Abstract
Cow antibodies are unusual in having an exceptionally long third complementarity determining region of the heavy chain (CDR H3). These CDR H3s have a multitude of cysteines and form a distinct domain characterized by a β-ribbon “stalk” and disulfide bonded “knob”. Cows appear to utilize somatic hypermutation of a single VDJ rearrangement to produce an astounding variety of distinct CDR H3 sequences with different disulfide bonding patterns within the knob. Thus, cows may be unique amongst vertebrates in evolving an antibody system with both a different scaffold for binding antigen as well as an unusual diversity creating process.
Introduction
Combinatorial diversity in antibody genes is driven by the multitude of V, D, and J gene segments encoded in germline DNA. Cows have limited genome encoded combinatorial diversity potential, as they possess only three known functionally rearranging VH region genes which are all highly homologous [1,2]. However, the third complementarity determining region of the heavy chain (CDR H3) is unusually long in cows, and a subset of the repertoire (the “ultralong” repertoire, representing 10–15% of heavy chains) can reach up to nearly 70 amino acids in length. These are much longer than the 8–16 amino acid CDR H3s of typical human antibodies. The identification of these unusual ultralong CDR H3s was made several years ago in studies of bovine B-cells transformed with bovine leukemia virus [4]. While these initial sequences were found in IgM, further work revealed ultralong CDR H3s in all five immunoglobulin isotypes [5]. Recently, the ultralong CDR H3 repertoire has been found to be incredibly diverse, and structural studies have shown that the ultralong CDR H3 is comprised of two unique domain substructures: (1) a β-ribbon “stalk” that protrudes out from the antibody surface which supports (2) a disulfide bonded “knob” domain [3]. Antigen binding appears to reside entirely within the knob, which is uniquely diversified in both amino acid content as well as disulfide bonded loop patterns. The paradigm for both antibody diversity and antigen binding is an unusual alternative to conventional antibodies; diversity occurs in part by wholesale changes of loop structures through disulfide mutations, and the scaffold that binds antigen is a unique “knob” that is distinct from the CDR loops of a traditional immunoglobulin domain. While the biological significance of these antibodies is unclear, they may enable binding of concave types of epitopes as a result of their novel protruding CDR H3 [6].
Antibody Diversity
In most species, antibodies display a wide range of diversity within the context of the immunoglobulin fold. The CDR H3 is the most varied portion of the antibody molecule and is derived from DNA rearrangement of variable (V), diversity (D), and junctional (J) gene segments [7–9]. Somatic hypermutation results in additional point mutations in the variable regions after antigen exposure [10,11]. Although humans and mice possess a large pool of VDJ genes, certain species such as chicken [12], pig [13], sheep [14], and cattle [1,2] have limited combinatorial diversity potential. Specifically, cows have a very limited number of V gene segments [1,15–17]; however, they are perfectly capable of mounting a robust adaptive immune response.
Combinatorial diversity also includes the multitude of potential heavy chain/light chain pairings that can occur. Interestingly, the VHH antibodies from camelids and the IgNAR of sharks contain bivalent heavy chain domains without light chains, thus their combinatorial diversity all rests within the heavy chain [18,19]. Both of these species still utilize the immunoglobulin scaffold, and thus, their heavy chain CDR loops to bind antigen. Cow ultralong CDR H3 antibodies appear to pair with a single germline light chain; therefore, VH/VL pairing also may not contribute significantly to repertoire diversity. Thus, with limited combinatorial diversity generated through VDJ recombination and limited VH/VL pairing, the majority of the diversity in the repertoire is concentrated in CDR H3.
The mechanisms for creating antibody diversity in the cow ultralong CDR H3 antibody repertoire are not well studied. Deep sequencing of ultralong CDR H3s has shown that these structures contain a complexity of cysteines, which initially indicated that disulfide-bonded mini-domains may form during repertoire development [3]. Sequence analysis suggested that diversity arises from somatic hypermutation of a single ultralong DH region, DH2. This germline encoded DH appears to have a bias towards codons that can mutate to cysteine with only one nucleotide change. Interestingly, somatic hypermutation is active early in B-cell development in cows, enabling somatic hypermutation to create diversity in the primary repertoire as opposed to the secondary repertoire as in many other species [3,20]. Examination of the crystal structures of two cow antibodies revealed that these CDR H3s form an unusual architecture comprised of a β-strand “stalk” that supports a structurally distinct, disulfide-bonded, “knob” domain [3]. Current research on the cow antibody system suggests that these animals produce an antibody repertoire composed of ultralong CDR H3s that fold into a multitude of knob mini-domains that are generated through somatic hypermutation which enables formation of different disulfide bonding patterns. Thus, somatic hypermutation creates diversity both through amino acid side change alterations as well as by changing loop patterns produced by disulfide bonds.
