Abstract
Introduction
GATA3 is a critical transcription factor in maintaining the differentiated state of luminal mammary epithelial cells. We sought to determine the prognostic and predictive roles of GATA3 genotypes for breast cancer.
Patients and Methods
Twelve single nucleotide polymorphisms (SNPs) were genotyped in two breast cancer cohorts, including SWOG S8897 trial where patients were treated with adjuvant chemotherapy (CAF vs. CMF) or untreated, and the observational Pathways Study.
Results
In the S8897 trial, rs3802604 and rs568727 were associated with disease-free survival (DFS) and overall survival (OS) in the treated group, regardless of chemotherapy regimen. The GG genotype of rs3802604 conferred poorer OS (adjusted HR=2.45, 95% CI: 1.48-4.05) and DFS (adjusted HR=1.95, 95% CI: 1.27-2.99) compared to the AA genotype. Similar associations were found for rs568727. In contrast, no association with either SNP was found in the untreated group. Subgroup analyses indicated that these two SNPs more strongly influenced outcomes in the patients who also received tamoxifen. However, the associations in the subgroup with tamoxifen treatment were not replicated in the Pathways Study, possibly due to substantial differences between the two patient cohorts, such as chemotherapy regimen and length of followup. Results from joint analyses across these two cohorts were marginally significant, driven by the results in S8897. Bioinformatic analyses support potential functional disruption of the GATA3 SNPs in breast tissue.
Conclusions
The present study provides some evidence for the predictive value of GATA3 genotypes for breast cancer adjuvant therapies. Future replication studies in appropriate patient populations are warranted.
Keywords: GATA3, SNPs, breast cancer, chemotherapy, outcomes
MicroAbstract
In an ancillary study to a completed clinical trial, we identified GATA3 genotypes predictive of survival after adjuvant chemotherapy, particularly among those subsequently treated with tamoxifen. Although the replication effort in a second cohort proved to be futile due to substantial differences between the discovery and the replication cohorts, mechanistic exploration of the identified variants supported their functional significance.
Introduction
GATA binding protein 3 (GATA3) is among the most important and well-studied transcription factors in breast cancer (1–3). GATA3 and its downstream target FOXA1 are critical for maintaining the luminal differentiation status of mammary epithelial cells (4, 5). It has a strong and specific presence in estrogen receptor (ER) positive luminal subtype tumors, which is gradually lost as tumor cells acquire a stem cell-like status. Moreover, GATA3 is the third most commonly-mutated gene in breast cancer (11%), following TP53 and PIK3CA, with the mutations occurring mostly in luminal subtypes (6).
Because GATA3 suppresses the epithelial to mesenchymal transition of tumor cells and thus inhibits metastatic dissemination, in part, via regulating microRNAs (7), the expression of GATA3 gene and protein has been investigated and shown to be prognostic in breast cancer; although the evidence has not been definitive. In an early genome-wide gene expression meta-analysis, GATA3 was identified as one of the top genes wherein low expression was related to poor breast cancer outcomes (8). Similar results were reported with IHC expression of GATA3, with the effects dependent on ER status in some studies (9–13). Several studies have also evaluated IHC expression of GATA3 with treatment response, and found it predictive of favorable endocrine therapy outcomes (14–16). In a neoadjuvant study, GATA3 negativity was an independent predictor for pathologic complete response (17), whereas in the case of chemoresistant triple-negative breast cancer, a GATA3-containing luminal-like gene signature was associated with better prognosis (18).
Despite mounting evidence of GATA3 as a critical transcription factor with potential prognostic and predictive value in breast cancer, few studies have investigated the inherited genetic variations with breast cancer outcomes. This becomes an even more warranted research question to address given the existing high-level evidence that two GATA3 single nucleotide polymorphisms (SNPs), rs3824662 and rs3781093, have been identified as the top hits in genome-wide association studies (GWAS) of risk and relapse of childhood acute lymphoblastic leukemia (ALL) (19–21) and another SNP with risk of Hodgkin lymphoma (22). It is also notable that GATA3 plays an important role in T-cell differentiation reminiscent of that in mammary epithelial cells (23). Thus, using a cross-validating study design, we sought to investigate the prognostic and predictive roles of GATA3 SNPs in two cohorts of breast cancer patients.
Materials and Methods
Patient Populations
Data and DNA samples for this study were obtained from two breast cancer patient populations: A completed phase III cooperative group trial SWOG S8897 (INT-0102) and a cohort of breast cancer survivors in the Pathways Study. Study schemas of the two populations are shown in Supplemental Figure 1 and descriptive characteristics of the two patients populations are shown in Table 1.
Table 1.
Descriptive characteristics of patient populations in S8897 trial and Pathways Study
| Characteristic | S8897 T reated Group |
S8897 Untreated Group |
Pathways Chemotherapy Group |
Pathways No Chemotherapy Group |
|---|---|---|---|---|
| n = 441 (%) | n = 799 (%) | n = 515 (%) | n = 483 (%) | |
| Age, years | ||||
| < 40 | 66 (15) | 70 (9) | 93 (19) | 21 (4) |
| 40-49 | 165 (37) | 225 (28) | 211 (44) | 125 (26) |
| 50-59 | 105 (24) | 211 (26) | 118 (24) | 127 (26) |
| 60-69 | 76 (17) | 199 (25) | 74 (15) | 121 (25) |
| ≥ 70 | 29 (7) | 94 (12) | 19 (4) | 89 (18) |
| Median(Range), years | 49 (27-85) | 54 (24-89) | 47 (25-76) | 56 (23-89) |
| Race | ||||
| White (non-Hispanic) | 391 (89) | 750 (93) | 270 (52) | 298 (62) |
| Black (non-Hispanic) | 27 (6) | 30 (4) | 37 (7) | 37 (8) |
| Hispanic | 12 (3) | 12 (2) | 93 (18) | 57 (12) |
| Other | 11 (2) | 7 (1) | 115 (23) | 91 (19) |
| Menopausal Status | ||||
| Premenopausal | 231 (52) | 306 (38) | 350 (68) | 213 (44) |
| Postmenopausal | 210 (48) | 493 (62) | 165 (32) | 270 (56) |
| Overall Survival Event | ||||
| Yes | 329 (75) | 577 (72) | 45 (9) | 32 (7) |
| No | 112 (25) | 222 (28) | 470 (91) | 451 (93) |
| Disease Free Survival Event | ||||
| Yes | 289 (66) | 482 (60) | 80 (16) | 71 (15) |
| No | 152 (34) | 317 (40) | 435 (84) | 412 (85) |
As previously described, women diagnosed with T1-T3a node-negative invasive breast cancer were eligible for S8897 (24). Patients were initially assigned to high risk (tumor size ≥ 2 cm, or hormonal receptor negative), low risk (tumor size ≤ 1 cm), or uncertain risk (tumor size between 1-2 cm or hormonal receptor positive) groups. The last group was subsequently classified as either high or low risk based on the S-phase fraction by flow cytometry. Patients in the high-risk group were randomly assigned to one of two adjuvant chemotherapy treatment arms to receive either an anthracycline-based regimen (CAF) consisted of 6 cycles of oral cyclophosphamide, intravenous doxorubicin, and 5-fluorouracil, or the non-anthracycline regimen (CMF) consisted of 6 cycles of oral cyclophosphamide, intravenous methotrexate, and 5-fluorouracil. After receiving chemotherapy, women in the treated group were randomly assigned to receive or to not receive tamoxifen endocrine therapy for 5 years. Women in the low-risk group were followed for observation only and did not receive either adjuvant chemotherapy or endocrine therapy. The trial enrolled a total of 3,965 patients between 1989-1993. After a median of 10.8 years follow-up, the study concluded with slightly better overall survival but not disease-free survival in patients treated with CAF vs. CMF (24). For this ancillary study, genomic DNA was required. Because lymph node tissues were not collected from patients in the initial high-risk group in S8897, only those from the initially low-risk group (n=490) and the initially uncertain-risk group (n=1,029) who were subsequently assigned to high- and low-risk groups for adjuvant therapy (“treated group”, n=516) or no adjuvant therapy (“untreated group”, n=513), respectively, were included.
