Abstract
The reject water of anaerobic digestors still contains high levels of methane and ammonium that need to be treated before these effluents can be discharged to surface waters. Simultaneous anaerobic methane and ammonium oxidation performed by nitrate/nitrite-dependent anaerobic methane-oxidizing(N-damo) microorganisms and anaerobic ammonium-oxidizing(anammox) bacteria is considered a potential solution to this challenge. Here, a stable coculture of N-damo archaea, N-damo bacteria, and anammox bacteria was obtained in a sequencing batch reactor fed with methane, ammonium, and nitrite. Nitrite and ammonium removal rates of up to 455 mg N-NO2− L−1 day−1 and 228 mg N-NH4+ L−1 were reached. All nitrate produced by anammox bacteria (57 mg N-NO3− L−1 day−1) was consumed, leading to a nitrogen removal efficiency of 97.5%. In the nitrite and ammonium limited state, N-damo and anammox bacteria each constituted about 30–40% of the culture and were separated as granules and flocs in later stages of the reactor operation. The N-damo archaea increased up to 20% and mainly resided in the granular biomass with their N-damo bacterial counterparts. About 70% of the nitrite in the reactor was removed via the anammox process, and batch assays confirmed that anammox activity in the reactor was close to its maximal potential activity. In contrast, activity of N-damo bacteria was much higher in batch, indicating that these bacteria were performing suboptimally in the sequencing batch reactor, and would probably be outcompeted by anammox bacteria if ammonium was supplied in excess. Together these results indicate that the combination of N-damo and anammox can be implemented for the removal of methane at the expense of nitrite and nitrate in future wastewater treatment systems.
Keywords: Anaerobic methane oxidation, Anammox, Wastewater treatment, Methylomirabilis, Methanoperedens
Introduction
Aerobic wastewater treatment systems are energy-intensive and costly, as aeration for the removal of organic carbon and oxidation of ammonium requires large amounts of oxygen (Jetten et al. 1997). Furthermore, for complete nitrogen removal by denitrification, the addition of an external electron donor is needed (Kartal et al. 2010). In this respect, anaerobic treatment systems offer many advantages such as biogas production in the form of methane, which can be used to generate electricity. In addition, as anaerobic microbes grow slower, the sludge production in these systems is lower compared with that in aerobic treatment systems. However, the reject water of anaerobic treatment systems is still rich in reduced compounds, such as methane and ammonium. While there are several established processes that can be used to remove ammonium from the effluents of these anaerobic systems, currently there are no established biological methods to remove dissolved methane, resulting in the release of this potent greenhouse gas into the atmosphere (van Kessel et al. 2018).
In the past few decades, anaerobic microbial processes consuming ammonium and methane that depend on nitrite and/or nitrate reduction have been discovered and described (Mulder et al. 1995; Haroon et al. 2013; Raghoebarsing et al. 2006). The anaerobic ammonium oxidation (anammox) process is performed by microorganisms that use nitrite as their terminal electron acceptor, which produce N2 and nitrate (Strous et al. 1999). Combined with partial nitrification, this process is already applied worldwide in wastewater treatment and has shown to be an efficient and cost-effective system for nitrogen removal (Kartal et al. 2010; Lackner et al. 2014). Nitrate/nitrite-dependent anaerobic methane oxidation (N-damo) by a consortium of bacteria belonging to the NC10 phylum and methanotrophic archaea related to the ANME-2d clade was first described in 2006 (Raghoebarsing et al. 2006). The N-damo bacteria, named “Candidatus Methylomirabilis oxyfera,” were shown to use nitrite as an electron acceptor for methane oxidation, while the N-damo archaea prefer nitrate (Ettwig et al. 2008; Haroon et al. 2013; Hu et al. 2011). These ANME-2d clade archaea, named “Candidatus Methanoperedens nitroreducens,” reduce nitrate to ammonium via nitrite and oxidize methane via reverse methanogenesis to carbon dioxide (Arshad et al. 2015; Ettwig et al. 2016; Haroon et al. 2013; Vaksmaa et al. 2017; Gambelli et al. 2018). The production of nitrite and ammonium makes these archaea suitable partners for both Ca. Methylomirabilis and anammox bacteria (Arshad et al. 2015; Haroon et al. 2013; Meng et al. 2016; Shen et al. 2017). Indeed, studies have shown that anammox and Ca.Methylomirabilis-like bacteria and Ca.Methanoperedens-like archaea can be cultured together in laboratory-scale membrane reactors, together removing methane, ammonium, nitrite, and nitrate from the influent (Ding et al. 2017; Lu et al. 2017; Shi et al. 2013; Xie et al. 2017a, b). In addition to cooperation, these organisms may also compete with each other under substrate limitation (Luesken et al. 2011a, 2011b; Arshad et al. 2017; Guerrero-Cruz et al. 2019; Welte et al. 2016). Ca. Methylomirabilis sp. and Ca. Methanoperedens sp. both use methane, while Ca. Methylomirabilis sp. and anammox bacteria are both dependent on nitrite. In previous studies of the N-damo process, methane was added via a membrane system. In most anaerobic wastewater treatment systems, methane will be present as dissolved gas in the bulk liquid. In addition, biofilm growth on the membranes needs to be carefully monitored and controlled, to optimize the N and C removal capacity of the system (Chen et al. 2015; Wu and Zhang 2017). This might pose a challenge to control such a system when applied for full-scale wastewater treatment, as influent concentrations of substrates and temperatures vary considerably over time. Therefore, other biomass retention systems might be more practical in full-scale use.
Here, we describe an anammox/N-damo coculture in a laboratory-scale sequencing batch reactor (SBR) system that we used to study the microbial interactions between the three involved functional clades of Ca. Methanoperedens, Ca. Methylomirabilis, and anammox bacteria. Specific consumption rates of ammonium, nitrite, nitrate, and methane were determined using stable 15N isotopes in reactor and ex situ batch assays to determine the contribution of the different microbial groups to substrate removal. Community composition was monitored with fluorescence in situ hybridization (FISH) with specific probes for the three different groups.
Materials and methods
Establishment of an anaerobic methane- and ammonium-oxidizing coculture
An anaerobic methane-oxidizing enrichment culture (1.6 L; Ettwig et al. 2008) was used to inoculate a 3-L sequencing batch reactor (SBR) equipped with pH and dissolved oxygen sensors and connected to an ADI1010 bio-controller (Applikon Biotechnology BV, Schiedam, The Netherlands). Every SBR cycle comprised 10 h and 40 min of constant medium supply, 20 min of settling, and 1 h of removal of excess liquid. During the filling period, the culture was kept anoxic by continuous flushing with CH4/CO2 (95%/5%, 10 mL min−1). The gas effluent was discharged via a 5-L bottle, which acted as a “gas buffer” to further minimize air incursion from the atmosphere. At the start, synthetic medium containing 0.5 mM NH4Cl, 1.5 mM NaNO3, and 10 mM NaNO2 was added continuously. The medium further contained 0.3 g KH2PO4, 0.2 g CaCl2·2H2O, 0.2 g MgSO4·7H2O, 0.14 mg ZnSO4·7H2O, 0.06 mg CoCl2·6H2O, 0.1 mg MnCl2·4H2O, 0.4 mg CuSO4, 0.09 mg NiCl2·6H2O, 0.007 mg H3BO3, 0.01 mg SeO2, 0.01 mg Na2WO4·2H2O, 0.05 mg Na2MoO4, 0.01 mg CeCl2, and 2.5 mg FeSO4·7H2O per liter. Trace elements were added using NTA-chelated stock solutions. The medium vessel was continuously flushed with Ar/CO2 (95%/5%, 10 mL min−1). The pH was maintained at 7.3 by KHCO3 addition. The influent flow rate was adjusted based on the nitrite removal capacity of the culture. The SBR was stirred at 80 rpm by means of a turbine stirrer, and the temperature was kept constant at 30 °C with a heating blanket. During the removal period, the liquid volume (1.6 L) in the bioreactor was maintained by a level-controlled effluent pump. After reaching stable performance, the culture volume was reduced to 1.1 L upon settling of the biomass. Subsequently, on day 111, 500 mL of Kuenenia stuttgartiensis enrichment culture was added (Kartal et al. 2010). Ammonium and nitrite in the medium were gradually increased (up to 20 mM NH4+ and 40 mM NO2−) following the increasing consumption of these substrates by the biomass of the culture. Influent and effluent samples were taken on a regular basis and centrifuged for 5 min at maximal speed, and the supernatant was stored at −20 °C until further analysis.