Ultralong CDR H3 Antibody Structures
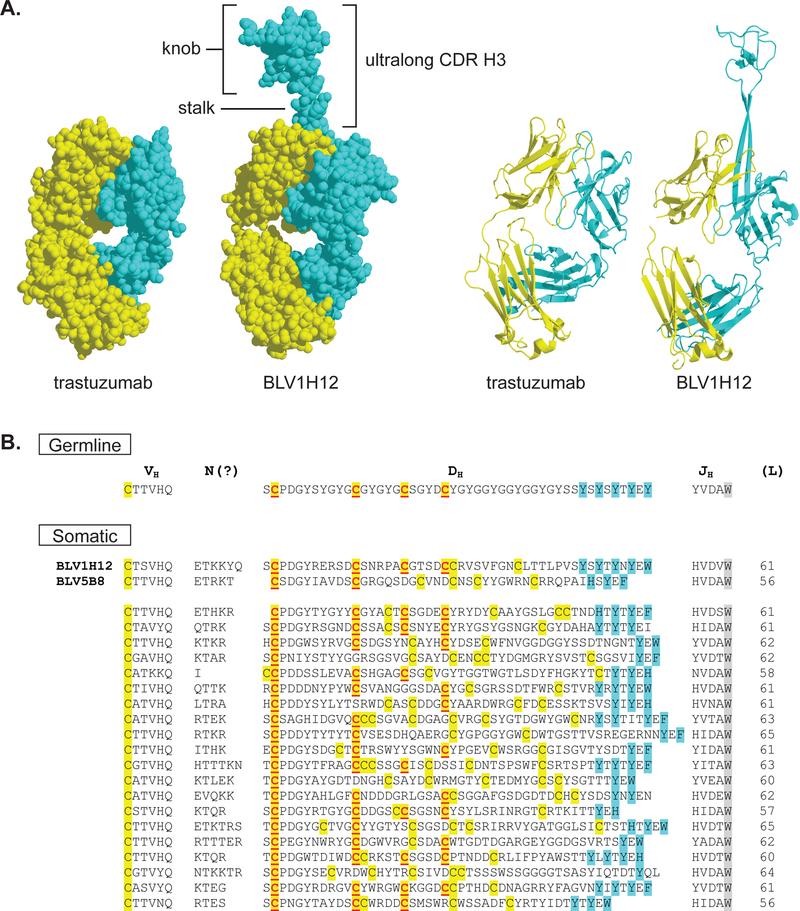
Antibody diversity and antigen binding in vertebrates is often restricted to the complementarity determining region loops of the immunoglobulin fold. A flat or undulating binding surface is usually formed by the CDRs of the heavy and light chains for antigen recognition. In humans, the CDR H3 is generally 8–16 amino acids in length; although, some longer CDR H3 loops, which protrude from the antibody surface, have been described. These longer CDR H3s contribute to important functions such as virus neutralization [21–26]. A variety of CDR H3 lengths exist across different species. Specifically, cow antibodies have the longest CDR H3 regions known with a 10–15% subset of ultralong CDR H3s that ranges in length from 50 to 69 amino acids (Figure 1) [1,4,15–17]. The heavy chains of these ultralong CDR H3s are thought to pair with a restricted set of lambda light chains [1]. Additionally, they have multiple yet an even number of cysteines, which indicates they likely participate in disulfide bonds [4]. The distinctive aspects of cow CDR H3s like the restricted VH/VL pairing, potential for multiple disulfide bonds, and unusually long length initially suggested that these structures might not be loops in the conventional sense but that they may have a unique and well-defined structural fold.
Figure 1.
