the gilts representing late age at puberty) were also genotyped with SowPro90 (Thermo Fisher Scientific Inc., Waltham, MA). Moreover, 2,309 animals from the two commercial populations including Landrace, Yorkshire, and maternal crossbred animals were genotyped with SowPro90.
Genomic Approaches for Novel Genetic Variants Identification
RNA sequencing. The RNA sequencing data were obtained from various swine populations and tissues. These include the hypothalamic arcuate nucleus from prepubertal (n = 12) and postpubertal gilts (n = 25) that expressed puberty at different ages (early and late) originating from the UNL population (Wijesena et al., 2017) and peripheral blood from commercial maternal crossbred (Large White × Landrace) pigs that expressed high and low levels of viremia following an experimental infection with Porcine circovirus 2b (PCV2b; n = 8, Walker et al., 2018).
The SNP detection was carried out using Genome Analysis Toolkit (GATK, version 3.1, DePristo et al., 2011) and Picard tools (version 2.1.1, Wysoker et al., 2013). Briefly, a sequence dictionary was created for the Sscrofa 10.2 reference genome (http://support.illumina.com/sequencing/sequencing_software/ igenome.html – [accessed March 7, 2016]) using CreateSequenceDictonary tool in Picard. RNA sequence BAM files were processed using Picard tools – AddOrReplaceReadGroups, MarkDuplicates, and ReorderSam. The sequence reads were split into exons and any leftover intronic regions were hard clipped using GATK SplitNCigarReads tool. The variants were called using the HaplotypeCaller tool and filtered using VariantFiltration tool in GATK (FisherStrand > 30.0 and QualitybyDepth < 2.0; Van der Auwera et al., 2013). The individual VCF files generated for each sample containing high-quality variant calls were then merged within each data set using GATK CombineVariant tool.
Genome sequencing. Landrace sires (n = 20) from the UNL population representing both ends of the distribution for average genomic prediction values for their daughters’ age at puberty were selected for whole-genome sequencing (Wijesena et al., 2017). Eleven of the sequenced sires represented early age at puberty, and nine sires represented late age at puberty. The sequence reads were mapped to Sscrofa 10.2 reference genome, and DNA variants were detected using default settings in the multiallelic and rare-variant option of BCFtools (Wijesena et al., 2017). Seventy-two founders in a U.S. Meat Animal Research Center (USMARC) experimental swine herd (12 Duroc and 12 Landrace boars and 48 Yorkshire × Landrace composite sows) were also sequenced to identify putative functional variants across the swine genome such as loss function, nonsynonymous, and regulatory SNP (Keel et al., 2017). Variant calling and filtering was performed as described in Keel et al. (2017).
Design of the SowPro90 SNP Array
The SowPro90 SNP array was designed and manufactured based on Affymetrix Axiom myDesign technology (Thermo Fisher Scientific Inc.) and contained 103,476 SNPs. The SNPs were obtained from sources mentioned above, including transcriptomic and genome sequencing and also scaffold SNP from the Porcine SNP60 BeadArray.
Briefly, transcriptomic and genomic sequence data were used to identify SNP located in genes and their proximal promoters (±2 kb region flanking the transcription start site) that overlapped the top 1% of QTL for age at puberty discovered by GWAS in the UNL (Wijesena et al., 2017) and USMARC (Nonneman et al., 2016b) resource populations. In the UNL population, the genes were identified in major 1-Mb windows extended by 500 kb in both directions (n = 42 windows) that explained the largest proportion of genetic variance for age at puberty (Wijesena et al., 2017). In the USMARC population, the genes overlapping QTL were identified in the five-SNP QTL windows extended by 300 kb in both directions (n = 222 windows) that explained the largest proportion of genetic variance for age at puberty (Nonneman et al., 2016b). Another portion of the array included SNP located in genes with ontologies associated with innate and adaptive immunity, and also SNP known to affect viral disease susceptibility (Walker et al., 2018). The immunity-related gene ontology terms were obtained from Ensemble BioMart tool (https://may2017.archive.ensembl.org/biomart/martview/– [accessed May 2, 2017]). Additionally, SNP in the proximal promoter of differentially expressed genes between gilts that expressed puberty at different ages, their upstream regulatory genes (e.g., transcription factors), genes overlapping selection sweep regions for litter size traits, and genes associated with structural soundness were included in SowPro90. A large majority of SNP incorporated in the array were gene based, located in coding (e.g., nonsynonymous, synonymous, splice region, stop gained, and stop lost) and untranslated regions (5′ and 3′) of positional candidate genes. The position of the genes was identified based on Sscrofa 10.2 reference genome annotation. The potential SNP consequences were obtained using Ensemble Variant Effect Predictor tool (https://may2017.archive.ensembl.org/info/docs/tools/vep/index.html – [accessed May 25, 2017]).
The SNP array also consisted of potential loss-of-function SNP (Keel et al., 2017) as well as DNA markers for age at puberty identified in the USMARC studies (Nonneman et al., 2016a). The scaffold SNP incorporated in the SowPro90 obtained from the Porcine SNP60 BeadArray had a minor allele frequency > 0.05 in the UNL maternal crossbred data sets used for sow reproductive (Wijesena et al., 2017) and viral disease (Walker et al., 2018) research. The SowPro90 was also supplemented with SNP included in the Neogen Porcine GGPHD Array (Neogen Genomics, Lincoln, NE) if they overlapped the top 1% of the QTL for age at puberty (Wijesena et al., 2017) and SNP from the Affymetrix Axiom Pig High Density (PigHD) Array (Groenen, 2015; Thermo Fisher Scientific Inc. Waltham, MA) located in the Swine Leukocyte Antigen complex II locus (Table 1). The SowPro90 array is commercially available and the array content can be found in the supplementary material (Supplementary Table S1).
