Abstract
Supplementation of appropriate probiotics can improve the health and productivity of ruminants while mitigating environmental methane production. Hence, this study was conducted to determine the effects of Enterococcus faecium SROD on in vitro rumen fermentation, methane concentration, and microbial population structure. Ruminal samples were collected from ruminally cannulated Holstein–Friesian cattle, and 40:60 rice straw to concentrate ratio was used as substrate. Fresh culture of E. faecium SROD at different inclusion rates (0, 0.1%, 0.5%, and 1.0%) were investigated using in vitro rumen fermentation system. Addition of E. faecium SROD had a significant effect on total gas production with the greatest effect observed with 0.1% supplementation; however, there was no significant influence on pH. Supplementation of 0.1% E. faecium SROD resulted in the highest propionate (P = 0.005) but the lowest methane concentration (P = 0.001). In addition, acetate, butyrate, and total VFA concentrations in treatments were comparatively higher than control. Bioinformatics analysis revealed the predominance of the bacterial phyla Bacteroidetes and Firmicutes and the archaeal phylum Euryarchaeota. At the genus level, Prevotella (15–17%) and Methanobrevibacter (96%) dominated the bacterial and archaeal communities of the in vitro rumen fermenta, respectively. Supplementation of 0.1% E. faecium SROD resulted in the highest quantities of total bacteria and Ruminococcus flavefaciens, whereas 1.0% E. faecium SROD resulted in the highest contents of total fungi and Fibrobacter succinogenes. Overall, supplementation of 0.1% E. faecium SROD significantly increased the propionate and total volatile fatty acids concentrations but decreased the methane concentration while changing the microbial community abundance and composition.
Keywords: Bar-coded pyrosequencing, Enterococcus faecium, In vitro rumen fermentation, Methane concentration, Microbial diversity
Introduction
Probiotics are beneficial live microorganisms that are used as feed supplements for improving the intestinal microbial balance as well as growth performance in livestock. The term “probiotics” has been amended by the Food and Agriculture Organization/World Health Organization to “live microorganisms, which, when administered in adequate amounts, confer a health benefit on the host” (Fuller 1989). Accordingly, probiotics have been used to modulate the balance and activities of the gastrointestinal microbiota and have been developed as functional foods (Uyeno et al. 2015) as well as growth promoters to replace the widely used antibiotic and synthetic chemical-based feed supplements (Fuller 1989).
The rumen microbiome is composed of complex and diverse groups of microorganisms, which are responsible for converting fibrous plant materials into energy used by the ruminants. These microorganisms thus play an important role in animal health and productivity, food safety, and the environment. The rumen microbial community, diversity, and quantity vary depending on the host’s dietary composition and dry matter intake. Hence, supplementation of specific probiotics to ruminants can increase microbial diversity and enhance the proportion of beneficial microbes in the community. Probiotics belong to a wide variety of yeasts, Bacillus, and lactic acid bacteria (Lactobacillus, Bifidobacterium, and Enterococcus), which are now commonly used for human as well as animal consumption.
Enterococcus is one of the main genera of lactic acid bacteria, which has been used as probiotics for decades. Enterococci are ubiquitous and facultative anaerobes, which means that they can be easily cultivated and proliferate under aerobic conditions during production as well as under the anaerobic conditions found inside the rumen. Moreover, enterococci are resistant to gastric juices and bile salts (Rossi et al. 2003; Li et al. 2019) and produce useful enzymes (Sarantinopoulos et al. 2001), vitamin B12 (Li et al. 2017) and enterocin (an antimicrobial compound) (Yang et al. 2012, 2018), and inhibit harmful microorganisms (Arena et al. 2018; Mansour et al. 2018). Enterococcus faecium helps in maintaining the activity of lactate-utilizing bacteria and stimulates the growth of rumen microbes, which can, in turn, increase the glucogenic propionate energy supply for host ruminants (Pang et al. 2014), while also producing antimicrobial agents (Lauková et al. 1993; Wang et al. 2018).
