Abstract
The aim of the present work is to provide an approach to safely deliver silver nanoparticles (AgNPs) as cytotoxic agents into cancer cells, and to provide a deeper insight into the cellular mechanisms affected by such targeted delivery. The use of mesoporous silica nanoparticles (MSNs) as nanovehicles decorated with transferrin (Tf, targeting agent) provides a nanoplatform for the nucleation and immobilization of AgNPs (MSNs-Tf-AgNPs). We have performed the physico-chemical characterization of the nanosystems and evaluated their therapeutic potential using bioanalytical strategies to estimate the efficiency of the targeting, the degree of cellular internalization in two cell lines with different TfR expression, and the cytotoxic effects of the delivered AgNPs. In addition, cellular localization of the nanosystems in cells has been evaluated by a transmission electron microscopy analysis of ultrathin sections of Human hepatocarcinoma (HepG2) cells exposed to MSNs-Tf-AgNPs. The in vitro assays demonstrate that only the nanosystem functionalized with Tf is able to transport the AgNPs inside the cells which overexpress transferrin receptors. Therefore, this novel nanosystem is able to deliver AgNPs specifically to cancer cells overexpressing Tf receptors and offers the possibility of a targeted therapy using reduced doses of silver nanoparticles as cytotoxic agents. Then, a quantitative proteomic experiment validated through the analysis of gene expression has been performed to identify the action molecular mechanisms associated with the chemotherapeutic potential of the MSNs-Tf-AgNPs nanocarriers.
Introduction
The rise of nanoscience in biomedical applications and the need for targeted therapies is a challenge in the development of nanosystems that allows reduced doses of therapeutic agents as well as their specific release in targeted cells. Nowadays, nanoparticles are a field in increasing development due to its diverse applications in consumer products, technology, industry or medicine. These materials exhibit excellent properties, such as their reduced size or improved physico-chemical properties compared to higher scale materials, and have recently allowed significant advances in therapeutic and preventive nanomedicine.1,2
It is well known that silver nanoparticles (AgNPs) have different applications based on their efficient and broad-spectrum of antibacterial activity and also exhibit plasmonic properties.3,4 Recently, AgNPs are also receiving considerable attention as potential anticancer therapeutic agents.5–7 In this case, it is important to point out that the use of AgNPs to inhibit angiogenesis or cell growth and their use in diagnosis is limited by their tendency to aggregate, as well as by the different mechanisms of toxicity that can be induced in the organism, since its administration could also provoke cytotoxic effects on healthy cells.8,9 In fact, several studies have shown the different effects of silver nanoparticles in cancer cell lines.3,10–12 Hence, the growing studies of AgNPs as therapeutic agents in cancer disease generates the need to avoid non-specific delivery and toxicity in healthy cells.13 Additionally, the AgNPs tendency to aggregate makes it necessary to improve their way of transport and selective intracellular release in cancer cells.
On the other hand, mesoporous silica nanoparticles (MSNs) possess, besides its biocompatibility, key features that make them versatile and very attractive for nanomedicine.14–16 MSNs usually show particle diameters in the 50-200 nm range and narrow pore size distributions of 3-6 nm. Their highly ordered pore network is used to store drug molecules,17,18 and their silanol rich surface provides covalent attachment of organic moieties to the MSNs to impart stealth, targeting19–21 or smart functionalities.22–24 Hence, several studies demonstrate their use in drug delivery,25–27 diagnosis28 or vaccines,29,30 among others.
Anticancer applications of nanoparticles are highly motivated by the enhanced permeability and retention (EPR) effect, which entails a preferential accumulation of the NPs in the solid tumor microenvironment, also known as passive targeting.31 For the active uptake, the differential characteristics of tumor cells, such as the over-expression of transferrin or folic acid receptors, allows the selective and specific recognition by these cells of nanomaterials presenting the corresponding ligands.32,33 An advantage of targeting with transferrin (Tf), in comparison to other signaling ligands, is its lack of complicated downstream signaling effects, because its internalization is mainly based on a nutrient uptake mechanism.34–36
Based on the above, immobilization of AgNPs in a supporting matrix together with transferrin as a cancer cell targeting ligand, would avoid AgNPs aggregation while maintaining the possibility of its selective delivery to specific cells. For this reason, the aim of this study is the synthesis and evaluation of a hybrid nanosystem consisting of metallic AgNPs well dispersed on the external surface of MSNs decorated with transferrin for cancer cell targeting. This new nanosystem exploits a biological entity, the transferrin protein, as nucleation agent to externally modify MSNs with silver nanoparticles and, at the same time, the transferrin protein works as a targeting agent in the final nanosystem. In this manuscript we explore the selective transport of silver nanoparticles to tumor cells with the aim to achieve a therapeutic delivery useful for anticancer therapy. The effectiveness of the nanosystem is confirmed by the development of a counterpart nanosystem with bovine serum albumin (BSA) as control protein. The anticancer potential of these nanosystems has been assayed in HepG2 and MC3T3-E1 cell lines with clear difference in transferrin receptor (TfR) expression, including a TEM study of ultrathin sections of HepG2 cells exposed to the nanosystem. Furthermore, a quantitative proteomics study validated by gene expression analysis has been performed to evaluate the differential nuclear proteins expression of HepG2 cells after exposure to the effective nanosystem. The results lead to the identification of key protein targets and molecular mechanisms associated with the potential antitumoral properties of the designed nanosystem.
Experimental
Reagents and equipment
Fluorescein isothiocynate (FITC), tetraethylorthosilicate (TEOS), cetyltrimethylammonium bromide (CTAB), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC.HCl), 2-morpholinoethanesulfonic acid (MES), bovine serum albumin (BSA), transferrin human (Tf), citric acid, silver nitrate 99.9% and water (HPLC grade) were purchased from Sigma-Aldrich. 3-Aminopropyltriethoxysilane 97% (APTS), 3-(triethoxysilyl)propylsuccinic anhydride 94% (TESPSA) were purchased from ABCR GmbH & Co.KG. All other chemicals (ammonium nitrate, absolute EtOH, dry toluene, NaOH, etc.) were of the highest quality commercially available and used as received.
The analytical methods used to characterize the synthesized compounds were as follows: thermogravimetric and differential thermal analysis (TGA), chemical microanalyses, solid state magic angle spinning (MAS) NMR and cross polarization (CP) MAS NMR spectroscopy, low-angle powder X-ray diffraction (XRD), N2 adsorption porosimetry, electrophoretic mobility measurements to calculate the values of zeta-potential (ζ), dynamic light scattering (DLS), scanning electron microscopy (SEM), energy dispersive X-ray spectroscopy (EDS) and transmission electron microscopy (TEM). The equipment and conditions used are described in the Electronic Supplementary Information (ESI).
Materials synthesis
MSNs and MSNs-COOHext materials were synthetized as published,37 see details in the Electronic Supplementary Information (Figure SI.1).
MSNs-Protein
A starting MSNs-COOHext material with a nominal value of –COOH groups of 1.22 × 10−3 mol/g SiO2 was used for the protein anchorage. Prior to the conjugation of proteins, the -COOH groups on the surface of MSNs-COOHext were activated. For this activation, EDC·HCl (820 mg, 10 equiv per nominal –COOH groups) was dissolved in water and added to a vigorously stirred suspension of MSNs-COOHext (0.4 g) well dispersed in water (150 mL, HPLC grade). The mixture was stirred at RT for 3 h in the dark. Then, the solid was centrifuged and rinsed with water to remove the residuals of EDC. Then, activated MSNs-COOHext was re-dispersed in MES monohydrate (50 mM, pH depending on the protein) under gently stirring. After that, each protein (BSA or Tf) was dissolved in MES monohydrate 50 mM (pH 4.8 or pH 6, respectively) and added over the material suspension. The mixture was stirred overnight in the dark, centrifuged at 11000 rpm for 20 min, the solid exhaustively washed with water and finally dried. Materials were denoted as MSNs-BSA and MSNs-Tf, respectively.
Silver nanoparticles nucleation onto MSNs-Protein
For the nucleation of AgNPs, silver nitrate was used as silver ions source. The procedure was optimized depending on the reduction agent used and the isoelectric point of each protein.
MSNs-BSA-AgNPs
5 mL of a silver nitrate solution (1 mg/mL) were added over a dispersion of 10 mg of MSNs-BSA in water and the mixture stirred 1 h at RT in the dark. After this, a solution of citric acid (Ag+/citric acid, 1:2) was added dropwise and stirred overnight in the dark at RT. The obtained solid was washed with water and dried.
MSNs-Tf-AgNPs
10 mL of a silver nitrate solution (1 mg/mL) were added over a dispersion of 10 mg of MSNs-Tf in HEPES buffer 20 mM (pH 8.2) and the mixture stirred 1 h at RT in the dark. After this, water was added to obtain a good dispersion that was stirred overnight in the dark at RT. The obtained solid was washed with water and dried.
In vitro cell assays
Cell culture
Human hepatocellular carcinoma cell line (HepG2) and mouse osteoblast-like cell line (MC3T3-E1) purchased from ATCC were used to evaluate internalization and biocompatibility of the nanomaterials. HepG2 cells were maintained in Dulbecco´s modified Eagle´s medium (DMEM) and MC3T3-E1 in alpha modified Eagle´s medium (a-MEM). Both culture media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C and 5% CO2.
Cytotoxicity assay
For evaluating the cell viability, HepG2 cells were seeded on 96-well plates 24 h prior to the experiment. After cell attachment, they were exposed to 10, 50 and 100 μg/mL of different materials for 24, 48 and 72 h of contact times. Then, 20 μL of 3-(4,5-dimethyl-thiazol-2-yl)2,5-diphenyl tetrazolium bromide (MTT, 5 mg/mL) were added to each well and incubated for 5 h at 37 °C. Then, the MTT solution was removed and 100 μL of dimethyl sulfoxide were added to dissolve the insoluble purple formazan products. The absorbance at a 595 nm was measured using a microplate reader (TECAN) and the cell viability calculated through the relation between the absorbance of treated-cells and the absorbance of control cells.
Live-dead staining assay
For calculating the percentage of dead cells, HepG2 cells were seeded on 6-well plates 24 h prior to the experiment. After cell attachment, they were exposed to 10, 50 and 100 μg/mL of different materials (MSNs-Tf and MSNs-Tf-AgNPs) for 24 h. Then, cells were harvested and stained with trypan blue. The percentages of live and dead cells were obtained using an automated cell counter (Countess II, Fisher Scientific).
Cell uptake assays
Two different assays were carried out to evaluate the cell uptake of MSN-Protein and MSNs-Tf-AgNPs materials. In order to confirm the preferential uptake of transferrin targeted-material, MC3T3-E1 and HepG2 cells were seeded on 6-wells plates and exposed to 100 μg/mL of MSNs-BSA and MSNs-Tf materials for 24 h. To evaluate the effect of metallic nanoparticles after cell uptake, several concentrations (10, 25, 50 μg/mL) were tested for different exposure times (24 and 48 h). In both cases, the procedure was the following: after the contact time, the medium was aspirated and the cells were washed with PBS and harvested using a 0.25% trypsin-EDTA solution. After 10 or 5 min, depending on the cell line, the reaction was stopped with culture medium and the cells were centrifuged at 289 g for 5 min and resuspended in PBS for analysis by flow cytometry. Trypan Blue (0.4%) was added at that time to quench the fluorescence of the MSNs adhered to the outside membrane of the cells. The percentage of cells that had internalized MSNs was quantified as the fraction of fluorescein positive cells among the number of total cells. Then, the intensity of the green fluorescence of the positive cells was measured.
