Abstract
Gut microbiota have profound effects on bile acid metabolism by promoting deconjugation, dehydrogenation, and dehydroxylation of primary bile acids in the distal small intestine and colon. High-fat diet-induced dysbiosis of gut microbiota and bile acid dysregulation may be involved in the pathology of steatosis in patients with non-alcoholic fatty liver disease. Epigallocatechin-3-gallate (EGCG), the most abundant polyphenolic catechin in green tea, has been widely investigated for its inhibitory or preventive effects against fatty liver. The aim of the present study was to investigate the effects of EGCG on the abundance of gut microbiota and the composition of serum bile acids in high-fat diet-fed mice and determine the specific bacterial genera that can improve the serum bile acid dysregulation associated with EGCG anti-hepatic steatosis action. Male C57BL/6N mice were fed with the control diet, high-fat diet, or high-fat diet + EGCG at a concentration of 0.32% for 8 weeks. EGCG significantly inhibited the increases in weight, the area of fatty lesions, and the triglyceride content in the liver induced by the high-fat diet. Principal coordinate analysis revealed significant differences in microbial structure among the groups. At the genus level, EGCG induced changes in the microbiota composition in high-fat diet-fed mice, showing a significantly higher abundance of Adlercreutzia, Akkermansia, Allobaculum and a significantly lower abundance of Desulfovibrionaceae. EGCG significantly reversed the decreased population of serum primary cholic acid and β-muricholic acid as well as the increased population of taurine-conjugated cholic acid, β-muricholic acid and deoxycholic acid in high-fat diet-fed mice. Finally, the correlation analysis between bile acid profiles and gut microbiota demonstrated the contribution of Akkermansia and Desulfovibrionaceae in the improvement of bile acid dysregulation in high-fat diet-fed mice by treatment with EGCG. In conclusion, the present study suggests that EGCG could alter bile acid metabolism, especially taurine deconjugation, and suppress fatty liver disease by improving the intestinal luminal environment.
Keywords: Akkermansia, dysbiosis, epigallocatechin-3-gallate, high-fat diet, taurine-conjugated bile acids
Introduction
Non-alcoholic fatty liver disease (NAFLD) has become an important public health issue because of its high prevalence.(1) The spectrum of NAFLD ranges from simple hepatic steatosis, commonly associated with obesity, to non-alcoholic steatohepatitis, which can progress to fibrosis, cirrhosis, and hepatocellular carcinoma.(2) Numerous factors are complexly involved in the pathology of NAFLD, including inflammatory factors such as lipopolysaccharides, cytokines, lipid overload, and genetic predisposition, but their mechanisms have not been clarified. Studies are gradually revealing the abnormalities of gut microbiota and their metabolites in patients with NAFLD,(3,4) as well as in animal models induced by a high-fat diet (HFD).(5) Hildebrandt et al.(5) first reported dysbiosis characterized by decreases in Proteobacteria and Firmicutes and a decrease in the Bacteroidetes in HFD-fed mice. This result has since been reproduced in many experiments. Recent metagenomic analysis by next generation sequencing has demonstrated that the Proteobacteria phylum is closely involved in hepatic fibrosis in NAFLD patients(6) and that Bilophila wadsworthia, a member of the Proteobacteria phylum, could aggravate an HFD-induced inflammatory response.(7)
In addition, when considering the mechanism by which dysbiosis of the resident microbiota could modify the pathology of NAFLD, several studies have revealed an important role of metabolites produced by microbiota using mass spectrometer (MS) analysis. Gut microbes synthesize many products such as short-chain fatty acids,(8) trimethylamine,(9) ammonia,(10) and hydrogen.(11) A recent study reported evidence of a link between hepatic steatosis/fibrosis and the gut microbiome-derived metabolite 3-(4-hydroxyphenyl) lactate, which is a product of aromatic amino acid metabolism,(12) through a prospective cohort study. On the other hand, numerous human clinical trials of bile acids targeting gastrointestinal disease have been carried out.(13–15) In recent years, several metabolites of bile acids (BAs) have been shown to regulate lipid and carbohydrate metabolism as well as energy homeostasis, in both hepatic and extrahepatic tissues, through positive and negative regulation of the activation of BA-specific receptors, farnesoid X receptor (FXR), and transmembrane G protein-coupled receptor (TGR)-5. The primary BAs synthesized in the human liver are cholic acid (CA) and chenodeoxycholic acid (CDCA). They are conjugated with glycine or taurine (Glyco-CA, Glyco-CDCA, Tauro-CA, and Tauro-CDCA), and then excreted into the bile. In the terminal ileum and the colon, bile salt hydrolase is expressed in various bacteria that deconjugate glycine and taurine. The hydroxyl group at the C-7α position of the deconjugated BAs is then dehydroxylated to form secondary BAs, deoxycholic acid (DCA), and lithocholic acid (LCA) by multi-step reactions of specific bacteria.(16) In addition, hydroxyl groups at the C-3α, 7α, and 12α positions of both conjugated and unconjugated BAs can be dehydrogenated to carbonyl groups and further epimerized to 3β-, 7β-, and 12β-hydroxyl groups by intestinal bacteria. In mice, α-muricholic acid (MCA) and β-MCA are produced in the liver from CDCA as a primary BA, and these BAs were also conjugated by glycine and taurine and dehydroxylated to form ω-MCA as a secondary BA. A highly sensitive and specific method for the quantification of many conjugated/unconjugated primary and secondary BAs in serum and feces has been developed based upon a stable isotope dilution technique by liquid chromatography-tandem mass spectrometry (LC-MS/MS).(16,17) However, it is poorly understood whether and to what extent gut microbiota are involved in the regulation of BAs and energy homeostasis, especially in the pathology of NAFLD.
Epigallocatechin-3-gallate (EGCG), the most abundant polyphenolic catechin in green tea, has been widely investigated in terms of its health benefits, including anti-inflammatory, anti-cancer, and anti-steatotic effects on the liver.(18–23) Studies have demonstrated that treatment with EGCG suppressed fat deposition in the liver in high-fat diet (HFD)-fed mice, a murine model of human NAFLD, via the regulation of intracellular second messengers, signal transduction pathways, transcriptional activation, and autophagy pathway.(24–27) In addition to these activities, the possibility that the functionality of EGCG could be derived from its influence on gut microbiota and intestinal environment has recently attracted attention, since catechins including EGCG have extremely low absorbability from the intestinal tract. It has been reported that EGCG reduced the abundance of Clostridium spp. and tends to increase the abundance of Bacteroides in rats.(28) Recently, Sheng et al.(29) reported that EGCG increases the abundance of Akkermansia muciniphila and promotes the release of glucagon-like peptide (GLP)-1 from the intestinal tract via the activation of TGR-5 in mice fed a Western diet.
To further investigate the concept that the gut microbiota-BA interaction may affect the development of NAFLD, we studied the influence of modulation of gut microbiota by EGCG on the serum composition of BAs and hepatic steatosis. Using a mouse model of NAFLD, we investigated the effects of EGCG on the abundance of gut microbiota by 16S rRNA sequencing and also determined the composition of serum BAs and the association between these two aspects. Finally, we determined the specific bacterial genera that improve the serum BA dysregulation associated with anti-hepatic steatosis after treatment with EGCG.
Materials and Methods
Animals
Five-week-old male C57BL/6N mice were obtained from Shimizu Laboratory Supplies (Kyoto, Japan). Mice were kept at 18–24°C and 40–70% relative humidity, with a 12 h light/dark cycle. They were allowed free access to water and food (CE-2; CLEA Japan, Tokyo, Japan) during a 1 week acclimatization period. Experimental procedures were conducted in accordance with the National Institutes of Health guidelines for the use of experimental animals. All experimental protocols were approved by the Animal Care Committee of Kyoto Prefectural University of Medicine (Permission #M27-474).
After acclimatization, the mice were divided into three groups of eight mice each. They were fed one of the following diets: control (CE-2; CLEA Japan), an HFD (HFD32; CLEA Japan), or an HFD with EGCG for 8 weeks. EGCG (Sunphenon®, Taiyo Kagaku Co., Ltd., Mie, Japan) was supplemented in the food at a concentration of 0.32%. In our study, the dose of EGCG used (0.32%) corresponded to consumption of approximately 10 cups of green tea per day in humans,(30) based on an allometric scale.(31) The CE-2 diet provided 32% protein, 59% carbohydrate, and 12% fat with a caloric value of 339 kcal/100 g. The HFD32 contained 25% protein, 29% carbohydrates, and 32% fat (with saturated, monounsaturated, and polyunsaturated fatty acids at 7, 22, and 4 g/100 g chow, respectively). HFD32 has a caloric value of 507 kcal/100 g. After 8 weeks of feeding, fresh stool samples were obtained from each mouse. Thereafter, the mice were sacrificed under anesthesia. Their blood, liver, epididymal adipose tissue, and cecal contents were collected immediately.