Table 1.
Number of SNP and overlapping genes included in SowPro90
| SNP category | Number of SNP | Number of genes |
|---|---|---|
| SNP in genes and regulatory regions (RNA and genome sequencing) | ||
| 42 QTL for age at puberty (UNL) | 11,474 | 788 |
| 222 QTL for age at puberty (USMARC) | 21,490 | 1,500 |
| Adaptive and immunity genes | 16,271 | 1,015 |
| Differentially expressed genes in hypothalamic arcuate nucleus | 107 | 17 |
| Upstream regulatory genes of differentially expressed genes | 308 | 31 |
| 11 selection sweep regions for litter size | 1,286 | 220 |
| Structural soundness genes | 607 | 224 |
| Predicted loss–of-function SNP | 617 | 376 |
| SNP from commercial genotyping platforms | ||
| Illumina Porcine SNP60 BeadArray | 49,710 | |
| Neogen Porcine GGPHD Array | 1,012 | |
| Affymetrix Axiom PigHD Array | 594 | |
| Total | 103,476 | 4,171 |
UNL = University of Nebraska-Lincoln; USMARC = U.S. Meat Animal Research Center.
The SowPro90 Genotype Quality Evaluation
There were 49,710 Porcine SNP60 BeadArray SNP included in the SowPro90 design. The genotype quality of SowPro90 was evaluated by assessing the genotype concordance defined as proportion of identical genotypes for common SNP between SowPro90 and Porcine SNP60 BeadArray using 277 UNL animals genotyped with both platforms.
The initial set of common SNP present in both platforms was generated using an SNP and sample call rate ≥80%. For SowPro90, the CEL files from all genotyped samples (n = 2,586) were imported into SNPolisher tool in Axiom Analysis Suite (AxAS; Thermo Fisher Scientific Inc.,) together with library files and diploid threshold parameter settings while the rest of the parameters were set at default levels. The optimum threshold for SNP call rate was obtained by analyzing the distribution of the data quality and genotype concordance across platforms at 2% SNP call rate increments from 80% to 100%. The sample call rate threshold was obtained by analyzing the data quality at different sample call rates (80%, 90%, 93%, and 97%) defined based on the distribution of data. Finally, the SowPro90 data (n = 103,476) were re-analyzed using all genotyped animals (n = 2,586) and the newly established optimum SNP and sample call rates to generate the genotypes for downstream analysis. The SowPro90 and Porcine SNP60 BeadArray SNP were mapped to the Sscrofa 11.1 reference genome assembly (https://support.illumina.com/sequencing/sequencing_software/igenome.html – [accessed August 10, 2018]) to understand the genome-wide distribution of SNP in two platforms.
RESULTS AND DISCUSSION
Development of the SowPro90 SNP Array
Reproductive longevity is a composite trait with a low heritability (h2 = 0.04; Tart et al., 2013), including many fertility traits, and expressed late in life. Previous research found that age at puberty is a trait with moderate heritability (h2 = 0.37) and an early indicator of reproductive longevity (Tart et al., 2013). Understanding the pleiotropic sources influencing phenotypic variation of age at puberty and other fertility traits could help in the development of a reliable approach to improve genomic prediction for sow reproductive longevity. Genetic variants (SNP) overlapping QTL for age at puberty and fertility traits as well as other economically important traits such as susceptibility to viral diseases were integrated into SowPro90, a custom Axiom myDesign SNP array (Thermo Fisher Scientific Inc.). The SowPro90 incorporated 103,476 SNPs overlapping 4,171 transcribed genes. Similar custom SNP panels targeting economically important traits have been developed in cattle industry to aid in genomic selection. For example, Mullen et al. (2013) and Boichard et al. (2018) developed custom SNP panels for dairy and beef cattle to screen for quantitative traits, lethal recessive, and congenital disorders. These panels included SNP from low-density Illumina BovineLD BeadChip and causative variants such as loss of function and nonsynonymous polymorphisms.
Approximately 50% of the SowPro90 SNP (n = 51,463) were identified using transcriptomic and genomic sequencing (Table 1). Of these, 32,964 SNPs were located in 2,288 genes overlapping major QTL for age at puberty discovered in prior studies (Nonneman et al., 2016b; Wijesena et al., 2017). The SNP array was also supplemented with 16,271 SNPs located in 1,015 genes involved in innate and adaptive immunity including genes overlapping the Swine Leukocyte Antigen complex II locus and other genes influencing viral disease susceptibility (Kreikemeier et al., 2015; Walker et al. 2018; Table 1). The rest of the SNPs identified were located in differentially expressed genes (including their proximal promoters) in the hypothalamic arcuate nucleus of gilts that expressed puberty early vs. late as well as their upstream regulatory genes or upstream trans modulators (Wijesena et al., 2017), SNP in genes overlapping 11 selective sweep regions for litter size traits (Trenhaile et al., 2016), and SNP in genes associated with structural soundness (Fan et al., 2009, 2011). Additionally, 565 SNPs located in 504 genes characterized by potential loss of function (Keel et al., 2017) as well as previously reported DNA markers for age at puberty (Nonneman et al., 2016a) were included. The remaining ~50% of the array content (n = 51,316) was comprised of scaffold SNP obtained from Porcine SNP60 BeadArray and SNP overlapping the top 1% of QTL for age at puberty and immunity genes obtained from other commercially available platforms (e.g., Neogen Porcine GGPHD Array and Affymetrix PigHD array; Table 1).