Structure and diversity of cow ultralong CDR H3 regions. (A) The Fab fragments for trastuzumab (PDB: 1N8Z) and BLV1H12 (PDB: 4K3D) are shown in space fill (left) and ribbon diagrams (right). The heavy chains are colored cyan and the light chains yellow. The ultralong CDR H3 in the cow Fab BLV1H12 is comprised of a β-ribbon “stalk” and disulfide bonded “knob” domain that protrude far from the typical antibody paratope. (B) Sequences of ultralong CDR H3s are shown, with the germline VHBUL, DH2 and JH illustrated at the top and several somatic sequences at the bottom. The sequences encoding the two Fabs whose crystal structures have been solved (BLV1H12 and BLV5B8) are indicated. Cysteines are highlighted in yellow, with those encoded by the germline DH2 underlined and in red. Aromatic residues expected to form the descending strand of the β-ribbon stalk are in cyan, and the conserved tryptophan in the JH region is greyed. Note the conservation of the first cysteine encoded by DH2, with the decrease in conservation proceeding C-terminally.
Two published crystal structures of Fab fragments, derived from the antibodies BLV1H12 and BLV5B8 [1,2] which contain CDR H3s of 56 and 61 amino acids, were recently solved at 1.9 Å and 2.0 Å resolution, respectively. BLV1H12 has the longest known complementarity determining region whose structure has been determined [3]. The ultralong CDR H3s of both antibodies form a previously undescribed structure that includes two anti-parallel β strands as a “stalk” and a disulfide-bonded “knob” which rests atop the stalk. This structure is analogous to a mushroom with a stalk and cap (Figure 1). This long anti-parallel β-ribbon (10 residues in BLV5B8 and 12 in BLV1H12) is rigidified using 8 backbone β-sheet hydrogen bonds and serves to link the “knob” domain with the main antibody scaffold. It is unusual to find a β-ribbon linking two folded domains, and the cow antibody architecture may be unique among all protein structures in this regard. The side chains of the ascending β-strand are largely hydrophilic, while the descending β-strand has an alternating aromatic pattern (e.g. YXYXY). The stacking of these aromatic amino acid side chains may contribute to the stability of this long solvent exposed two-stranded β-sheet. The base of the CDR H3 stalk interacts with complementarity determining regions H1, H2, L1, and L3. The base of the ascending strand is formed by a “CTTVHQ” motif that is encoded by the VH region and appears to only exist in cow antibodies with ultralong CDR H3s. This motif interacts with a “DKAVG” motif in CDR H1, which is also highly conserved in cow antibodies with ultralong CDR H3s [3]. The 34-amino-acid knob domain of BLV1H12 is folded such that the three disulfide bonds adopt a 1–4, 2–6, 3–5 pattern, whereas for BLV5B8 the pattern is 1–3, 2–4, 5–6. While the two Fab fragments share similar overall structures, including the immunoglobulin frameworks of the heavy and light chains and the stalk and knob features of the CDR H3, significant differences exist in the CDR H3 region. The β-ribbon stalk of BLV5B8 is two residues shorter than BLV1H12, which leads to a stalk orientation shift. This changes the position as well as the shape of the “knob” domain. The amino acid sequences of the knobs are highly dissimilar, which contribute to different surface potentials and different disulfide bonding patterns. Thus, the fold, surface contour, and surface potential of the “knob” domain of the two Fabs are distinct despite both CDR H3s containing the key structural features of “stalk” and “knob”.
Recently, Stanfield, et.al. solved three additional crystal structures of ultralong CDR H3 antibodies [27]. All of these contained the “stalk” and “knob” architecture; however, additional conserved features were identified. Remarkably, each knob contained a very short three-stranded β-sheet, despite having very little sequence conservation in this region. This conserved β-sheet is structurally similar to other small disulfide bonded domains, with the closest structural match being Kalata B1, which, interestingly, plays a role in pathogen protection in plants. A conserved disulfide was also identified, which utilized a germline encoded cysteine but whose cysteine bonding partner was not conserved. Furthermore, the loops between the strands were diverse in length and amino acid content, suggesting that the short loops within CDR H3 may be functionally analogous to the CDRs of a traditional antibody.