Localization of MSNs-Tf-AgNPs inside HepG2 cells
Cells were exposed to MSNs-Tf-AgNPs at two concentrations (10 and 25 μg/mL) and incubated at 37 °C in 5% CO2 for 24 h. Afterwards, the cells were harvested, rinsed with PBS and fixed with 2.5% glutaraldehyde and 4% p-formaldehyde in PBS for 4 h at 4 °C. After this time, cells were washed twice with PBS and incubated in PBS at 4 °C overnight. After incubation with 1% OsO4 in Milli-Q water for 1 h at RT, the samples were washed, dehydrated in a graded series of acetone and embedded in resin for 72 h at 67 °C. Ultrafine sections were cut with ultramicrotome, deposited on copper grids, stained with uranyl acetate and chrome citrate and analyzed by transmission electron microscopy (JEOL 2100, CNME).
Quantitative proteomics
Metabolic labelling
HepG2 cells were maintained in DMEM medium supplemented with 10% dialyzed FBS, 100 units per mL of penicillin/streptomycin and either naturally-occurring isotope abundances (“light”) or stable isotope-labeled (“heavy”) 13C6 arginine and 13C6 lysine amino acids. Culture media were refreshed when 100% (1×107 cells) plate confluence was reached and the cells were grown for at least 6 doublings to allow full incorporation of the labeled amino acids. Six large-scale SILAC replicates (3×107 cells per condition) were performed. The full incorporation of labeled amino acids was verified by MS analysis of a protein digest (data not shown).
Protein extraction, SDS-page and in gel digestion
After differential labeling, control and cells exposed to 25 μg/mL MSN-Tf-AgNPs for 48 h were mixed in a 1:1 ratio. Nucleic acid binding proteins fraction was extracted by following the extraction protocol of Qproteome Nuclear Protein Kit (QIAGEN®). Protein extracts were separated by SDS-PAGE on 10% SDS-polyacrylamide gels, visualized by Coomassie blue staining and the gel lanes were cut horizontally into 20 sections. Excised gel bands were de-stained firstly in 25 mM ammonium bicarbonate followed by a 50:50 25 mM ammonium bicarbonate/acetonitrile solution and then were dried. Gel pieces were rehydrated with 12.5 ng/μL trypsin solution in 25 mM ammonium bicarbonate and incubated overnight at 37 °C. Peptides were extracted using acetonitrile and 5% formic acid, dried by vacuum centrifugation and reconstituted in 12 μL 2% acetonitrile 0.1% formic acid.
Mass spectrometry analysis: protein identification and quantification
Peptide mixtures were analyzed using nanoflow LC-MS/MS (Eksigent). Peptides were loaded onto a 0.3 × 10 mm C18 precolumn (SGE) and separated by a reverse-phase column (75 μm × 15 cm fused silica capillary C18 HPLC PepMap column, 3 μm, 100 Å, Thermo) with linear gradient of 5-95% acetonitrile in 0.1% aqueous solution of formic acid. The samples were delivered over 120 min at a flow rate of 200 nL/min through the analytical column to a stainless steel nanobore emitter (Proxeon). The peptides were scanned and fragmented with an LTQ XL linear ion trap mass spectrometer (Thermo Scientific) operated in data-dependent ZoomScan and MS/MS switching mode using the three most intense precursors detected in a survey scan from 400 to 1600 u (three μscans). ZoomScan mass window was set to 12 Da enabling monitoring of the entire 12C/13C isotopic envelope of double and triple charged peptides; single charged ions were excluded for MS/MS analysis. Normalized collision energy was set to 35% and dynamic exclusion was applied during 3 min periods to avoid repetitive fragmentation ions. The generated .raw files were converted to .mgf files for MASCOT data search. A database containing the NCBInr Homo sapiens sequences containing 113620 entries (31/03/18) was searched using MASCOT software (version 2.3 MatrixScience) for protein identification. Oxidation of methionine and 13C6-Arg and 13C6-Lys were specified as variable modifications, trypsin as the specific enzyme and one missed cleavage allowed. Minimum precursor and fragment-ion mass accuracies of 1.2 and 0.3 Da were used. A requirement of at least one bold (unique) red peptide (i.e. the highest scoring peptide matches to the protein with the highest total score) was required for protein identification and at least two bold red (unique) peptides were required for quantification. Cut-off values for MASCOT scores of peptides and proteins were set to 39 (p < 0.05) and 46 (p < 0.01), respectively. The false positive rate was calculated by searching the same spectra against the NCBInr Homo sapiens decoy database. Relative quantitation ratios of identified proteins were calculated using QuiXoT (version 1.4.02). SILAC ratios were defined by the area of the heavy peptides (13C) divided by the area of light (12C). Protein ratios obtained by QuiXoT were manually verified for all peptides. As observed in previous studies, a proportion of 13C6-Arg was converted to 13C5-Pro, leading to a reduction in the intensity of the isotope labeled peptide peak; this was corrected for all peptides containing one or more proline residues. Molecular and cellular functions of the proteins found to be deregulated by SILAC were assigned based on the biological knowledge available in Gene Ontology (GO) annotations.
RNA extraction and RT-PCR analysis
Total RNA was isolated from HepG2 cells after treatment with MSN-Tf-AgNPs (25 µg/mL), using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The quantity of extracted RNA was measured by Nanodrop One (Thermo Fisher Scientific). Synthesis of cDNA with integrated removal of genomic DNA contamination was performed by Quantitect reverse transcription kit (Quiagen) using 1 μg of RNA. RT-PCR analysis was carried out using TaqMan gene expression assays (Thermo Fisher Scientific) and TaqMan Fast advance master mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. The references of TaqMan gene expression assays used are listed in Table 1. All reactions were performed in a final volume of 10 μL. The reaction protocol was 2 min at 50 °C, 10 min at 95 °C for activating polymerase and 40 cycles for 15 seconds at 95 °C and 1 min at 60 °C. Relative expression of genes was normalized using GADPH as the endogenous control. Gene expression in each sample was calculated as 2−ΔΔCt.
Table 1. References of TaqMan gene expression assays used for the RT-PCR analysis.
| Gene | RefSeq | Assay ID |
|---|---|---|
| RCF3 | RCF3 | NM_002915.3 |
| CHERP | CHERP | NM_006387.5 |
| DDX3X | DDX3X | NM_001193416.2 |
| SHMT2 | SHMT2 | NM_001166356.1 |
| Ciclina E | CCNE1 | NM_001238.3 |
| Cdk2 | CDK2 | NM_001290230.1 |
| GADPH | GADPH | NM_001256799.2 |
Results and discussion
Proteins grafting to mesoporous silica nanoparticles
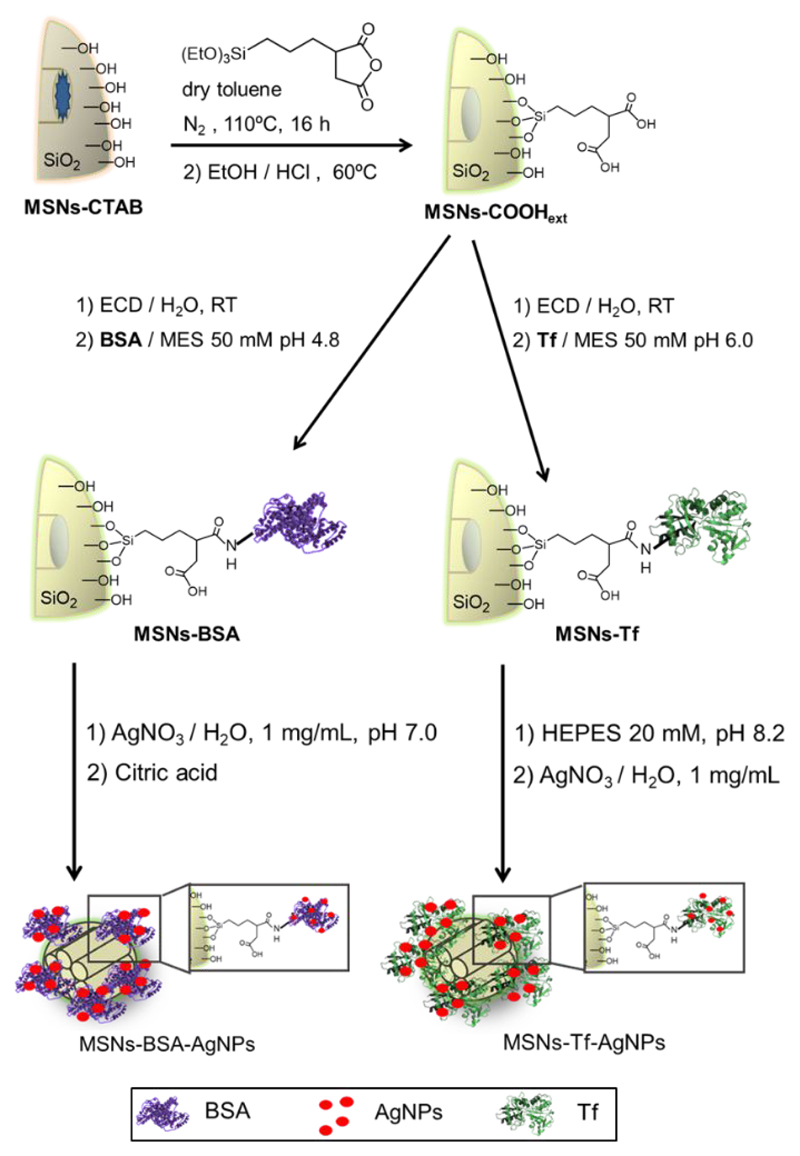
The whole synthetic approach to obtain hybrid materials is shown in Scheme 1. To provide anchoring points for the proteins, the external surface of the MSNs was functionalized with carboxylic acid groups in a first step, using a post-synthesis method.37 In a second step, amide bonds were formed for the covalent anchorage of proteins to the MSNs-COOHext material. To achieve this, some of the free primary amino groups on the BSA or Tf proteins, present in lysine and arginine amino acids, were condensed with the carboxylic acid groups in the external MSNs surface through carbodiimide chemistry. The isoelectric points of the proteins and the pH of the condensation reaction play an important role in the protein anchorage to the MSNs. Therefore, the pH of the reaction was fixed below the isoelectric point of the proteins (IEPBSA 5.87 ‡,38 and IEPTf 6.97 ‡‡,39) to avoid electrostatic repulsion of the negative surface of the silica nanoparticles (MSNs-COOHext) with the negatively charged proteins above the isoelectric point.40 Due to steric hindrance effects that occur when macromolecules are employed to functionalize the silica surface, the amount of protein used in these syntheses was previously optimized by using an excess of protein for the reaction. Hence, the amount of anchored protein was estimated from TG measurements and set up as the optimal or maximum amount of protein able to be anchored. The obtained molar relation between carboxylic acid groups to anchored protein (nCOOH/nProt) was 1 to 1.4 × 10−3 and it was then used for the rest of reactions.
Scheme 1.
Synthesis of hybrid MSN materials functionalized with BSA or Tf proteins and silver nanoparticles.