We previously isolated an E. faecium SROD strain (KCCM11098P) (Kim et al. 2016) that showed promise as a fumarate reductase-producing enterococci bacterium, and also resulted in enhanced production of total volatile fatty acids (VFAs) while decreasing the concentration of methane during rumen fermentation in vitro. However, the effects of E. faecium SROD on the ruminal microbiome remain unclear. Therefore, in the present study, we applied molecular techniques of quantitative real-time polymerase chain reaction (qRT-PCR) and pyrosequencing to determine these effects and gain a better general understanding of rumen microbes’ symbiosis and functions. In particular, we added E. faecium SROD at different inclusion rates to an in vitro rumen fermentation system and determined the effects on methane concentration, microbial diversity, and population structure.
Materials and methods
Cultivation of E. faecium SROD
Enterococcus faecium SROD KCCM11098P, which was previously isolated in our laboratory (Kim et al. 2016), was used in this study. A frozen stock culture of E. faecium SROD was thawed and re-cultivated at a 1% inoculum using deMan, Rogosa, and Sharpe broth (Becton–Dickinson and Company, Difco, Sparks, MD, USA), and then incubated in a horizontal shaking incubator (120 rpm) (Hanbaek Scientific Co., Korea) at 37 °C. E. faecium SROD was then subcultured three times on the same medium to ensure full activity. The cell growth was monitored based on an optical density value at 600 nm of approximately 1.50, which is equivalent to 7.0 × 108 colony-forming units (CFU)/mL.
Rumen fluid collection and in vitro fermentation
All animal care procedures conducted for this study followed protocols approved by the Sunchon National University Committee on Animal Care. Three ruminally cannulated Holstein–Friesian cows with body weights of 600 ± 47 kg that were fed twice daily with feed concentrate (NongHyup Co., Anseong, Korea) and rice straw (2:8 ratio) were used in this study. Three hours after morning feeding, the ruminal contents were collected, strained through four layers of surgical gauze, placed in amber bottles, maintained at 39 °C, and then immediately transported to the laboratory. Asanuma buffer used in this study was composed of (per L) 0.45 g K2HPO4, 0.45 g KH2PO4, 0.9 g (NH4)2SO4, 0.12 g CaCl2·2H20, 0.19 g MgSO4·7H2O, 1.0 g trypticase peptone (BBL; Becton–Dickinson), 1.0 g yeast extract (Difco Laboratories, MI, USA), and 0.6 g cysteine HCl (Asanuma et al. 1999). It was prepared, autoclaved at 121 °C for 15 min, maintained in a 39 °C water bath, flushed with N2 for 30 min and continuously flushed until it was transferred to serum bottles. The pH was adjusted to 6.9 using 10 N NaOH. The mixed buffered rumen fluid (1:3 rumen fluid:buffer ratio) was anaerobically transferred (100 mL) to 160 mL serum bottles containing the substrates. The substrates were grinded and sieved to 1 mm particle size and then 1 g dry matter of rice straw and concentrate at 40:60 ratio was put into the serum bottles. The following inocula treatments were conducted under a stream of O2-free N2: no addition (control; Con), and 0.1% (T1), 0.5% (T2), and 1.0% (T3) supplementation of E. faecium SROD (7.0 × 108 colony-forming units (CFU)/mL). The bottles were then sealed and incubated at 39 °C (Mamuad et al. 2014). Five replicates were established for all treatments and incubation times.
In vitro rumen fermentation parameters
In vitro rumen parameters were sampled in each serum bottle at the end of each incubation period. Total gas was measured using a press and sensor machine (Laurel Electronics, Inc., Costa Mesa, CA, USA), and the pH was determined using a Pinnacle series M530p meter (Schott Instruments, Mainz, Germany). Gas samples were collected for determination of methane concentration, and the in vitro rumen fermenta was collected for ammonia–nitrogen (NH3–N), VFA, and molecular analyses. One millilitre of gas sample contained in vacuum tubes was used to determine the methane concentration through gas chromatography (GC; HP 5890 system, Agilent Technologies, Foster City, CA, USA) with a thermal conductivity detector and an HP stainless packed GC Porapak Q 80/100 outer dimension 1/8 in × inner dimension 2.0 mm, length 3.05 m (10 ft) with 200 °C inlet, 200 °C detector, 40 °C oven temperature, and 3 mL/min N2 carrier gas flow. Estimation of the amount of methane produced was conducted using the formula described by Ørskov and McDonald (1979). Peaks identification and standards analyses were performed using the procedure described by Kim et al. (2016). Gas standards of known composition were used to identify the peaks. Standards with R2 = 0.999 were prepared prior to sample analysis.