Reactor assay—anaerobicmethane-oxidizing activity
On day 105, before the addition of K. stuttgartiensis, the nitrite and methane consumption rate of the anaerobic methane-oxidizing enrichment culture was determined. The SBR cycle, influent flow, and CH4 inflow were stopped. The reactor was flushed with Ar/CO2 until the CH4 concentration in the headspace was approximately 8% (v/v). Subsequently, the headspace was sealed and the reactor was supplemented with 0.4 mM NO2−. Methane and nitrite samples were taken every hour.
Reactor assay—substrate turnover and dinitrogen and nitrous oxide gas production
Turnover rates of ammonium, nitrite, and nitrate in the anaerobic methane- and ammonium-oxidizing coculture were determined during two reactor assays on day 520 and day 524. After the removal phase of the SBR cycle, methane inflow was stopped and the headspace of the reactor was sealed. Inflow of medium containing 20 mM NH4+, 40 mM NO2−, and 1.5 mM NO3− was continued during the assay. After 1 h, the medium was replaced with medium containing 15NO2− (all substrate concentrations were as described above). Gas and liquid samples were taken every 30 min.
Batch assay—potentialmethane- and nitrogen-removing activity
The headspace of 60-mL serum bottles containing 25 mL medium (see the above section) with substrates (Table 1) supplemented with 5 mM 3-morpholinopropanesulfonic acid (MOPS) buffer (pH 7.4) was exchanged with argon by 5 cycles of vacuum/gassing followed by 10 min flushing with argon. Subsequently, 5 mL biomass from the reactor was added. The headspace was evacuated and exchanged with argon twice to remove any remaining methane from the liquid. CH4 and CO2 were added to achieve a final concentration of 4% (v/v, Table 1. All incubations were performed in duplicate. Bottles were incubated in a shaking incubator at 30 °C and 135 rpm. Gas and liquid samples were taken every hour.
Table 1.
Specification of substrate and gas additions in the batch assays to determine the potential activity of the culture
| Treatment | NO2− (mM) | NH4+ (mM) | NO3− (mM) | CH4 (%) | CO2 (%) |
|---|---|---|---|---|---|
| NO2− + CH4 | 0.5 | – | – | 4 | 4 |
| NO3− + CH4 | – | – | 0.5 | 4 | 4 |
| NO2− | 0.5 | – | – | – | 4 |
| NO3− | – | – | 0.5 | – | 4 |
| NH4+ + NO2− | 1 | 1 | – | – | 4 |
| NH4+ + NO2− + CH4 | 0.5 | 0.5 | – | 4 | 4 |
Analytical methods
Liquid samples taken during activity tests were centrifuged for 1 min at maximal speed, and the supernatant was stored at −20 °C until further analysis. Ammonium was determined colorimetrically using a modified orthophataldialdehyde assay and nitrite by the Griess reaction (Ettwig et al. 2008). Nitrate was measured with the NOA280i nitric oxide analyzer (GE Analytical Instruments, Manchester, UK) after nitrite reduction to NO with acidic VCl3 at 95 °C. To measure the protein content of the biomass, 1.5 mL biomass samples was taken in triplicate and centrifuged for 5 min at maximal speed. The supernatant was removed, and subsequently the pellet was homogenized in 0.5 M NaOH, boiled at 90 °C for 30 min, and neutralized with 0.5 M HCl. Next, protein content was determined by the bicinchoninic acid (BCA) assay (Pierce, USA). Methane was measured using gas chromatography (HP 5890 gas chromatograph equipped with a Porapak Q column and a flame ionization detector). N2, O2, CO2, and N2O production was analyzed using gas chromatography (Agilent 6890, Porapak Q column, 80 °C) in combination with mass spectrometry (Agilent 5975c, quadrupole inert MS) (Arshad et al. 2017)
Fluorescence in situ hybridization
Biomass samples (2 mL) from the coculture were taken on days 85, 136, and 464 and centrifuged for 5 min at maximum speed. The pellet was washed with 1 mL phosphate-buffered saline (10 mM Na2HPO4/NaH2PO4 pH 7.5 and 130 mM NaCl) and fixed using paraformaldehyde. Subsequently, FISH was performed as described before (Ettwig et al. 2008). A mixture of three probes (EUB338, EUB338II, and EUB338III, targeting most bacteria) was used to visualize the general bacterial population (Amann et al. 1990; Daims et al. 1999). A general probe targeting most archaea (ARCH0915) was used to visualize archaea (Stahl and Amann 1991). To visualize anammox bacteria, Ca.Methylomirabilis-like bacteria, and Ca.Methanoperedens-like archaea, AMX820, DBACT1043, and DBACT1043b mix and DARCH0641 probes were used, respectively (Schmid et al. 2001, Raghoebarsing et al. 2006, Ettwig et al. 2009, Schubert et al. 2011). Probes were Cy3, Cy5, or FLUOS labeled. All samples were counterstained with the DNA stain 4′,6-diamidino-2-phenylindole (DAPI). Slides were examined and images obtained by utilization of a Zeiss Axioplan 2 epifluorescence microscope equipped with a digital camera, in combination with the AxioVision software package (Zeiss, Germany).