Silver nanoparticles nucleation onto proteins functionalized MSNs materials
To obtain the hybrid materials with proteins and metallic nanoparticles, two synthetic routes were followed depending on the protein attached onto the MSNs (Scheme 1). Silver nitrate was the metallic ion precursor and the pH was fixed above the IEP of each protein in both cases. Thus, the proteins present residues of glutamate (Glu) and aspartate (Asp) acids negatively charged which must be involved in the site-specific localization of positive silver ions.41–43 For the nucleation of silver nanoparticles (AgNPs) onto MSNs-BSA, silver ions were allowed to be complexed first at pH 7 and, subsequently, citric acid was used as reducing agent carrying out the reaction in a slightly acid pH but higher than the isoelectric point of the protein (IEPBSA 5.87).44 On the other hand, a HEPES solution was used to nucleate AgNPs onto MSNs-Tf. The structure of transferrin possesses less amount of negative residues Asp and Glu than BSA and the isoelectric point is higher (IEPTf 6.97). For these reasons it was necessary a basic medium for the synthesis. The HEPES solution was used as buffer to set the pH at 8.2 during the synthesis, and also as a reducing agent, because it has been reported in previous studies for the preparation of gold and silver nanoparticles.45–48 Nevertheless, it is important to mention that only a small proportion of the carboxylic acid groups on the external surface of the MSNs have been used for protein attachment. Therefore, there are carboxylate residues also acting as nucleating sites for the Ag+ at the pH established for the syntheses, since carboxylic acid group in hydrocarbon chains with three to six carbon long have a pKa around 4.8.49
Materials characterization
The incorporation of the alkoxysilane derivative and the BSA or Tf proteins was followed by quantification of the organic content of the MSNs materials by TGA and elemental chemical analyses (Table 2). The results confirm the expected successive increase of the organic content after functionalization of the MSNs with the alkoxysilane in the first step and the proteins attached in the second step. Sulphur and nitrogen content in MSNs-BSA and MSNs-Tf samples confirm the presence of amino acids from the anchored proteins.
Table 2. Organic content and elemental composition from thermogravimetric and chemical analysis of MSNs and functionalized MSNs materials.
| Material | Theor. Org. (wt%) | Org. Content (wt%) | %C | %N | %S |
|---|---|---|---|---|---|
| MSNs | - | 4.53 | 3.64 | 0.05 | 0.03 |
| MSNs-COOHext-CTAB | - | 36.27 b | 30.86 | 1.85 | 0.02 |
| MSNs-COOHext | 9.73 | 7.81 b | 10.11 | 0.10 | 0.01 |
| MSNs-BSA | 19.54 a | 13.69 b | 14.27 | 2.93 | 0.26 |
| MSNs-Tf | 19.55 a | 13.67 b | 13.94 | 2.86 | 0.26 |
Theoretical organic content (wt%) is calculated without considering the loss of water molecules which are produced in the condensation reaction for the protein anchorage.
Organic content (wt%) is determined from the TGA weight losses, excluding the weight loss due to the desorption of water (up to 125 °C) and further corrected by the weight loss of the remaining alkoxysilanes after the solgel reaction (surfactant extracted unmodified MSNs).
Zeta-potential (ζ) values and hydrodynamic sizes were measured to evaluate the functionalization process (Table 3). The grafting of the alkoxysilane TESPSA produces a more negative ζ-potential compared to the bare MSNs, due to the co-existence of negative-charged –SiO− groups of silica in water plus -COO− groups from the new carboxylic acid functionalities. The subsequent introduction of the proteins onto MSNs-COOHext, provokes a change towards a less negative ζ-potential. These results are consistent with the fact that only a small proportion of the –COOH groups are converted into amide bonds due to the expected steric hindrance of the proteins. Also, in the water media the proteins must be above or close to their isoelectric point, i.e., negatively charged or close to neutral, therefore not influencing the final value.
Table 3. ζ-potential values and hydrodynamic particle size in water medium of MSNs materials.
| Material | ζ-potential (mV) a | Hydrodynamic size a,b (nm) |
|---|---|---|
| MSNs | -25 ± 8 | 199 ± 8 |
| MSNs-COOHext | -29 ± 5 | 209 ± 15 |
| MSNs-BSA | -21 ± 3 | 166 ± 33 |
| MSNs-Tf | -20 ± 4 | 166 ± 16 |
| MSNs-BSA-AgNPs | -13 ± 7 | 197 ± 21 |
| MSNs-Tf-AgNPs | -8 ± 3 | 196 ± 12 |
Notes.
samples were measured in quintuplicate (n = 5).
maximum of the size distribution measured by DLS.
All MSNs exhibit high enough negative ζ-potential values to be in the colloidal stability zone50 and dynamic light scattering (DLS) measurements show monomodal hydrodynamic size distributions between 130-250 nm for all the MSNs materials (See Figure SI.2 for MSNs-Tf material as a representative example). The maximum of the size distribution was not significantly altered during the alkoxysilane and protein grafting processes. Nevertheless, the MSNs-BSA and MSNs-Tf materials possess a smaller hydrodynamic size than bare MSNs and the functionalized MSNs-COOHext, reflecting a decrease in the magnitude of the aggregates of nanoparticles in water media. Therefore, the presence of organic macromolecules attached to the external surface of the MSNs in the hybrid materials contributes as well to a steric repulsion that decreases the size of the aggregates in solution.
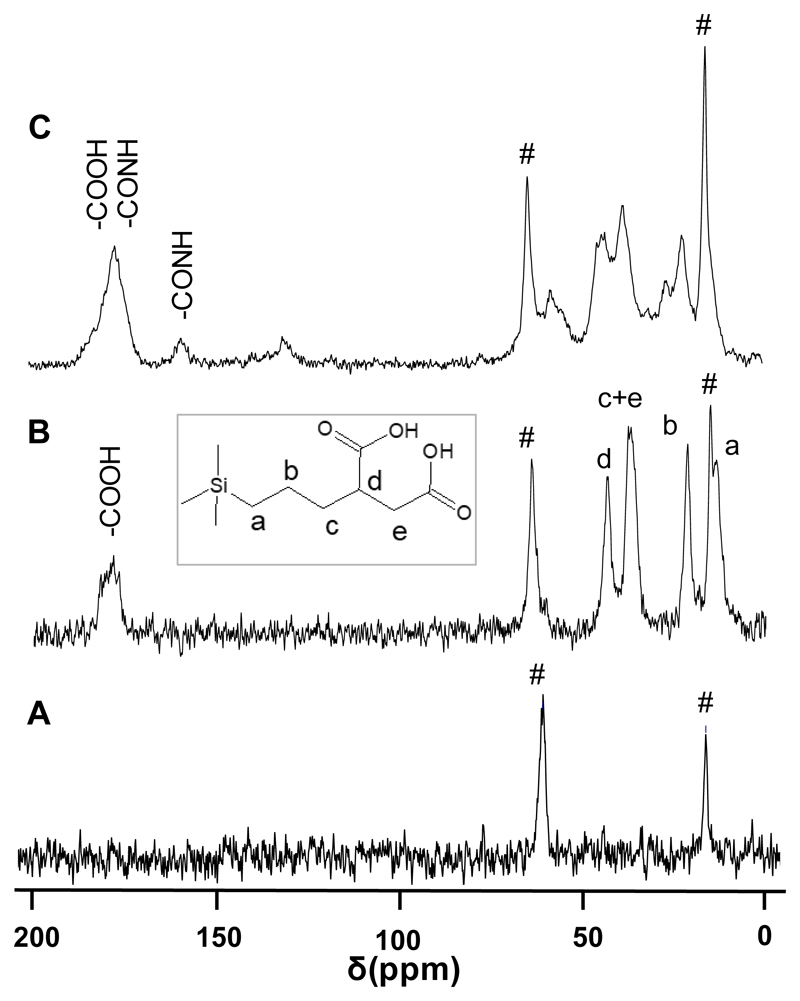
Figure 1 shows the 13C{1H} CP MAS NMR spectra of materials recorded to follow the incorporation of organic matter on the silica surface. The two peaks at 64 and 15 ppm, which are common in all the spectra, correspond to the methylene and methyl groups, respectively, from ethoxy groups due to incomplete hydrolysis and condensation of the tetraethylorthosilicate precursor during the sol-gel synthesis of MSNs.51 These ethoxy groups represent around a 5% of organic content in the TG analysis of extracted MSNs, which does not correspond to the remaining surfactant since the %N is negligible (see TG and AQE data in Table 2). The spectrum of MSNs-COOHext (Figure 1B), shows the signals of the hydrolyzed propylsuccinic acid present on the hybrid material. The broad signal at ca. 178 ppm corresponds to the carbonyl moiety from carboxylic acid groups. Then, the CH adjacent to the carboxylic acid group shows a peak at 43 ppm (signal d) and the peak at 37 ppm corresponds to the sum of CH2 groups close to the carboxylic acid group (signal c and e). The peak at 21 ppm matches with the methylene carbon in the chain (signal b) and, finally, methylene carbon directly attached to silicon atom shows a peak at 13 ppm (signal a). As a representative example for both proteins, Figure 1C shows the spectrum of MSNs-BSA which confirms the covalent attachment of BSA protein to the carboxylic acid groups on the surface of MSNs-COOHext. Chemical shifts in this spectrum are coincident with those of the respective BSA protein in the solution NMR spectrum of the free protein (see Figure SI.3 in ESI). The carbonyl signal at ca. 178 ppm is broadened with respect to the MSNs-COOHext material due to the presence of carbonyl groups of carboxylic acids and amide bonds in the protein structure itself. However, a more intense signal shows up at 158 ppm supporting the formation of amide bonds between the –COOH from TESPSA and –NH2 from some lysine and arginine amino acids of the protein.
Figure 1.
13C {1H} CP MAS NMR spectra of materials: MSNs (A), MSNs-COOHext (B) and MSNs-BSA (C). Peaks designated with # correspond to ethoxy groups due to incomplete hydrolysis and condensation.
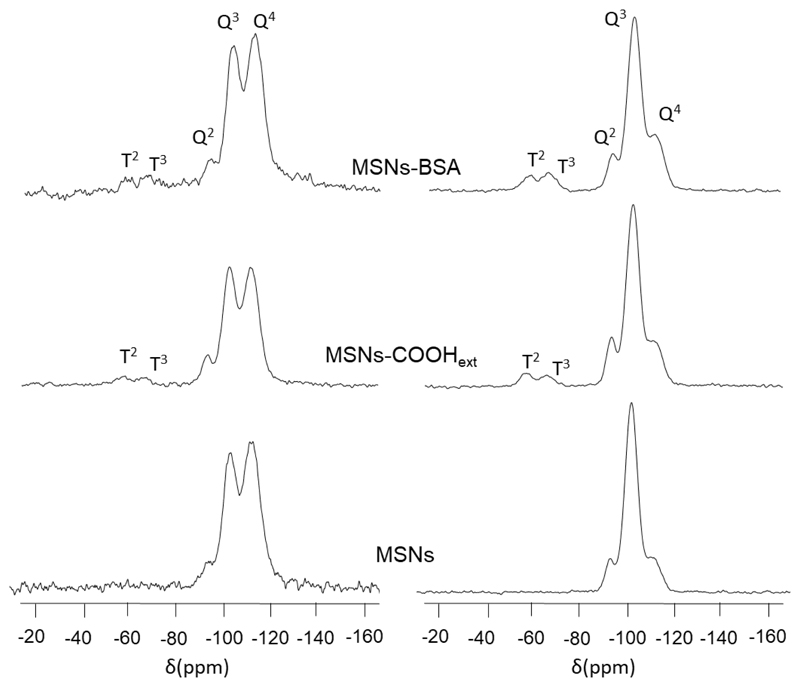
Solid state 29Si MAS NMR spectroscopy was used to further analyze the functionalization of mesoporous silica nanoparticles. Figure 2 (left) compares the quantitative spectra from the direct polarization method obtained for the bare MSNs material with those obtained for the functionalized materials MSNs-COOHext and MSNs-BSA. In addition, 29Si CP MAS NMR spectra were also registered (Figure 2, right) to assess the existence of T units [R-Si(OSi)n(OX)3–n] (X = H, C). This method uses cross-polarization from the nearby protons and yields unquantitative experiments, but confirms the presence of the functionalizing trialkoxysilane in the materials.
Figure 2.
29Si MAS NMR spectra (left) and 29Si CP MAS NMR spectra (right) of MSNs, MSNs-COOHext and MSNs-BSA materials.