Samples were measured from each of the serum bottles at different incubation times. Two 1-mL in vitro rumen fermenta from each serum bottles were immediately centrifuged after sampling at 16,609×g for 10 min at 4 °C using a Micro 17TR centrifuge (Hanil Science Industrial Co. Ltd., Incheon, Korea). Then, supernatant and pellet were separated, kept in 1.5 Eppendorf tubes and stored at − 80 °C until subjected to NH3–N, VFA, and molecular analyses. The supernatant was used for determination of NH3–N (Chaney and Marbach 1962) and VFA concentrations (Kim et al. 2016) with spectrophotometry (Libra S22 spectrophotometer, Biochrom Ltd., Cambridge, UK) and high-performance liquid chromatography (HPLC; Agilent Technologies 1200 series, USA), respectively. The samples for VFA analysis were filtered through 0.2-μm Millipore filters. HPLC had a UV detector set at 210 and 220 nm while MetaCarb 87H (300 × 7.8 mm; Agilent, Germany) column was used in the determination of fermentation products with the application of 0.0085 N H2SO4 solvent as a buffer at a flow rate of 0.6 mL/min and a column temperature of 35 °C. This was done according to the methods of Tabaru et al. (1988) and Han et al. (2005). Standard was made at 0.999 (R2) before analysis. Standards with R2 = 0.999 were prepared prior to sample analysis. The VFA concentration in mM was calculated in ppm divided by the molecular weight.
qRT-PCR
Total genomic deoxyribonucleic acid (DNA) from the rumen pellets was extracted using a Fast-DNA spin kit (MPbio) according to the manufacturer instructions. The general bacteria, general fungi, methanogens, protozoa, Fibrobacter succinogenes, and Ruminococcus flavefaciens were enumerated using qRT-PCR on the DNA extracted from the in vitro rumen fermenta using the primers reported in Denman and McSweeney (2006) and Denman et al. (2007) (Table 1). Amplification was performed in triplicates using the Eco Real-Time PCR System (Illumina, USA) with QuantiSpeed SYBR no-Rox Kit (PhileKorea, Korea) in final reaction volumes of 20 μL.
Table 1.
Real time PCR primers used for the quantification of microbial population
| Target gene | Primer sequence | Length | Initial denaturation | Denaturation | Annealing | Extension | Cycles | Reference |
|---|---|---|---|---|---|---|---|---|
| General bacteria | Denman and McSweeney (2006) | |||||||
| F sequence (5′-3′) | CGGCAACGAGCGCAACCC | 130 |
95 °C 2 min |
95 °C 15 s |
60 °C 60 s |
72 °C 30 s |
40 | |
| R sequence (5′-3′) | CCATTGTAGCACGTGTGTAGCC | |||||||
| General anaerobic fungi | ||||||||
| F sequence (5′-3′) | GAGGAAGTAAAAGTCGTAACAAGGTTTC | 120 |
95 °C 2 min |
95 °C 15 s |
60 °C 60 s |
72 °C 30 s |
40 | |
| R sequence (5′-3′) | CAAATTCACAAAGGGTAGGATGATT | |||||||
| Methanogens | ||||||||
| F sequence (5′-3′) | TTCGGTGGATCDCARAGRGC | 140 |
95 °C 2 min |
95 °C 15 s |
60 °C 60 s |
72 °C 30 s |
40 | |
| R sequence (5′-3′) | GBARGTCGWAWCCGTAGAATCC | |||||||
| Fibrobacter succinogenes | ||||||||
| F sequence (5′-3′) | GTTCGGAATTACTGGGCGTAAA | 121 |
95 °C 2 min |
95 °C 15 s |
60 °C 60 s |
72 °C 30 s |
40 | |
| R sequence (5′-3′) | CGCCTGCCCCTGAACTATC | |||||||
| Ruminococcus flavefaciens | ||||||||
| F sequence (5′-3′) | CGAACGGAGATAATTTGAGTTTACTTAGG | 132 |
95 °C 2 min |
95 °C 15 s |
60 °C 60 s |
72 °C 30 s |
40 | |
| R sequence (5′-3′) | CGGTCTCTGTATGTTATGAGGTATTACC | |||||||
Bar-coded pyrosequencing, PCR, and data analysis