The Pathways Study is an observational prospective cohort study of breast cancer survivors established at Kaiser Permanente Northern California (KPNC) to investigate factors associated with recurrence and mortality (25, 26). Between 2006 and 2013, a total of 4,505 eligible patients diagnosed with incident invasive breast cancer were enrolled typically within 2 months of diagnosis. Baseline in-person interviews were conducted to query demographic, health history, and extensive lifestyle and other epidemiological information potentially related to breast cancer risk and prognosis; anthropometric measures were also taken. Blood and/or saliva samples were obtained shortly thereafter. Regular follow-ups on lifestyle and other factors were conducted via mailed or telephone questionnaires at 6, 24, and 72 months post-baseline, and health outcomes and comorbidities at 12, 24, 48, 72, and 96 months post-baseline. Complete clinical data including initial diagnosis and treatment were obtained from the KPNC Cancer Registry and KPNC electronic clinical and administrative databases. For the replication cohort, we selected a sub-cohort of patients enrolled in Pathways who received adjuvant tamoxifen therapy as the replication cohort.
Written informed consent was obtained from all patients for the use of their tissues in future research studies. This study was approved by the Institutional Review Boards at Roswell Park Comprehensive Cancer Center and KPNC.
Biospecimens and genotyping
In S8897, three 5-micron paraffin-embedded, formalin-fixed (FFPE) slides of normal lymph node tissue were used for extraction of genomic DNA. Adequate DNA for genotyping was obtained from 1,253 eligible patients, including 446 in the treated group and 807 in the untreated group. An Illumina GoldenGate 384-plex custom panel was designed for genotyping, which included 12 tagSNPs selected from GATA3 based on HapMap CEU data. Two SNPs shown to be significant in the previous GWAS for childhood ALL, rs3824662 and rs3781093, were forced in during the tagSNPs selection process. For genotyping QC purpose, 2% duplicates were randomly selected and blinded in across genotyping plates and an in-house trio set was also included in each plate. The overall successful genotyping call rate was 98.3%, with complete concordance between duplicate pairs. Thirteen samples with a call rate <85% were removed from analysis, resulting in a final sample size of 441 in the treated group and 799 in the untreated group.
In the Pathways Study, genomic DNA was extracted from blood and/or saliva samples and genotyping of the same set of 12 GATA3 SNPs were conducted using the Agena Bioscience MassARRAY, with the same percent of blind duplicate samples and in-house trio samples included. The overall genotyping call rate was 98.4%, and 10 samples with a call rate <85% were removed from analysis, resulting in a final sample size of 1,000 patients.
None of the SNPs in either study had Mendelian error or violated Hardy Weinberg Equilibrium (HWE) (P > 0.001). The DNA preparation work was performed at Roswell park Data Bank and Biorepository (DBBR) resource and genotyping performed at Roswell Park Genomics Shared Resource (GSR).
Statistical analysis
Analytical endpoints were overall survival (OS), defined as the time from registration to death from any cause (censored at the last date of contact), and disease-free survival (DFS), defined as the time from registration to disease recurrence, new second primary, or death. OS and DFS probabilities were plotted with Kaplan-Meier survival curves by genotypes of each SNP separately in the treated and untreated groups in S8897, with P-values derived from log-rank test. Bonferroni correction was used to adjust for multiple testing, giving a significance threshold of P ≤ 0.001 after correcting for testing 12 SNPs, 2 endpoints in 2 subgroups (0.05/48). Using a co-dominant model of inheritance, Cox proportional hazards regression was used to estimate hazards ratios (HRs) and 95% confidence intervals (CIs) for each SNP in treated and untreated groups separately, before and after adjusting for covariates including age at diagnosis, race and ethnicity, and treatment arm in the treated group. An additive model of inheritance was used in Cox regression analysis to estimate HRs and accompanying 95% CIs per risk allele for each variant. As GATA3 plays an important role in regulating estrogen signaling in breast cancer, which may have an impact on treatment response to tamoxifen, the 12 SNPs were further analyzed among patients who received tamoxifen following adjuvant chemotherapy and those who did not. Similar approaches were used to analyze data from the Pathways Study cohort. All analyses were conducted in R program.
Bioinformatic analysis
To explore the potential functional impact of GATA3 SNPs, bioinformatic analyses were conducted based on publicly available data resources using HaploReg v4.1 (27). The program comprehensively interrogates chromatin state (promoter histone marks, enhancer histone marks, DNAse sensitivity), protein binding, transcription factor motif, expression quantitative trait loci (eQTL), and evolutionary sequence conservation across mammals.