Initial Quality Evaluation of the SowPro90
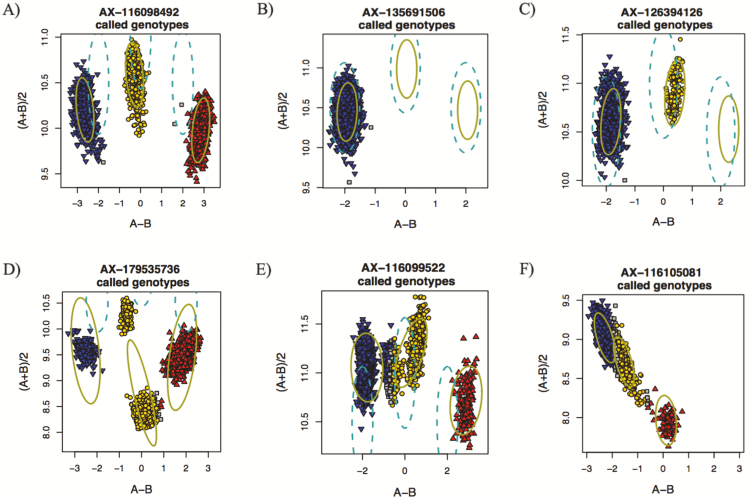
The AxAS software classified SNP into six quality classes (Figure 1) including (i) polymorphic SNP with three genotype clusters that passed all the quality control parameters (poly high resolution), (ii) SNP that were monomorphic (mono high resolution), (iii) SNP with only one homozygote and heterozygote genotype clusters (no minor homozygotes), (iv) SNP with more than one heterozygote cluster or the average signal for heterozygote cluster much lower than for the homozygote clusters (off target SNP), (v) SNP with genotype call rate below the threshold (e.g., <80%), and (vi) SNP that failed one or more quality control parameters (other). Polymorphic and monomorphic high-resolution SNP and SNP lacking minor homozygotes were recommended for downstream analysis.
Figure 1.
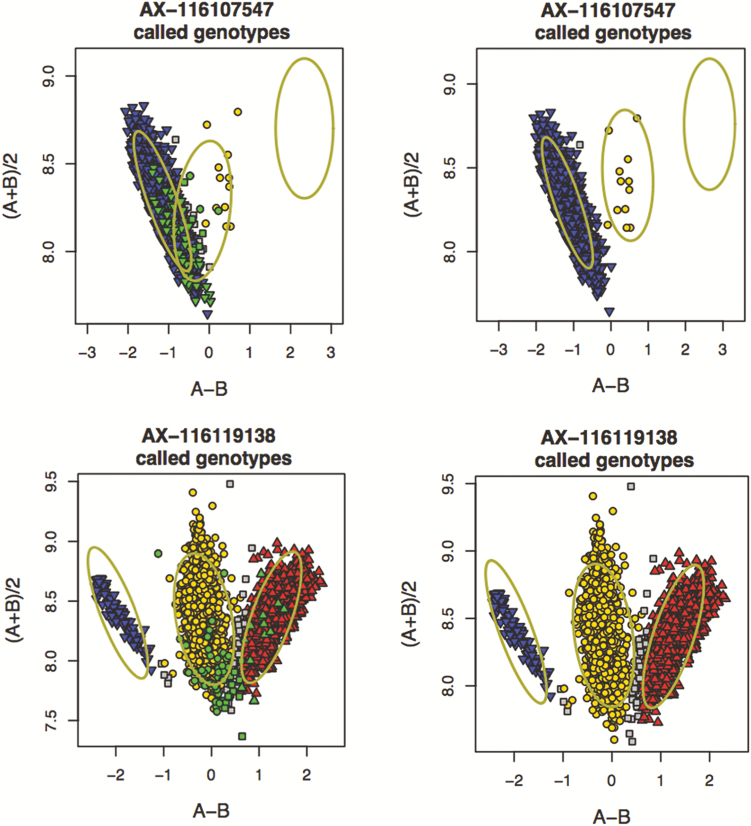
Example of SNP classification based on cluster properties by Axiom Analysis Suite. (A) poly high resolution, (B) mono high resolution, (C) no minor homozygote, (D) off target variant, (E) call rate below threshold, and (F) other. Red: AA genotype, yellow: AB genotype, blue: BB genotype, and grey: no genotype call. SNP with three genotype clusters (poly high resolution), monomorphic SNP (mono high resolution), and SNP lacking minor homozygotes (no minor homozygotes) were recommended for downstream analysis.