Therefore, T2 [R-Si(OSi)2(OX)] and T3 [R-Si(OSi)3] units show signals at −57 and −66 ppm, respectively, in the CP spectra of the functionalized MSNs-COOHext and MSNs-BSA materials. Table 4 shows the chemical shifts and populations of these silicon environments from the 29Si MAS NMR. All spectra showed three resonances at around −93,−102 and −113 ppm for the Q2 [Si(OSi)2(OX)2], Q3 [Si(OSi)3(OX)] and Q4 [Si(OSi)4] silicon sites, respectively (X = H, C). The grafting of alkoxysilane species on the silica surface provokes a decrease in the Q2 and Q3 peak areas and an increase in the Q4 population due to the conversion from Si-OH to fully condensed Si-O-Si species. As shown in Table 4, a decrease in Q2 and Q3 area peaks and an increase in Q4 confirm the covalent grafting of TESPSA in MSNs-COOHext. The relative ratio of partially to fully condensed silicon sites, (Q2 + Q3)/Q4, confirms that the inner surface of the channels was preserved from functionalization, since this step is performed before the surfactant extraction stage. These findings are also verified by the N2 sorption studies (see ESI and Figure SI.4). As expected, the populations of Qn environments in MSNs-BSA are maintained as compared to the previous material, MSNs-COOHext, because the protein is attached through the carboxylic acid groups. The relation of Qn/Tn environments is also maintained when MSNs-COOHext is compared with MSNs-BSA, supporting the anchorage of proteins over carboxylic acids groups without involving silicon sites.
Table 4. Chemical shifts and populations (%) of the silicon Qn and Tn environments and peak area relations, (Q2 + Q3)/Q4 and Qn/Tn units, on the basis of the deconvolution of 29Si MAS NMR spectra of MSNs materials.
| Material | δ, ppm (peak area, %) | δ (ppm) | peak area | ||||
|---|---|---|---|---|---|---|---|
| Q2 | Q3 | Q4 | (Q2 + Q3)/Q4 | T2 | T3 | Q/T | |
| MSNs | -93.2 (6.2) | -102.3 (44.6) | -112.0 (49.1) | 1.0 | --- | --- | --- |
| MSNs-COOHext | -93.6 (3.2) | -102.7 (36.5) | -112.0 (60.2) | 0.6 | -57.8 | -66.9 | 15.3 |
| MSNs-BSA | -92.5 (3.7) | -102.8 (32.1) | -112.5 (64.0) | 0.5 | -58.3 | -66.4 | 15.7 |
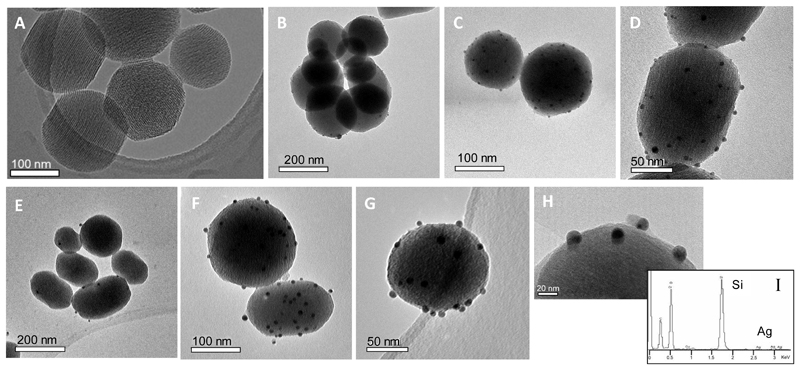
Figure 3 shows TEM micrographs of the materials at different magnifications (see also Figure SI.5). A well dispersion of the silver nanoparticles onto the surface of both MSNs-BSA-AgNPs (Figure 3B-D) and MSNs-Tf-AgNPs (Figure 3E-G) can be observed. The metallic nanoparticles present homogeneous sizes in both materials, which are around 15 nm for MSNs-Tf-AgNPs (Figure 3H) and smaller for the MSNs-BSA-AgNPs. In addition, free AgNPs away from the MSNs surface were not found in the TEM analysis, indicating that the nucleation and growth of the AgNPs takes place on the outer MSNs surface covalently functionalized with the proteins. These results are also verified by DLS measurements of the hydrodynamic size of the materials (Table 3) where only a population around 196 nm was found, corresponding to the whole nanosystem (MSNs-Protein-AgNPs). Moreover, the absence of a population around 20 nm indicates that in water suspension the AgNPs are not released from the protein modified-mesoporous silica nanosystem. AgNPs containing nanosystems have the maximum of the hydrodynamic size distribution shifted towards a slightly higher value than the corresponding MSNs-Protein materials. This effect is probably due because the MSNs-Protein-AgNPs present a ζ-potential value less negative than the formers, i.e., away from the zone of colloidal stability.
Figure 3.
TEM images of the materials: MSNs (A), MSNs-BSA-AgNPs (B, C and D) and MSNs-Tf-AgNPs (E, F, G and H). EDX analysis of MSNs-Tf-AgNPs (I).
The atomic percentages for silver and silicon were measured by EDS analysis registered at low magnifications of AgNPs containing materials, finding 0.40% Ag and 99.60% Si for MSNs-BSA-AgNPs and 0.53% Ag and 99.47% Si for MSNs-Tf-AgNPs. These values represent a 0.004 molar ratio of Ag/Si for the MSNs-BSA-AgNPs material and 0.005 Ag/Si molar ratio for MSNs-Tf-AgNPs material, thus being comparable and representing a similar content in silver in both materials, independent of the attached protein. These results also support the participation of the carboxylic acid groups on the MSN-Protein silica external surface in the localization of silver cations besides the protein itself for their subsequent reduction and nucleation, as discussed above.
In addition, attempts to nucleate silver nanoparticles by using the free protein as template did not lead to the formation of silver nanoparticles. Instead, inhomogeneous silver clusters were obtained with this synthetic approach (see Figure SI.6 in ESI). This fact highlights the need to use the mesoporous silica nanosystem functionalized with -COOH groups and decorated with a protein as a platform for the nucleation and synthesis of silver nanoparticles, as well as a platform for the vectorization of those nanoparticles, in the case of the transferrin protein.
In vitro cell studies
Evaluation of the cellular uptake in HepG2 and MC3T3-E1 cell lines
The enhanced cellular uptake of MSNs materials functionalized with Tf protein as targeting agents was assessed in a first step in two cell lines. HepG2, hepatocarcinoma cells, and MC3T3-E1, osteoblast-like cells, were selected due to their clear different expression of transferrin receptor (TfR)52,53. This receptor resides in the external cell membrane and cycles in acid endosomes within the cell in a clatrin-dependent way to finally release iron and return to the cell membrane.34,35 Cells were incubated with 100 μg/mL of the bare MSNs and protein decorated MSNs for 24 h. Analysis by flow cytometry was used to evaluate the degree of materials internalization by quantifying the intensity of the living cells that exhibited green fluorescence.
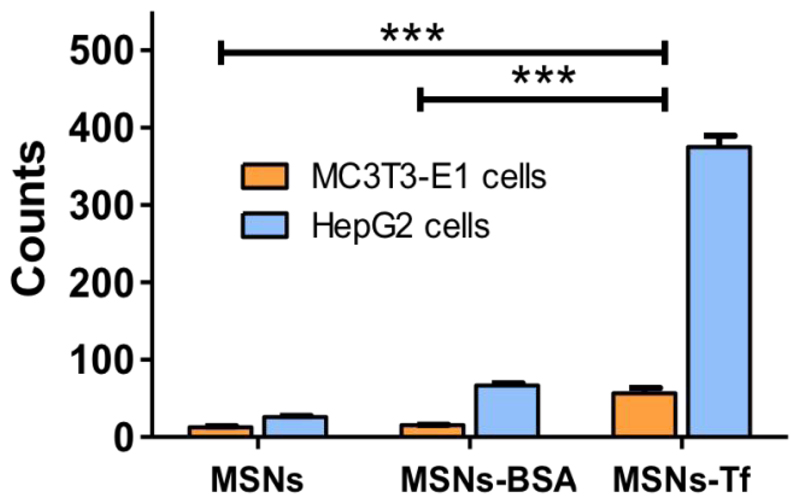
As shown in Figure 4, functionalization with Tf (MSNs-Tf) enhances the uptake of materials in HepG2 cells when compared with the bare MSNs and BSA functionalized MSNs (MSNs-BSA), then supporting the transferrin receptor (TfR) mediated cellular uptake.54
Figure 4.
Cellular uptake of MSNs materials evaluated on HepG2 and MC3T3 cells exposed to 100 μg/mL of MSNs, MSNs-BSA and MSNs-Tf for 24 h. Statistical significance: ***p < 0.001.
In addition, internalization of MSNs-Tf in MC3T3-E1 cells was significantly lower as compared to HepG2 cells. This is consistent with the fact that HepG2 overexpressed TfR while MC3T3-E1 cells have low TfR expression. Thus, demonstrating the selective internalization of MSNs-Tf towards cancer cells overexpressing TfR. On the other hand, the slightly higher uptake of BSA functionalized materials compared with the bare MSNs in both cells lines, may be ascribed to a better dispersion of the nanosystems in the cell culture media due to the higher stabilization once MSNs are externally functionalized with the proteins, as reflected by the decrease of the maximum of the hydrodynamic size distribution (Table 3).
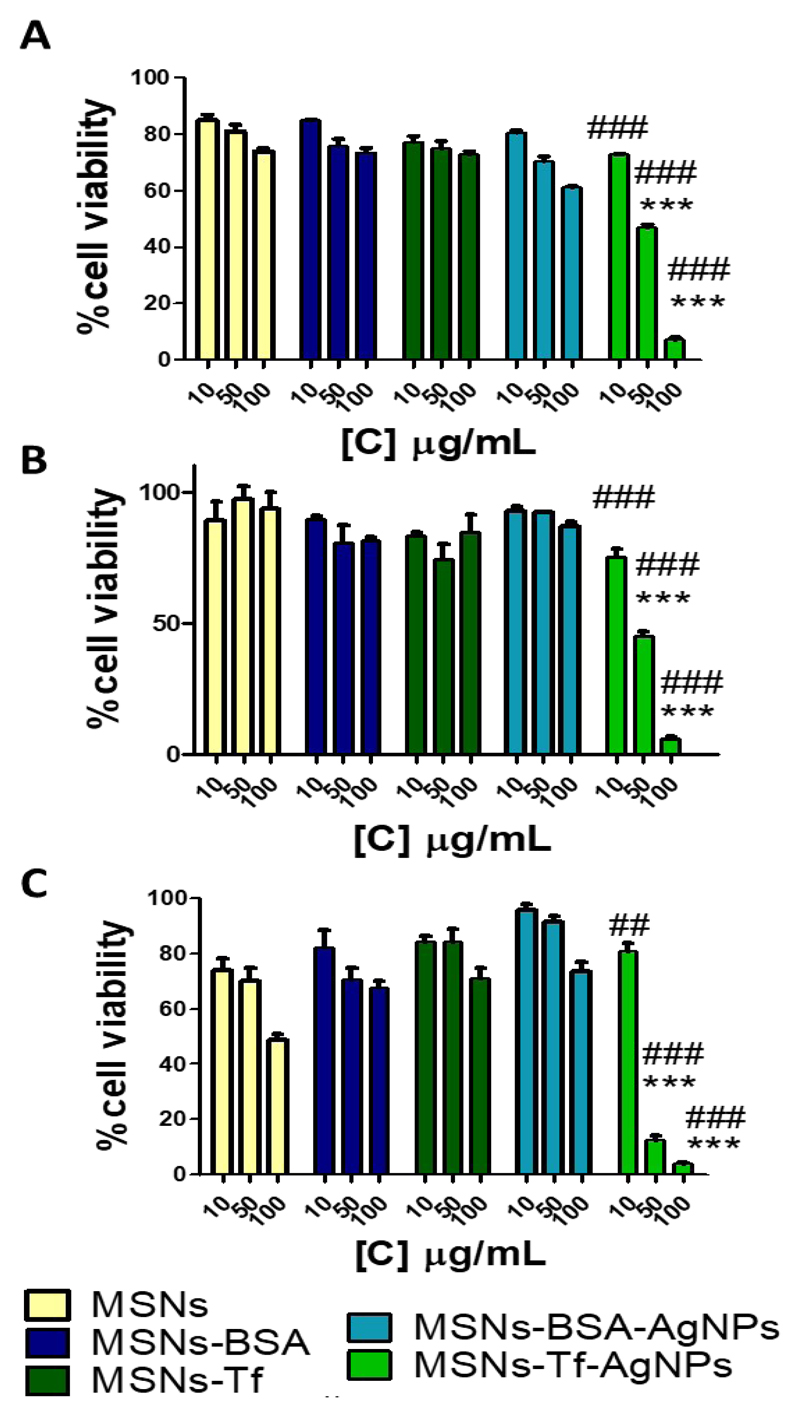
Cell viability
Once tested the efficacy of the targeting, we evaluated the toxicity of the nanosystems containing silver nanoparticles (MSNs-Protein-AgNPs) in comparison with the bare MSNs and protein functionalized MSNs (MSNs-Protein). This study was carried out by evaluating the viability of cells exposed to different concentrations of materials at different times (24, 48 and 72 h) (Figure 5). When HepG2 cells were exposed to different concentrations of bare MSNs or MSNs functionalized with BSA or Tf proteins (MSNs-Protein), the viability at every time assayed was sustained without relevant changes, especially for the shortest times. However, cells treated with silver nanoparticles supported on MSNs-Tf (MSNs-Tf-AgNPs) showed a significant concentration-dependent reduction in viability when compared to the other tested materials (Figure 5). Indeed, MSNs-Tf-AgNPs caused cell death to a greater extent than the analogous material decorated with BSA (MSNs-BSA-AgNPs). Bearing in mind that EDX analyses of both materials showed similar Si/Ag ratios, this fact is explained by the increased cellular uptake of materials when using transferrin as targeting ligand as compared to the material decorated with BSA, which is in agreement with the results of the internalization assay (Figure 4).