Results
GATA3 SNPs and breast cancer outcomes in the treated vs. untreated groups in S8897
Descriptive characteristics of S8897 patients with successful genotyping results are summarized in Table 1. Patients in the treated group were younger with a median age of 49 years, compared to patients in the untreated group with a median age of 54 years (P <0.05), and were also more likely to be premenopausal (P <0.05). A majority of patients (92%) self-reported as non-Hispanic Whites, 5% non-Hispanic Blacks, 2% Hispanics, and 1% others.
Figure 1 depicts the results of log-rank tests for OS and DFS in the treated and untreated groups, along with the linkage disequilibrium (LD) map. Of the twelve GATA3 SNPs tested, four were associated with both OS and DFS in the treated group with a nominal significance (P <0.05). After correction for multiple testing, two SNPs, rs3802604 and rs568727 in relatively strong LD (r2=0.79), remained statistically significant (threshold P=0.001) in the treated group; whereas the other two SNPs, rs568727 and rs10905284, became non-significant.
Figure 1.
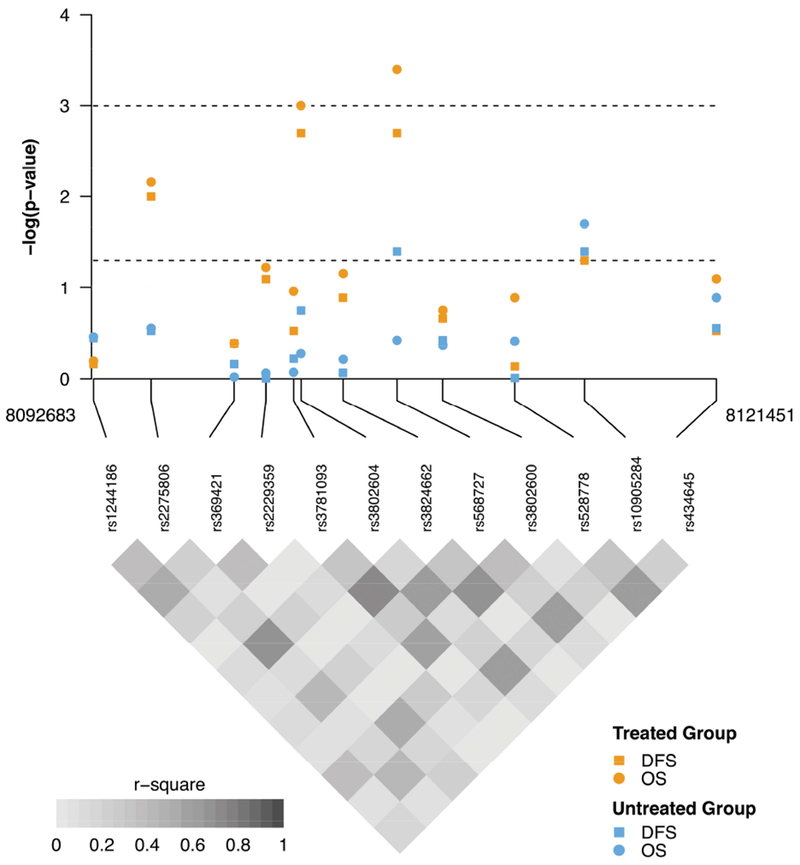
Genotyped single nucleotide polymorphisms in GATA3 and breast cancer outcomes in S8897 trial
P-values from log-rank test of single nucleotide polymorphisms with breast cancer outcomes, including disease-free survival (DFS) and overall survival (OS), are log10-transformed and plotted along with linkage disequilibrium map. Analyses were stratified by the treated and untreated group and results from each subgroup analysis are indicated by colors and shapes shown above. The lower and upper dashed lines indicate nominal significance threshold (0.05) and that after correction for multiple testing (0.001), respectively.
Kaplan-Meier curves of OS by the top two SNPs are shown in Figure 2 and curves of DFS shown in Supplemental Figure 2. Among women treated with adjuvant therapy, those carrying the GG genotype of rs3802604 had poorer OS compared to the AA genotype (HR=2.45, 95% CI: 1.48-4.05, P=0.0005), in a dose-dependent manner (per G allele: HR=1.57, 95% CI: 1.21-2.03, P=0.0007) (Table 2). Similarly, those in the treated group carrying the AA genotype of rs568727 had poorer OS compared to the CC genotype (HR=2.57, 95% CI: 1.43-4.30, P=0.0003), also in a dose-dependent manner (per A allele: HR=1.59, 95% CI: 1.23-2.06, P=0.0005). Analyses of those 2 SNPs with DFS showed results consistent with OS and the results were similar in subgroups defined by CAF vs. CMF regimen. Neither of those two SNPs were associated with breast cancer outcomes in the untreated group of patients who did not receive adjuvant therapy (Table 2; for complete results of all 12 SNPs, see Supplemental Tables 1 and 2). Interaction tests of the two SNPs between the treated and untreated groups did not reach statistical significance (P>0.05).
Figure 2.
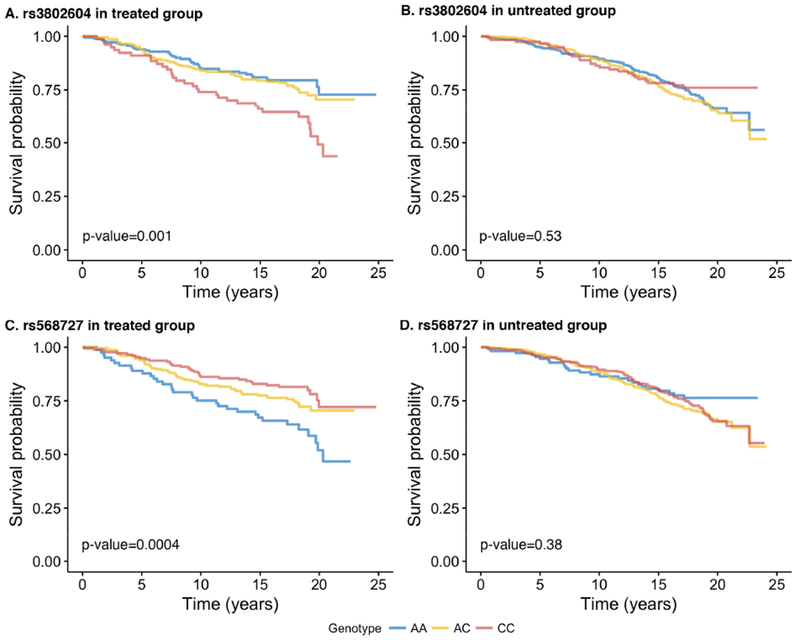
Kaplan-Meier survival curves of overall survival by genotypes of the top two single nucleotide polymorphisms in GATA3 in S8897 trial
Kaplan-Meier curves of overall survival by the genotypes of rs3802604 and rs568727, the two single nucleotide polymorphisms in GATA3 remained significant after correcting for multiple comparison, are plotted separately for the treated and untreated group in S8897 trial. P-values are from log-rank test.