Blood biochemical analysis
Blood supernatants were obtained as serum, after centrifugation at 1,500 × g for 10 min. Concentrations of triacylglyceride (TG), total cholesterol (T-Cho), non-esterified fatty acid (NEFA), aspartate amino-transferase (AST), alanine aminotransferase (ALT), high-density lipoprotein cholesterol (HDL-Cho) and low-density lipoprotein cholesterol (LDL-Cho) in serum were measured at SRL, Inc. (Tokyo, Japan).
Histological analysis
The liver tissue samples were fixed in 20% formalin and embedded in paraffin. Sections (4 µm thick) were cut and stained with hematoxylin-eosin (HE). Histological features and the fat area in liver tissue sections were determined using a BZ-X710 fluorescence microscope (Keyence, Osaka, Japan). In each HE-stained section, the severity of hepatic histological steatosis was assessed and the ratio of fatty lesions was calculated as fatty area/(total area-vascular area) × 100 (%) using computer-assisted image analysis with All-in-One analysis software (KEYENCE SOFTWARE Co., Osaka, Japan), which were performed blind to prevent observer bias.
Lipid analysis
TG concentration in the liver was measured enzymatically with a test kit (Triglyceride E Test Wako; Wako Pure Chemical Co. Ltd., Osaka, Japan). Lipids within the liver were extracted, using previously described methods.(32)
Collection of cecal contents and extraction of bacterial genomic DNA
Fresh cecal contents were collected, placed into tubes, and kept at −80°C until further use. Bacterial genomic DNA was extracted from the cecal content samples, as previously described.(33) Briefly, whole bacterial DNA was extracted from cecal content by using the QuickGene DNA Tissue kit SII (Kurabo, Osaka, Japan), with a nucleic acid extraction machine (QuickGene-Mini80; Kurabo).
Library preparation and DNA sequencing
Preparation of the library for DNA sequencing was conducted as previously described(34) using a MiSeq desktop sequencer (Illumina, San Diego, CA). Briefly, the V3–4 region of the 16S rRNA genes in each sample was amplified using the KAPA HiFi HotStart Ready Mix (Kapa Biosciences, Wilmington, MA), with primers 341F and 805R that contained a 5' overhang adapter sequence. The amplicon was purified by NucleoFast 96 PCR plates (Takara Bio Inc., Shiga, Japan). A second PCR was carried out using the KAPA HiFi HotStart Ready Mix to attach a unique combination of dual indices (I5 and I7 index) and Illumina sequencing adapters to each sample. The amplicon of the second PCR was purified, and the concentration was normalized with a SequalPrep Normalization Plate Kit (Life Technologies Japan, Tokyo, Japan). Each of the normalized amplicons was then evenly pooled and concentrated using AMPure XP beads (Beckman Coulter, Tokyo, Japan). From the library, 11 pM were combined with phiX Control (v3, Illumina; expected 20%) and sequenced using a 300 bp paired-end strategy on the MiSeq (Illumina), as per the manufacturer’s instructions.
Sequence data analysis
Sequence data analysis was performed using USEARCH v8.0(35) and QIIME v1.9.0(36) as previously described.(34) Statistical differences (p<0.05) in the relative abundance of bacterial phyla and genera between groups were evaluated using Student’s paired t test. The observed Chao1 and Shannon phylogenetic diversity indices were calculated using the R “phyloseq” package(37) and were statistically analyzed using a Kruskal-Wallis test followed by the Steel-Dwass post hoc test. The β-diversity was estimated using the UniFrac metric to calculate distances between the samples and visualized by principal coordinate analysis (PCoA); it was statistically examined using permutational multivariate analysis of variance (PERMANOVA). The final figures were generated using QIIME (ver. 1.9.0).
Determination of the serum BA profile
Serum BA profiles were detected using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) system, as previously described.(17,38) Briefly, 20 µl of mouse serum was diluted 100-fold with 2H-labelled internal standards and 0.5 M potassium phosphate buffer (pH 7.4). The mixture was injected into a Bond Elut C18 cartridge (200 mg; Agilent Technologies, Santa Clara, CA). Target molecules were eluted in water/ethanol (1:9, vol/vol). The eluate was evaporated under nitrogen until dry and then dissolved in 20 mM ammonium acetate buffer (pH 7.5)/methanol (1:1, vol/vol). An aliquot of the resulting sample solution was injected into the LC-MS/MS system for analysis. Chromatographic separation was performed using a Hypersil GOLD column (150 × 2.1 mm, 3 µm; Thermo Fisher Scientific, Waltham, MA). A mixture of 20 mM ammonium acetate buffer (pH 7.5), acetonitrile, and methanol (70:15:15, vol/vol/vol) was used for the initial mobile phase and was gradually changed to 30:35:35 (vol/vol/vol) over 30 min.
Statistical analysis
All data are presented as mean ± SEM. Comparisons among the three groups were performed using one-way analysis of variance (ANOVA) with Tukey’s post hoc tests. Body weight gain curves were analyzed with an ANOVA, followed by Tukey’s multiple comparison tests. These statistical analyses were performed using statistical software (Graph Pad Prism, ver. 6.07; Graph Pad Software, San Diego, CA). The Pearson’s rank correlation coefficient was used to analyze the correlation between gut microbiota and BA. The correlation and outcome analysis was performed using the JMP statistical software package ver. 14. (SAS Institute, Inc., Cary, NC). All results were considered statistically significant at p<0.05.
Results
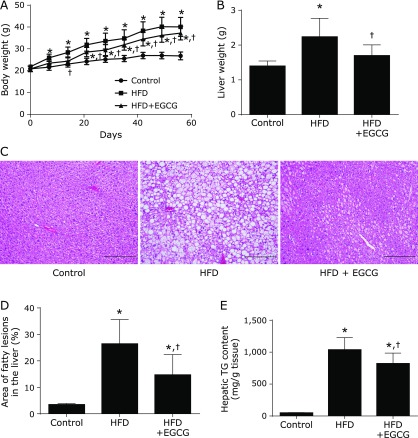
EGCG inhibited body weight increase and lipid accumulation in HFD-fed mice
Six-week-old mice were fed for 8 weeks with either the control diet, HFD, or HFD supplemented with 0.32% EGCG. Body weight and food consumption were monitored weekly throughout the experiment. As shown in Fig. 1A, body weight was significantly higher in the HFD group than in the control group after 1 week of treatment (i.e., at 7 weeks of age). This difference was maintained through the end of the study. EGCG treatment significantly suppressed the body weight gain of mice during weeks 3–8 (Fig. 1A). During the experiment, the average food intake of mice in the HFD group was significantly lower than the control group. However, the food consumption of mice between the HFD group and the HFD + EGCG group was not different during the experiment (Supplemental Table 1*). The HFD-induced body weight gain was suppressed in the HFD + EGCG group. The final liver weight at the end of the experiment was significantly increased in the HFD group compared to the control group, and this increase was significantly inhibited by treatment with EGCG (Fig. 1B). Although the histological liver sections of the control group were free of lipid droplets, increased accumulation of lipid droplets was observed in the HFD group, leading to a condition of hepatic steatosis, and this increase was suppressed in the HFD + EGCG group (Fig. 1C). Consistent with the histological findings, the area of fatty lesions in the liver was significantly increased in the HFD group compared to the control, and this increase was significantly inhibited by treatment with EGCG (Fig. 1D). In addition, the content of TG in the liver was significantly increased in the HFD group, and this increase was also inhibited by treatment with EGCG (Fig. 1E).
Fig. 1.
Inhibitory effect of EGCG on the HFD-induced obese phenotype and hepatic triacylglycerol accumulation. (A) Time-course of mouse body weight. Body weight of male mice receiving an EGCG-containing diet for 8 weeks. (B) Liver weight. (C) Hematoxylin-eosin (HE)-stained liver sections. (D) Area of fatty lesions in the liver. (E) Hepatic triacylglycerol (TG) content. C57BL/6N mice were fed a CE-2 diet (control), a high-fat diet (HFD), or an HFD supplemented with 0.32% EGCG (HFD + EGCG) for 8 weeks. Values are expressed as the means and SEM of eight mice in each group. Significant differences compared with the control group are denoted * (p<0.05), and those with the HFD group are denoted † (p<0.05). Photographs are of HE staining of liver sections from representative mice of each group (scale bar = 500 µm).