Figure 5.
Cell viability of HepG2 cells exposed to different concentrations (10, 50, 100 μg/mL) of MSNs, MSNs-BSA, MSNs-Tf, MSNs-BSA-AgNPs and MSNs-Tf-AgNPs for 24 (A), 48 (B) and 72 (C) hours (n=5). Data were analyzed by ANOVA followed by Bonferroni´s multiple-comparison test. ***p < 0.001 versus MSNs-Tf. #p < 0.05. ##p < 0.01 ###p < 0.001 versus MSNs-BSA-AgNPs.
Furthermore, when the viability of cells exposed to MSNs-Tf-AgNPs is compared to the viability of cells exposed to the same material but without AgNPs (MSNs-Tf), a significant difference is evident for all the times assayed, especially at the higher concentrations tested. This fact confirms that the decrease in cell viability observed is provoked by the effect of AgNPs selectively transported inside the cells and can not be due to the nanocarrier used.
To confirm that the nucleation of AgNPs onto MSNs-Tf material does not affect the enhanced uptake of these materials, another internalization assay was carried out, comparing bare MSNs and MSNs-Tf-AgNPs (see Figure SI.7 in ESI). The uptake of bare MSNs was negligible while for MSNs-Tf-AgNPs was significantly higher, supporting that the nucleation of AgNPs on transferring decorated MSNs does not provoke alterations in the receptor mediated internalization process.
Cell death
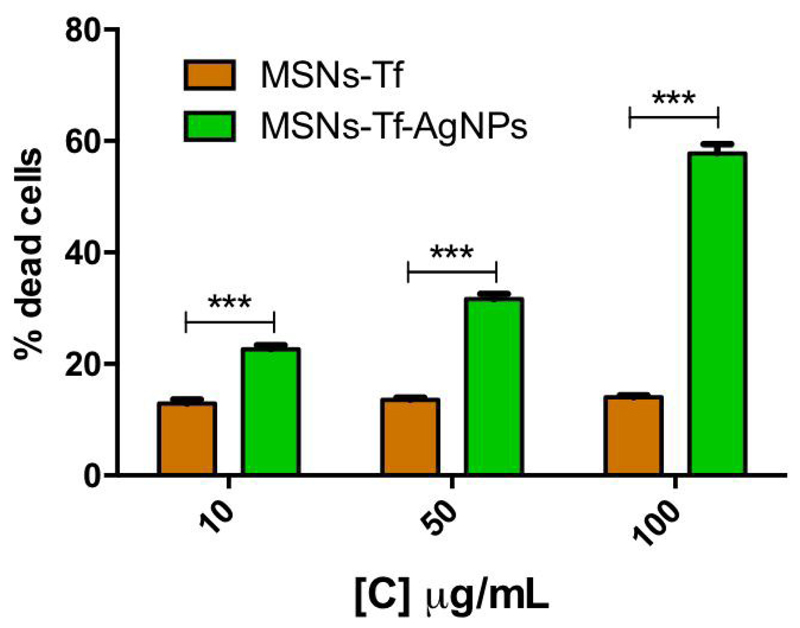
In order to evaluate whether or not the decreased observed in cell viability is consequence of MSNs-Tf-AgNPs-induced cell death, and to confirm that such effect is exclusively due to the released of Ag+ from the AgNPs, we evaluated the percentage of dead cells after exposure for 24 h to different concentrations of MSNs-Tf-AgNPs (10, 50 and 100 μg/mL) as compared to the same material but without AgNPs (MSNs-Tf). The results (Figure 6) demonstrated that at all the tested concentrations, the percentage of dead cells was significantly higher in cells exposed to MSNs-Tf-AgNPs as compared to cells exposed to MSNs-Tf; thus, sustaining the cytotoxic role of AgNPs in the proposed nanosystem.
Figure 6.
Percentage of HepG2 dead cells after exposure for 24h to different concentrations of MSNs-Tf-AgNPs and MSNs-Tf (10, 50 and 100 μg/mL). Statistical significance: ***p < 0.001.
MSNs intracellular localization assay by TEM
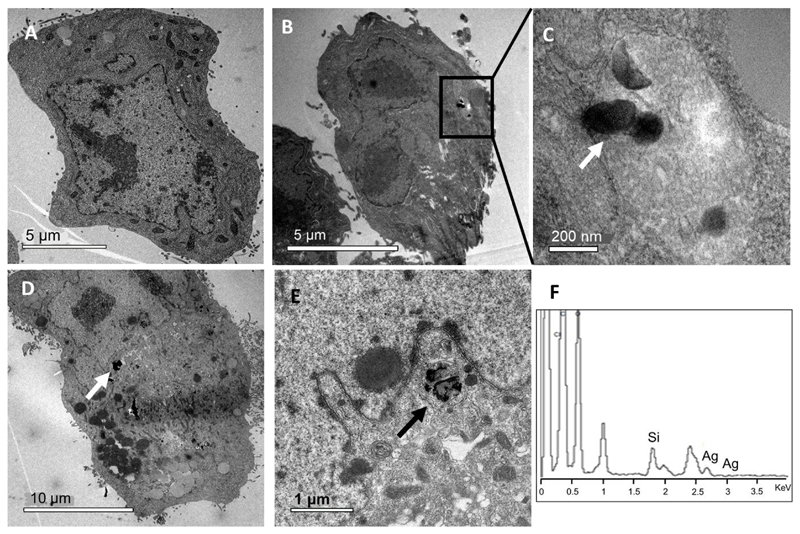
To better examine the effect of AgNPs carried inside the cells, transmission electron microscopy (TEM) analysis of cell sections was used to obtain a deeper insight into the internalization and localization of MSNs-Tf-AgNPs in exposed HepG2 cells. Transmission electron micrographs of ultrathin sections of HepG2 cells exposed to 25 and 50 μg/mL of MSNs-Tf-AgNPs are shown in Figure 7 and Figure SI.9 in ESI. Differences between control cells (Figure 7A) and cells treated with the nanosystem (Figure 7B-7E) can be observed. While control (untreated) cells show no abnormalities, cells treated with the nanosystem exhibit large endosomes with engulfed nanoparticles and thus, increased degree of vacuolization. This effect is in accordance with the previous observed greater degree of internalization and reduction of cell viability as the nanosystem concentration and exposure times increase (See Figure 5 and Figures SI.7 and SI.8 in ESI).
Figure 7.
TEM images of ultrathin sections of HepG2 cells exposed to MSNs-Tf-AgNPs. Control cells (A), HepG2 cells exposed to 25 μg/mL for 24 h (B and C) and 50 μg/mL for 48 h (D and E) of MSNs-Tf-AgNPs. EDX analysis of treated cells (F). Arrows indicate the localization of MSNs.
The nanosystems were mainly located in lysosomes (Figure 7 B and C) due to the transferrin receptor (TfR) mediated cellular uptake mechanism proposed for their cell internalization as well as because this nanosystem was not provided with a rapid endosomal escape mechanism.55,56 Moreover, considering the size of the nanosystem MSNs could not penetrate the nuclear membrane (Figure 7E).57 In addition, the action of lysosomal proteases are likely to degrade the Tf proteins anchored on the external surface of the nanosystem, and therefore both AgNPs and MSNs would be losing their protective coating.58,59 However, AgNPs were not observed inside the cells, neither free nor supported on the nanocarrier, so dissolution of the metal nanoparticles must have been produced. This fact can be explained with the “lysosome-enhanced Trojan horse effect” mechanism,60 in which metallic nanoparticles internalized by active endocytosis mechanisms can release toxic ions due to the degradation of nanoparticles promoted by the acidic conditions of the lysosomal environment (pH 4.5-5.0). As reported in previous studies about toxicity of silver nanoparticles, one of the chemical reactions that may be implicated in this process also involves the reaction with reactive oxygen species (ROS) that are also caused by the presence of silver nanoparticles and silver ions, i.e., 2Ag + H2O2 + 2H+ → 2Ag+ + 2H2O (Ec0 = 0.98 V). §,61–63 For these reasons, we can hypothesized that the toxic effects of silver nanoparticles are based on the metal ions (Ag+) released as effectors of the activation cascades responsible for damage of the mitochondrial membranes, disturbances of the redox state and increased ROS production, apoptosis, cell cycle deregulation, and DNA damage, among others.62 The presence of these silver ions inside the cells has been confirmed by energy-dispersive X-ray spectroscopy (EDX) analysis of cells exposed to the nanosystem (Figure 7F).60,64,65
On the other hand, the observed MSNs present signals of degradation, such as loss of their spherical shape (Figure 7E), as a function of the assayed time, i.e., the time elapsed within the lysosome. Silica can be dissolved releasing silicic acid in the conditions of the lysosomal pH, although its dissolution is much slower than in the case of AgNPs.59,60 The EDX analysis also confirms that the nanoparticles found inside the lysosomes contain mainly Si in their composition.
Quantitative proteomics
To gain a deeper insight in the molecular mechanisms involved in the interaction of the developed nanosystem with HepG2 cells, a SILAC experiment was carried out to evaluate the differential nuclear protein expression after MSNs-Tf-AgNPs exposure (Figure SI.10.A and B in ESI). A total of 822 proteins were identified by mass spectrometry analysis with at least one unique peptide and a false discovery rate of 0.12%, estimated from the number of hits against reverse or randomized sequence (decoy database)/total hits ratio at p > 0.01. However, only 451 proteins passed the selected criteria for protein quantitation (at least two unique peptides). From the quantified proteins, those with a SILAC ration (RSILAC) higher than 1.5 or below −1.5 were considered significantly deregulated. According to these criteria, 15 proteins were found up-regulated and 17 were inhibited after MSNs-Tf-AgNPs exposure (Table 5). As for the SILAC ratio distribution, most of the quantified proteins were within a SILAC ratio close to 1, as expected for a 1:1 mixture (Figure SI.10.C). The molecular and cellular functions of the altered proteins were classified using the Gene Ontology (GO) database.