Table 2.
GATA3 single nucleotide polymorphisms and breast cancer outcomes in S8897 trial
| Genotype | Treated Group (n=441) |
Untreated Group (n=799) |
||||
|---|---|---|---|---|---|---|
| # events/patients | Adjusted HR (95% CI) | Adjusted P | # events/patients | Adjusted HR (95% CI) | Adjusted P | |
| A. Overall survival | ||||||
| rs3802604 | ||||||
| AA | 31/164 | 1.00 | 96/337 | 1.00 | ||
| GA | 51/206 | 1.36 (0.87-2.14) | 0.18 | 99/342 | 1.07 (0.81-1.42) | 0.64 |
| GG | 30/70 | 2.45 (1.48-4.05) | 0.0005 | 27/166 | 0.78 (0.51-1.20) | 0.27 |
| Per risk allele | 1.57 (1.21-2.03) | 0.0007 | 0.94 (0.78-1.13) | 0.48 | ||
| rs568727 | ||||||
| CC | 28/160 | 1.00 | 95/328 | 1.00 | ||
| AC | 52/204 | 1.50 (0.94-2.39) | 0.09 | 102/357 | 1.03 (0.78-1.36) | 0.86 |
| AA | 32/77 | 2.57 (1.43-4.30) | 0.0003 | 24/111 | 0.74 (0.47-1.16) | 0.19 |
| Per risk allele | 1.59 (1.23-2.06) | 0.0005 | 0.91 (0.75-1.10) | 0.32 | ||
| B. Disease-free survival | ||||||
| rs3802604 | ||||||
| AA | 49/164 | 1.00 | 134/337 | 1.00 | ||
| GA | 66/206 | 1.08 (0.74-1.56) | 0.69 | 145/342 | 1.15 (0.91-1.45) | 0.25 |
| GG | 37/70 | 1.95 (1.27-2.99) | 0.002 | 38/116 | 0.80 (0.56-1.15) | 0.24 |
| Per risk allele | 1.38 (1.10-1.73) | 0.005 | 0.96 (0.82-1.12) | 0.62 | ||
| rs568727 | ||||||
| CC | 45/160 | 1.00 | 131/328 | 1.00 | ||
| AC | 68/204 | 1.22 (0.83-1.79) | 0.31 | 153/357 | 1.14 (0.90-1.44) | 0.28 |
| AA | 39/77 | 2.02 (1.31-3.12) | 0.001 | 32/111 | 0.71 (0.48-1.04) | 0.08 |
| Per risk allele | 1.41 (1.13-1.76) | 0.003 | 0.92 (0.79-1.08) | 0.33 | ||
Footnote: Hazard ratios (HRs), 95% confidence intervals (CIs) and p-values were derived from Cox hazard regression models with adjustment for age at enrollment, race and ethnicity, and treatment arms. Estimates of per risk allele were derived from an additive genetic model by treating genotypes as 0, 1 and 2 according to the copy number of risk allele. P-values ≤ 0.001 were considered statistically significant to account for multiple testing for 12 genetic variants in GATA3.
GATA3 SNPs and breast cancer outcomes in the treated group by tamoxifen therapy in S8897
Because GATA3 is critical for the estrogen signaling pathways in breast cancer, the analyses in the treated group in S8897 were further stratified by whether or not patients received adjuvant tamoxifen therapy following chemotherapy. As shown in Table 3 for the top two SNPs, the associations of rs3802604 and rs568727 with both OS and DFS remained nominally significant among patients treated tamoxifen, with the associations restricted to the homogenous variant genotypes, although the p-values did not meet the cutoff of significance level after correction for multiple comparison (P>0.001). Among those not treated with tamoxifen, the associations, particularly with DFS, were weaker and non-significant after correction for multiple testing. Interaction tests of the two SNPs between the group treated with and without tamoxifen did not reach statistical significance (P>0.05).
Table 3.
GATA3 single nucleotide polymorphisms and breast cancer outcomes among patients in the treated group by tamoxifen therapy in S8897 trial
| Genotype | Tamoxifen Treated Group (n=220) |
No Tamoxifen Treated Group (n=221) |
||||
|---|---|---|---|---|---|---|
| # events/patients | Adjusted HR (95% CI) | Adjusted P | # events/patients | Adjusted HR (95% CI) | Adjusted P | |
| A. Overall survival | ||||||
| rs3802604 | ||||||
| AA | 14/78 | 1.00 | 17/85 | 1.00 | ||
| GA | 19/105 | 1.09 (0.54-2.20) | 0.82 | 31/100 | 1.56 (0.86-2.81) | 0.14 |
| GG | 17/35 | 2.66 (1.31-5.43) | 0.007 | 13/35 | 2.23 (1.08-4.60) | 0.03 |
| Per risk allele | 1.66 (1.13-2.44) | 0.009 | 1.50 (1.05-2.14) | 0.03 | ||
| rs568727 | ||||||
| CC | 13/76 | 1.00 | 14/82 | 1.00 | ||
| AC | 19/105 | 1.02 (0.50-2.06) | 0.96 | 33/99 | 1.97 (1.05-3.71) | 0.04 |
| AA | 18/37 | 2.89 (1.40-5.96) | 0.004 | 14/40 | 2.28 (1.08-4.80) | 0.03 |
| Per risk allele | 1.71 (1.15-2.54) | 0.008 | 1.51 (1.07-2.14) | 0.02 | ||
| B. Disease-free survival | ||||||
| rs3802604 | ||||||
| AA | 21/78 | 1.00 | 28/85 | 1.00 | ||
| GA | 27/105 | 0.95 (0.53-1.70) | 0.87 | 38/100 | 1.16 (0.71-1.89) | 0.56 |
| GG | 22/35 | 2.59 (1.41-4.73) | 0.002 | 15/35 | 1.53 (0.81-2.87) | 0.19 |
| Per risk allele | 1.62 (1.16-2.26) | 0.005 | 1.22 (0.90-1.67) | 0.2 | ||
| rs568727 | ||||||
| CC | 20/76 | 1.00 | 24/82 | 1.00 | ||
| AC | 28/105 | 0.98 (0.55-1.74) | 0.94 | 40/99 | 1.43 (0.86-2.39) | 0.17 |
| AA | 22/37 | 2.60 (1.41-4.82) | 0.002 | 17/40 | 1.65 (0.88-3.07) | 0.12 |
| Per risk allele | 1.60 (1.14-2.24) | 0.007 | 1.29 (0.96-1.75) | 0.09 | ||
Footnote: Hazard ratios (HRs), 95% confidence intervals (CIs) and p-values were derived from Cox hazard regression models with adjustment for age at enrollment, race and ethnicity, and treatment arms. Estimates of per risk allele were derived from an additive genetic model by treating genotypes as 0, 1 and 2 according to the copy number of risk allele. P-values ≤ 0.001 were considered statistically significant to account for multiple testing for 12 genetic variants in GATA3.