The sequence and structural analysis of bovine antibodies with ultralong CDR H3s reveals a potential mechanism that enables a small amount of germline-encoded genetic material to generate substantial sequence and structural diversity by generating disulfide crosslinked hypervariable mini-folds de novo. There are a multitude of small disulfide bonded peptides with diverse functions in nature. The bovine ultralong CDR H3 system may serve as a model for diversification of small minifolds containing disulfide bonds. In this vein, the covalent disulfide bonds may enable stabilization of small folds where other molecular interactions like hydrogen bonds or hydrophobic interactions are either too weak, or take too long in evolutionary time to optimize. The ultralong bovine CDR H3 domain not only provides a novel scaffold to study an unusual disulfide-based structural diversity paradigm but also potentially allows for the engineering of unique therapeutics, including creating functional proteins with different folds fused into the CDR H3 region or selecting novel disulfide bonded structures from molecular libraries. These investigational and applied aspects of antibody diversity could be of significant biomedical importance as recombinant antibodies are now an important class of therapeutics for multiple indications.
Ultralong CDR H3 Formation
The structural and sequence information available for ultralong CDR H3 antibodies suggests mechanisms for how they arise in vivo. The “CTTVHQ” motif at the base of the ascending strand of the stalk is a highly unusual sequence. Most antibody variable region genes encode CAR or CAK in this region, with the arginine or lysine often forming a salt bridge with a conserved aspartate encoded by the J region. The relative conservation of CTTVHQ in ultralong sequences suggested that it was encoded in the germline, and indeed, a new variable region gene termed VHBUL (variable heavy bovine ultralong) was identified in the Bos taurus genome [3]. VHBUL appears to be preferentially used in cow antibodies with ultralong CDR H3s. There is only one germline DH region (DH2) capable of encoding an ultra-long CDR H3 in cattle, and this DH encodes four cysteines [28]. As only one JH is used in cattle, all ultralong sequences appear to use the same VDJ recombination germline gene segments. Despite the combinatorial diversity limitation by using single V, D, and J segments, junctional diversity can additionally add or remove nucleotides between V-D and D-J segments through the action of terminal deoxynucleotidyl transferase (TdT) and exonuclease during the process of V(D)J recombination. Of particular note, the V-D and D-J joints encode regions of the ascending and descending strands of the stalk, respectively. Thus, variation in the stalk sequence and length can likely occur solely through junctional diversity. Rearranged ultralong CDR H3 genes are somewhat unusual as extensive insertion appears to occur at the V-D junction, where multiple amino acids are added and even appear somewhat conserved. These inserted amino acids are often positively charged, and a threonine is usually found in the second position. Typically, insertions occur during V(D)J recombination through the action of TdT or through palindromic-based insertions (P-nucleotides) at the coding ends. TdT-based N-nucleotide insertions are not usually as long as the insertions seen in ultralong V-D junctions, and analysis of whether P-nucleotide additions or gene conversion could account for these insertions at the V-D junction have not yielded a clear explanation. However, a novel “nucleotide capture” mechanism has been proposed to account for the mechanism behind these insertions [4]. Given the structural conservation of the ascending stalk, clearly only certain residues could be accommodated in this region.
The knob region appears to be entirely encoded by DH2. As DH2 is the only ultralong D region, and it is one of approximately ten DH regions in the germline, the fact that the ultralong antibody repertoire is present at about 10% of the repertoire can be entirely explained by the usage of DH2 in 10% of antibody rearrangements. Deep sequencing analysis of ultralong CDR H3s did not reveal any evidence for an alternative D region; however, given the extensive diversity in the knob region, one cannot completely rule out the presence of alternative DHs or alternative mechanisms like gene conversion in participating in repertoire diversification. Interestingly, of the four cysteines encoded by the germline DH2, the first appears nearly completely conserved in somatically mutated knob sequences (Figure 1, B). This cysteine participates in a disulfide bond at the base of the knob. While this cysteine is conserved, its disulfide partner appears variable, as the conservation of the remaining three cysteines is not particularly high. In fact, it appears as though cysteine conservation decreases as one moves from the N- to the C- terminus of the knob amino acid sequence. As described above, somatic hypermutation is hypothesized to generate the substantial diversity in CDR H3, and its role includes generating mutations that alter amino acid side chains but also alter disulfide patterns and their associated loops. The combination of somatic hypermutation and selection acting on DH2 seems to produce the remarkable diversity in the knob; however, the first cysteine remains conserved, as does the overall architecture of the stalk and knob. Further structural and genetic research should yield important details on the mechanisms behind formation and diversification of these unique CDR H3s.