Table 5. Deregulated proteins obtained in the SILAC study after HepG2 cells exposure to the MSNs-Tf-AgNPs nanosystem.
| gi number | Protein name | Common name | RSILAC | RSDSILAC |
|---|---|---|---|---|
| 194394237 | Villin-1 [Homo sapiens] | VIL1 | -3.34 | 13.73 |
| 119226260 | Calcium homeostasis endoplasmic reticulum protein [Homo sapiens] | CHERP | -2.85 | 4.69 |
| 261862348 | Serine hydroxymethyltransferase, mitochondrial isoform 3 [Homo sapiens] | SHMT2 | -2.26 | 17.30 |
| 4506489 | Replication factor C subunit 3 isoform 1 [Homo sapiens] | RFC3 | -1.91 | 18.71 |
| 4557469 | AP-2 complex subunit beta isoform b [Homo sapiens] | AP2B1 | -1.90 | 8.30 |
| 56237027 | Insulin-like growth factor 2 mRNA-binding protein 1 isoform 1 [Homo sapiens] | IGF2BP1 | -1.78 | 9.63 |
| 301171467 | ATP-dependent RNA helicase DDX3X isoform 2 [Homo sapiens] | DDX3X | -1.77 | 19.17 |
| 98986457 | Host cell factor 1 [Homo sapiens] | HCFC1 | -1.67 | 11.74 |
| 4506701 | 40S ribosomal protein S23 [Homo sapiens] | RPS23 | -1.65 | 10.25 |
| 11321585 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 [Homo sapiens] | GNB1 | -1.61 | 14.35 |
| 5453740 | Myosin regulatory light chain 12A [Homo sapiens] | MYL12A | -1.61 | 18.81 |
| 116256489 | Septin-9 isoform c [Homo sapiens] | SEPT9 | -1.59 | 18.39 |
| 4505917 | Exosome component 10 isoform 2 [Homo sapiens] | EXOSC10 | -1.59 | 11.80 |
| 15011936 | 40S ribosomal protein S26 [Homo sapiens] | RPS26 | -1.56 | 3.97 |
| 4504897 | Importin subunit alpha-2 [Homo sapiens] | KPNA2 | -1.56 | 19.75 |
| 189491630 | Upstream-binding protein 1 isoform LBP-1b [Homo sapiens] | UBP1 | -1.54 | 19.34 |
| 24497603 | Nuclear pore glycoprotein p62 [Homo sapiens] | NUP62 | -1.50 | 19.29 |
| 10800130 | Histone H2A type 1-D [Homo sapiens] | HIST1H2AD | 1.50 | 10.32 |
| 5453994 | Double-strand-break repair protein rad21 homolog [Homo sapiens] | RAD21 | 1.57 | 19.53 |
| 118601081 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 [Homo sapiens] | HNRNPUL2 | 1.60 | 4.51 |
| 4505343 | Nuclear cap-binding protein subunit 1 [Homo sapiens] | NCBP1 | 1.60 | 12.42 |
| 10863903 | E3 ubiquitin-protein ligase TRIP12 [Homo sapiens] | TRIP12 | 1.62 | 1.16 |
| 4503571 | Alpha-enolase isoform 1 [Homo sapiens] | ENO1 | 1.68 | 2.87 |
| 89179321 | Protein unc-45 homolog A isoform 3 [Homo sapiens] | UNC45A | 1.68 | 11.73 |
| 283436222 | ATPase family AAA domain-containing protein 3A isoform 2 [Homo sapiens] | ATAD3A | 1.71 | 6.41 |
| 4503529 | Eukaryotic initiation factor 4A-I isoform 1 [Homo sapiens] | EIF4A1 | 1.71 | 19.97 |
| 4506675 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 precursor [Homo sapiens] | RPN1 | 1.78 | 0.75 |
| 19923653 | PC4 and SFRS1-interacting protein isoform 1 [Homo sapiens] | PSIP1 | 1.93 | 16.95 |
| 101943240 | General transcription factor 3C polypeptide 1 [Homo sapiens] | GTF3C1 | 2.12 | 15.46 |
| 14589866 | Aspartyl/asparaginyl beta-hydroxylase isoform a [Homo sapiens] | ASPH | 2.21 | 14.65 |
| 45593130 | Guanine nucleotide-binding protein-like 3 isoform 1 [Homo sapiens] | GNL3 | 2.58 | 8.80 |
| 148839305 | Structural maintenance of chromosomes flexible hinge domain-containing protein 1 [Homo sapiens] | SMCHD1 | 2.66 | 7.37 |
Cell cycle and cellular proliferation
Most of the proteins found de-regulated upon MSNs-Tf-AgNPs exposure are involved in processes related to cell proliferation and cell cycle. The protein GNL3 is related with the regulation of cellular growth and proliferation through the control of the tumor suppressor p53.66 Overexpression of GNL3 (RSILAC = 2.58) impairs cell cycle progression, thus precluding cell proliferation67 and inducing apoptosis.68 On another hand, overexpression of TRIP12 (RSILAC = 1.62) produces inhibition of USP7, which is an ubiquitin specific processing protease whose depletion induce cell cycle arrest at the S phase.69 The inhibition of a big part of the proteins found altered in the study also has effects on cell proliferation. Different studies carried out in hepatocarcinoma cells such as Huh-7 or HepG2 (cells employed in our work) have shown that an inhibition of SHMT2 (RSILAC = −2.26) or RFC3 (RSILAC = −1.91), is translated into an inhibition of cell proliferation and tumorigenicity70 by stabilization of p21, p53 and p57, and the inhibition of the cyclin A. All these effects induce cell cycle arrest on the S phase,71 which is consistent with the overexpression of TRIP12 commented above and the inhibition of NUP62 (RSILAC = −1.50) and RPL34 (RSILAC = −1.68), as it has been observed in ovarian and lung cancer cells.72,73 Other proteins found downregulated after exposure to MSNs-Tf-AgNPs that are also involved in alteration of the cell cycle by stabilization of p53 are RPS23 (RSILAC = −1.65) and RPS26 (RSILAC = −1.37). In addition, the inhibition of RPS26 and RPS6 (RSILAC = −1.37) induce downregulation of RPL11 (RSILAC = −1.22), another key protein involved in cell cycle and progression.74 That is also the case of KPNA2 (RSILAC = -1.56), which promotes the cell cycle transition G1/S by increasing c-Myc, Akt and cyclin D1 expression, and inhibiting FOXO3a, p21 and p27. This protein has been found to be overexpressed in numerous cancer types. In fact, when this protein is downregulated, as it has been observed in our study (RSILAC = −1.56), inhibition of the tumorigenicity and proliferation of cells occur by decreasing c-Myc and increasing FOXO3a activity.75 This result is in agreement with the downregulation of IGF2BP1 (RSILAC = −1.77) found in our SILAC experiment, which also affects cell proliferation and induce apoptosis76 through c-Myc and Ki-67, as it has been observed in hepatocellular carcinoma and melanoma cells.77 Finally, downregulation of CHERP (RSILAC = −2.85) that impairs Ca2+ mobility through the stimulation of the T lymphocyte receptor by stimulating the G-protein-linked thrombin receptor,78 and DDX3X (RSILAC = −1.77), which prevents the transition G1/S,79 has also been linked with decreased cyclin D1 expression and cell proliferation. In the case of CHERP, its downregulation has also been demonstrated to induce cellular apoptosis through the ATF4/CHOP/DR5 pathway, producing inhibition of the AKT/mTOR signaling pathway;80 while inhibition of DDX3X also produces depletion of Rac-1 and β-catenin, which enhance cellular adhesion, compromise cellular mobility, and therefore decrease the metastatic charge, as shown in breast and lung cancer.79
The presence of MSNs-Tf-AgNPs inside lysosomes, detected by TEM (Figure 7) could also be related with the mechanisms involved in the inhibition of cell proliferation. In fact, several proteins related to lysosomes were found altered in the proteomic experiment. That is the case of KPNA2, PSIP1 or SEPT9. Downregulation of SEPT9 (RSILAC = -1.59), is particularly significant because SEPT9 belongs to a family of proteins called septins, that are modulators of endo-lysosomal membrane trafficking.81 Specifically, SEPT9 participates indirectly in the degradation of tyrosine kinase receptors, such as EGFR. Diesenberg et al.82 demonstrated that depletion of SEPT9 resulted in a significant reduction of the expression of EGFR in the cell membrane.83 This receptor is related to the activation of different signalling pathways including those involved in cell proliferation, regulation of apoptosis and tumorigenesis. These results help to explain the mechanisms associated to the observed antiproliferative potential of the proposed nanosystem.
DNA damage repair and stress response
If most proteins found inhibited in our SILAC experiments are related to cell cycle regulation and cell proliferation, most of the proteins found up-regulated after exposure to the MSNs-Tf-AgNPs nanosystem, are involved in DNA repair and stress response mechanisms. SMCHD1, which takes part in gene silencing and DNA damage related processes, and usually appears down-regulated in hepatic cancer.84 The overexpression and accumulation of this protein in areas where DNA damage has been produced indicates a high degree of DNA repair.85 A similar role is played by HNRNPUL2 and RAD21, which also accumulate in DNA damaged areas, where they promote effective DNA repair.86,87 In our study, all three proteins were found overexpressed after MSNs-Tf-AgNPs exposure: SMCHD1 (RSILAC = 2.66), HNRNPUL2 (RSILAC = 1.60) and RAD21 (RSILAC = 1.57). These results suggest the activation of the DNA repair machinery after exposing HepG2 cells to the designed MSNs-Tf-AgNPs nanosystem. In addition to DNA damage response, there are some proteins that appear overexpressed as a response to stress generated in the cells. One of these proteins is ENO1 which is overexpressed and impaired in tumor cells contributing to the Warburg effect.88 In addition, ENO1 has also been demonstrated to be upregulated in hypoxic conditions in cancer cells. Considering this, ENO1 acts as a stress response protein allowing the cells to overcome these situations through the activation of the anaerobic metabolism.89 ENO1 was found to be overexpressed in our experiment (RSILAC = 1.68), which is consistent to the existence of hypoxic conditions generated by oxygen reactive species (ROS) induced by AgNPs. Similarly, the protein PSIP1 (LEDGF/p75), which is also involved in oxidative stress,90 was also found overexpressed (RSILAC = 1.93), which support the hypothesis that the designed nanosystem is inducing oxidative stress in cancer cells.