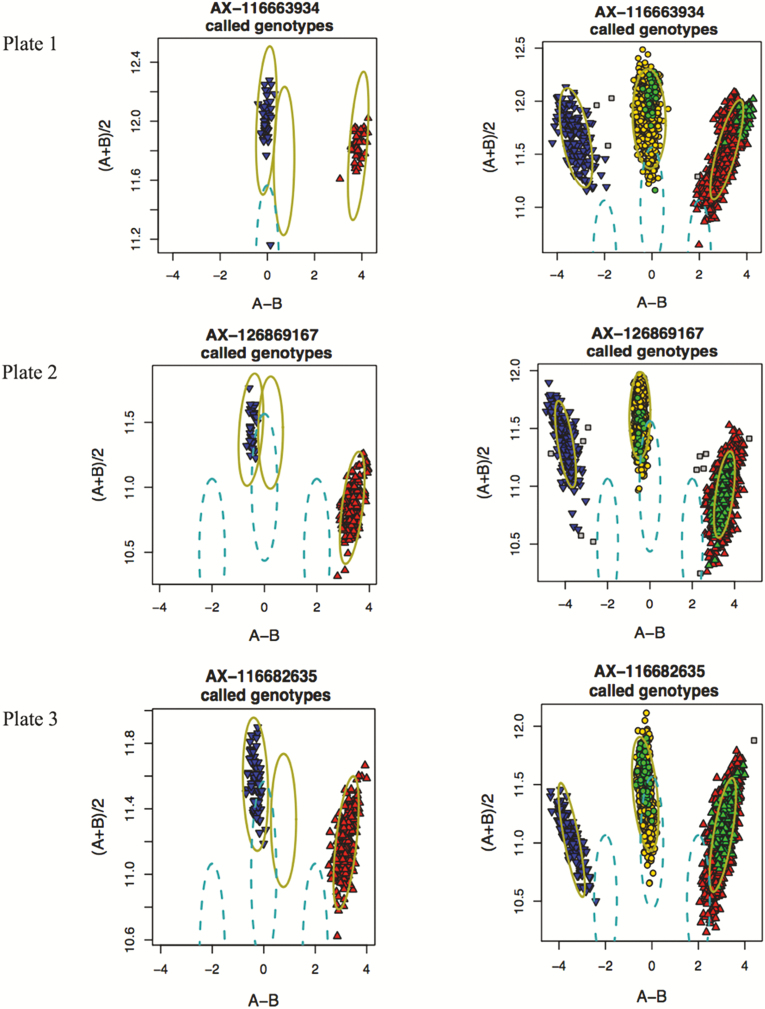
The genotype quality of SowPro90 was evaluated by merging genotype data from the three populations (n = 2,586), rather than evaluating each population or plate (96 well) separately to achieve an optimum genotype clustering and reliable genotype calls. For example, the number of recommended SNP at default call rates (97% SNP and 94% sample call rate) ranged from 62,145 to 94,428 when genotype quality was evaluated in 16 separate plates and only 36,897 (36%) SNPs were consistently recommended across all plates. We hypothesize that the variation in data is a result of limited genetic diversity present in single plates. Plates do not usually include randomly assigned samples from different genetic backgrounds, but rather batches of samples of similar genetics. Some of the nonrecommended SNP could therefore result from a lack of genetic diversity within a plate (or within genetic line). For example, SNPs were identified that appeared to lack heterozygote genotypes but both homozygote genotypes (AA and BB) being present. The proportion of this group of SNP varied from 3% to 5% when samples from three genetic lines were allocated to three separate plates and analyzed individually. When the genotype data from the three populations were merged (n = 2,586), 20% to 86% of these SNPs were recommended and “rescued” since heterozygote calls were previously miscalled as one of the homozygote calls (AA or BB) (Figure 2). The absence or rarity of one of the homozygote classes in samples of similar genetics (or single plates) could limit the power to distinguish homozygote from heterozygote clusters. This problem does not exist when a large and diverse set of samples is analyzed together, and all three genotypes are expected to be present.
Figure 2.
Genotype calling in three individual plates, each representing a genetic line (left) generated non recommended SNP that appeared to lack heterozygote genotypes but both homozygote genotypes being present (AA and BB). When the three populations were merged (right), these SNPs were recommended since one of the homozygote calls (AA or BB) was miscalled in individual plates when they were actually heterozygotes. Each data point represents the genotype of an animal. Red: AA genotype, yellow: AB genotype, blue: BB genotype, and gray: no genotype call. The green dots represent the animals in each individual plate.
The Genotype Concordance Rate Per SNP Between SowPro90 and SNP60 BeadArray
The evaluation of the genotype concordance was performed using animals (n = 277) genotyped with both SowPro90 and Porcine SNP60 BeadArray. There were 49,710 common SNPs in both platforms. Of those, 44,708 SNPs with ≥80% call rate in each platform were selected for evaluation of the genotype concordance. This call rate is generally considered acceptable in high-density genotyping (Tart et al., 2013).
The mean genotype concordance rate per SNP across the 44,708 SNPs was 98.4%. A small proportion (~0.65%) of the homozygote genotypes in one platform was called as alternate homozygote genotypes in the other platform (Table 2A). A subset of discordant SNP with <90% genotype concordance rate (n = 2,418) exhibited higher incidences of calling heterozygote variants as homozygotes and calling homozygote genotypes in one platform as alternate homozygote genotypes in the other platform. Approximately 13% of the heterozygous Porcine SNP60 BeadArray genotypes were called homozygous in SowPro90 and ~11% of the homozygous Porcine SNP60 BeadArray genotypes were called alternate homozygotes in SowPro90 (Table 2B). Selecting for SNP with ≥90% genotype concordance rate (n = 42,290) increased the overall mean genotype concordance to 99.5% (Table 2C).
Table 2.
Genotype occurrence (%) in Porcine SNP60 BeadArray and SowPro90 using common genotyped animals (n = 277).
| A | Porcine SNP60 BeadArray | SowPro90 | ||
|---|---|---|---|---|
| AA | AB | BB | ||
| AA | 98.39 | 0.96 | 0.66 | |
| AB | 0.90 | 98.22 | 0.88 | |
| BB | 0.64 | 0.70 | 98.66 | |
| B | Porcine SNP60 BeadArray | SowPro90 | ||
| AA | AB | BB | ||
| AA | 81.69 | 6.90 | 11.41 | |
| AB | 13.39 | 73.34 | 13.27 | |
| BB | 11.00 | 5.24 | 83.75 | |
| C | Porcine SNP60 BeadArray | SowPro90 | ||
| AA | AB | BB | ||
| AA | 99.32 | 0.65 | 0.03 | |
| AB | 0.2 | 99.61 | 0.19 | |
| BB | 0.03 | 0.46 | 99.51 |
(A) All SNPs (n = 44,708), (B) SNP with <90% genotype concordance (n = 2,418), and (C) SNP with ≥90% genotype concordance (n = 42,290).