The biochemical parameters of the serum obtained from mice at the end of the experiment are shown in Table 1. Serum levels of ALT and AST in the HFD group tended to increase compared to the control, and these increased levels tended to decrease in the HFD + EGCG group. However, these changes were not significant. Compared with the control, mice fed with the HFD exhibited significantly higher plasma NEFA, T-Cho, HDL-Cho, and LDL-Cho levels. EGCG supplementation had no effect on the HFD-mediated up-regulation of these parameters.
Table 1.
Effect of HFD and EGCG on serum metabolic parameters at 8 weeks of treatment
| Control | HFD | HFD + EGCG | |
|---|---|---|---|
| TG (mg/dl) | 68 ± 6 | 35 ± 4* | 25 ± 2* |
| NEFA (mEQ/L) | 1,198 ± 119 | 1,514 ± 80* | 1,250 ± 52 |
| T-Cho (mg/dl) | 82 ± 5 | 216 ± 8* | 197 ± 9* |
| AST (IU/L) | 67 ± 10 | 105 ± 21 | 72 ± 5 |
| ALT (IU/L) | 27 ± 3 | 48 ± 20 | 33 ± 5 |
| HDL-Cho (mg/dl) | 54 ± 3 | 80 ± 3* | 70 ± 2* |
| LDL-Cho (mg/dl) | 8 ± 1 | 29 ± 3* | 30 ± 3* |
C57BL/6N mice were fed with the control CE-2 diet (control), a High-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for 8 weeks. Values are expressed as the means ± SEM of eight mice in each group; *p<0.05 compared with the control group. TG, triacylglycerol; NEFA, non-esterified fatty acid; mEQ, milliequivalent; T-Cho, total cholesterol; AST, aspartate amino-transferase; ALT, alanine amino-transferase; HDL-Cho, high-density lipoprotein cholesterol; LDL-Cho, low-density lipoprotein cholesterol.
Effects of EGCG on HFD-induced gut microbiota dysbiosis
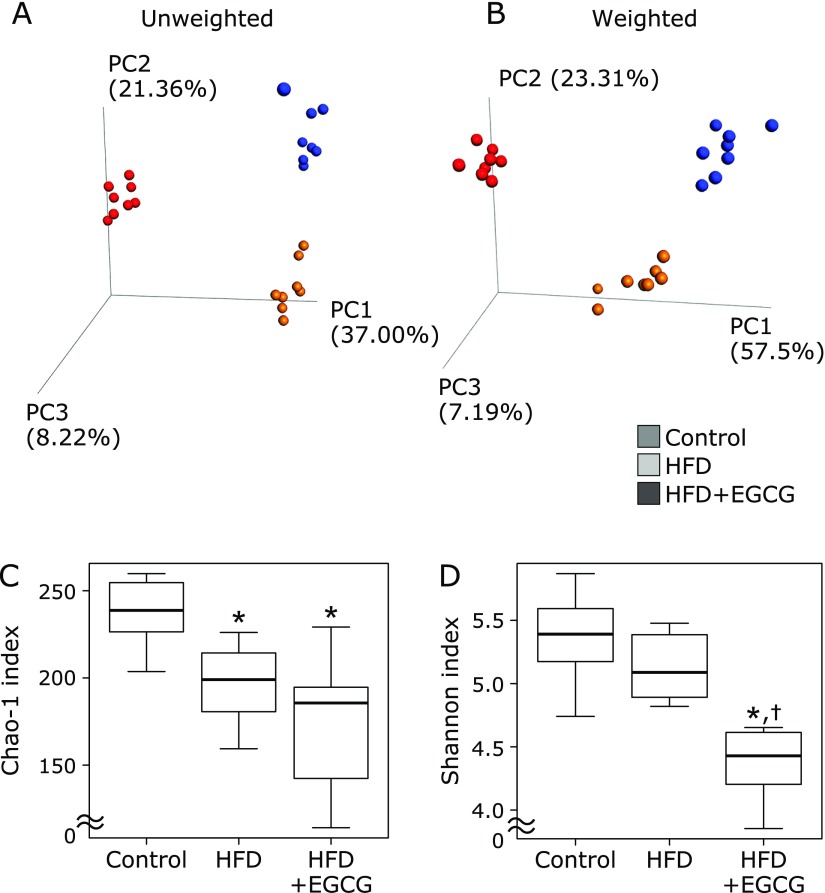
In total, 412 OTUs were detected from Illumina high-quality sequence reads. Initially, the overall structure of the cecal microbiota among the control, HFD, and HFD + EGCG groups using β-diversity indices were calculated for unweighted and weighted UniFrac distances. PCoA revealed that there were significant microbial structural differences among the control, HFD and HFD + EGCG groups in unweighted (PERMANOVA, p = 0.0001) and weighted (PERMANOVA, p = 0.0001) UniFrac distances (Fig. 2A and B). Subsequently, we compared α-diversity among the control, HFD and HFD + EGCG groups using different indices the observed species and Chao 1 index (OTU richness estimation), and the Shannon index (OTU evenness estimation). The Chao 1 index was decreased in the HFD group compared to the control, and the decrease was not affected by EGCG (Fig. 2C). The Shannon index was significantly decreased in the HFD + EGCG group compared to the control, as well as to the HFD group (Fig. 2D).
Fig. 2.
Effects of EGCG administration on gut microbiota in α and β diversity indices. (A) Principal coordinate analysis (PCoA) plot of unweighted UniFrac data. (B) PCoA plot of weighted UniFrac data. (C) The Chao 1 index (OTU richness estimation). (D) The Shannon index (OTU evenness estimation). C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for eight weeks. The composition of cecal content was analyzed using Illumina-based 16S rRNA sequencing. Values are expressed as the means and SEM of six mice in each group. Significant differences compared with the control group are denoted * (p<0.05), and those with the HFD group are denoted † (p<0.05).
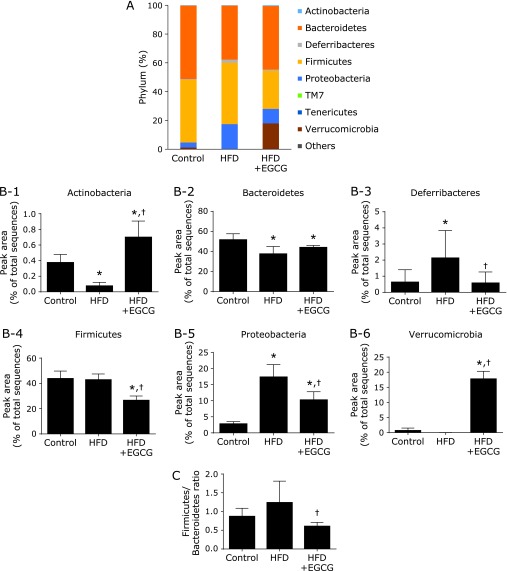
Compared to the control, HFD-fed mice had a lower abundance of phyla Actinobacteria and Bacteroidetes and had a higher abundance of phyla Deferribacteres and Proteobacteria (Fig. 3 and Supplemental Table 2*). Treating mice with EGCG induced the changes in microbiota composition in HFD-fed mice, showing a significantly higher abundance of the phyla Verrucomicrobia and Actinobacteria, and significantly lower abundance of the phyla Deferribacteres, Proteobacteria, and Firmicutes (Fig. 3B-1–6 and Supplemental Table 2*). The Firmicutes/Bacteroidetes ratio, an index of dysbiosis, was significantly decreased in the HFD + EGCG group compared to the HFD group (Fig. 3C).
Fig. 3.
Effects of EGCG administration on gut microbiota at the phylum level. (A) The microbial composition at the phylum level. (B) At the phylum level of microbiota (calculated as the percentage of total microbiota). (C) Firmicutes/Bacteroidetes ratio. C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for eight weeks. The composition of cecal content was analyzed using Illumina-based 16S rRNA sequencing. Values are expressed as the means and SEM of six mice in each group. Significant differences compared with the control group are denoted * (p<0.05), and those with the HFD group are denoted † (p<0.05).