Validation of the results obtained by SILAC by gene expression analysis
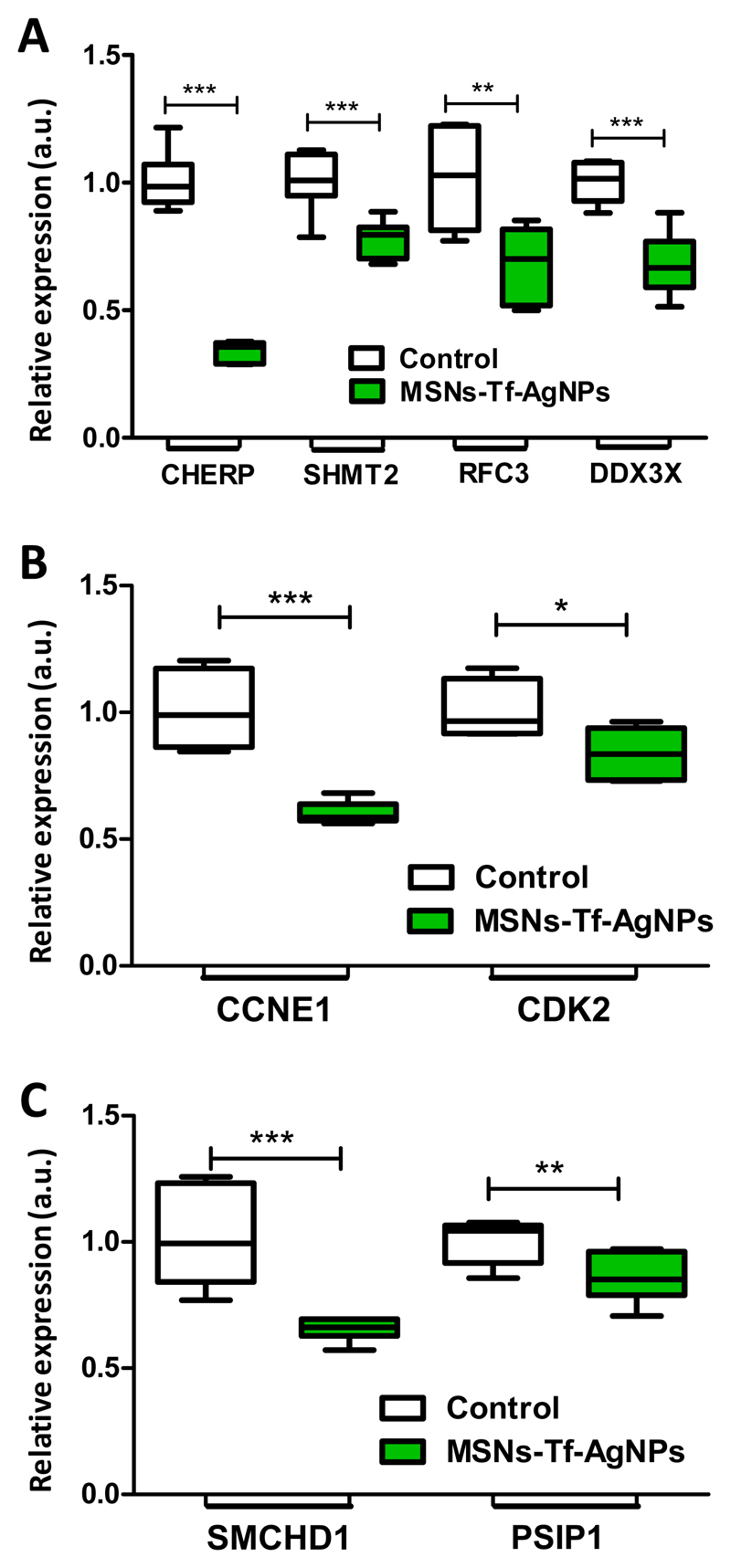
In order to validate the results obtained by quantitative proteomics, gene expression analysis was carried out for selected mRNAs. We first evaluated the expression of 4 mRNA related to 4 proteins that were found significantly altered in our SILAC experiment: CHERP, SHMT2, RFC3 and DDX3X. We selected these mRNAs since they are well-known targets for cancer therapy. Thus, it seemed crucial to validate if the designed nanosystem affects such relevant targets. The results obtained (Figure 8A) confirm the inhibition of the 4 tested transcripts after exposure to the MSNs-Tf-AgNPs nanosystem (Table 5). As commented before, this inhibition implies a reduction in cell proliferation. In particular, it has been shown that the inhibition of any of these proteins, which have been studied in numerous types of cancer, leads to a failure in the synthesis and replication of DNA inducing a cell cycle arrest at the G1/S transition.91 This cell cycle checkpoint is regulated by the action of a complex formed by Cyclin E and Cdk2. For this reason, and in order to obtain a deeper knowledge about the effect of the designed nanosystem at the molecular level, the mRNA expression of Cyclin E and Cdk2 were also evaluated. The results obtained (Figure 8B) also showed the inhibition of these transcripts upon exposure to the MSNs-Tf-AgNPs nanosystem, and thus demonstrate its effect on arresting the cell cycle at the G1/S transition. Finally, the mRNA expression levels of two of the most upregulated proteins (SMCHD1 and PSIP1) found in the SILAC experiment were also evaluated. The results demonstrated an inhibition in the expression of the two transcripts (Figure 8C), which is not consistent with the results obtained for the corresponding proteins (Table 5). However, it is important to consider that while the protein expression was evaluated only considering the nuclear fraction, the mRNA expression analysis was carried out with total lysates. Therefore, these results suggest that the increase protein expression observed by SILAC for SMCHD1 and PSIP1, might be caused by an accumulation of these proteins in the nucleus, rather than by a general overexpression in the cell. These results are consistent with the fact that both, SMCHD1 and PSIP1 tend to accumulate in DNA damage sites to facilitate the repair of double-strand breaks,85,92 which support the previous finding related to the DNA damage induced by the MSNs-Tf-AgNPs nanosystem.
Figure 8.
A) Levels of mRNA of the different altered genes: CHERP, SHMT2, RFC3 and DDX3X. Statistical significance when compared to the control: **p<0.01 and ***p<0.001. B) Levels of mRNA of the proteins that form the G1/S transition regulatory complex (Cyclin E and Cdk2). Statistical significance when compared to the control: *p<0.05 and ***p<0.001. C) Levels of mRNA of SMCHD1 and PSIP1. Statistical significance when compared to the control: **p<0.01 and ***p<0.001.
Conclusions
In conclusion, a novel nanosystem able to transport AgNPs selectively to cancer cells has been developed. In a first stage, mesoporous silica nanoparticles externally decorated with carboxylic acid groups and proteins such as BSA and Tf act as effective platform for the nucleation of AgNPs homogeneous in size of approximately 15 nm. After physico-chemical characterization, in vitro assays in hepatocarcinoma cells (HepG2) and osteoblast-like cells (MC3T3-E1) demonstrate that only the nanosystem functionalized with Tf is able to selectively transport the AgNPs inside the cells overexpressing transferrin receptors (HepG2), therefore causing a reduction in cell viability and an increased in cell death. Due to the receptor mediated endocytic mechanism for the internalization of the nanosystem, the transported AgNPs dissolve in toxic Ag+ ions during the retention time within the lysosomes, following the “lysosome-enhanced Trojan horse effect”. Therefore, this novel nanomaterial is able to deliver AgNPs specifically to cancer cells overexpressing TfR, affecting key proteins and transcripts involved in cell cycle regulation, cell proliferation and DNA damage, as it has been demonstrated by quantitative proteomics and validated by qPCR. Thus, the present nanosystem offers the possibility of a targeted therapy using reduced doses of silver nanoparticles as cytotoxic agent. Furthermore, the silica matrix of the presented nanosystem could be used to host another drug in the pore network which would enhance its cytotoxic properties against cancer cells, transforming this material into a multidrug delivery device for a combined therapy. Due to the selectivity and effectiveness of the designed nanosystem in reducing cancer cell proliferation and in targeting relevant cancer-related proteins and transcripts, on-going work is aiming at exploring the suitability of this hybrid nanosystem in clinical applications.
Supplementary Material
Acknowledgements
This work was supported by Ministerio de Economía y Competitividad (MINECO) grants CTQ2017-85673-R, MAT2015-64831-R and MAT2016-75611-R AEI/FEDER; European Research Council, ERC-2015-AdG (VERDI) Proposal No. 694160; Instituto de Salud Carlos III (ISCIII) grant PI15/00978, and FEDER funds of the European Union. CIBER is a public research consortium created by ISCIII whose actions are co-funded by the European Regional Development Fund. S. Montalvo-Quiros acknowledges Ministry of Education, Culture and Sports from the Spanish Government for a predoctoral fellowship (FPU13/00723).
Footnotes
Conflicts of interest
There are no conflicts to declare.
IEPBSA calculated from the protein sequence NCBI GenBank (BSA, CAA76847.1) and the Expasy program (https://web.expasy.org/protparam/).
IEPTf calculated from the protein sequence NCBI GenBank (Human apotransferrin Tf, AAB22049.1) and the Expasy program (https://web.expasy.org/protparam/).
H2O2 + 2H+ + 2e− → 2H2O (E0 = 1.78 V) ; Ag → Ag+ + e−(E0 = 0.80 V)
Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available.
References
- 1.Riehemann K, Schneider SW, Luger TA, Godin B, Ferrari M, Fuchs H. Angew Chem Int Ed. 2009;48:872–897. doi: 10.1002/anie.200802585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. Chem Soc Rev. 2012;41:2740–2779. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei L, Lu J, Xu H, Patel A, Chen Z-S, Chen G. Drug Discov Today. 2015;20:595–601. doi: 10.1016/j.drudis.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krajczewski J, Kołątaj K, Kudelski A. RSC Adv. 2017;7:17559–17576. [Google Scholar]
- 5.Ong C, Lim JZZ, Ng C-T, Li JJ, Yung L-YL Bay B-H. Curr Med Chem. 2013;20:772–781. [PubMed] [Google Scholar]
- 6.Liang J, Zeng F, Zhang M, Pan Z, Chen Y, Zeng Y, Xu Y, Xu Q, Huang Y. RSC Adv. 2015;5:43733–43740. [Google Scholar]
- 7.Locatelli E, Naddaka M, Uboldi C, Loudos G, Fragogeorgi E, Molinari V, Pucci A, Tsotakos T, Psimadas D, Ponti J, Franchini MC. Nanomedicine. 2014;9:839–849. doi: 10.2217/nnm.14.1. [DOI] [PubMed] [Google Scholar]
- 8.Uygur B, Craig G, Mason MD, Ng AK. ACS Nano. 2009;3:279–290. [Google Scholar]
- 9.Luque-Garcia JL, Sanchez-Díaz R, Lopez-Heras I, Camara C, Martin P. Trends Anal Chem. 2013;43:254–268. [Google Scholar]
- 10.Gurunathan S, Lee KJ, Kalishwaralal K, Sheikpranbabu S, Vaidyanathan R, Eom SH. Biomaterials. 2009;30:6341–6350. doi: 10.1016/j.biomaterials.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Jang SJ, Yang IJ, Tettey CO, Kim KM, Shin HM. Mater Sci Eng C. 2016;68:430–435. doi: 10.1016/j.msec.2016.03.101. [DOI] [PubMed] [Google Scholar]
- 12.Faedmaleki F, Shirazi FH, Salarian A. Iran J Pharm Res. 2014;13:235–242. [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Muñiz MN, Muñoz-Olivas R, Luque-Garcia JL. Nanotoxicology. 2019 doi: 10.1080/17435390.2019.1579374. In press. [DOI] [PubMed] [Google Scholar]
- 14.Castillo RR, Baeza A, Vallet-Regí M. Biomater Sci. 2017;5:353–377. doi: 10.1039/c6bm00872k. [DOI] [PubMed] [Google Scholar]
- 15.Castillo RR, Colilla M, Vallet-Regí M. Expert Opin Drug Deliv. 2017;14:229–243. doi: 10.1080/17425247.2016.1211637. [DOI] [PubMed] [Google Scholar]
- 16.Manzano M, Vallet-Regí M. J Mater Sci Mater Med. 2018;29:65. doi: 10.1007/s10856-018-6069-x. [DOI] [PubMed] [Google Scholar]
- 17.Vallet-Regí M, Rámila A, del Real RP, Pérez-Pariente J. Chem Mater. 2001;13:308–311. [Google Scholar]
- 18.Martínez-Carmona M, Colilla M, Ruiz-González ML, González-Calbet JM, Vallet-Regí M. Microporous Mesoporous Mater. 2016;225:399–410. [Google Scholar]
- 19.López V, Villegas MR, Rodríguez V, Villaverde G, Lozano D, Baeza A, Vallet-Regí M. ACS Appl Mater Interfaces. 2017;9:26697–26706. doi: 10.1021/acsami.7b06906. [DOI] [PubMed] [Google Scholar]
- 20.Baeza A, Vallet-Regí M. Current Drug Targets. 2018;19:213–224. doi: 10.2174/1389450117666160603023037. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Carmona M, Lozano D, Colilla M, Vallet-Regí M. Acta Biomater. 2018;65:393–404. doi: 10.1016/j.actbio.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Colilla M, González B, Vallet-Regí M. Biomater Sci. 2013;1:114–134. doi: 10.1039/c2bm00085g. [DOI] [PubMed] [Google Scholar]
- 23.Croissant JG, Fatieiev Y, Almalik A, Khashab NM. Adv Healthcare Mater. 2018;7 doi: 10.1002/adhm.201700831. 1700831. [DOI] [PubMed] [Google Scholar]