Results
Anaerobic methane-oxidizing culture
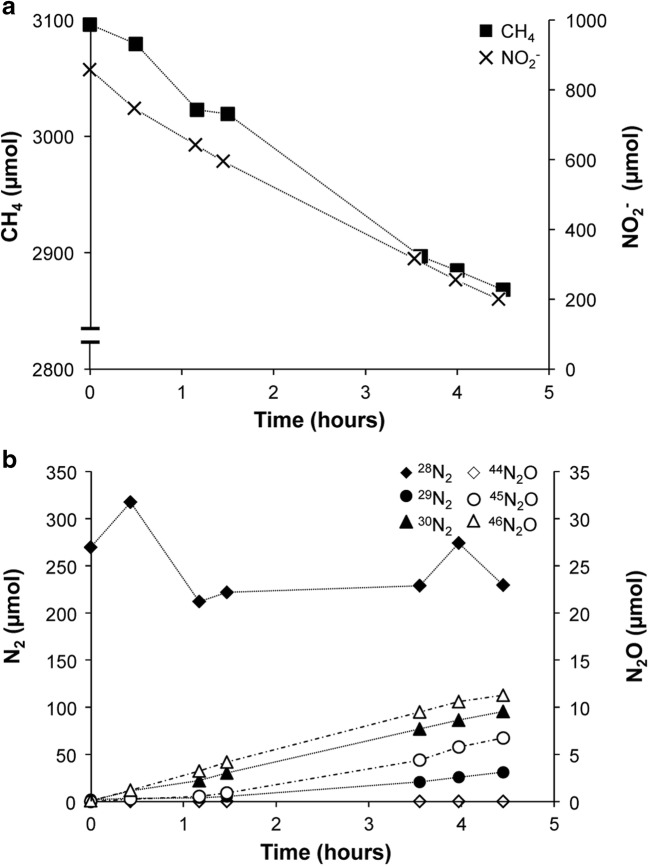
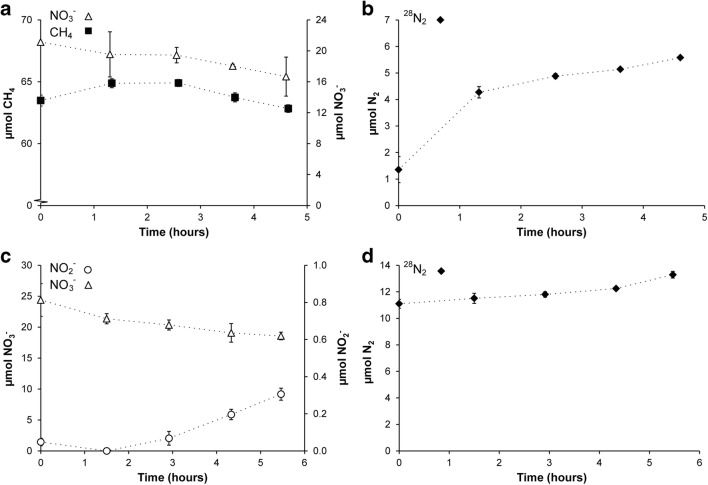
A 3-L laboratory-scale SBR was started with a culture that contained both nitrate- and nitrite-dependent anaerobic methane-oxidizing(N-damo) microorganisms (Ettwig et al. 2008). In the first 110 days, the culture was supplemented with nitrate, nitrite, and methane until a stable consumption rate was achieved. Ammonium was only present as a nitrogen source. During this period, nitrite was consumed at an average rate of 63 ± 12 mg N-NO2− L−1 day−1 (Fig. 1b), while no significant nitrate consumption was detected (Fig. 1c). On day 85, Ca.Methylomirabilis-like bacteria were the most abundant bacteria in the culture. Hardly any Ca.Methanoperedens-like archaea were visible with FISH, indicating that they had disappeared below the detection of less than a few percent of the biomass. At this phase of cultivation, also no anammox bacteria could be detected. On day 105, reactor activity was determined by supplementing the culture with methane and 15N-labeled nitrite. Nitrate (1.1 ± 0.04 mM) was still present in the reactor. Methane and nitrite were consumed at average rates of 53 nmol h−1 mg protein−1 and 143 nmol h−1 mg protein−1, respectively (Fig. 2a). Nitrate consumption was initially 70 nmol h−1 mg protein−1, but the consumption decreased after 1.5 h. No anaerobic ammonium consumption was observed. Consumption of 14N-nitrate and 15N-nitrite led to the production of both 29N2 and 30N2. Next to the production of dinitrogen gas, nitrous oxide production comprised about 13% of the total nitrogenous gas production (Fig. 2b).
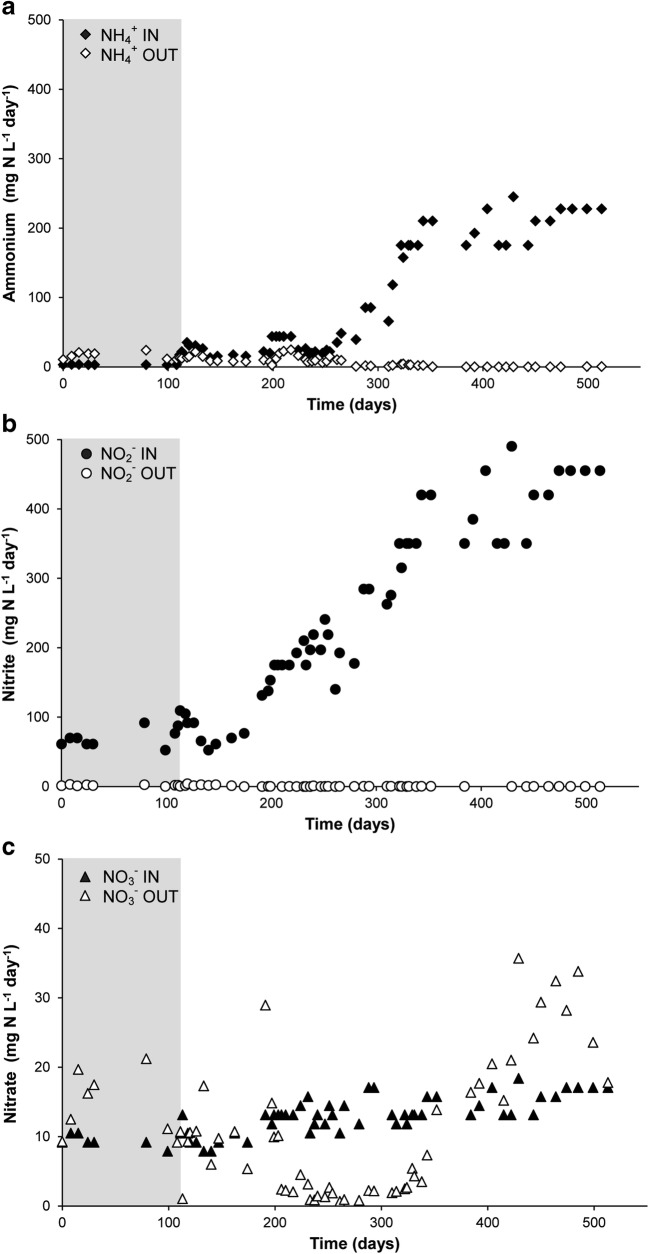
Fig. 1.
Ammonium (a), nitrite (b), and nitrate (c) consumption in the reactor. Closed symbols represent substrate per day fed to the reactor, open symbols show the amount of substrate per day in the effluent. In the period of t = 0 till t = 111 days (gray area), the reactor was operated as an anaerobic methane-oxidizing culture; from t = 111 days onwards, it was operated as an anaerobic methane- and ammonium-oxidizing coculture
Fig. 2.
Reactor activity assay (t = 105 days). a Methane (closed squares) and nitrite (crosses) consumption. Total protein of the reactor was 9.6 ± 0.9 g. b Production of dinitrogen gas (closed symbols) and nitrous oxide (open symbols), 28N2/44N2O (diamonds), 29N2/45N2O (circles), and 30N2/46N2O (triangles)
Anammox/N-damo coculture establishment
To establish a culture that would consume both methane and ammonium at the expense of nitrite and nitrate, 500 mL of K. stuttgartiensis biomass was added to the SBR on day 111 (Kartal et al. 2010). During the first 8 months after the addition, ammonium concentration in the medium could be gradually increased up to 20 mM. A stable performing culture was obtained within 363 days after the addition of the anammox cells, consuming up to 455 mg N-NO2− L−1 day−1 and up to 228 mg N-NH4+ L−1 day−1(Fig. 1a). Nitrate consumption increased over time to 16 mg N-NO3− L−1 day−1, but from around day 390 onwards, the activity of anammox bacteria resulted in net production of nitrate. As most of the ammonium was consumed via the anammox process, following the anammox stoichiometry (1:1.3:0.3, ammonium to nitrite to nitrate, Strous et al. 1997), an ammonium consumption of 228 mg N-NH4+ L−1 day−1 (t = 474 – 513 days) would correspond to a theoretical nitrite consumption and nitrate production by anammox bacteria of 296 mg N-NO2− L−1 day−1 and 68 mg N-NO3− L−1 day−1, respectively. Taking this and the nitrate in the medium feed into account, it was calculated that the total nitrate consumption by the culture was about 66 mg N-NO3− L−1 day−1 by the end of the bioreactor operation.