In addition to the above two SNPs, six other GATA3 SNPs showed some evidence of association with OS and/or DFS among patients treated with tamoxifen, and one of them, rs369421, remained significant after correction for multiple testing (P<0.001; Supplemental Tables 4 and 5). Interestingly, the two GWAS SNPs previously associated with childhood ALL risk and relapse, rs3781093 and rs3824662, were associated with OS, and the latter also with DFS, at a nominal significance level among patients with no tamoxifen therapy. The directions of the associations were consistent with those from the ALL GWAS.
Replication of GATA3 SNPs with breast cancer outcomes in the Pathways Study
Because the associations of GATA3 SNPs with breast cancer outcomes appeared to be stronger in patients treated with adjuvant chemotherapy followed by tamoxifen, consistent with its putative roles in estrogen signaling pathways in breast cancer, the replication analysis was conducted in the Pathways Study among 1,000 patients with early-stage breast cancer who received adjuvant tamoxifen treatment. Descriptive characteristics of the replication cohort are summarized in Table 1. The median follow-up time was 6.3 years, with an OS rate of 7.7% and a DFS rate of 15.1%. In Cox regression models, none of the twelve SNPs, including rs3802604 and rs568727, were associated with either OS and DFS. The results remained unchanged when stratified by whether patients received adjuvant chemotherapy (Supplemental Table 6), aromatase inhibitors, menopausal status, or when the analyses were restricted to non-Hispanic White women (data not shown). In a joint analysis in which data were pooled from the SWOG and Pathways Study cohorts, the AA genotype of rs568727 was associated with both OS and DFS at a nominal significance level (P<0.05), and the GG genotype of rs3802604 showed a marginal association with OS and DFS (Table 4). However, the test for study heterogeneity between the two cohorts was significant (P<0.05).
Table 4.
Joint analysis of GATA3 single nucleotide polymorphisms and breast cancer outcomes in the S8897 trial and Pathways Study
| Genotype | # events/patients | Adjusted HR (95% CI) | Adjusted P |
|---|---|---|---|
| A. Overall survival | |||
| rs3802604 | |||
| AA | 34/261 | 1.00 | |
| GA | 44/368 | 0.98 (0.62-1.53) | 0.92 |
| GG | 23/122 | 1.59 (0.94-2.71) | 0.09 |
| Per risk allele | 1.23 (0.93-1.63) | 0.15 | |
| rs568727 | |||
| CC | 39/290 | 1.00 | |
| AC | 37/364 | 0.71 (0.45-1.11) | 0.13 |
| AA | 25/96 | 1.86 (1.11-3.12) | 0.02 |
| Per risk allele | 1.26 (0.94-1.68) | 0.13 | |
| B. Disease-free survival | |||
| rs3802604 | |||
| AA | 52/261 | 1.00 | |
| GA | 71/368 | 0.96 (0.67-1.38) | 0.84 |
| GG | 33/122 | 1.49 (0.96-2.30) | 0.08 |
| Per risk allele | 1.18 (0.94-1.49) | 0.15 | |
| rs568727 | |||
| CC | 57/290 | 1.00 | |
| AC | 66/364 | 0.85 (0.59-1.21) | 0.37 |
| AA | 33/96 | 1.80 (1.16-2.79) | 0.009 |
| Per risk allele | 1.25 (0.98-1.59) | 0.07 | |
Footnote: Hazard ratios (HRs), 95% confidence intervals (CIs) and p-values were derived from Cox hazard regression models with adjustment for study, age at enrollment, race and ethnicity, and treatment arms. Estimates of per risk allele were derived from an additive genetic model by treating genotypes as 0, 1 and 2 according to the copy number of risk allele.
Potential functional impact of GATA3 SNPs
As summarized in Table 5, there is ample bioinformatic evidence supporting that those SNPs may alter the functionality of GATA3. For rs3802604, the most significant SNP in our study, it was predicted to disrupt chromatin state in a variety of tissues, including breast; alter motifs for multiple transcription factors, including estrogen receptor alpha central in the ER signaling pathway; and perturb the binding of proteins in Chip-Seq experiments. Moreover, this SNP has also been identified as a cis-eQTL associated with GATA3 expression in peripheral blood monocytes (P=1.7e-6) (28).
Table 5.