- 24.Paris JL, Manzano M, Cabañas MV, Vallet-Regí M. Nanoscale. 2018;10:6402–6408. doi: 10.1039/C8NR00693H. [DOI] [PubMed] [Google Scholar]
- 25.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Acc Chem Res. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doadrio AL, Sánchez-Montero JM, Doadrio JC, Salinas AJ, Vallet-Regí M. Eur J Pharm Sci. 2017;97:1–8. doi: 10.1016/j.ejps.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Vallet-Regí M, Colilla M, Izquierdo-Barba I, Manzano M. Molecules. 2018;23:47. doi: 10.3390/molecules23010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh RK, Patel KD, Leong KW, Kim H-W. ACS Appl Mater Interfaces. 2017;9:10309–10337. doi: 10.1021/acsami.6b16505. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Dong K, Zhang Y, Ju E, Chen Z, Ren J, Qu X. Chem Commun. 2015;51:15975–15978. doi: 10.1039/c5cc06794d. [DOI] [PubMed] [Google Scholar]
- 30.Mahony D, Cavallaro AS, Stahr F, Mahony TJ, Qiao SZ, Mitter N. Small. 2013;9:3138–3146. doi: 10.1002/smll.201300012. [DOI] [PubMed] [Google Scholar]
- 31.Maeda H, Nakamura H, Fang J. Adv Drug Deliv Rev. 2013;65:71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Adv Drug Deliv Rev. 2013;65:121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byrne JD, Betancourt T, Brannon-Peppas L. Adv Drug Deliv Rev. 2008;60:1615–1626. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Daniels TR, Delgado T, Rodriguez JA, Helguera G, Penichet ML. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA, Holler E, Ljubimova JY, Helguera G, Penichet ML. Biochim Biophys Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang AA, Lu J, Tamanoi F, Zink JI. Small. 2015;11:319–328. doi: 10.1002/smll.201400765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez A, Fuentes-Paniagua E, Baeza A, Sánchez-Nieves J, Cicuéndez M, Gómez R, de la Mata FJ, González B, Vallet-Regí M. Chem Eur J. 2015;21:15651–15666. doi: 10.1002/chem.201501966. [DOI] [PubMed] [Google Scholar]
- 38.Ge S, Kojio K, Takahara A, Kajiyama T. J Biomater Sci Polym Ed. 1998;9:131–150. doi: 10.1163/156856298x00479. [DOI] [PubMed] [Google Scholar]
- 39.Gomme PT, McCann KB. Drug Discov Today. 2005;10:267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- 40.Luo Z, Hu Y, Xin R, Zhang B, Li J, Ding X, Hou Y, Yang L, Cai K. J Biomed Mater Res Part A. 2014;102:3781–3794. doi: 10.1002/jbm.a.35049. [DOI] [PubMed] [Google Scholar]
- 41.Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. Nano Lett. 2003;3:413–417. [Google Scholar]
- 42.Nam KT, Lee YJ, Krauland EM, Kottmann ST, Belcher AM. ACS Nano. 2008;2:1480–1486. doi: 10.1021/nn800018n. [DOI] [PubMed] [Google Scholar]
- 43.Kittler S, Greulich C, Gebauer JS, Diendorf J, Treuel L, Ruiz L, Gonzalez-Calbet JM, Vallet-Regí M, Zellner R, Köller M, Epple M. J Mater Chem. 2010;20:512–518. [Google Scholar]
- 44.Rodríguez-Galván A, Contreras-Torres FF, Basiuk EV, Heredia A, Basiuk VA. Can J Chem Eng. 2013;91:264–270. [Google Scholar]
- 45.Ahsan H, Masaaki T, Guang WY. Bull Chem Soc Jpn. 2005;78:262–269. [Google Scholar]
- 46.Sun RW-Y, Chen R, Chung NP-Y, Ho C-M, Lin C-LS, Che C-M. Chem Commun. 2005:5059–5061. doi: 10.1039/b510984a. [DOI] [PubMed] [Google Scholar]
- 47.Chen R, Wu J, Li H, Cheng G, Lu Z, Che C-M. Rare Met. 2010;29:180–186. [Google Scholar]
- 48.Tan S, Erol M, Attygalle A, Du H, Sukhishvili S. Langmuir. 2007;23:9836–9843. doi: 10.1021/la701236v. [DOI] [PubMed] [Google Scholar]
- 49.Serjeant EP, Dempsey B, Perrin DD. pKa Prediction for Organic Acids and Bases. Springer Science & Business Media; 1981. [Google Scholar]
- 50.Rosenholm JM, Sahlgren C, Lindén M. Nanoscale. 2010;2:1870–1883. doi: 10.1039/c0nr00156b. [DOI] [PubMed] [Google Scholar]
- 51.González B, Colilla M, Díez J, Pedraza D, Guembe M, Izquierdo-Barba I, Vallet-Regí M. Acta Biomater. 2018;68:261–271. doi: 10.1016/j.actbio.2017.12.041. [DOI] [PubMed] [Google Scholar]
- 52.Tang J, Wang Q, Yu Q, Qiu Y, Mei L, Wan D, Wang X, Li M, He Q. Acta Biomater. 2019;83:379–389. doi: 10.1016/j.actbio.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 54.Hwang AA, Lu J, Tamanoi F, Zink JI. Small. 2015;11:319–328. doi: 10.1002/smll.201400765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Martens TF, Remaut K, Demeester J, De Smedt SC, Braeckmans K. Nano Today. 2014;9:344–364. [Google Scholar]
- 57.Barua S, Mitragotri S. Nano Today. 2014;9:223–243. doi: 10.1016/j.nantod.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Setyawati MI, Yuan X, Xie J, Leong DT. Biomaterials. 2014;35:6707–6715. doi: 10.1016/j.biomaterials.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Paris JL, Colilla M, Izquierdo-Barba I, Manzano M, Vallet-Regí M. J Mater Sci. 2017;52:8761–8771. [Google Scholar]
- 60.Sabella S, Carney RP, Brunetti V, Malvindi MA, Al-Juffali N, Vecchio G, Janes SM, Bakr OM, Cingolani R, Stellacci F, Pompa PP. Nanoscale. 2014;6:7052–7061. doi: 10.1039/c4nr01234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar C. Nanomaterials-Toxicity, Health and Environmental Issues. Vol. 5 Wiley-VCH Verlag GmbH & Co; Weinheim, Germany: 2006. [Google Scholar]
- 62.AshaRani PV, Mun GLK, Hande MP, Valiyaveettil S. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 63.Loza K, Diendorf J, Sengstock C, Ruiz-Gonzalez L, Gonzalez-Calbet JM, Vallet-Regí M, Köllerb M, Epple M. J Mater Chem B. 2014;2:1634–1643. doi: 10.1039/c3tb21569e. [DOI] [PubMed] [Google Scholar]
- 64.Park EJ, Yi J, Kim Y, Choi K, Park K. Toxicol In Vitro. 2010;24:872–878. doi: 10.1016/j.tiv.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Miyayama T, Matsuoka M. J Occup Med Toxicol. 2016;11:1–6. doi: 10.1186/s12995-016-0090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asadi MH, Derakhshani A, Mowla SJ. Tumor Biol. 2014;35:7177–7185. doi: 10.1007/s13277-014-1966-1. [DOI] [PubMed] [Google Scholar]
- 67.Dai M-S, Sun X-X, Lu H. Mol Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai R, McKay R. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Yang X, Li Y, Zhao S, Li C, Ma P. FEBS Lett. 2016;1:4213–4222. doi: 10.1002/1873-3468.12471. [DOI] [PubMed] [Google Scholar]
- 70.Woo CC, Chen WC, Teo XQ, Radda GK, Hock Lee PT. Oncotarget. 2016;7:53005–53017. doi: 10.18632/oncotarget.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao Z, Hu K, Huang HE, Xu S, Wang Q. Int J Mol Med. 2015;36:1393–1399. doi: 10.3892/ijmm.2015.2350. [DOI] [PubMed] [Google Scholar]
- 72.Kinoshita Y, Kalir T, Rahaman J, Dottino P, Kohtz DS. Am J Pathol. 2012;180:375–389. doi: 10.1016/j.ajpath.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang S, Cui J, Yang Y, Liu Z, Yan H, Tang C, Wang H, Qin H, Li X, Li J, Wang W, et al. Gene. 2016;576:421–428. doi: 10.1016/j.gene.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 74.Cui D, Li L, Lou H, Sun H, Ngai SM, Shao G, Tang J. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 75.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. Cell Death Dis. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gutschner T, Hämmerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, Heim A, Gro M, Hofmann N, Geffers R, Skawran B, et al. Hepatology. 2014;59:1900–1911. doi: 10.1002/hep.26997. [DOI] [PubMed] [Google Scholar]
- 77.Kim TW, Havighurst T, Kim KM, Albertini M, Xu YG, Spiegelman VS. Mol Carcinog. 2018;57:678–683. doi: 10.1002/mc.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Rourke Fa, LaPlante JM, Feinstein MB. Biochem J. 2003;373:133–143. doi: 10.1042/BJ20030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bol GM, Xie M, Raman V. Mol Cancer. 2015;14:188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang D, Wang F, Pang Y, Ke X, Zhu S, Zhao E. Oncotarget. 2017;8:80956–80970. doi: 10.18632/oncotarget.20898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song K, Russo G, Krauss M. Front Cell Dev Biol. 2016;4:124. doi: 10.3389/fcell.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diesenberg K, Beerbaum M, Fink U, Schmieder P, Krauss M. J Cell Sci. 2015;128:397–407. doi: 10.1242/jcs.162206. [DOI] [PubMed] [Google Scholar]
- 83.Nyati MK, Morgan MA, Feng FY, Lawrence TS. Nat Rev Cancer. 2006;6:876–885. doi: 10.1038/nrc1953. [DOI] [PubMed] [Google Scholar]
- 84.Tang M, Li Y, Zhang X, Deng T, Zhou Z, Ma W, Zhou S. J Biol Chem. 2014;289:34024–34032. doi: 10.1074/jbc.M114.601179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coker H, Brockdorff N. J Cell Sci. 2014;127:1869–1874. doi: 10.1242/jcs.140020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, Dobner T, et al. Molecular Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deb S, Xu H, Tuynman J, George J, Yan Y, Li J, Ward RL, Mortensen N, Hawkins NJ, McKay MJ, Ramsay RG, et al. Br J Cancer. 2014;110:1606–1613. doi: 10.1038/bjc.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song Y, Luo Q, Long H, Hu Z, Que T, Zhang X, Li Z, Wang G, Yi L, Liu Z, Fang WY, et al. Molecular Cancer. 2014;13:1–12. doi: 10.1186/1476-4598-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang QL, Huang ZG, Li GN, Du JL, Ou YP, Zhang XN, Chen TT. Int J Clin Exp Med. 2015;8:231–239. [PMC free article] [PubMed] [Google Scholar]
- 90.Basu A, Rojas H, Banerjee H, Cabrera IB, Perez KY, de León M, Casiano CA. PLoS ONE. 2012;7:26–29. doi: 10.1371/journal.pone.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sørensen CS, Petersen NHT, Sorensen PHB, Lukas C, Bartek J, Lukas J, et al. Nat Struct Mol Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.