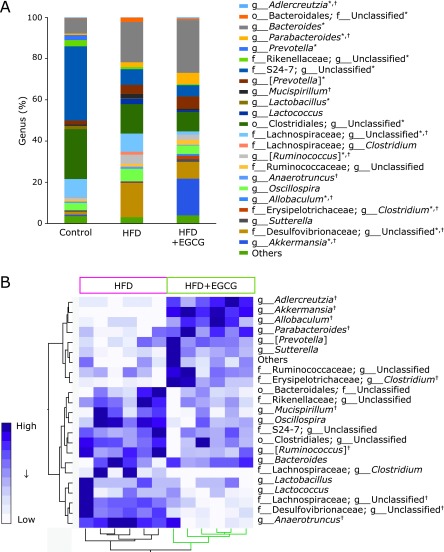
Changes in gut microbiota induced by a HFD were also observed at the genus level (Fig. 4 and Table 2). Compared to the control, HFD-fed mice had a significantly lower abundance of 6 genera and had a significantly higher abundance of 8 genera shown in Fig. 4A and Table 2. Treating mice with EGCG induced the changes in microbiota composition in HFD-fed mice, showing significantly higher abundance of the genera Adlercreutzia, Akkermansia, Allobaculum, Parabacteroides, f_Erysipelotrichaceae; g_Clostridium which clustered together, and a significantly lower abundance of the genera Mucispirillum, [Ruminococcus], f_Lachnospiraceae; g_Unclassified, f_Desulfovibrionaceae; g_Unclassified, and Anaerotruncus which clustered together (Fig. 4A, B and Table 2).
Fig. 4.
Effects of EGCG administration on gut microbiota at the genus level. (A) The microbial composition at the genus level. (B) Heat map of 16S rRNA gene sequencing analysis of cecal content at the genus level. The scale reflects the data as follows: violet indicates high values whereas white indicates low values for the percentage of reads that were classified at that rank. C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for eight weeks. The composition of cecal content was analyzed using Illumina-based 16S rRNA sequencing. Significant differences compared with the control group are denoted * (p<0.05), and those with the HFD group are denoted † (p<0.05).
Table 2.
Average abundance of genus-level OTUs in the C57BL/6N mice treated with control, HFD, or HFD + EGCG
| (%) | |||||||
|---|---|---|---|---|---|---|---|
| Taxon |
Control | HFD | HFD + EGCG | ||||
| Phylum | Class | Order | Family | Genus | |||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Adlercreutzia | 0.36 ± 0.11 | 0.08 ± 0.05* | 0.70 ± 0.21*,† |
| Bacteroidetes | Bacteroidia | Bacteroidales | Unclassified | Unclassified | 0.11 ± 0.12 | 2.06 ± 1.70* | 0.69 ± 0.33* |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 7.41 ± 2.54 | 19.68 ± 10.43* | 25.44 ± 3.30* |
| Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Parabacteroides | 0.60 ± 0.14 | 2.23 ± 2.46 | 5.58 ± 2.60*,† |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 2.49 ± 0.87 | 0.00 ± 0.00* | 0.00 ± 0.00* |
| Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Unclassified | 3.01 ± 0.45 | 1.08 ± 0.23* | 0.55 ± 0.28* |
| Bacteroidetes | Bacteroidia | Bacteroidales | S24-7 | Unclassified | 35.96 ± 3.56 | 7.61 ± 1.95* | 5.65 ± 2.03* |
| Bacteroidetes | Bacteroidia | Bacteroidales | [Paraprevotellaceae] | [Prevotella] | 2.08 ± 1.46 | 4.34 ± 2.73 | 5.80 ± 2.06* |
| Deferribacteres | Deferribacteres | Deferribacterales | Deferribacteraceae | Mucispirillum | 0.63 ± 0.78 | 2.13 ± 1.71* | 0.58 ± 0.69† |
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | 1.54 ± 1.24 | 0.16 ± 0.16* | 0.08 ± 0.07* |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Lactococcus | 0.00 ± 0.00 | 2.66 ± 2.85* | 0.84 ± 0.56 |
| Firmicutes | Clostridia | Clostridiales | Unclassified | Unclassified | 24.20 ± 0.89 | 14.28 ± 0.42* | 9.49 ± 1.03* |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Unclassified | 8.98 ± 2.30 | 8.91 ± 3.02 | 1.55 ± 1.04*,† |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Clostridium | 0.23 ± 0.18 | 1.35 ± 1.86 | 0.01 ± 0.04 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | [Ruminococcus] | 0.88 ± 0.42 | 4.51 ± 0.97* | 2.35 ± 1.20*,† |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Unclassified | 0.93 ± 0.19 | 1.25 ± 0.65 | 2.50 ± 0.75 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Anaerotruncus | 0.55 ± 0.23 | 1.14 ± 0.29* | 0.31 ± 0.38† |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | 3.68 ± 1.58 | 6.06 ± 1.42* | 4.05 ± 1.92 |
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Allobaculum | 0.00 ± 0.00 | 0.18 ± 0.21 | 0.91 ± 0.56*,† |
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Clostridium | 0.00 ± 0.00 | 0.26 ± 0.16 | 1.58 ± 1.46*,† |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.70 ± 0.33 | 0.40 ± 0.21 | 1.54 ± 1.63 |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Unclassified | 1.38 ± 0.51 | 16.48 ± 3.66* | 8.11 ± 1.03*,† |
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Verrucomicrobiaceae | Akkermansia | 0.66 ± 0.85 | 0.00 ± 0.04 | 17.76 ± 2.54*,† |
| Others (<0.01) | 3.65 ± 0.17 | 3.16 ± 0.10 | 3.95 ± 0.20 | ||||
C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for 8 weeks. Values are expressed as the means ± SEM (%) of eight mice in each group; *p<0.05 compared with the control group. †p<0.05 compared with the HFD group.
Effects of EGCG on serum levels of unconjugated/conjugated BAs
To investigate the effects of changes in gut microbiota on BA metabolism, we conducted a panel of BAs in the serum. A total of 17 unconjugated and 13 conjugated BAs, shown in Table 3, were quantified. Data from the serum BA profiles showed that HFD significantly decreased the level of total free BAs, and the reduction was significantly reversed by EGCG supplementation. Among free BAs, CA, ω-MCA, ursodeoxycholic acid (UDCA), ursocholic acid (UCA), hyodeoxycholic acid (HDCA), and 7-oxodeoxycholic acid (7-oxo DCA) were significantly decreased in the HFD group compared to the control group. The HFD-induced decrease in CA, a predominant primary BA in mice, was significantly reversed by treatment with EGCG. The level of free deoxycholic acid (DCA), a predominant secondary BA in mice, was significantly increased in the HFD group, but this increase was not affected by the EGCG treatment (Table 3).
Table 3.