The amplification of bacterial and archaeal 16S rRNA genes for bar-coded pyrosequences and subsequent data analysis were performed according to the procedure described by Lee et al. (2012). The primer sets used for amplification were Bac9F (5′-adaptor B-AC-GAG TTT GAT CMT GGC TCA G-3′)/Bac541R (5′-adaptor A-X-AC-WTT ACC GCG GCT GG-3′) and Arc21F (5′-adaptor B-GA-TCC GGT TGA TCC YGC CGG-3′)/Arc519R (5′-adaptor A-X-GA-GGT DTT ACC GCG GCK GCT G-3′) (Delong 1992; Sørensen and Teske 2006; Roesch et al. 2007; Chun et al. 2010). Unique 7–11 bp barcode sequences, denoted as “X” in the primer sequences above, were inserted between the 454 Life Sciences adaptor A sequence and the common linkers AC and GA. The polymerase chain reaction (PCR) products were purified and quantified using a PCR purification kit (Solgent, Korea) and an enzyme-linked immunosorbent assay reader equipped with a Take3 multivolume plate, respectively. Equal amounts of purified PCR amplicons from each sample were prepared as a composite DNA sample. The samples were sent to Macrogen (Korea) for pyrosequencing using a 454 GS-FLX Titanium system (Roche, Germany), and the sequencing data were analyzed using the RDP pyrosequencing pipeline (http://pyro.cme.msu.edu/) (Cole et al. 2009). The aligned sequences were clustered into operational taxonomic units (OTUs), defined at 97% similarity, using the complete-linkage clustering tool. The Shannon–Weaver index (Shannon and Weaver 1964), Chao 1 biodiversity indices (Chao 1987), and evenness index and rarefaction analyses were determined using the RDP pyrosequencing pipeline. In addition, the processed sequences were taxonomically classified using the RDP naive Bayesian rRNA classifier (Wang et al. 2007) based on an 80% confidence threshold.
Statistical analyses
Data were statistically evaluated using Proc Glimmix for a complete randomized design. The experiment was done twice and the control and treatments were conducted in five replicates. Least square means was used to identify differences among control and treatments. Orthogonal contrasts were used to examine the differences between the control and treatment groups. The linear and quadratic effects of E. faecium SROD supplementation were analyzed using orthogonal polynomial coefficients to describe the functional relationships among the control and treatment levels. P ≤ 0.05 indicated statistical significance. All analyses were carried out using Statistical Analysis Systems (SAS) software version 9.4 (SAS Institute 2012).
Results
Effects of E. faecium SROD supplementation on rumen fermentation in vitro
The volume of total gas produced was found highest (P = 0.017) in supplementation of 0.1% E. faecium SROD with 59.45 mL and lowest in control with 55.15 mL (Table 2). Although the addition of E. faecium SROD at increasing inclusion rates tended to result in a lower pH value than the control, the difference was not statistically significant. NH3–N concentrations were also comparable among the control and treatment groups. However, the methane concentration was linearly correlated (P = 0.001) with E. faecium SROD addition, and was lowest (P = 0.001) with supplementation of 0.1%, followed by 0.5% E. faecium SROD, and was highest in the control and 1.0% E. faecium SROD groups with no significant difference between them. Addition of 0.1% E. faecium SROD resulted in the lowest carbon dioxide (P = 0.053) with 3.70 mM/mL but the highest (P < 0.001, P = 0.005) concentrations of total VFAs and propionate with 55.40 mM and 14.15 mM, respectively (Table 3). Acetate concentration increased (P < 0.001) with increasing inclusion rate of E. faecium SROD with linearly (P < 0.020) and quadratically (P < 0.043) correlation of the concentration and inclusion rate. Butyrate (P < 0.018) and total VFA (P < 0.001) concentrations were comparatively higher than control while propionate (P < 0.005) concentration was found the highest in addition of 0.1% E. faecium SROD.