Similar mean genotype concordance rates were reported by other studies in livestock and humans. An evaluation of 49,859 SNPs in sheep samples (n = 84) genotyped by Illumina and Affymetrix platforms reported a 98.1% mean genotype concordance rate per SNP (Berry et al., 2016). This study reported that only a small proportion (~0.3%) of homozygous genotypes in one platform was called as alternate homozygous in the other platform. In humans, a comparison between six technical replicates genotyped with both Illumina and Affymetrix platforms reported a mean genotype concordance of 98.8% (Hong et al., 2012). In a simulation study, Hong et al. (2012) reported that using genotypes with lower concordance in GWAS could affect the research outcome. Jiang et al. (2013) evaluated the within sample genotype concordance between Illumina and Affymetrix platforms in humans for 146,885 SNPs and reported a mean genotype concordance of 99.9%.
A potential source of limited genotype concordance across SNP is represented by minor allele frequency (MAF). Across the 44,708 SNPs used for genotype concordance evaluation, the number of SNP in different MAF categories ranged from 369 (0.83%) that were monomorphic to 5,608 (12.5%) with MAF between 0.45 and 0.50 (Table 3). The lowest mean genotype concordance (61.6%) was observed for monomorphic SNP while the highest (98.89%) was observed for SNP with MAF > 0.05 to ≤ 0.10 (Table 3). The study of Berry et al. (2016) observed similar results.
Table 3.
Number and genotype concordance of SNP in each minor allele frequency (MAF) category
| MAF category | Common SNP used for genotype concordance evaluation (44,708 SNPs) | Mean genotype concordance (%, 44,708 SNPs) | SNPs in SowPro90 at ≥97% SNPs and ≥ 93% sample call rates (89,040 SNPs) |
|---|---|---|---|
| 0 | 369 | 61.63 | 9,293 |
| >0 to ≤0.05 | 2,593 | 98.06 | 9,269 |
| >0.05 to ≤0.1 | 3,338 | 98.89 | 7,643 |
| >0.1 to ≤0.15 | 3,713 | 98.76 | 7,149 |
| >0.15 to ≤0.2 | 4,111 | 98.83 | 7,298 |
| >0.2 to ≤0.25 | 4,439 | 98.68 | 7,835 |
| >0.25 to ≤0.3 | 4,910 | 98.73 | 7,856 |
| >0.3 to ≤0.35 | 5,116 | 98.78 | 8,134 |
| >0.35 to ≤0.4 | 5,055 | 98.73 | 8,015 |
| >0.4 to ≤0.45 | 5,456 | 98.76 | 8,241 |
| >0.45 to ≤0.5 | 5,608 | 98.73 | 8,307 |
Evaluation of Optimal SNP Call Rate
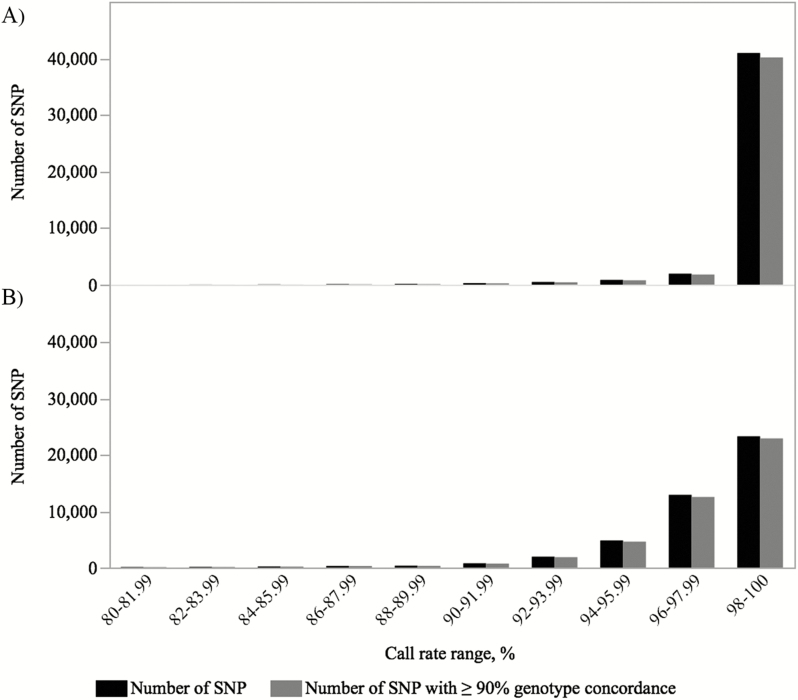
Concordance between the genotypes across platforms was evaluated within 2% SNP call rate ranges starting from 80% up to 100% (Figure 3). For SowPro90, the SNP call rates of the majority of SNP (n = 40,939, 91.6%,) were distributed between 98% and 100% (Figure 3A). In this range, there were 40,155 SNPs (98.1%) with ≥90% genotype concordance between the platforms and 35,767 of these SNP (89.1%) had ≥99% genotype concordance. Based on the distribution of SNP genotype call rates and the number of SNP with ≥ 90% genotype concordance, an SNP call rate ≥97% was considered to be the optimal threshold for SowPro90 quality evaluation which allowed retention of a maximum number of SNP (n = 42,151, 94.3%). The mean genotype concordance of the SNP with ≥97% SNP call rate was 98.7%.