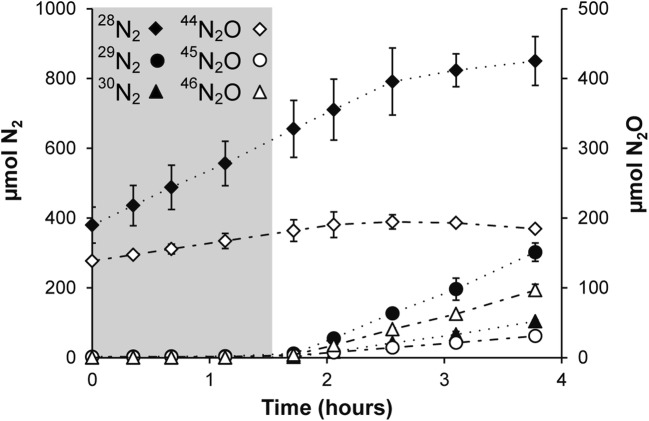
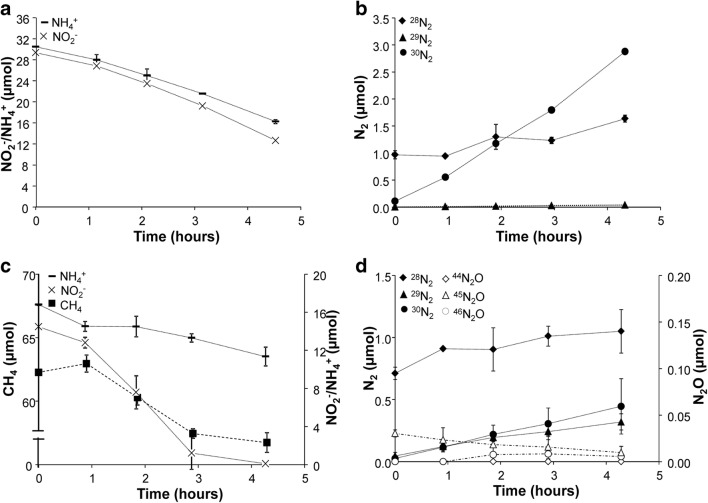
In order to analyze the contribution of different processes to nitrogen removal within this SBR system, two reactor activity assays (on days 520 and 524) were performed in which medium supplemented with isotopically labeled 15N nitrite was continuously fed to the culture. The headspace was closed to measure the production of labeled nitrogen gas. Oxygen levels in the headspace remained below 30 ppm. Ammonium was consumed at a rate of 355 ± 55 nmol mg protein−1 h−1, while nitrite consumption was 664 ± 125 nmol mg protein−1 h−1. Nitrate concentration did not change throughout the experiments (results not shown). These results indicate that anammox bacteria removed about 70% of nitrite, while other processes, including N-damo, accounted for the remaining 30%. Similar results were obtained from off-gas analysis (Fig. 3), as about 75% of the produced labeled dinitrogen gas was 29N2, originating from combining labeled nitrite with unlabeled substrate (either ammonium or nitrate). Besides labeled dinitrogen gas, both 45N2O and 46N2O were produced, together accounting for 20% of the total nitrogenous gas production.
Fig. 3.
Dinitrogen gas and nitrous oxide production during activity assays (t = 520 days and t = 524 days). The closed symbols represent N2 and the open symbols N2O (diamonds: 28N2/44N2O, circles: 29N2/45N2O, triangles: 30N2/46N2O). The gray box indicates the period that medium with 14N-nitrite was fed to the reactor
FISH analysis
Community composition was studied by means of FISH on days 136 and 464, 25 and 354 days after the addition of K. stuttgartiensis cells, respectively (Fig. 4). Already 25 days after the addition of anammox biomass, clusters of anammox bacteria were visible in the biomass that was dominated (70–80%) by Ca.Methylomirabilis-like bacteria (Fig. 4a). In addition, archaeal biomass had increased up to about 5% of the biomass and mainly consisted of Ca.Methanoperedens-like archaea (Fig. 4b). At day 354, Ca.Methylomirabilis-like bacteria were no longer the dominant group and represented about 40% of the biomass. Both anammox bacteria and the Ca.Methanoperedens-like archaea had increased in numbers, representing 40% and 10–20% of the total biomass, respectively (Fig. 4c, d). Furthermore, upon close inspection of the biomass during the settling and removal period of the SBR, two types of biomass could be detected. The bottom layer consisted of quickly settling, dense, gray granules, which mainly consisted of Ca.Methylomirabilis-like bacteria and Ca.Methanoperedens-like archaea (Fig. 4e). In contrast, the biomass in the top layer was fluffier, less compact, and more floc-like. The flocs settled much slower but were comparable in size. Anammox bacteria comprised about 80% of this biomass (Fig. 4f).
Fig. 4.
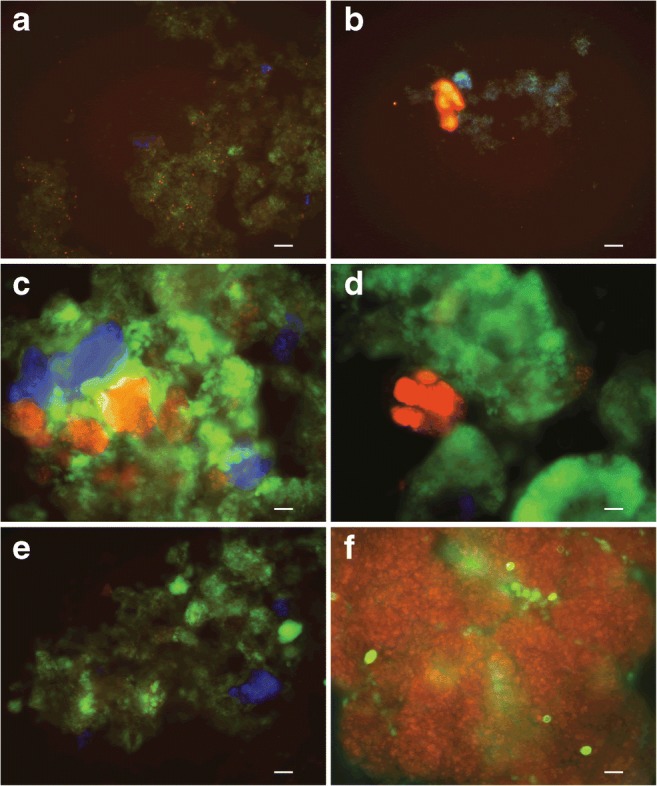
Fluorescence in situ visualization of N-damo/anammox coculture after 25 days (a, b) and 354 days (c–f) after the addition of K. stuttgartiensis. a Anammox bacteria are visible in red (Cy3, AMX820), N-damo bacteria in green (FLUOS, DAMOBACT1027), and archaea in blue (Cy5, ARCH0915). b 25 days after K. stuttgartiensis addition, most archaea were N-damo archaea (red, Cy3, DAMOARCH0641). N-damo bacteria are visible in light blue (FLUOS, DAMOBACT1027) and all bacteria in dark blue (Cy5, EUBmix). c 354 days after the addition of K. stuttgartiensis, anammox bacteria comprised 40% of the total biomass (red, Cy3, AMX820), as did N-damo bacteria (green, FLUOS, DAMOBACT1027). Archaea visible in blue (Cy5, ARCH0915) represented about 10–20% of the biomass. d N-damo archaea (red, Cy3, DAMOARCH0641) were the dominant archaea in the culture. N-damo bacteria are shown in green (FLUOS, DAMOBACT1027) and anammox bacteria in blue (Cy5, AMX820). e Of the two layers that were visible in the reactor during the settling and removal period, the bottom layer, consisting of dense granules, constitutes mainly N-damo bacteria (green, FLUOS, DAMOBACT1027) and archaea (blue, Cy5, ARCH0915). f In the top layer, with more floc-like biomass, anammox bacteria (red, Cy3, AMX820) were dominant. In green, some N-damo bacteria (FLUOS, DAMOBACT1027) are visible. Scale bars represent 20 μm
Potential activity batch assays
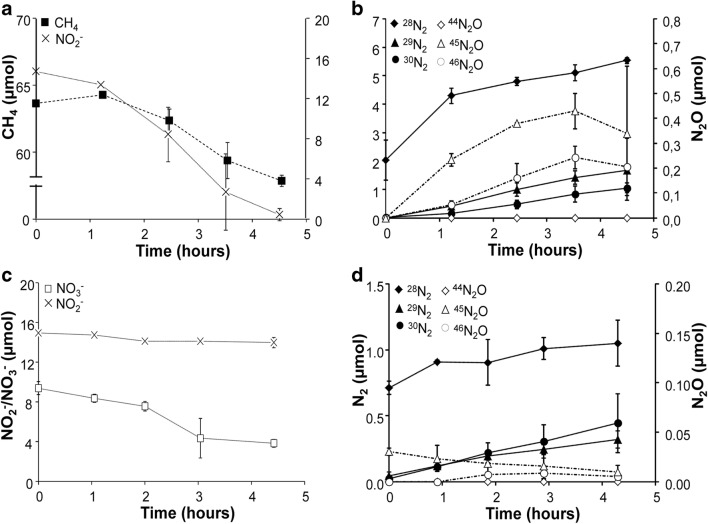
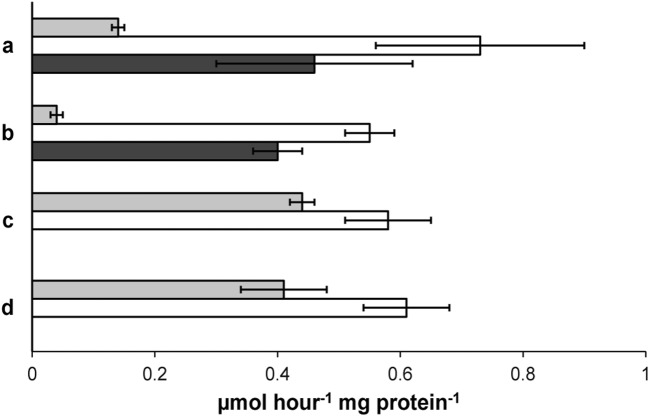