Table 2.
Total gas, pH, ammonia–nitrogen, methane, and carbon dioxide concentrations during in vitro rumen fermentation (12 h)
| Parameters | Treatments | SEM | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Con | T1 | T2 | T3 | Treatment | Linear | Quadratic | ||
| Total gas (mL) | 55.15b | 59.45a | 56.36ab | 58.16ab | 0.869 | 0.017 | 0.535 | 0.419 |
| pH | 5.43 | 5.39 | 5.38 | 5.38 | 0.083 | 0.083 | 0.037 | 0.734 |
| Ammonia–nitrogen (mM) | 22.28 | 22.63 | 20.94 | 23.16 | 0.790 | 0.386 | 0.271 | 0.663 |
| Methane (mM/mL) | 11.20a | 9.12b | 9.93b | 10.27ab | 0.238 | 0.001 | 0.001 | 0.074 |
| Carbon dioxide (mM/mL) | 4.18ab | 3.70b | 4.09ab | 4.30a | 0.145 | 0.053 | 0.035 | 0.555 |
Con control (no addition), T1 0.1% E. faecium, T2 0.5% E. faecium, T3 1.0% E. faecium, SEM standard error of the mean
Different superscript letters indicate a statistically significant difference. P-value, calculated probability
Table 3.
Volatile fatty acid concentrations during in vitro rumen fermentation (12 h)
| Parameters | Treatments | SEM | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Con | T1 | T2 | T3 | Treatment | Linear | Quadratic | ||
| Acetate (mM) | 32.52c | 33.91b | 34.97ab | 35.66a | 0.286 | < 0.001 | 0.020 | 0.043 |
| Propionate (mM) | 9.97b | 14.15a | 10.83b | 10.39b | 0.180 | 0.005 | 0.382 | 0.013 |
| Butyrate (mM) | 5.87b | 7.20a | 7.41a | 7.38a | 0.295 | 0.018 | 0.042 | 0.142 |
| A/P ratio (mM) | 3.11 | 2.98 | 3.48 | 3.21 | 0.211 | 0.474 | 0.627 | 0.680 |
| Total VFA (mM) | 49.19b | 55.40a | 55.38a | 54.17a | 0.583 | < 0.001 | 0.006 | 0.039 |
Con control (no addition), T1 0.1% E. faecium, T2 0.5% E. faecium, T3 1.0% E. faecium, SEM standard error of the mean
Different superscript letters indicate a statistically significant difference. P-value, calculated probability
Effects of E. faecium SROD supplementation on the in vitro rumen microbial community composition and abundance
Comparable quantities of general bacteria were observed among the control and treatment groups (Table 4). Between the cellulolytic bacteria determined, there were more log copies of F. succinogenes than R. flavefaciens. However, supplementation of 0.1% E. faecium SROD resulted in the significantly highest quantities of general fungi (P = 0.026), F. succinogenes (P = 0.010), and R. flavefaciens (P = 0.008). The control group had the lowest quantities of general fungi (P = 0.026) and F. succinogenes (P = 0.010) but the highest log copy numbers of methanogens (P = 0.048), which showed a significant linear decrease with increasing supplementation of E. faecium SROD. In addition, supplementation of 0.1% E. faecium SROD resulted in lower quantities of methanogen (P = 0.048) than control.
Table 4.