Bioinformatic evidence of potential functionality of GATA3 SNPs
| Variant | Promoter histone marks |
Enhancer histone marks |
DNAse sensitivity | Proteins bound in Chip-Seq experiment |
Motifs changed | eQTL | Evolutionary conservation |
|---|---|---|---|---|---|---|---|
| rs1244186 | 19 tissues (including BRST) | 9 tissues (including BRST) | 35 tissues (including BRST) | FOXA1,FOXA2,HNF4A,P300,TCF4,HAE2F1 | NANOG,TATA | GATA3 in lymphoblastoids | Yes |
| rs2275806 | 15 tissues (including BRST) | 14 tissues (including BRST) | 36 tissues (including BRST) | CTCF,RAD21,SMC3,YY1,CTCFL,HAE2F1,POL2,ZNF143 | GR,ZBRK1 | ||
| rs369421 | 15 tissues (including BRST) | 5 tissues (including BRST) | 26 tissues (including BRST) | BATF,NFKB,PU1,POL2,CTBP2 | EBF,ETS,PU1,STAT,TATA | GATA3 in whole blood | Yes |
| rs2229359 | 6 tissues (including BRST) | 9 tissues (including BRST) | 11 tissues (including BRST) | CMYC,POL2 | EGR1,LBP1,NRSF,PAX4 | Yes | |
| rs3781093 | 10 tissues (including BRST) | 9 tissues (including BRST) | 15 tissues (including BRST) | ZNF263 | CEMP1 and FCGR2B in breast tumors | ||
| rs3802604 | 14 tissues (including BRST) | 10 tissues (including BRST) | 34 tissues (including BRST) | BATF,EBF1,POL2,SIX5,NFKB,P300,RAD21,ZNF263 | BP1,ESR1,EWSR1-FL1,GR,NRSF,PU1,RAD21,SMC3,SPZ1,TATA,WT1,ZNF263,ZNF143 | GATA3 in blood monocytes | Yes |
| rs3824662 | BRST | 13 tissues (including BRST) | 12 tissues (including BRST) | PU1,HAE2F1 | NF1,NRSF,PAX6,ZBTB3 | XAGE1B in blood mononcytes | |
| rs568727 | ESC, KID | 10 tissues (including BRST) | 9 tissues (including BRST) | POL2 | MAF,NR2F2 | ||
| rs3802600 | 4 tissues | BLD | GATA2 | GR,HSF | |||
| rs528778 | BRN | 6 tissues (including BRST) | BRST,BRN,SKIN | POL2 | POU5F1 | ||
| rs10905284 | ESDR,THYM,VAS | DMRT2,YY1 | AOC2 in blood monocytes |
Discussion
There are strong biological rationales for a potential predictive value of GATA3 SNPs for outcomes after breast cancer adjuvant chemotherapy, particularly among patients who were subsequently treated with tamoxifen. Results from the S8897 cohort support this hypothesis, and in silico bioinformatic prediction of SNP functionality also support these findings.
However, the observations in the S8897 cohort were not replicated in the independent Pathway Study cohort, with null results. We considered several reasons for the lack of a cross-validation success between the two cohorts. First, the length of follow-up time differed, with a median 18 years in S8897 and only 6 years in the Pathways Study. Indeed, Kaplan-Meier curves of OS by rs3802604 genotypes did not start to separate until approximately 7 years after enrollment in S8897. Nevertheless, the separation of survival curves of DFS and by the other top SNP rs568727 did become apparent early during the follow-up. Second, because of the varying lengths of follow-up time, the event rates in the Pathways Study were much lower than those in S8897. However, HRs from regression models in the Pathways Study were closer to null than any suggestive trends consistent with those from S8897. Thus, it is unclear whether simply extending the follow-up time and accumulating more events in the second cohort would result in convergence of findings with that in the first cohort. Third, in the treated group in S8897, all patients received CAF vs. CMF; while in the Pathways Study, chemotherapy regimens were more heterogeneous, with a majority of patients receiving CA (39%), CT (23%), or C monotherapy (19%) and CAF and CMF only occasionally prescribed (<1% each). Because the findings related to GATA3 SNPs in S8897 were observed primarily in the treated group but not in the untreated group without adjuvant chemotherapy, we speculated the associations to be predictive for chemotherapy efficacy. If the SNP effects of GATA3 were specific to 5-fluorouracil, which is no longer commonly used in the adjuvant settings for breast cancer, instead of cyclophosphamide or cytotoxic chemotherapy in general, we would not expect to replication of associations in the second cohort.
With the first cohort from a clinical trial conducted in the early 1990’s where treatment regimens were closely prescribed, and the second cohort from a more contemporary observational cohort based in a large integrated healthcare system which reflects a community-based clinical setting, these cohort differences are logical reasons for the inconsistent findings. If the findings had been replicated, heterogeneity between the discovery and the validation cohorts would have underscored the robustness of the GATA3 SNPs as biomarkers of potential clinical utility outside the tightly controlled “experimental” setting of clinical trials. However, failure to replicate suggests that these SNPs do not play important roles in breast cancer under current practice.
Alternatively, the lack of replication in the second cohort in our study could reflect the true lack of an impact of GATA3 genetic variations on breast cancer outcomes. The two GWAS index SNPs with ALL susceptibility and relapse, rs3824662 and rs3781093, were not significant in the treated or untreated group in S8897, but only showed associations at a nominal significance level in the subgroup of patients who received adjuvant chemotherapy without tamoxifen. Although the direction of the associations in our study of breast cancer outcomes was consistent with the GWAS findings in ALL, it is in contrast to our hypothesis of a more important role of GATA3 for endocrine therapy. There was some evidence of rs3 824662 being an eQTL and causal SNP responsible for the GWAS signal (19), and rs3824602 as an eQTL associated with GATA3 expression in monocytes, yet definitive evidence from functional studies still await, and it remains to be determined whether the functional impact is specific to leukocytes or applicable to other cell types including mammary cells.
To date, genetic association studies for breast cancer prognosis and prediction of drug efficacy and toxicity, candidate-gene based or genome-wide design alike, have not produced many bona fide risk variants, which is in stark contrast with studies of breast cancer susceptibility, where more than 100 loci have been identified through GWAS based on ever increasing sample sizes (29). It was hypothesized that the effect size of common genetic variants for toxicity and survival outcomes might be larger than that for cancer susceptibility due to lack of evolutionary selection pressure, which presumably would facilitate the power of discovery (30). However, this advantage may have been more or less dampened by the complexity of cancer outcomes as study phenotypes. Unlike binary cancer susceptibility (yes vs. no), cancer outcomes are time-varying and under influences such as cytotoxic cancer therapy, the effects of which may be overwhelming and mask any moderate genetic effects. Also, due to complex outcome phenotypes and multifaceted heterogeneity in patient characteristics and treatment modalities, it is more challenging to pool and harmonize data collected under different settings and from various sources, despite the necessity and proven value of pooling across studies in cancer susceptibility GWAS. Our present study highlights those challenges and calls for large collaborative efforts to overcome them.
In addition to the heterogeneities between the discovery cohort and the replication cohort as discussed above, another limitation should also be noted. S8897 was designed to randomize patients deemed at high risk to CMF vs. CAF with or without tamoxifen, whereas those at low risk were not randomized and did not received chemotherapy. Because achieved normal lymph nodes were not available from patients at high risk based on large tumor size or hormonal receptor negativity, they were not included in this ancillary study. Thus, the patient population in this study represented those with smaller tumor or with hormonal receptor positive cancer. This limits the generalizability of the study findings to low risk patients.