The potential activity of the culture to remove nitrite, nitrate, ammonium, and methane was tested by batch assays approximately 1 year after K. stuttgartiensis addition. Incubations supplemented with nitrite (15NO2−) and methane showed high consumption rates (Fig. 5a), with maximum rates of 532 ± 179 nmol NO2− mg protein−1 h−1 and 289 ± 62 nmol CH4 mg protein−1 h−1, respectively. The ratio of consumed nitrite to methane (8:4.3) was close to, but not identical to, the expected stoichiometry of nitrite-dependent methane oxidation (8:3, nitrite to methane). No ammonium was detected and background nitrate remained stable over the course of the experiment. However, 29N2, 30N2, and nitrous oxide was produced (Fig. 5b), suggesting that part of the nitrite and nitrate was consumed by N2O-producing microorganisms. Control incubations with only nitrite (15NO2−) and nitrate indeed gave a background nitrite and nitrate reduction rate of 35 ± 14 nmol NO2− mg protein−1 h−1 and 198 ± 24 nmol NO3− mg protein−1 h−1, respectively. Although nitrite consumption in these control incubations was low, production of labeled 29N2 and 30N2 as well as 45N2O and 46N2O could be observed (Fig. 5c, d). Incubations containing methane and nitrate as substrates showed that nitrate was also consumed in combination with methane (Fig. 6a), with average rates of 97 ± 27 nmol NO3− mg protein−1 h−1 and 70 ± 5.8 nmol CH4 mg protein−1 h−1. Methane consumption exceeded the nitrate consumption based on the described stoichiometry for Ca. Methanoperedens archaea (methane to nitrate ratio, 2.9:4 as opposed to 1:4), suggesting that the nitrite produced in this process was most likely immediately consumed by Ca. Methylomirabilis bacteria. Incubations with nitrate as the sole electron acceptor only produced dinitrogen gas, and no nitrous oxide production was detected (Fig. 6b). Incubations with only nitrate did show accumulation of nitrite, but not of ammonium (Fig. 6c). Like in the incubations containing both methane and nitrate, only dinitrogen gas was produced (Fig. 6d). When ammonium (15NH4+) and nitrite were available as substrates, these were consumed at rates of 438 ± 23 nmol NH4+ mg protein−1 h−1 and 583 ± 65 nmol NO2− mg protein−1 h−1 (Fig. 7a), respectively, nicely fitting the anammox stoichiometry of 1:1.3 (ammonium to nitrite). The fact that mainly 29N2 was produced (Fig. 7b) also indicated that the substrate consumption in these incubations was most likely catalyzed by the anammox process. Remarkably, the addition of methane, ammonium, and nitrite (15NO2−) as substrates resulted in much lower ammonium consumption rates (140 ± 11 nmol NH4+ mg protein−1 h−1) compared with incubations where ammonium was the only electron donor (Fig. 7c). In contrast, nitrite and methane consumption rates were similar to those in incubations without ammonium, namely 726 ± 166 nmol NO2− mg protein−1 h−1 and 237 ± 80 nmol CH4 mg protein−1 h−1, respectively (Fig. 7c). In addition, also in these incubations, both 29N2 and 30N2 and 45N2O and 46N2O were produced (Fig. 7d). Apparently, in the presence of methane and excess nitrite, anaerobic ammonium oxidation is somewhat inhibited. This effect was even stronger at lower ammonium concentrations, while using lower ammonium concentrations in the absence of methane did not affect the consumption rate (Fig. 8). Methane itself, when added to a final concentration of 4%, does not inhibit anammox activity.
Fig. 5.
Batch incubations to determine the production of dinitrogen gas and nitrous oxide with methane and/or 15N-labeled nitrite as substrates. a Methane (closed squares) and nitrite (crosses) consumption b Dinitrogen gas and nitrous oxide production in incubations with methane (4%) and nitrite (0.5 mM 15NO2−). The closed symbols represent N2 and the open symbols N2O (diamonds: 28N2/44N2O, circles: 29N2/45N2O, triangles: 30N2/46N2O). c Nitrate (open squares) and nitrite (crosses) consumption in batch incubations. d Dinitrogen gas and nitrous oxide production in incubations with 15NO2− as a substrate. The closed symbols represent N2 and the open symbols N2O (diamonds: 28N2/44N2O, circles: 29N2/45N2O, triangles: 30N2/46N2O)
Fig. 6.
Production of nitrogenous gasses during batch incubations with methane and/or 15N-labeled nitrate as substrates. a Methane (closed squares) and nitrate (open triangles) consumption in incubations. b Only dinitrogen gas was formed in incubations with methane and nitrate. c Nitrate consumption (open triangles) and nitrite (open circles) production in incubations with nitrate (0.7 mM). d Only production of dinitrogen gas was detected in incubations with only nitrate
Fig. 7.