Relative bile acid profiles of the serum
| (%) | ||||
|---|---|---|---|---|
| Control | HFD | HFD + EGCG | ||
| Free- | Cholic acid | 19.66 ± 3.32 | 9.83 ± 2.46* | 22.97 ± 5.45† |
| ω-Muricholic acid | 11.90 ± 4.29 | 3.08 ± 0.87* | 13.44 ± 3.58† | |
| α-Muricholic acid | 2.09 ± 0.75 | 0.91 ± 0.72 | 1.47 ± 1.03 | |
| β-Muricholic acid | 8.70 ± 2.73 | 10.19 ± 5.25 | 9.47 ± 5.76 | |
| Chenodeoxycholic acid | 2.49 ± 1.20 | 2.51 ± 0.71 | 4.35 ± 1.52*,† | |
| Deoxycholic acid | 24.01 ± 5.82 | 38.83 ± 6.07* | 33.16 ± 9.47 | |
| Lithocholic acid | 5.46 ± 2.44 | 2.92 ± 1.47 | 3.95 ± 1.40 | |
| Ursodeoxycholic acid | 3.17 ± 1.81 | 1.12 ± 0.64* | 0.85 ± 0.50* | |
| Ursocholic acid | 0.35 ± 0.17 | 0.08 ± 0.10* | 0.09 ± 0.11* | |
| Hyocholic acid | 0.03 ± 0.04 | 0.01 ± 0.02 | 0.09 ± 0.19 | |
| Murideoxycholic acid | 0.14 ± 0.11 | 0.33 ± 0.14 | 0.42 ± 0.20* | |
| Hyodeoxycholic acid | 2.45 ± 0.60 | 1.18 ± 0.75* | 1.14 ± 0.33* | |
| Isodeoxycholic acid | 1.65 ± 0.71 | 1.12 ± 0.88 | 0.84 ± 0.23 | |
| 3-Dehydrocholic acid | 0.13 ± 0.11 | 0.03 ± 0.03 | 0.21 ± 0.25 | |
| 7-Oxodeoxycholic acid | 4.60 ± 1.85 | 0.87 ± 0.57* | 1.00 ± 0.46* | |
| 3-Dehydrodeoxycholic acid | 0.03 ± 0.03 | 0.08 ± 0.06 | 0.16 ± 0.15 | |
| 12-Oxolithocholic acid | 1.52 ± 0.88 | 0.80 ± 0.50 | 1.01 ± 0.34 | |
| Free-bile acid | 88.38 ± 7.12 | 73.90 ± 3.10* | 94.62 ± 2.10† | |
| Glyco- | Cholic acid | 0.09 ± 0.04 | 0.30 ± 0.65 | 0.04 ± 0.04 |
| Chenodeoxycholic acid | 0.08 ± 0.06 | 0.02 ± 0.02 | 0.05 ± 0.07 | |
| Deoxycholic acid | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.15 ± 0.20 | |
| Lithocholic acid | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.01 | |
| Ursodeoxycholic acid | 0.02 ± 0.04 | 0.00 ± 0.00 | 0.01 ± 0.02 | |
| Glyco-bile acid | 0.22 ± 0.09 | 0.32 ± 0.64 | 0.27 ± 0.26 | |
| Tauro- | Cholic acid | 6.33 ± 4.05 | 16.80 ± 6.42* | 1.97 ± 1.98† |
| ω-Muricholic acid | 2.09 ± 2.04 | 0.59 ± 0.62 | 0.64 ± 0.43 | |
| α-Muricholic acid | 0.59 ± 0.72 | 0.55 ± 0.52 | 0.34 ± 0.62 | |
| β-Muricholic acid | 2.09 ± 1.47 | 4.84 ± 3.23 | 0.90 ± 1.24† | |
| Chenodeoxycholic acid | 0.05 ± 0.04 | 0.13 ± 0.05 | 0.20 ± 0.20 | |
| Deoxycholic acid | 0.16 ± 0.20 | 2.73 ± 1.88* | 0.87 ± 0.83† | |
| Lithocholic acid | 0.03 ± 0.03 | 0.03 ± 0.04 | 0.04 ± 0.06 | |
| Ursodeoxycholic acid | 0.08 ± 0.08 | 0.11 ± 0.14 | 0.13 ± 0.17 | |
| Tauro-bile acid | 11.41 ± 7.05 | 25.78 ± 3.28* | 5.11 ± 2.16† | |
C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for 8 weeks. Values are expressed as the means ± SEM (%) of six mice in each group; *p<0.05 compared with the Control group. †p<0.05 compared with the HFD group.
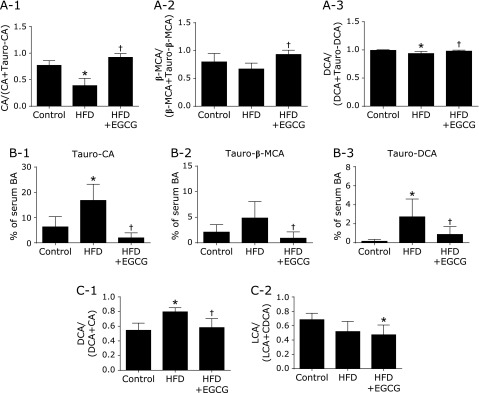
In all 3 groups, glycine-conjugated BAs were almost below the detection limit in the mouse serum, but taurine-conjugated forms of primary and secondary BAs were detected. Contrary to the changes in free BAs, HFD increased the total level of taurine-conjugated BAs, and this increase was significantly inhibited by treatment with EGCG. Among taurine-conjugated BAs, Tauro-CA and Tauro-DCA were significantly increased in the HFD group compared to the control, and these increases were significantly inhibited by EGCG (Fig. 5B-1). The level of Tauro-β-MCA was also significantly decreased in the HFD + EGCG group compared to the HFD group (Fig. 5B-2).
Fig. 5.
Determination of BA profile and calculation of BA transformation activities by gut microbiota. (A) Deconjugation. (B) Taurine-conjugated BAs. (C) 7α-Dehydroxylation. C57BL/6N mice were fed with the control CE-2 diet (control), a high-fat diet (HFD), or the HFD supplemented with 0.32% EGCG (HFD + EGCG) for eight weeks. Values are expressed as the means and SEM of six mice in each group. Significant differences compared with the control group are denoted * (p<0.05), and those with the HFD group are denoted † (p<0.05). CA, free cholic acid; Tauro-CA, taurine-conjugated cholic acid; β-MCA, free β-muricholic acid; Tauro-β-MCA, taurine-conjugated β-muricholic acid; DCA, free deoxycholic acid; Tauro-DCA, taurine-conjugated deoxycholic acid; BA, bile acid; LCA, free lithocholic acid; CDCA, free chenodeoxycholic acid.
To estimate the activities of BA transformation by gut microbiota and the effects of EGCG on transformation of BAs by deconjugation or 7α-dehydroxylation, we calculated the product/(product + substrate) ratio for each reaction. Deconjugation was calculated, and the CA/(CA + Tauro-CA) ratio and DCA/(DCA + Tauro-DCA) ratios were significantly decreased in the HFD group (Fig. 5A-1). These HFD-induced decreases in the deconjugation ratio were significantly reversed by treatment with EGCG. The β-MCA/(β-MCA + Tauro-β-MCA) ratio (β-MCA being a mouse-specific primary BA) was significantly increased in the HFD + EGCG group only (Fig. 5A-2). 7α-Dehydroxylation by gut microbiota was calculated by the DCA/(DCA + CA) ratio or LCA/(LCA + CDCA) ratio. The ratio of DCA/(DCA + CA) was significantly increased in the HFD group compared to the control, and this increase was significantly inhibited by treatment with EGCG (Fig. 5C-1). There was no significant difference in the LCA/(LCA + CDCA) ratio between the HFD and the HFD + EGCG group (Fig. 5C-2).
Correlation between gut microbiota and BA composition
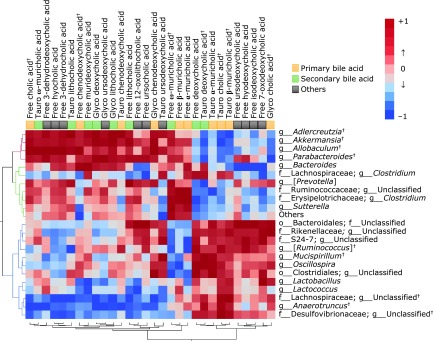
To clarify the relationship between gut microbiota (22 genera and others) and BAs (17 unconjugated and 13 conjugated BAs), we used Pearson’s correlation test (Fig. 6 and Supplemental Table 3*). The heat map data revealed a positive correlation between serum predominant primary CA and CDCA and Adlercreutzia, Akkermansia, or Allobaculum which clustered closely. A similar positive correlation was also seen between mouse-specific primary BAs (α-MCA and β-MCA) and f_Erysipelotrichaceae; g_Clostridium, or f_Ruminococcaceae; g_Unclassified, which clustered together. In addition, Desulfovibrionaceae; g_Unclassified, Anaerotruncus, and Lachnospiraceae; g_Unclassified, which clustered together, were correlated negatively and positively to the composition of CA and CDCA and to those of taurine-conjugated BAs (Tauro-CA, Tauro-α-MCA Tauro-β-MCA, or Tauro-DCA), respectively.
Fig. 6.
A heat map generated by Spearman’s correlation analysis reveals the relationship between the abundance of gut microbiota at the genus level and BA profile. Correlation heat map demonstrating the association between the indicated gut microbiota taxonomic genera and BAs. Red denotes a positive association, blue a negative association, and white no association. C57BL/6N mice were fed with a high-fat diet (HFD), or a HFD supplemented with 0.32% EGCG (HFD + EGCG) for eight weeks.