The biological function of ultralong CDR H3 antibodies remains unknown. The heavy chain locus of the cow is unusual compared with other mammals, with a duplication that results in two IgM genes [29]. Interestingly, ultralong antibodies seem to be completely associated to rearrangement to IgM2. Any differential roles in immunity of the two IgM genes remain to be determined.
Conclusions
The underlying structure and diversity mechanisms driving humoral immunity are not entirely evolutionarily conserved across vertebrate species. Jawless fish utilize a non-antibody variable lymphocyte receptor scaffold that utilizes unique leucine rich repeat modules to create a novel paratope [30–32]. Camelids and sharks have evolved novel antigen combining sites in antibodies that only utilize a heavy chain; such paratopes have longer CDR H3s that replace the light chain interface and allow a smaller protruding binding site that may be able them to interact with concave surfaces [6,33]. Cows, on the other hand, have evolved an ultralong CDR H3 that protrudes far from the typical antibody binding site. By creating structural diversity within ultralong CDR H3s encoded only by a single VHBUL, DH2, and JH paired with a limited number of lambda light chains, cattle tremendously expand their limited combinatorial repertoire. It is tempting to speculate about the evolutionary purpose for developing this unique humoral immune system. Cows are capable of mounting a very robust immune response to infectious agents. Cattle are ruminants, which are characterized by digestive systems that include a rumen, reticulum, omasum, abomasum, and a large cecum. The rumen, in particular, functions in fermenting plant-based feed stuff and maintains an expansive gut microbiome comprised of both prokaryotic and eukaryotic microorganisms [34]. One possible purpose for the cow’s unique antibody system is their necessity in regulating the gut microbiome and general mucosal immunity. Additional suppositions concentrate on ruminant reproductive immunity. Current understanding of fetal development in the cow indicates that immunoglobulins are not transported across the placenta as in other species. This failure requires the neonate to ingest these macromolecules through colostrum before the intestinal cells are unable to absorb them within the first few hours of life [35]. It is possible that cows have developed this unique antibody system to ensure immunological coverage of the species’ neonates. Further research in this specialized field will shed light on the function and evolutionary drivers behind these unusual ultralong CDR H3 cow antibodies.
Highlights.
Cows have a limited combinatorial antibody repertoire.
Cows have an unusual ultralong CDR H3 comprising “stalk” and “knob” features.
Diversity may occur through a DH region with codons primed to mutate to cysteine.
A rich variety of sequence and cysteine content is found in cow ultralong CDR H3s.
Acknowledgements
This work was supported in part by the National Institute of General Medical Sciences (R01GM105826–01A1).
References
- 1.Saini SS, Farrugia W, Ramsland PA, Kaushik AK: Bovine IgM antibodies with exceptionally long complementarity-determining region 3 of the heavy chain share unique structural properties conferring restricted VH + Vlambda pairings. Int Immunol 2003, 15:845–853. [DOI] [PubMed] [Google Scholar]
- 2.Sinclair MC, Gilchrist J, Aitken R: Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol 1997, 159:3883–3889. [PubMed] [Google Scholar]
- **3.Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, et al. : Reshaping antibody diversity. Cell 2013, 153:1379–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]; Description of the first structures of ultralong CDR H3 antibodies, as well as diversity generating mechanisms. This study also defined the “knob” domain of CDR H3 as interacting with antigen in a model antibody.
- 4.Saini SS, Allore B, Jacobs RM, Kaushik A: Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol 1999, 29:2420–2426. [DOI] [PubMed] [Google Scholar]
- **5.Walther S, Czerny C-P, Diesterbeck US: Exceptionally long CDR3H are not isotype restricted in bovine immunoglobulins. PLoS One 2013, 8:e64234. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mapping of the bovine immunoglobulin heavy chain locus and identification of ultralong CDR H3 sequences in all antibody isotypes.