Consumption of substrates and production of dinitrogen gas and nitrous oxide during batch assays to determine the nitrite, nitrate, ammonium, and methane removal potential of the culture. a Ammonium and nitrite consumption in incubations with ammonium (1 mM 15NH4+) and nitrite (1 mM). b Production of 28N2 and 29N2 in incubations with ammonium (1 mM 15NH4+) and nitrite (1 mM). No 30N2 was formed. c Consumption of ammonium, nitrite, and methane in incubations with methane (4%), ammonium (0.5 mM), and nitrite (0.5 mM 15NO2−). Total protein was 8.3 ± 2.7 mg. d Dinitrogen gas and nitrous oxide production in incubations with methane (4%), ammonium (0.5 mM), and nitrite (0.5 mM 15NO2−)
Fig. 8.
Specific consumption rates of ammonium (light gray bars), nitrite (white bars), and methane (dark gray bars) during batch assays to determine the nitrite, nitrate, ammonium, and methane removal potential of the culture. Conditions: (a) 4% methane, 0.5 mM ammonium, and 0.5 mM nitrite; (b) 4% methane, 0.25 mM ammonium, and 0.5 mM nitrite; (c) 1 mM ammonium and 1 mM nitrite; (d) 0.5 mM ammonium and 0.5 mM nitrite
Discussion
By complementing an enrichment culture containing both N-damo bacteria and archaea with K. stuttgartiensis single cells, a stable coculture of anammox bacteria, Ca.Methylomirabilis-like bacteria, and Ca.Methanoperedens-like archaea was established. Together, these microorganisms removed methane, ammonium, nitrite, and nitrate from the supplied medium via a network of interactions (Fig. 9). Interestingly, prior to the addition of anammox cells, the N-damo enrichment culture showed somewhat lower specific anaerobic methane-oxidizing rates than N-damo enrichments reported before (Ettwig et al. 2009; Luesken et al. 2011a). After the addition of 500 mL K. stuttgartiensis cells, nitrite and ammonium consumption of the culture rapidly increased up to 455 mg N L−1 day−1 and 228 mg N L−1 day−1, respectively. Once a stable population was obtained, nearly all nitrate produced by anammox bacteria was consumed by Ca.Methanoperedens-like archaea at a rate of 57 mg N L−1 day−1 and the total nitrogen removal efficiency of the culture was as high as 97.5%. The rates for ammonium and nitrite removal reported here are amongst the higher rates for anammox/N-damo cultures (Allegue et al. 2018; Liu et al. 2019; Luesken et al. 2011a, 2011b; Shi et al. 2013; Xie et al. 2017a, b). Yet, as in these previous studies, it was shown that in a mixed anammox/N-damo culture, the presence of Ca.Methylomirabilis-like bacteria in large numbers does not necessarily correspond to similar contributions to nitrite consumption (Luesken et al. 2011a; Guerrero-Cruz et al. 2019). Here, Ca.Methylomirabilis-like and anammox bacteria both constituted about 30–40% of the culture, yet about 70% of the nitrite was removed via the anammox process similar to earlier reports (Luesken et al. 2011a). In a recent study mimicking estuarine conditions with methane, sulfide, and ammonium as electron donors for nitrate conversion, a similar biomass composition and contribution of anammox bacteria, Ca. Methylomirabilis, and Ca. Methanoperedens were observed (Arshad et al. 2017).
Fig. 9.
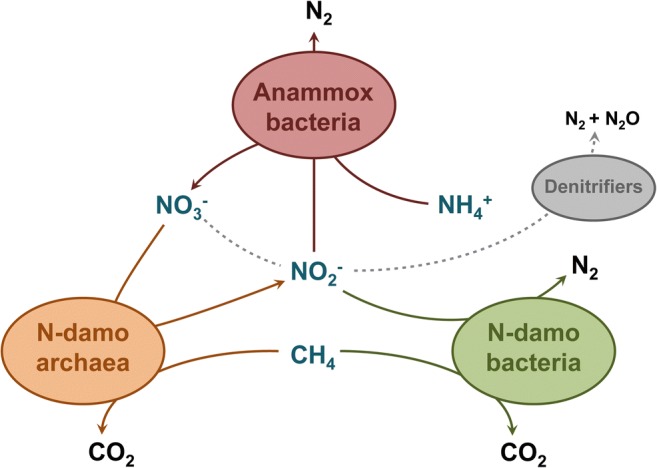
Main interactions within the SBR reactor system removing methane, ammonium, nitrite, and nitrate
In full-scale wastewater treatment, influent concentrations of substrates and temperatures are varying considerably over time. In anaerobic wastewater treatment systems, methane will be present as dissolved gas in the bulk liquid. To mimic this, we used continuous gas supply resulting in saturated methane concentration in the reactor liquid. As a proof of principle, we show that an active nitrogen/methane-removing community can be established, but we realize that for applications the supply will be through the water. Previous studies used methane supply via a membrane system (Allegue et al. 2018; Chen et al. 2015; Wu and Zhang 2017). These systems may be good alternatives if biofilm growth on the membranes is carefully controlled.
In addition, ex situ batch incubations with ammonium and nitrite as substrates showed similar specific anammox activity as the activity observed in situ in the reactor. This suggests that the activity of anammox bacteria in the reactor was close to their maximum potential activity. In contrast, the specific nitrite consumption rate of Ca.Methylomirabilis-like bacteria in the reactor was about two times lower than the specific rates measured in the batch incubations with nitrite and methane. The batch test activities performed in this study support the previous in situ observations that these bacteria loose the competition for nitrite from anammox bacteria under nitrite limitation (Luesken et al. 2011a; Winkler et al. 2015). For future work, it may be beneficial to perform both in situ and ex situ measurements on these systems in order to better understand the microbial interactions. In addition, community analysis with FISH showed two different types of biomass layers that were visible upon settling of the biomass, indicating a physical separation of anammox bacteria and Ca.Methylomirabilis-like bacteria. Separation of microbiological guilds in different aggregates has been seen before (Nielsen et al. 2005). They proposed that the separation of aerobic ammonium oxidizers and anammox bacteria within a partial-nitrification anammox bioreactor into respectively smaller and bigger granules is an effect of the oxygen sensitivity of anammox bacteria. In the current system, oxygen is most likely not the cause of the separation into flocs and granules. Possibly, the formation of dense granules by N-damo bacteria and archaea prevents biomass washout (Luesken et al. 2011a, 2011b; He et al. 2015).
During batch incubations, nitrous oxide production was observed. Nitrous oxide production within N-damo cultures has been described before, and it was speculated that it originated from the denitrifying community members (Etttwig et al. 2010). In this study, only batch incubations fed with nitrite showed N2O production, while no N2O production was detected in incubations with only nitrate as an electron acceptor. In other studies, similar effects on N2O production by cultures after adding pulses of nitrite have been observed (Allegue et al. 2018; Itokawa et al. 2001). This might explain the observations in batch incubations, where the microbial community is subjected to pulse feeding of substrates. In addition, since the exact functions of the putative quinol-dependent NO reductases found in Ca. Methylomirabilis bacteria are still unknown, it cannot be excluded that these bacteria produce N2O under certain conditions (Etttwig et al. 2010; Wu et al. 2011; Zhu et al. 2017; Graf et al. 2018; Versantvoort et al. 2018).
As observed in previous studies and here, competition for nitrite is an important regulator of anammox/N-damo systems, but also other chemical compounds, such as trace elements, could play a role in this. Recently, it has been reported that the concentration of available iron and copper might play a role in the competition between anammox and Ca. Methylomirabilis bacteria (He et al. 2015; Lu et al. 2018). Concentrations of iron and copper used in the current study were 10 μM and 2.5 μM, respectively. More research is needed to determine how the use of medium containing more iron and copper could influence the community and the competition for substrates in an anammox/N-damo coculture. Remarkably, also the presence or absence of rare earth elements may determine which Ca. Methylomirabilis bacteria will become dominant in the culture. Recently it was shown that inclusion of cerium in the trace elements of a Ca. Methylomirabilis enrichment culture resulted in the appearance of a new species named Ca. Methylomirabilis lanthanidiphila that only had a xoxF-type methanol dehydrogenase encoded in its genome (Versantvoort et al. 2018).