Figure 3.
Distribution of SNP call rates at 80% SNP and 80% sample call rates. (A) SowPro90 SNP and (B) Porcine SNP60 BeadArray SNP. Black: total number of SNP and gray: number of SNP with ≥90% concordance.
The SNP call rates for most of the SNP (n = 41,043, 91.8%) on the Porcine SNP60 BeadArray were distributed between 94% and 100% and in this range there were 40,085 SNPs (97.7%) with ≥90% genotype concordance between platforms (Figure 3B). Based on call rate distribution and concordance data, a SNP call rate ≥94% was considered to be the optimal threshold for Porcine SNP60 BeadArray (Figure 3B). The mean genotype concordance of the SNPs with ≥94% SNPs call rate was 98.6%.
Evaluating the Optimal Sample Call Rate
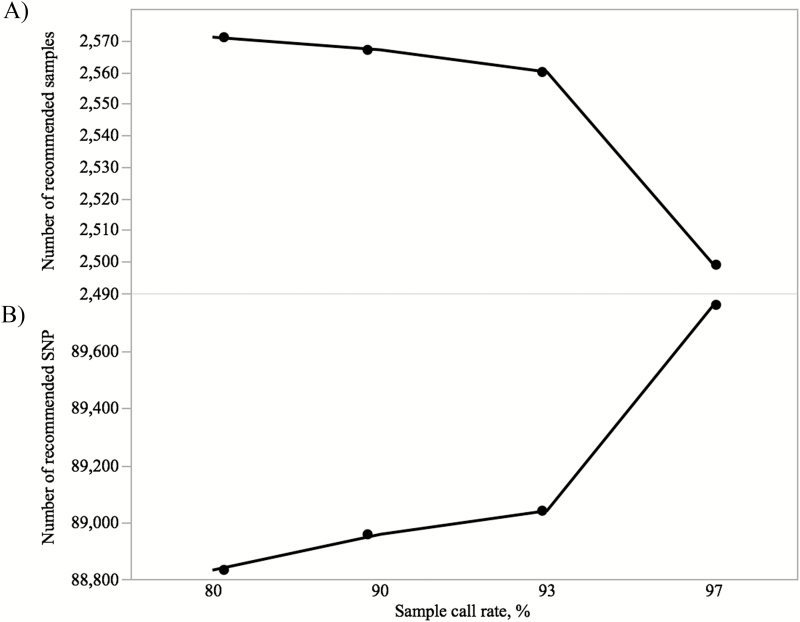
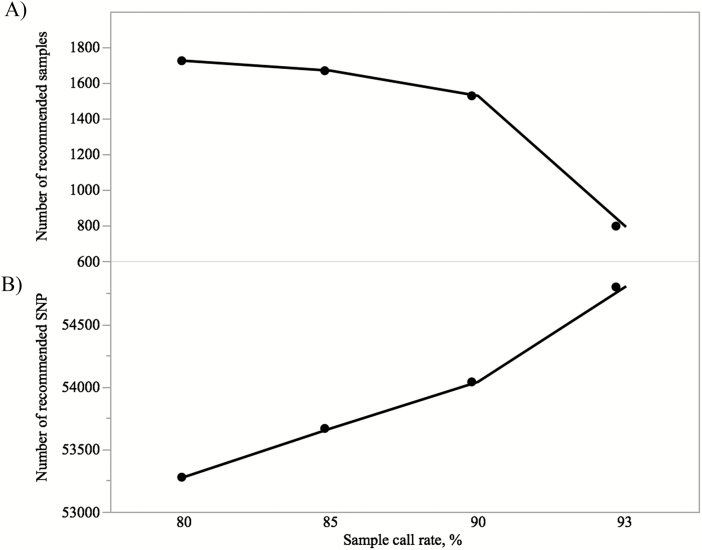
To identify the optimal sample call rate for SowPro90 quality evaluation, the SNP array (n = 103,476) was re-analyzed at an SNP call rate ≥97% and different sample call rates (e.g., 80%, 90%, 93%, and 97%) using all the genotyped animals (n = 2,586). The largest number of genotyped animals (n = 2,571) was retained at ≥80% sample call rate (Figure 4A). An increase in sample call rate (and removing low-quality samples) led to an improvement in genotype clustering and a larger number of SNP that passed the filtering criteria (Figure 5). For example, at ≥97% sample call rate there were 932 additional SNP (+1%) retained compared with >80% sample call rate (Figure 4B) while 72 animals (−2.9%) failed the threshold parameters (Figure 4A). Therefore, to retain the maximum number of animals with highest quality genotypes, a less stringent ≥93% sample call rate was considered to be the optimal threshold for SowPro90. In this case, there were 308 additional SNP retained with only 11 animals failing this threshold parameter compared with ≥80% sample call rate.
Figure 4.
A) Number of recommended samples and (B) number of recommended SNP at 97% SNP call rate and different sample call rates for SowPro90.
Figure 5.
An increase in sample call rate improved the overall genotype clustering as illustrated by two examples. The genotype clustering was compared at 97% SNP and 80% (left) and 97% (right) sample call rates. Each data point represents the genotype of an animal. Red: AA genotype, yellow: AB genotype, blue: BB genotype, and gray: no genotype call. The green dots represent the 72 animals that were removed when the sample call rate increased from 80% to 97%.