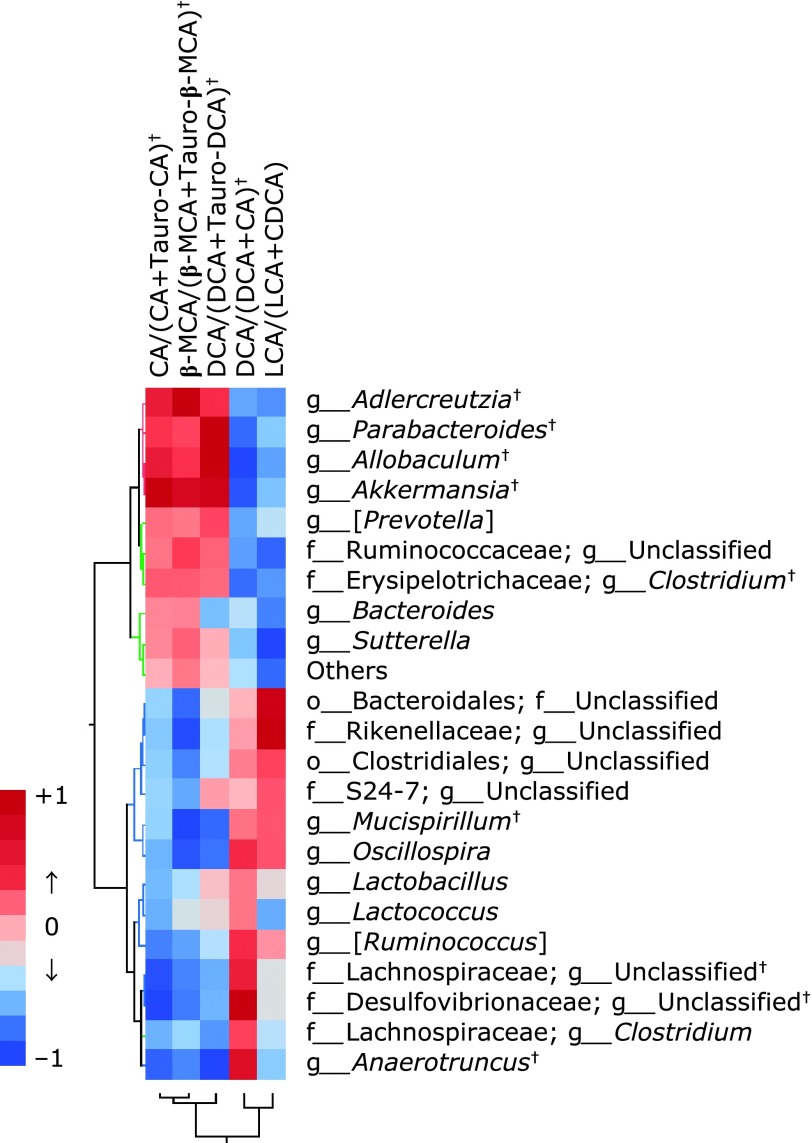
Finally, we investigate the correlation between BA transformation index and the abundance of gut microbiota. As shown in Fig. 7 and Supplemental Table 4*, significant positive correlations were observed between serum CA/(CA + Tauro-CA) and Akkermansia, Allobaculum, Adlercreutzia, or Parabacteroides, and between DCA/(DCA + CA) and f_Desulfovibrionaceae; g_Unclassified, Anaerotruncus, f_Lachnospiraceae; g_Unclassified, or [Ruminococcus].
Fig. 7.
Correlation heat map demonstrating the association between the indicated gut microbiota taxonomic genera and BA composition. Red denotes a positive association, blue a negative association, and white no association. CA, free cholic acid; Tauro-CA, taurine-conjugated cholic acid; β-MCA, free β-muricholic acid; Tauro-β-MCA, taurine-conjugated β-muricholic acid; DCA, free deoxycholic acid; Tauro-DCA, taurine-conjugated deoxycholic acid; LCA, free lithocholic acid; CDCA, free chenodeoxycholic acid.
Discussion
In the present study, we investigated the effect of oral administration of EGCG on the gut microbiota and serum BA profile in mice fed a HFD by 16S rRNA sequencing analysis and quantitative systematic LC-MS/MS, respectively. We found EGCG significantly inhibited the increase in histological fatty deposit and TG accumulation in the liver induced by an HFD and also improved gut dysbiosis. In addition, EGCG significantly reversed the decrease in the serum level of primary BAs and the increases in those of secondary BAs and taurine-conjugated BAs in mice fed an HFD. Finally, we determined crucial genera of gut microbiota correlated with BA fluctuation in serum. These results demonstrate a novel anti-fatty liver action of EGCG, which is hardly absorbed from the intestinal tract, by modulation of BA metabolism to improve gut microbiota.
A number of findings indicate that dysbiosis of gut microbiota occurs in NAFLD in humans, and novel treatments targeting microbiota and their metabolites have recently been developed to address this issue.(3,39) The mouse fatty liver model through HFD used in this study has been used previously(40) and the abnormality of gut microbiota determined by 16sRNA metagenomic analysis has already been reported.(5) In the present study, PCoA showing β-diversity clearly distinguished control, HFD and HFD + EGCG groups. At the phylum level, the relative abundance of Actinobacteria was significantly decreased and those of Deferribacteres and Proteobacteria were significantly increased in the HFD group as compared to the control group; EGCG significantly reversed these changes. The Firmicutes/Bacteroidetes ratio, an index of dysbiosis, tended to increase in the HFD group as compared to the control, and this increase was significantly inhibited in the HFD + EGCG group. These results suggest that EGCG improved dysbiosis of gut microbiota induced by HFD and, as a result, this improvement may be involved in the anti-fatty liver action of EGCG.
One key finding in this study is that the abundance of Proteobacteria phylum increased markedly in the HFD group compared with the control group, and this increase was significantly suppressed in the HFD + EGCG group. Recently, Bilophila wadsworthia belonging to p_Proteobacteria; f_Desulfovibrionaceae; g_Bilophila was identified as a bacterium exacerbating metabolic disorder caused by HFD, and it has been demonstrated that Bilophila wadsworthia bacteria aggravates the metabolic dysfunction in HFD-fed mice by enhancing intestinal mucosal permeability, promoting inflammatory immune response, and altering BA metabolism.(7) Although g_Bilophila was hardly detected in this study, a related genus, p_Proteobacteria; f_Desulfovibrionaceae; g_Unclassified, increased more than 10-fold in the HFD group as compared with the control, and EGCG significantly inhibited this increase. These results suggest that f_Desulfovibrionaceae may play a crucial role in the development of HFD-induced fatty liver in this rodent model.
p_Actinobacteria contain important beneficial bacteria which comprise about 10% of the Japanese gut microbiota,(41) and administration of Bifidobacterium, which is a major genus in this phylum, could ameliorate HFD-induced fatty liver in mice.(42,43) In the present study, the abundance of p_Actinobacteria was significantly decreased in HFD-fed mice compared to control, and EGCG reversed this decrease and the levels increased more in the EGCG-treated group as compared to the control. However, in this experimental study, increased bacteria at Actinobacteria phylum by EGCG was f_Coriobacteriaceae; g_Adlercreutzia, but not g_Bifidobacterium. Some Adlercreutzia genera could metabolize and decompose EGCG into smaller molecular weight catechins (epicatechin, catechin, etc.), like Adlercreutzia equolifaciens.(44,45) In the future, when considering the function of EGCG, it may be necessary to analyze microbiota that could metabolize or decompose EGCG into smaller molecule catechin metabolites.
In the present study, the most striking feature was the significant increase of the Akkermansia genus in the Verrucomicobia phylum in the HFD + EGCG group, consistent with recent reports.(29,46) Recently, Sheng et al.(29) reported that Akkermansia genus is significantly increased by western diet with EGCG administration. Akkermansia muciniphila is the main genus classified in the Verrucomicrobia phylum and recent studies revealed the involvement of this bacteria in obesity, sugar metabolism, and intestinal immunity.(47) In previous studies, cranberry extracts,(48) concord grape polyphenols,(49) and apple procyanidin(50) have been interestingly reported to show an anti-obesity effect through an increased abundance of the Akkermansia genus.(51) In addition, these effects have also been reproduced by the administration of Akkermansia muciniphila to mice.(52) Such polyphenols, including EGCG, generally have low absorption in the intestinal tract, and they reach the large intestine without being digested and absorbed, but the details of the mechanism by which the abundance of Akkermansia increases have not been elucidated. It is thought that Akkermansia muciniphila produces short-chain fatty acids such as acetic acid by feeding intestinal mucin and supplies energy to goblet cells that produce mucin. This study suggested that the marked increase in Akkermansia by EGCG treatment may be involved in hepatoprotection via various mechanisms. Akkermansia genus in feces may be one of the good surrogate markers to consider the functionality of EGCG. A human clinical trial is necessary to confirm the functionality of EGCG with respect to the gut microbiome.