In addition, it remains unclear why anammox bacteria seemed not active in batch incubations with ammonium, nitrite, and 4% methane, while winning the competition for nitrite under methane saturated conditions in the reactor. Batch incubations with the K. stuttgartiensis biomass in the presence of 4% methane did not show any inhibition of anammox activity, indicating that N-damo microorganisms might influence anammox activity when nitrite is not limiting. Also, to apply an anammox/N-damo system in anaerobic wastewater treatment plants, nitrite will have to be produced from ammonium via partial nitrification, as is the case in partial nitritation-anammox systems (van Kessel et al. 2018). Next to ammonium oxidizers, other aerobic microorganisms like aerobic methanotrophs and heterotrophs might start to grow, while N-damo and anammox might be inhibited (Castro-Barros et al. 2017; Guerrero-Cruz et al. 2018; Luesken et al. 2012; Wu and Zhang 2017). Several studies have explored the feasibility of such a combined nitritation-anammox-N-damo system by modeling (Bhattacharjee et al. 2016; Castro-Barros et al. 2017; Chen et al. 2015; Winkler et al. 2015). Now the next step would be to test the potential of such a system in laboratory-scale reactors.
Acknowledgments
We would like to thank Katinka van de Pas-Schoonen and Guylaine Nuijten for their help with the bioreactor maintenance.
Funding information
M.A.H.J.v.K, K.S., and S.G.C. were funded by the Technology Foundation STW (grant 13146). M.A.H.J.v.K. is further supported by an NWO Veni Grant (016.veni.192.062). H.J.M.O.d.C. is supported by the European Research Council (ERC 669371) and M.S.M.J by the European Research Council (ERC 339880) and by the Netherlands Organization for Scientific Research (NWO SIAM 024002002). BK was supported by the European Research Council (ERC 640422).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allegue T, Arias A, Fernandez-Gonzalez N, Omil F, Garrido JM. Enrichment of nitrite-dependent anaerobic methane oxidizing bacteria in a membrane bioreactor. Chem Eng J. 2018;347:721–730. doi: 10.1016/j.cej.2018.04.134. [DOI] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Deveruex R, Stahl RA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad A, Speth DR, de Graaf RM, Op den Camp HJM, Jetten MSM, Welte CU (2015) A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 6. 10.3389/fmicb.2015.01423 [DOI] [PMC free article] [PubMed]
- Arshad A, Dalcin Martins P, Frank J, Jetten MSM, Op den Camp HJM, Welte CU. Mimicking microbial interactions under nitrate-reducing conditions in an anoxic bioreactor: enrichment of novel Nitrospirae bacteria distantly related to Thermodesulfovibrio. Environ Microbiol. 2017;19:4965–4977. doi: 10.1111/1462-2920.13977. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee AS, Motlagh AM, Jetten MSM, Goel R. Methane dependent denitrification- from ecosystem to laboratory-scale enrichment for engineering applications. Water Res. 2016;99:244–252. doi: 10.1016/j.watres.2016.04.070. [DOI] [PubMed] [Google Scholar]
- Castro-Barros CM, Ho LT, Winkler MKH, Volcke EIP. Integration of methane removal in aerobic anammox-based granular sludge reactors. Environ Tech. 2017;39:1615–1625. doi: 10.1080/09593330.2017.1334709. [DOI] [PubMed] [Google Scholar]
- Chen XM, Guo J, Xie G-J, Liu Y, Yuan Z, Ni B-J. A new approach to simultaneous ammonium and dissolved methane removal from anaerobic digestion liquor: a model-based investigation of feasibility. Water Res. 2015;85:295–303. doi: 10.1016/j.watres.2015.08.046. [DOI] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann RI, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Ding Z-W, Lu Y-Z, Fu L, Ding J, Zeng RJ. Simultaneous enrichment of denitrifying anaerobic methane-oxidizing microorganisms and anammox bacteria in a hollow-fiber membrane biofilm reactor. Appl Microbiol Biotechnol. 2017;101:437–446. doi: 10.1007/s00253-016-7908-7. [DOI] [PubMed] [Google Scholar]
- Etttwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S., Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen TA, Luesken FA, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M (2010)Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–550 [DOI] [PubMed]
- Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJM, Jetten MSM, Strous M (2008) Denitrifying bacteria anaerobically oxidize methane in the absence of archaea. Environ Microbiol 10:3164–3173 [DOI] [PubMed]
- Ettwig KF, van Alen TA, van de Pas-Schoonen KT, Jetten MSM, Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol. 2009;75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Zhu B, Speth DR, Keltjens JT, Jetten MSM, Kartal B. Archaea catalyze iron-dependent anaerobic oxidation of methane. PNAS. 2016;113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambelli L, Guerrero-Cruz S, Mesman RJ, Cremers G, Jetten MSM, Op den Camp HJM, Kartal B, Lueke C, van Niftrik L. Community composition and ultrastructure of a nitrate-dependent anaerobic methane-oxidizing enrichment culture. Appl Environ Microbiol. 2018;17:e02186–e02117. doi: 10.1128/AEM.02186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf JS, Mayr MJ, Marchant HK, Tienken D, Hach PF, Brand A, Schubert KMMM, Milucka J. Bloom of a denitrifying methanotroph, ‘Candidatus Methylomirabilis limnetica’, in a deep stratified lake. Environ Microbiol. 2018;20:2598–2614. doi: 10.1111/1462-2920.14285. [DOI] [PubMed] [Google Scholar]
- Guerrero-Cruz S, Cremers G, van Alen TA, Op den Camp HJM, Jetten MSM, Rasigraf O, Vaksmaa A. Response of the anaerobic methanotroph “Candidatus Methanoperedens nitroreducens” to oxygen stress. Appl Environ Microbiol. 2018;84:online first. doi: 10.1128/AEM.01832-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Cruz S, Stultiens K, van Kessel MAHJ, Versantvoort W, Jetten MSM, Op den Camp HJM, Kartal B. Key physiology of nitrite-dependent methane oxidizing enrichment culture. Appl Environ Microbiol. 2019;85:e00124–e00119. doi: 10.1128/AEM.00124-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- He Z, Geng S, Pan Y, Cai C, Wang J, Wang L, Liu S, Zhen P, Xu X, Hu B. Improvement of the trace metal composition of medium for nitrite-dependent anaerobic methane oxidation bacteria: iron (II) and copper (II) make a difference. Water Res. 2015;85:235–243. doi: 10.1016/j.watres.2015.08.040. [DOI] [PubMed] [Google Scholar]
- Hu S, Zeng RJ, Keller J, Lant PA, Yuan Z. Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ Microbiol Rep. 2011;3:315–319. doi: 10.1111/j.1758-2229.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- Itokawa H, Hanaki K, Matsuo T. Nitrous oxide production in high-loading biological nitrogen removal process under low COD/N ratio condition. Wat Res. 2001;35:657–664. doi: 10.1016/S0043-1354(00)00309-2. [DOI] [PubMed] [Google Scholar]
- Jetten MSM, Horn SJ, van Loosdrecht MCM. Towards a more sustainable municipal wastewater treatment system. Water Sci Technol. 1997;35:171–180. doi: 10.2166/wst.1997.0341. [DOI] [Google Scholar]