Similar to above, the Porcine SNP60 BeadArray (n = 61,565) data were re-analyzed at ≥94% SNPs call rate and different sample call rates (e.g., 80%, 85%, 90%, and 93%) using 1,836 genotyped animals. The largest number of SNP was retained at 93% sample call rate with 43% of the animals failing these filtering criteria (Figure 6). To retain the maximum number of animals with highest quality genotypes, 85% was determined to be the optimal sample call rate for Porcine SNP60 BeadArray data, retaining 53,668 SNPs and 1,668 animals (Figure 6). This sample call rate was also suggested as the minimum by Purfield et al. (2016).
Figure 6.
A) Number of recommended samples and (B) number of recommended SNPs at 94% SNPs call rate and different sample call rates for Porcine SNP60 BeadArray.
Final Genotype Evaluation of the SowPro90
At ≥97% SNPs and ≥93% sample call rates, there were 89,040 (86%) recommended SNPs and 2,560 (98.7%) samples that passed the quality thresholds for SowPro90. The recommended SNP included 74,661 poly high resolution, 9,293 mono high resolution, and 5,086 SNPs without homozygotes for the minor allele. The monomorphic SNPs were presumably sequencing artifacts as the majority of these SNPs (94%) originated from transcriptome (73%) and genome (21%) sequencing. The average observed heterozygosity of polymorphic SNP was 0.35 and the average MAF of the recommended array content was 0.25. The number of recommended SNPs in different MAF categories (excluding monomorphic SNP) ranged from 7,149 (>0.1 to 0.15) to 9,269 (0 to 0.05, Table 3).
In the Porcine SNP60 BeadArray, there were 1,812 SNPs overlapping 42 QTL windows for age at puberty identified in the UNL population. These regions were enriched with 13,511 SNPs in SowPro90. The average distance between SNP in the enriched QTL regions was 5,150 bp for SowPro90 compared to 38,753 bp for Porcine SNP60 BeadArray. In the updated swine genome assembly (Sscrofa 11.1), the SNPs included in SowPro90 were distributed across the 18 autosomes and the X chromosome ranging from 1,669 (SSCX) to 8,413 SNPs (SSC7) per chromosome. At the genome-wide level, there were an average of 36 SNPs per 1-Mb window for SowPro90 compared with 21 SNPs for the Porcine SNP60 BeadArray.
CONCLUSION
Distribution of genotype quality across various platforms tends to differ, likely due to different chemistries and allelic detection approaches used for genotyping. For example, the majority of SowPro90 SNP (91.6%) had an SNP call rate ≥98% while for Porcine SNP60 BeadArray the majority of SNP (91.8%) had an SNP call rate ≥94% (Figure 2), suggesting that these platforms used different stringency levels when calling genotypes. For these specific ranges, a high genotype concordance rate (≥98.5%) between platforms was observed. Based on these observtions, it is not ideal to use the same threshold parameters for quality evaluations across different genotyping platforms. The approach used in this study, assessing genotype concordance between two genotyping platforms at different SNP and sample call rates, allowed identification of specific quality thresholds necessary to retain the maximum number of SNP and samples with high quality. This strategy will be helpful when integrating data from various genotyping sources for different applications such as genomic evaluations and genome-wide association.
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
This research was funded by Agriculture and Food Research Initiative Competitive (grant no. 2013-68004-20370) and Hatch Funds from the USDA National Institute of Food and Agriculture. Mention of a trade name, proprietary product, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. The USDA is an equal opportunity provider and employer. We thank the staff at the UNL Swine Research Facility for tissue and data collection and Thermo Fisher Scientific Inc. for designing and manufacturing the SowPro90.
LITERATURE CITED
- Berry D. P., O’Brien A., Wall E., McDermott K., Randles S., Flynn P., Park S., Grose J., Weld R., and McHugh N.. 2016. Inter- and intra-reproducibility of genotypes from sheep technical replicates on illumina and affymetrix platforms. Genet. Sel. Evol. 48:86. doi: 10.1186/s12711-016-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boichard D., Boussaha M., Capitan A., Rocha D., Hoze C., Sanchez M.-P., Tribout T., Letaief R., Croiseau P., Grohs C., Li W., Harland C., Charlier C., Lund M. S., Sahana G., Georges M., Barbier S., Coppieters W., Fritz S., and Guldbrandtsen B.. 2018. Experience from large scale use of the EuroGenomics custom SNP chip in cattle. Proc. World Congress Genetics Appl. Livest. Prod. Mol. Genet. 4:675 http://www.wcgalp.org/proceedings/2018/experience-large-scale-use-eurogenomics-custom-snp-chip-cattle [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M.,. et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle T. B., Jobman E. E., Moural T. W., McKnite A. M., Bundy J. W., Barnes S. Y., Davis E. H., Galeota J. A., Burkey T. E., Plastow G. S.,. et al. 2014. Variation in time and magnitude of immune response and viremia in experimental challenges with porcine circovirus 2b. BMC Vet. Res. 10:286. doi: 10.1186/s12917-014-0286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Onteru S. K., Du Z. Q., Garrick D. J., Stalder K. J., and Rothschild M. F.. 2011. Genome-wide association study identifies loci for body composition and structural soundness traits in pigs. PLoS One 6:e14726. doi: 10.1371/journal.pone.0014726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Onteru S. K., Mote B. E., Serenius T., Stalder K. J., and Rothschild M. F.. 2009. Large-scale association study for structural soundness and leg locomotion traits in the pig. Genet. Sel. Evol. 41:14. doi: 10.1186/1297-9686-41-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves K. L. 2015. Factors associated with puberty onset and reproductive performance of gilts [graduate theses and dissertations]. https://lib.dr.iastate.edu/etd/14580. [Google Scholar]