In conclusion, consistent with the established role of GATA3 as one of the most important transcription factors in breast tissue, our present study provides some indication for potential predictive values of GATA3 germline variants for breast cancer chemotherapy and endocrine therapy, and bioinformatic evidence supports these findings. However, these observations in one cohort were not replicated in another cohort. This may have been a result of substantial heterogeneity between the discovery and the replication cohorts, including differences in chemotherapy regimens and length of follow-up. Our findings call for future studies to continue investigating GATA3 SNPs in breast cancer treatment outcomes and whether they are relevant to current and evolving practice in breast cancer treatment.
Supplementary Material
Acknowledgement
The authors thank office and field staff for data collection, processing, and preparation. We thank all SWOG S8897 and Pathways Study participants for their numerous contributions to this study. Dr. Yao had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. No conflict of interest was declared by any coauthors. This study is supported in part by a Career Catalyst Award from Susan G. Komen Foundation to Dr. Yao. SWOG S8897 is supported by the National Cancer Institute at the National Institutes of Health (R01 CA167013, U10CA180888, U10CA180819, U10CA180801, UG1CA189821, and UG1 CA189830-04). The Pathways Study is supported by the National Cancer Institute at the National Institutes of Health (R01 CA105274, PI: Kushi LH; U01 CA195565, PIs: Kushi LH, Ambrosone CB; U24 CA171524, PI: Kushi LH). Biospecimens are processed and managed by the Roswell Park Comprehensive Cancer Center DataBank and BioRepository (DBBR) and genotyping assay was performed by the Genomics Shared Resource (GSR), both of which are Cancer Center Support Grant Shared Resource supported by P30 CA16056 (PI: Johnson CS). Drs. Hayes, Rae and Ambrosone are supported by the Breast Cancer Research Foundation. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies. The funders played no roles in the design and conduct of the study; in collection, management, analysis, and interpretation of the data; in preparation, review, or approval of the manuscript; or in decision to submit the manuscript for publication.
Grant support: Research reported in this publication was supported by the National Institutes of Health under Award Numbers R01 CA105274, R01 CA167013, U01 CA195565, U10CA180888, U10CA180819, U10CA180801, UG1CA189821, UG1 CA189830-04 and U24 CA171524; Susan G. Komen Foundation (S.Y.) and Breast Cancer Research Foundation (D.F.H., J.M.R. and C.B.A.). The RPCI DBBR and GSR are CCSG Shared Resources supported by NIH grant P30 CA016056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no potential conflicts of interest.
References:
- 1.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 2010;222(1):42–9. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takaku M, Grimm SA, Wade PA. GATA3 in Breast Cancer: Tumor Suppressor or Oncogene? Gene Expr. 2015;16(4):163–8. doi: 10.3727/105221615X14399878166113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asch-Kendrick R, Cimino-Mathews A. The role of GATA3 in breast carcinomas: a review. Hum Pathol. 2016;48:37–47. doi: 10.1016/j.humpath.2015.09.035. [DOI] [PubMed] [Google Scholar]
- 4.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23(1):12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127(5):1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15(2):201–13. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005;65(24):11259–64. doi: 10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 9.Yoon NK, Maresh EL, Shen D, Elshimali Y, Apple S, Horvath S, Mah V, Bose S, Chia D, Chang HR, Goodglick L. Higher levels of GATA3 predict better survival in women with breast cancer. Hum Pathol. 2010;41(12):1794–801. doi: 10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voduc D, Cheang M, Nielsen T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev. 2008;17(2):365–73. doi: 10.1158/1055-9965.EPI-06-1090. [DOI] [PubMed] [Google Scholar]
- 11.Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, Lunet N, Schmitt F. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11(3):R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciocca V, Daskalakis C, Ciocca RM, Ruiz-Orrico A, Palazzo JP. The significance of GATA3 expression in breast cancer: a 10-year follow-up study. Hum Pathol. 2009;40(4):489–95. doi: 10.1016/j.humpath.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Jacquemier J, Charafe-Jauffret E, Monville F, Esterni B, Extra JM, Houvenaeghel G, Xerri L, Bertucci F, Birnbaum D. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 2009;11(2):R23. doi: 10.1186/bcr2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parikh P, Palazzo JP, Rose LJ, Daskalakis C, Weigel RJ. GATA-3 expression as a predictor of hormone response in breast cancer. J Am Coll Surg. 2005;200(5):705–10. doi: 10.1016/jjamcollsurg.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Gulbahce HE, Sweeney C, Surowiecka M, Knapp D, Varghese L, Blair CK. Significance of GATA-3 expression in outcomes of patients with breast cancer who received systemic chemotherapy and/or hormonal therapy and clinicopathologic features of GATA-3-positive tumors. Hum Pathol. 2013;44(11):2427–31. doi: 10.1016/j.humpath.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Fang SH, Chen Y, Weigel RJ. GATA-3 as a marker of hormone response in breast cancer. J Surg Res. 2009;157(2):290–5. doi: 10.1016/j.jss.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Tominaga N, Naoi Y, Shimazu K, Nakayama T, Maruyama N, Shimomura A, Kim SJ, Tamaki Y, Noguchi S. Clinicopathological analysis of GATA3-positive breast cancers with special reference to response to neoadjuvant chemotherapy. Ann Oncol. 2012;23(12):3051–7. doi: 10.1093/annonc/mds120. [DOI] [PubMed] [Google Scholar]