- Kartal B, Kuenen JG, van Loosdrecht MC (2010) Engineering. Sewage treatment with anammox. Science 328:702–703 [DOI] [PubMed]
- Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM. Full-scale partial nitritation/anammox experiences – an application survey. Water Res. 2014;55:292–303. doi: 10.1016/j.watres.2014.02.032. [DOI] [PubMed] [Google Scholar]
- Liu T, Hu S, Guo J. Enhancing mainstream nitrogen removal by employing nitrate/nitrite-dependent anaerobic methane oxidation processes. Crit Rev Biotechnol. 2019;39:732–745. doi: 10.1080/07388551.2019.1598333. [DOI] [PubMed] [Google Scholar]
- Lu Y-Z, Li N, Ding Z-W, Fu L, Bai Y-N, Sheng G-P, Zeng RJ. Tracking the activity of the anammox-DAMO process using excitation emission matrix (EEM) fluorescence spectroscopy. Water. 2017;122:624–632. doi: 10.1016/j.watres.2017.06.036. [DOI] [PubMed] [Google Scholar]
- Lu Y-Z, Fu L, Li N, Ding J, Bai Y-N, Samaras P, Zeng RJ. The content of trace element iron is a key factor for competition between anaerobic ammonium oxidation and methane-dependent denitrification processes. Chemosphere. 2018;198:370–376. doi: 10.1016/j.chemosphere.2018.01.172. [DOI] [PubMed] [Google Scholar]
- Luesken FA, Sánchez J, van Alen TA, Snabria J, Op den Camp HJM, Jetten MSM, Kartal B. Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl Environ Microbiol. 2011;77:6802–6807. doi: 10.1128/AEM.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TLG, Zeeman G, Temmink H, Strous M, Op den Camp HJM, Jetten MSM. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol. 2011;92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MSM (2012) Effect of oxygen on the anaerobic methanotroph 'Candidatus Methylomirabilis oxyfera': kinetic and transcriptional analysis. Environ Microbiol 14:1024–1034 [DOI] [PubMed]
- Meng H, Wang YF, Chan HW, Wu RN, Gu JD. Co-occurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in subtropical acidic forest soils. Appl Microbiol Biotechnol. 2016;100:7727–7739. doi: 10.1007/s00253-016-7585-6. [DOI] [PubMed] [Google Scholar]
- Mulder A, van de Graaf AA, Robertson LA, Kuenen JG. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol. 1995;16:177–184. doi: 10.1111/j.1574-6941.1995.tb00281.x. [DOI] [Google Scholar]
- Nielsen M, Bollman A, Sliekers O, Jetten MSM, Schmid M, Strous M, Schmidt I, Larsen LH, Nielsen LP, Revsbech NP. Kinetics, diffusional limitation and microscale distribution of chemistry and organisms in a CANON reactor. FEMS Microbiol Ecol. 2005;51:247–256. doi: 10.1016/j.femsec.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra IC, Schouten S, Sinninghe Damsté JS, Op den Camp HJM, Jetten MSM, Strous M (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed]
- Schmid MC, Schmitz-Esser S, Jetten MSM, Wagner M (2001)16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ Microbiol 3:450–459 [DOI] [PubMed]
- Schubert CJ, Vazquez F, Lösekann-Behrens T, Knittel K, Tonolla M, Boetius A. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno) FEMS Micobiol Ecol. 2011;76:26–38. doi: 10.1111/j.1574-6941.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- Shen LD, Wu HS, Liu X, Li J. Cooccurrence and potential role of nitrite- and nitrate-dependent methanotrophs in freshwater marsh sediments. Water Res. 2017;123:162–172. doi: 10.1016/j.watres.2017.06.075. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hu S, Lou J, Lu P, Keller J, Yuan Z. Nitrogen removal from wastewater by coupling anammox and methane-dependent denitrification in a membrane biofilm reactor. Environ Sci Technol. 2013;47:11577–11583. doi: 10.1021/es402775z. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Amann RI. In: Development and application of nucleic acid probes. Nucleic acids techniques in bacterial systematics. Stackebrandt E, Goodfellow E, editors. Chichester: John Wiley & Sons; 1991. pp. 205–248. [Google Scholar]
- Strous M, van Gerven E, Zheng P, Kuenen JG, Jetten MSM. Ammonium removal from concentrated waste streams with the anaerobic ammonium oxidation (anammox) process in different reactor configurations. Water Res. 1997;31:1955–1962. doi: 10.1016/S0043-1354(97)00055-9. [DOI] [Google Scholar]
- Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van der Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449 [DOI] [PubMed]
- Vaksmaa A, Guerrero-Cruz S, van Alen TA, Cremers G, Ettwig KF, Lüke C, Jetten MSM. Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl Microbiol Biotechnol. 2017;101:7075–7084. doi: 10.1007/s00253-017-8416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MAHJ, Stultiens K, Slegers MFW, Guerrero-Cruz S, Jetten MSM, Kartal B, Op den Camp HJM (2018) Current perspectives on the application of N-damo and anammox in wastewater treatment. Curr Opin Biotechnol 50:222–227 [DOI] [PubMed]
- Versantvoort W, Guerrero-Cruz S, Speth DR, Frank J, Gambelli L, Cremers G, van Alen TA, Jetten MSM, Kartal B, Op den Camp HJM, Reimann J (2018) Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis lanthanidiphila. Front Microbiol 9. 10.3389/fmicb.2018.01672 [DOI] [PMC free article] [PubMed]
- Welte CU, Rasigraf O, Vaksmaa A, Versantvoort W, Arshad A, Op den Camp HJM, Jetten MSM, Lüke C, Reimann J. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ Microbiol Rep. 2016;8:941–955. doi: 10.1111/1758-2229.12487. [DOI] [PubMed] [Google Scholar]
- Winkler MKH, Ettwig KF, Vannecke TPW, Stultiens K, Bogdan A, Kartal B, Volcke EIP. Modelling simultaneous anaerobic methane and ammonium removal in a granular sludge reactor. Water Res. 2015;73:323–331. doi: 10.1016/j.watres.2015.01.039. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang Y. Evaluation of the impact of organic material on the anaerobic methane and ammonium removal in a membrane aerated biofilm reactor (MABR) based on the multispecies biofilm modeling. Environ Sci Pollut Res. 2017;24:1677–1685. doi: 10.1007/s11356-016-7938-9. [DOI] [PubMed] [Google Scholar]
- Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, van Niftrik L (2011) A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus 'Methylomirabilis oxyfera'. Biochem Soc Trans 39:243–248 [DOI] [PubMed]
- Xie G-J, Liu T, Cai C, Hu S, Yuan Z. Achieving high-level nitrogen removal in mainstream by coupling anammox with denitrifying anaerobic methane oxidation in a membrane biofilm reactor. Water Res. 2017;131:196–204. doi: 10.1016/j.watres.2017.12.037. [DOI] [PubMed] [Google Scholar]
- Xie G-J, Cai C, Hu S, Yuan Z. Complete nitrogen removal from synthetic anaerobic sludge digestion liquor through integrating anammox and denitrifying anaerobic methane oxidation in a membrane biofilm reactor. Environ Sci Technol. 2017;51:819–827. doi: 10.1021/acs.est.6b04500. [DOI] [PubMed] [Google Scholar]
- Zhu B, Bradford L, Huang S, Szalay A, Leix C, Weissbach M, Táncsics A, Drewes JE, Lueders T. Unexpected diversity and high abundance of putative nitric oxide dismutase (Nod) genes in contaminated aquifers and wastewater treatment systems. Appl Environ Microbiol. 2017;83(4):e02750–e02716. doi: 10.1128/AEM.02750-16. [DOI] [PMC free article] [PubMed] [Google Scholar]