- Groenen M. A. M. 2015. Development of a high-density Axiom® porcine genotyping array to meet research and commercial needs. Plant and Animal Genome Conference, January 10–14, 2015; San Diego, CA. [Google Scholar]
- Hong H., Xu L., Liu J., Jones W. D., Su Z., Ning B., Perkins R., Ge W., Miclaus K., Zhang L., Park K., Green B., Han T., Fang H., Lambert C. G., Vega S. C., Lin S. M., Jafari N., Czika W., Wolfinger R. D., Goodsaid F., Tong W., and Shi L.. 2012. Technical reproducibility of genotyping SNP arrays used in genome-wide association studies. PLoS One 7:e44483. doi: 10.1371/journal.pone.0044483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Willner D., Danoy P., Xu H., and Brown M. A.. 2013. Comparison of the performance of two commercial genome-wide association study genotyping platforms in Han Chinese samples. G3 (Bethesda). 3:23–29. doi: 10.1534/g3.112.004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel B. N., Nonneman D. J., and Rohrer G. A.. 2017. A survey of single nucleotide polymorphisms identified from whole-genome sequencing and their functional effect in the porcine genome. Anim. Genet. 48:404–411. doi: 10.1111/age.12557. [DOI] [PubMed] [Google Scholar]
- Kreikemeier C. A., Engle T. B., Lucot K. L., Kachman S. D., Burkey T. E., and Ciobanu D. C.. 2015. Genome-wide analysis of TNF-alpha response in pigs challenged with porcine circovirus 2b. Anim. Genet. 46:205–208. doi: 10.1111/age.12262. [DOI] [PubMed] [Google Scholar]
- Miller P. S., Moreno R., and Johnson R. K.. 2011. Effects of restricting energy during the gilt developmental period on growth and reproduction of lines differing in lean growth rate: responses in feed intake, growth, and age at puberty. J. Anim. Sci. 89:342–354. doi: 10.2527/jas.2010-3111. [DOI] [PubMed] [Google Scholar]
- Mullen M. P., McClure M. C., Kearney J. F., Waters S. M., Weld R., Flynn P., Creevey C. J., Cromie A. R., and Berry D. P.. 2013. Development of a custom SNP chip for dairy and beef cattle breeding, parentage, and research. In: INTERBULL, August 23–25, Nantes, France. Bulletin 47.
- Nonneman D. J., Lents C. A., Kalbfleisch T. S., Vallet J. L., and Rohrer G. A.. 2016a. Potential functional variants associated with age at puberty in a validation population of swine. Plant and Animal Genome Conference, January 9–13, 2016; San Diego, CA [Google Scholar]
- Nonneman D. J., Schneider J. F., Lents C. A., Wiedmann R. T., Vallet J. L., and Rohrer G. A.. 2016b. Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 17:50. doi: 10.1186/s12863-016-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D. C., McClure M., and Berry D. P.. 2016. Justification for setting the individual animal genotype call rate threshold at eighty-five percent. J. Anim. Sci. 94:4558–4569. doi: 10.2527/jas.2016-0802. [DOI] [PubMed] [Google Scholar]
- Rowland R. R., Lunney J., and Dekkers J.. 2012. Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance. Front. Genet. 3:260. doi: 10.3389/fgene.2012.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serenius T., and Stalder K. J.. 2006. Selection for sow longevity. J. Anim. Sci. 84(Suppl): E166–E171. doi: 10.2527/2006.8413_supple166x. [DOI] [PubMed] [Google Scholar]
- Tart J. K., Johnson R. K., Bundy J. W., Ferdinand N. N., McKnite A. M., Wood J. R., Miller P. S., Rothschild M. F., Spangler M. L., Garrick D. J.,. et al. 2013. Genome-wide prediction of age at puberty and reproductive longevity in sows. Anim. Genet. 44:387–397. doi: 10.1111/age.12028. [DOI] [PubMed] [Google Scholar]
- Trenhaile M. D., Petersen J. L., Kachman S. D., Johnson R. K., and Ciobanu D. C.. 2016. Long-term selection for litter size in swine results in shifts in allelic frequency in regions involved in reproductive processes. Anim. Genet. 47:534–542. doi: 10.1111/age.12448. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J.,. et al. 2013. From fastq data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 43:11.10.1–11.1033. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. R., Engle T. B., Vu H., Tosky E. R., Nonneman D. J., Smith T. P. L., Borza T., Burkey T. E., Plastow G. S., Kachman S. D.,. et al. 2018. Synaptogyrin-2 influences replication of porcine circovirus 2. PLoS Genet. 14:e1007750. doi: 10.1371/journal.pgen.1007750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesena H. R., Lents C. A., Riethoven J. J., Trenhaile-Grannemann M. D., Thorson J. F., Keel B. N., Miller P. S., Spangler M. L., Kachman S. D., and Ciobanu D. C.. 2017. GENOMICS SYMPOSIUM: using genomic approaches to uncover sources of variation in age at puberty and reproductive longevity in sows. J. Anim. Sci. 95:4196–4205. doi: 10.2527/jas2016.1334. [DOI] [PubMed] [Google Scholar]
- Wysoker A., Tibbetts K., and Fennel T.. 2013. Picard tools version 1.90 http://picard.sourceforge.net – [accessed December 14, 2016]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.