- 6.de Los Rios M, Criscitiello MF, Smider VV: Structural and genetic diversity in antibody repertoires from diverse species. Curr Opin Struct Biol 2015, 33:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG: The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 2000, 18:495–527. [DOI] [PubMed] [Google Scholar]
- 8.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T: An evolutionary view of the mechanism for immune and genome diversity. J Immunol 2012, 188:3559–3566. [DOI] [PubMed] [Google Scholar]
- 9.Smider V, Chu G: The end-joining reaction in V(D)J recombination. Semin Immunol 1997, 9:189–197. [DOI] [PubMed] [Google Scholar]
- 10.Di Noia JM, Neuberger MS: Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 2007, 76. [DOI] [PubMed] [Google Scholar]
- 11.Kocks C, Rajewsky K: Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc Natl Acad Sci U S A 1988, 85:8206–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynaud CA, Dahan A, Anquez V, Weill JC: Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell 1989, 59:171–183. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Kacskovics I, Brown WR, Butler JE: Expressed swine VH genes belong to a small VH gene family homologous to human VHIII. J Immunol 1994, 153:5618–5627. [PubMed] [Google Scholar]
- 14.Dufour V, Malinge S, Nau F: The sheep Ig variable region repertoire consists of a single VH family. J Immunol 1996, 156:2163–2170. [PubMed] [Google Scholar]
- 15.Berens SJ, Wylie DE, Lopez OJ: Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol 1997, 9:189–199. [DOI] [PubMed] [Google Scholar]
- 16.Lopez O, Perez C, Wylie D: A single VH family and long CDR3s are the targets for hypermutation in bovine immunoglobulin heavy chains. Immunol Rev 1998, 162:55–66. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Jackson SM, Aitken R: The bovine antibody repertoire. Dev Comp Immunol 2006, 30:175–186. [DOI] [PubMed] [Google Scholar]
- 18.Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L: A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 1999, 7:361–370. [DOI] [PubMed] [Google Scholar]
- 19.Stanfield RL, Dooley H, Flajnik MF, Wilson IA: Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004, 305:1770–1773. [DOI] [PubMed] [Google Scholar]
- 20.Liljavirta J, Niku M, Pessa-Morikawa T, Ekman A, Iivanainen A: Expansion of the preimmune antibody repertoire by junctional diversity in Bos taurus. PLoS One 2014, 9:e99808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collis AVJ, Brouwer AP, Martin ACR: Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol 2003, 325:337–354. [DOI] [PubMed] [Google Scholar]
- 22.Kwong PD, Wilson IA: HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol 2009, 10:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA: Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A 2010, 107:11483–11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA: Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 2001, 293:1155–1159. [DOI] [PubMed] [Google Scholar]
- 25.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. : Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011, 480:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. : Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489:526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Stanfield RL, Wilson IA, Smider V:Conservation and diversity in the ultralong CDR H3 of bovine antibodies. Science Immunology 2016, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structural analysis of five ultralong CDR H3 antibodies reveals a short conserved β-sheet within the “knob” domain, with connecting loops that are diverse in sequence and length.
- 28.Shojaei F, Saini SS, Kaushik AK: Unusually long germline DH genes contribute to large sized CDR3H in bovine antibodies. Mol Immunol 2003, 40:61–67. [DOI] [PubMed] [Google Scholar]
- *29.Ma L, Qin T, Chu D, Cheng X, Wang J, Wang X, Wang P, Han H, Ren L, Aitken R, et al. : Internal Duplications of DH, JH, and C Region Genes Create an Unusual IgH Gene Locus in Cattle. J Immunol 2016, doi: 10.4049/jimmunol.1600158. [DOI] [PubMed] [Google Scholar]; Description of the heavy chain loocus in cattle, and discovery of a duplication that gives rise to two IgM genes. Ultralong CDR H3 antibodies appear to be exclusively rearranged to IgM2.
- 30.Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD: Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430:174–180.15241406 [Google Scholar]
- 31.Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD: Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430:174–180.15241406 [Google Scholar]
- 32.Kanda R, Sutoh Y, Kasamatsu J, Maenaka K, Kasahara M, Ose T: Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module. PLoS One 2014, 9:e85875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyldermans S, Smider VV: Distinct antibody species: structural differences creating therapeutic opportunities. Curr Opin Immunol 2016, 40:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao S, Zhang M, Liu J, Zhu W: Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci Rep 2015, 5:16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staley TE, Bush LJ: Receptor mechanisms of the neonatal intestine and their relationship to immunoglobulin absorption and disease. J Dairy Sci 1985, 68:184–205. [DOI] [PubMed] [Google Scholar]