In mice, CA, CDCA, α-MCA and β-MCA are primary BAs. In this study, free forms of these primary BAs were detected in the serum of the control mice, and the most predominant one was CA (about 60% of primary BAs). In the present study, the proportion of CA in serum markedly decreased in the HFD group and this decrease was significantly recovered to the equivalent levels of the control by the treatment with EGCG. The proportion of CDCA also increased about twice in the EGCG group as compared with the HFD group. CA and CDCA are ligands that bind most strongly to BA receptor FXR (CA<CDCA), and it is suspected that FXR activity was promoted by treatment with EGCG. Recently, Sheng et al.(29) has reported that EGCG increased the concentration of FXR agonists, especially CDCA and CA, in serum and their regulated signaling-associated gene expression in the liver of mice, which supports our hypothesis. When FXR, a BA receptor present in the liver, is activated, SREBP1c is suppressed, TG synthesis is downregulated, and fatty liver is finally inhibited. These data suggest that EGCG increases serum levels of CA and CDCA through improving the intestinal environment, leading to the improvement of liver lipid metabolism and the inhibition of fatty liver. Furthermore, Tauro-CA increased markedly in the HFD group as compared with the control group, in contrast to the CA value, which markedly decreased in the HFD + EGCG group. In addition to Tauro-CA, the levels of other taurine-conjugated BAs (β-MCA, CDCA, and DCA) significantly increased in the HFD group as compared with the control, and these increases were markedly inhibited in the EGCG group. As Tauro-β-MCA is a natural antagonist of FXR,(53) the reduction of Tauro-β-MCA also contributes to the activation of FXR in EGCG treated mice. Thus, the reduction of serum taurine-conjugated BAs by EGCG appears to be another important mechanism explaining the anti-fatty liver action of EGCG.
From the correlation between microbiota abundance obtained by 16S rRNA metagenomic analysis and serum BA profiles obtained by quantitative LC-MS/MS, we further examined the functionality of EGCG from the standpoint of the interaction of these markers. Judging from the distribution of correlation coefficients by heat map analysis, the abundance of Akkermansia and Parabacteroides genus showed a positive correlation with CA and a negative correlation with Tauro-CA, significantly increased in the EGCG group as compared with the HFD group and was 1.0% or more. As we suspected that Tauro-CA was recovered in the EGCG group due to the deconjugation reaction of Tauro-CA by intestinal bacteria with the bile salt hydrolase (bsh) gene, we searched for genera positively correlated with the ratio of CA/CA + Tauro-CA, β-MCA/β-MCA + Tauro-β-MCA, and DCA/DCA + Tauro-DCA, as indices of deconjugation of BAs. As a result, the abundance of Akkermansia and Parabacteroides genera showed significant and strong correlation with the deconjugation indices. The above results indicate that the increased abundance of Akkermansia and/or Parabacteroides may be involved in promoting the taurine deconjugation reaction from taurine-conjugated BAs to free type of BAs. However, the deconjugation activity(54) of Akkermansia is not confirmed, and further investigation is required to confirm the same.
Since the production of secondary BA depends on the 7α-hydroxylation gene of gut microbiota, heat map analysis was also performed on the abundance of microbiota correlated with the ratio of DCA/DCA + CA and LCA/LCA + CDCA, as indices of 7α-hydroxylation of BAs. The present study showed that the ratio of DCA/DCA + CA was significantly increased in the HFD group compared with the control, and this increase was significantly inhibited by EGCG. Five genera were chosen as bacteria significantly correlated with the 7α-hydroxylation ratio; f_Lachnospiraceae; g_Unclassified, g_[Ruminococcus], and g_Oscillospira with moderate positive correlation and g_Anaerotruncus and f_Desulfovibrionaceae; g_Unclassified with strong positive correlation. Among the five genera, f_Desulfovibrionaceae; g_Unclassified is high abundance in the HFD group, suggesting the possibility that this bacterium was key in the production of DCA, but it is unknown how much it contributes to secondary BA production.
The present study has several limitations. This study was based on serum BA and gut microbiota analysis, without information on BA concentrations in the liver and intestinal tract, changes in the intestinal BA transport system, or the expression of BA synthesis gene in the liver. More information will be required to improve our understanding of the gut-microbiome-liver axis. Furthermore, since we used homogeneous C57BL/6N mice fed an HFD as a model of NAFLD, there are issues in the application of these findings to human beings. As we focused on the role of taurine-conjugated BAs associated with the development of fatty liver, glycine-conjugated BAs, a predominant form in humans, should be analyzed in future studies on patients with NAFLD.
Through quantitative systematic LC-MS/MS we found that serum taurine-conjugated BAs (Tauro-CA, Tauro-β-MCA, and Tauro-DCA) increased in HFD-fed mice and that the correlation analysis between BA profiles and gut microbiota demonstrated the contribution of Akkermansia and f_Desulfovibrionaceae; g_Unclassified in the modulation of BA metabolism induced by EGCG treatment. The present study suggests that EGCG could alter BA metabolism and suppress fatty liver by improving the intestinal luminal environment.
Author Contributions
YN and YI directed the paper’s conception. CU and YN wrote the paper. TT, KU, and YH provided intellectual support and modified the language. CU, KM, and YH performed the experiments. ZY and TO prepared EGCG. RI analyzed the sequence data. AH and YM analyzed the BA data. All authors read and approved the final manuscript for publication.
Data Availability Statement
The datasets generated for this study can be found in the DDBJ Sequence Read Archive Database (DDBJ accession number; PRJDB7523).
Funding
This work was supported by Grants-in-Aid for Scientific Research (B) (JSPS KAKENHI) to YN (Grant Number JP 16H05289) and by Grant of Industry-Academia-Government Collaboration of “Field for Knowledge Integration and Innovation” (FKII) to YN (No. 16824414) from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Abbreviations
- BA
bile acid
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- EGCG
epigallocatechin-3-gallate
- FXR
farnesoid X receptor
- HFD
high fat diet
- LCA
lithocholic acid
- MCA
muricholic acid
- NAFLD
non-alcoholic fatty liver disease
- PCoA
principal coordinate analysis
- Tauro-β-MCA
taurine-conjugated β-muricholic acid
- Tauro-CA
taurine-conjugated cholic acid
- Tauro-DCA
taurine-conjugated deoxycholic acid
- TG
triacylglyceride
Conflict of Interest
YN received scholarship funding from EA Pharma Co., Ltd. and collaboration research funding from Fujifilm Medical Co., Ltd. and has been paid lecture fees by Janssen Pharma K.K., Mylan EPD Co., Takeda Pharma. Co., Ltd., Mochida Pharma. Co., Ltd., EA Pharma Co., Ltd., Otsuka Pharma. Co., Ltd., Astellas Pharma Inc. and Miyarisan Pharma. Co., Ltd. ZY and TO are having the limited stock ownership as an employee of the company. No further financial interest is stated. The research was partly funded by these funds. Neither the funding agency nor any outside organization has participated in study design or have any competing of interest. These pharmaceutical companies had final approval of the manuscript. YI has an affiliation with a donation-funded department from Nichinichi Pharmaceutical Co., Ltd. The other authors have no conflicts of interest to disclose.