- 18.Yu KD, Zhu R, Zhan M, Rodriguez AA, Yang W, Wong S, Makris A, Lehmann BD, Chen X, Mayer I, Pietenpol JA, Shao ZM, Symmans WF, Chang JC. Identification of prognosisrelevant subgroups in patients with chemoresistant triple-negative breast cancer. Clinical Cancer Res. 2013;19(10):2723–33. doi: 10.1158/1078-0432.CCR-12-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, Xu H, Gastier-Foster J, E S, Lim JY, Chen IM, Fan Y, Devidas M, Borowitz MJ, Smith C, Neale G, Burchard EG, Torgerson DG, Klussmann FA, Villagran CR, Winick NJ, Camitta BM, Raetz E, Wood B, Yue F, Carroll WL, Larsen E, Bowman WP, Loh ML, Dean M, Bhojwani D, Pui CH, Evans WE, Relling MV, Hunger SP, Willman CL, Mullighan CG, Yang JJ. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nature Genet. 2013;45(12):1494–8. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Andreu V, Roberts KG, Xu H, Smith C, Zhang H, Yang W, Harvey RC, Payne-Turner D, Devidas M, Cheng IM, Carroll WL, Heerema NA, Carroll AJ, Raetz EA, Gastier-Foster JM, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Rowe JM, Luger SM, Tallman MS, Dean M, Burchard EG, Torgerson DG, Yue F, Wang Y, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Loh ML, Willman CL, Hunger SP, Mullighan CG, Yang JJ. A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood. 2015;125(4):680–6. doi: 10.1182/blood-2014-09-595744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, da Silva Filho MI, Vijayakrishnan J, Koehler R, Thomsen H, Irving JA, Allan JM, Lightfoot T, Roman E, Kinsey SE, Sheridan E, Thompson P, Hoffmann P, Nothen MM, Muhleisen TW, Eisele L, Zimmermann M, Bartram CR, Schrappe M, Greaves M, Stanulla M, Hemminki K, Houlston RS. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298–307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 22.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, van den Berg A, Olver B, Lloyd A, Dobbins SE, Lightfoot T, van Leeuwen FE, Forsti A, Diepstra A, Broeks A, Vijayakrishnan J, Shield L, Lake A, Montgomery D, Roman E, Engert A, von Strandmann EP, Reiners KS, Nolte IM, Smedby KE, Adami HO, Russell NS, Glimelius B, Hamilton-Dutoit S, de Bruin M, Ryder LP, Molin D, Sorensen KM, Chang ET, Taylor M, Cooke R, Hofstra R, Westers H, van Wezel T, van Eijk R, Ashworth A, Rostgaard K, Melbye M, Swerdlow AJ, Houlston RS. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nature Genet. 2010;42(12): 1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9(2):125–35. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchins LF, Green SJ, Ravdin PM, Lew D, Martino S, Abeloff M, Lyss AP, Allred C, Rivkin SE, Osborne CK. Randomized, controlled trial of cyclophosphamide, methotrexate, and fluorouracil versus cyclophosphamide, doxorubicin, and fluorouracil with and without tamoxifen for high-risk, node-negative breast cancer: treatment results of Intergroup Protocol INT-0102. J Clinical Oncol. 2005;23(33):8313–21. Epub 2005/11/19. doi: 23/33/8313 [pii] 10.1200/JCO.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 25.Kwan ML, Ambrosone CB, Lee MM, Barlow J, Krathwohl SE, Ergas IJ, Ashley CH, Bittner JR, Darbinian J, Stronach K, Caan BJ, Davis W, Kutner SE, Quesenberry CP, Somkin CP, Sternfeld B, Wiencke JK, Zheng S, Kushi LH. The Pathways Study: a prospective study of breast cancer survivorship within Kaiser Permanente Northern California. Cancer Causes Control. 2008;19(10):1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao S, Kwan ML, Ergas IJ, Roh JM, Cheng TD, Hong CC, McCann SE, Tang L, Davis W, Liu S, Quesenberry CP Jr., Lee MM, Ambrosone CB, Kushi LH. Association of Serum Level of Vitamin D at Diagnosis With Breast Cancer Survival: A Case-Cohort Analysis in the Pathways Study. JAMA Oncol. 2017;3(3):351–7. doi: 10.1001/jamaoncol.2016.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. Epub 2011/11/09. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6(12):e1 Epub 2009/02/19. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, Perkins BJ, Czene K, Eriksson M, Darabi H, Brand JS, Bojesen SE, Nordestgaard BG, Flyger H, Nielsen SF, Rahman N, Turnbull C, Bocs, Fletcher O, Peto J, Gibson L, dos-Santos-Silva I, Chang-Claude J, Flesch-Janys D, Rudolph A, Eilber U, Behrens S, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Khan S, Aaltonen K, Ahsan H, Kibriya MG, Whittemore AS, John EM, Malone KE, Gammon MD, Santella RM, Ursin G, Makalic E, Schmidt DF, Casey G, Hunter DJ, Gapstur SM, Gaudet MM, Diver WR, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Berg CD, Chanock SJ, Figueroa J, Hoover RN, Lambrechts D, Neven P, Wildiers H, van Limbergen E, Schmidt MK, Broeks A, Verhoef S, Cornelissen S, Couch FJ, Olson JE, Hallberg E, Vachon C, Waisfisz Q, Meijers-Heijboer H, Adank MA, van der Luijt RB, Li J, Liu J, Humphreys K, Kang D, Choi JY, Park SK, Yoo KY, Matsuo K, Ito H, Iwata H, Tajima K, Guenel P, Truong T, Mulot C, Sanchez M, Burwinkel B, Marme F, Surowy H, Sohn C, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Gonzalez-Neira A, Benitez J, Zamora MP, Perez JI, Shu XO, Lu W, Gao YT, Cai H, Cox A, Cross SS, Reed MW, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, kConFab I, Group A, Lindblom A, Margolin, Teo SH, Yip CH, Taib NA, Tan GH, Hooning MJ, Hollestelle A, Martens JW, Collee JM, Blot W, Signorello LB, Cai Q, Hopper JL, Southey MC, Tsimiklis H, Apicella C, Shen CY, Hsiung CN, Wu PE, Hou MF, Kristensen VN, Nord S, Alnaes GI Nbcs, Giles GG, Milne RL, McLean C, Canzian F, Trichopoulos D, Peeters P, Lund E, Sund M, Khaw KT, Gunter MJ, Palli D, Mortensen LM, Dossus L, Huerta JM, Meindl A, Schmutzler RK, Sutter C, Yang R, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Hartman M, Miao H, KS Chia, Chan CW, Fasching PA, Hein A, Beckmann MW, Haeberle L, Brenner H, Dieffenbach AK, Arndt V, Stegmaier C, Ashworth A, Orr N, Schoemaker MJ, Swerdlow AJ, Brinton L, Garcia-Closas M, Zheng W, Halverson SL, Shrubsole M, Long J, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Network G, Radice P, Peterlongo P, Manoukian S, Bernard L, Bogdanova NV, Dork T, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Devilee P, Tollenaar RA, Seynaeve C, Van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Huzarski T, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Slager S, Toland AE, Ambrosone CB, Yannoukakos D, Kabisch M, Torres D, Neuhausen SL, Anton-Culver H, Luccarini C, Baynes C, Ahmed S, Healey CS, Tessier DC, Vincent D, Bacot F, Pita G, Alonso MR, Alvarez N, Herrero D, Simard J, Pharoah PP, Kraft P, Dunning AM, Chenevix-Trench G, Hall P, Easton DF. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nature Genet. 2015;47(4):373–80. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nature Rev Genet. 2013;14(1):23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