Supplementary Material
References
- 1.Hashimoto E, Taniai M, Tokushige K. Characteristics and diagnosis of NAFLD/NASH. J Gastroenterol Hepatol 2013; 28: 64–70. [DOI] [PubMed] [Google Scholar]
- 2.Chu H, Duan Y, Yang L, Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut 2019; 68: 359–370. [DOI] [PubMed] [Google Scholar]
- 3.Mouzaki M, Bandsma R. Targeting the gut microbiota for the treatment of non-alcoholic fatty liver disease. Curr Drug Targets 2015; 16: 1324–1331. [DOI] [PubMed] [Google Scholar]
- 4.Saltzman ET, Palacios T, Thomsen M, Vitetta L. Intestinal microbiome shifts, dysbiosis, inflammation, and non-alcoholic fatty liver disease. Front Microbiol 2018; 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009; 137: 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab 2017; 25: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natividad JM, Lamas B, Pham HP, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 2018; 9: 2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell JM, Fahey GC Jr, Wolf BW. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J Nutr 1997; 127: 130–136. [DOI] [PubMed] [Google Scholar]
- 9.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A 2006; 103: 12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verbeke KA, Boobis AR, Chiodini A, et al. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr Res Rev 2015; 28: 42–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pimentel M, Mathur R, Chang C. Gas and the microbiome. Curr Gastroenterol Rep 2013; 15: 356. [DOI] [PubMed] [Google Scholar]
- 12.Caussy C, Hsu C, Lo MT, et al. Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD Hepatology 2018. DOI: 10.1002/hep.29892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki J, Suzuki H, Asakura K, et al. Gallstones increase the prevalence of Barrett’s esophagus. J Gastroenterol 2010; 45: 171–178. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki J, Suzuki H, Tsugawa H, et al. Bile acids increase levels of microRNAs 221 and 222, leading to degradation of CDX2 during esophageal carcinogenesis. Gastroenterology 2013; 145: 1300–1311. [DOI] [PubMed] [Google Scholar]
- 15.Nishizawa T, Suzuki H, Takahashi M, Kaneko H, Suzuki M, Hibi T. Effect of ursodeoxycholic acid and endoscopic sphincterotomy in long-term stenting for common bile duct stones. J Gastroenterol Hepatol 2013; 28: 63–67. [DOI] [PubMed] [Google Scholar]
- 16.Murakami M, Iwamoto J, Honda A, et al. Detection of gut dysbiosis due to reduced Clostridium subcluster XIVa using the fecal or serum bile acid profile. Inflamm Bowel Dis 2018; 24: 1035–1044. [DOI] [PubMed] [Google Scholar]
- 17.Higashimura Y, Naito Y, Takagi T, et al. Protective effect of agaro-oligosaccharides on gut dysbiosis and colon tumorigenesis in high-fat diet-fed mice. Am J Physiol Gastrointest Liver Physiol 2016; 310: G367–G375. [DOI] [PubMed] [Google Scholar]
- 18.Song WY, Aihara Y, Hashimoto T, Kanazawa K, Mizuno M. (–)-Epigallocatechin-3-gallate induces secretion of anorexigenic gut hormones. J Clin Biochem Nutr 2015; 57: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tipoe GL, Leung TM, Liong EC, Lau TY, Fung ML, Nanji AA. Epigallocatechin-3-gallate (EGCG) reduces liver inflammation, oxidative stress and fibrosis in carbon tetrachloride (CCl4)-induced liver injury in mice. Toxicology 2010; 273: 45–52. [DOI] [PubMed] [Google Scholar]
- 20.Kochi T, Shimizu M, Terakura D, et al. Non-alcoholic steatohepatitis and preneoplastic lesions develop in the liver of obese and hypertensive rats: suppressing effects of EGCG on the development of liver lesions. Cancer Lett 2014; 342: 60–69. [DOI] [PubMed] [Google Scholar]
- 21.Bose M, Lambert JD, Ju J, Reuhl KR, Shapses SA, Yang CS. The major green tea polyphenol, (–)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat-fed mice. J Nutr 2008; 138: 1677–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park HJ, DiNatale DA, Chung MY, et al. Green tea extract attenuates hepatic steatosis by decreasing adipose lipogenesis and enhancing hepatic antioxidant defenses in ob/ob mice. J Nutr Biochem 2011; 22: 393–400. [DOI] [PubMed] [Google Scholar]
- 23.Xiao J, Ho CT, Liong EC, et al. Epigallocatechin gallate attenuates fibrosis, oxidative stress, and inflammation in non-alcoholic fatty liver disease rat model through TGF/SMAD, PI3 K/Akt/FoxO1, and NF-kappa B pathways. Eur J Nutr 2014; 53: 187–199. [DOI] [PubMed] [Google Scholar]
- 24.Chen YK, Cheung C, Reuhl KR, et al. Effects of green tea polyphenol (–)-epigallocatechin-3-gallate on newly developed high-fat/Western-style diet-induced obesity and metabolic syndrome in mice. J Agric Food Chem 2011; 59: 11862–11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Farah BL, Sinha RA, et al. Epigallocatechin-3-gallate (EGCG), a green tea polyphenol, stimulates hepatic autophagy and lipid clearance. PLoS One 2014; 9: e87161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bae UJ, Park J, Park IW, et al. Epigallocatechin-3-gallate-rich green tea extract ameliorates fatty liver and weight gain in mice fed a high fat diet by activating the sirtuin 1 and AMP activating protein kinase pathway. Am J Chin Med 2018; 46: 617–632. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Liu Q, Liu L, Hu YY, Feng Q. Potential biological effects of (–)-epigallocatechin-3-gallate on the treatment of nonalcoholic fatty liver disease. Mol Nutr Food Res 2018; 62. DOI: 10.1002/mnfr.201700483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unno T, Sakuma M, Mitsuhashi S. Effect of dietary supplementation of (–)-epigallocatechin gallate on gut microbiota and biomarkers of colonic fermentation in rats. J Nutr Sci Vitaminol (Tokyo) 2014; 60: 213–219. [DOI] [PubMed] [Google Scholar]
- 29.Sheng L, Jena PK, Liu HX, et al. Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J 2018. DOI: fj201800370R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sae-Tan S, Grove KA, Kennett MJ, Lambert JD. (–)-Epigallocatechin-3-gallate increases the expression of genes related to fat oxidation in the skeletal muscle of high fat-fed mice. Food Funct 2011; 2: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider K, Oltmanns J, Hassauer M. Allometric principles for interspecies extrapolation in toxicological risk assessment--empirical investigations. Regul Toxicol Pharmacol 2004; 39: 334–347. [DOI] [PubMed] [Google Scholar]
- 32.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957; 226: 497–509. [PubMed] [Google Scholar]
- 33.Matsumoto M, Inoue R, Tsuruta T, Hara H, Yajima T. Long-term oral administration of cows’ milk improves insulin sensitivity in rats fed a high-sucrose diet. Br J Nutr 2009; 102: 1324–1333. [DOI] [PubMed] [Google Scholar]
- 34.Inoue R, Sawai T, Sawai C, et al. A preliminary study of gut dysbiosis in children with food allergy. Biosci Biotechnol Biochem 2017; 81: 2396–2399. [DOI] [PubMed] [Google Scholar]
- 35.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ando M, Kaneko T, Watanabe R, et al. High sensitive analysis of rat serum bile acids by liquid chromatography/electrospray ionization tandem mass spectrometry. J Pharm Biomed Anal 2006; 40: 1179–1186. [DOI] [PubMed] [Google Scholar]
- 39.Bashiardes S, Shapiro H, Rozin S, Shibolet O, Elinav E. Non-alcoholic fatty liver and the gut microbiota. Mol Metab 2016; 5: 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He C, Cheng D, Peng C, Li Y, Zhu Y, Lu N. High-fat diet induces dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front Microbiol 2018; 9: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takagi T, Naito Y, Inoue R, et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol 2019; 54: 53–63. [DOI] [PubMed] [Google Scholar]
- 42.Cano PG, Santacruz A, Trejo FM, Sanz Y. BifidobacteriumCECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity (Silver Spring) 2013; 21: 2310–2321. [DOI] [PubMed] [Google Scholar]
- 43.Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: molecular insights and therapeutic perspectives. Hepatology 2017; 65: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagaki A, Kato Y, Nanjo F. Isolation and characterization of rat intestinal bacteria involved in biotransformation of (–)-epigallocatechin. Arch Microbiol 2014; 196: 681–695. [DOI] [PubMed] [Google Scholar]
- 45.Takagaki A, Nanjo F. Bioconver. of (–)-epicatechin, (+)-epicatechin, (–)-catechin, and (+)-catechin by (–)-epigallocatechin-metabolizing bacteria. Biol Pharm Bull 2015; 38: 789–794. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z, Chen Z, Guo H, et al. The modulatory effect of infusions of green tea, oolong tea, and black tea on gut microbiota in high-fat-induced obese mice. Food Funct 2016; 7: 4869–4879. [DOI] [PubMed] [Google Scholar]
- 47.Zhou K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J Funct Foods 2017; 33: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anhê FF, Roy D, Pilon G, et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 2015; 64: 872–883. [DOI] [PubMed] [Google Scholar]
- 49.Roopchand DE, Carmody RN, Kuhn P, et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 2015; 64: 2847–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masumoto S, Terao A, Yamamoto Y, Mukai T, Miura T, Shoji T. Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci Rep 2016; 6: 31208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr 2018; 63: 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneeberger M, Everard A, Gómez-Valadés AG, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep 2015; 5: 16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 2013; 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 54.Martin G, Kolida S, Marchesi JR, Want E, Sidaway JE, Swann JR. In vitro modeling of bile acid processing by the human fecal microbiota. Front Microbiol 2018; 9: 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the DDBJ Sequence Read Archive Database (DDBJ accession number; PRJDB7523).