Quantification of general bacteria, general fungi, methanogens, Fibrobacter succinogenes, and Ruminococcus flavefaciens by real-time PCR
| Target genes | Treatments (log10 copies number) | SEM | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| Con | T1 | T2 | T3 | Treatment | Linear | Quadratic | ||
| General bacteria | 8.35 | 8.41 | 8.34 | 8.37 | 7.355 | 0.678 | 0.956 | 0.350 |
| General anaerobic fungi | 3.60c | 4.01a | 3.82bc | 3.97ab | 2.875 | 0.026 | 0.101 | 0.008 |
| Methanogens | 1.44a | 1.19b | 1.08b | 1.01b | 0.454 | 0.048 | 0.032 | 0.085 |
| F. succinogenes | 4.37c | 5.21a | 4.83bc | 4.93b | 3.958 | 0.010 | 0.028 | 0.005 |
| R. flavefaciens | 2.29b | 3.18a | 2.83b | 2.72b | 2.060 | 0.008 | 0.017 | 0.012 |
Con control (no addition), T1 0.1% E. faecium, T2 0.5% E. faecium, T3 1.0% E. faecium, SEM standard error of the mean
Different superscript letters indicate a statistically significant difference. P-value, calculated probability
The barcoded pyrosequencing results of 24 PCR amplicons (NCBI SRA accession PRJNA505970; NCBI Temporary Submission ID: SUB4770572) of the 16S rRNA genes for the bacterial and archaeal communities are shown in Tables 5 and 6, respectively. After filtering, quality control, and chimera removal, the average number of reads, number of operational taxonomic units (OTUs), Chao index, Shannon–Weaver index, evenness, and average read length were 4238.13, 2213.25, 6573.85, 7.15, 0.93, and 476.33 for bacterial communities, and were 4177.50, 116.92, 144.80, 2.93, 0.62, and 490.67 for archaeal communities, respectively (Tables 5 and 6). Rarefaction lines in all samples extended all the way to the right end of the axis.
Table 5.
Summary of the pyrosequencing data and statistical analysis of bacterial communities of Enterococcus faecium SROD
| Treatments | No. of reads | No. of OTUs | Chao | Shannon–Weaver index (H’) | Evenness | Avg. read length |
|---|---|---|---|---|---|---|
| Control | 4340.00 | 2228.50 | 6333.04 | 7.16 | 0.93 | 475.33 |
| 0.10% | 4137.00 | 2175.50 | 6498.96 | 7.13 | 0.93 | 478.00 |
| 0.50% | 3959.50 | 2056.50 | 6420.18 | 7.06 | 0.93 | 476.67 |
| 1.00% | 4516.00 | 2392.50 | 7043.22 | 7.24 | 0.93 | 475.33 |
OTU operational taxonomic units
OTUs were calculated by the RDP pipeline with a 97% OTU cut-off of the 16S rRNA gene sequences. Diversity indices of the microbial communities and numbers of phyla and genera were calculated using the RDP pyrosequencing pipeline based on the 16S rRNA gene sequences
Table 6.
Summary of the pyrosequencing data and statistical analysis of archaeal communities of Enterococcus faecium SROD
| Treatments | No. of reads | No. of OTUs | Chao | Shannon–Weaver index (H’) | Evenness | Avg read length |
|---|---|---|---|---|---|---|
| Control | 2723.33 | 102.00 | 125.95 | 2.92 | 0.63 | 490.67 |
| 0.10% | 5105.67 | 126.67 | 145.14 | 2.95 | 0.61 | 487.67 |
| 0.50% | 4866.33 | 122.67 | 164.85 | 2.94 | 0.61 | 493.33 |
| 1.00% | 4014.67 | 116.33 | 143.26 | 2.91 | 0.61 | 491.00 |
OTU operational taxonomic units
OTUs were calculated by the RDP pipeline with a 97% OTU cut-off of the 16S rRNA gene sequences. Diversity indices of the microbial communities and numbers of phyla and genera were calculated using the RDP pyrosequencing pipeline based on the 16S rRNA gene sequences
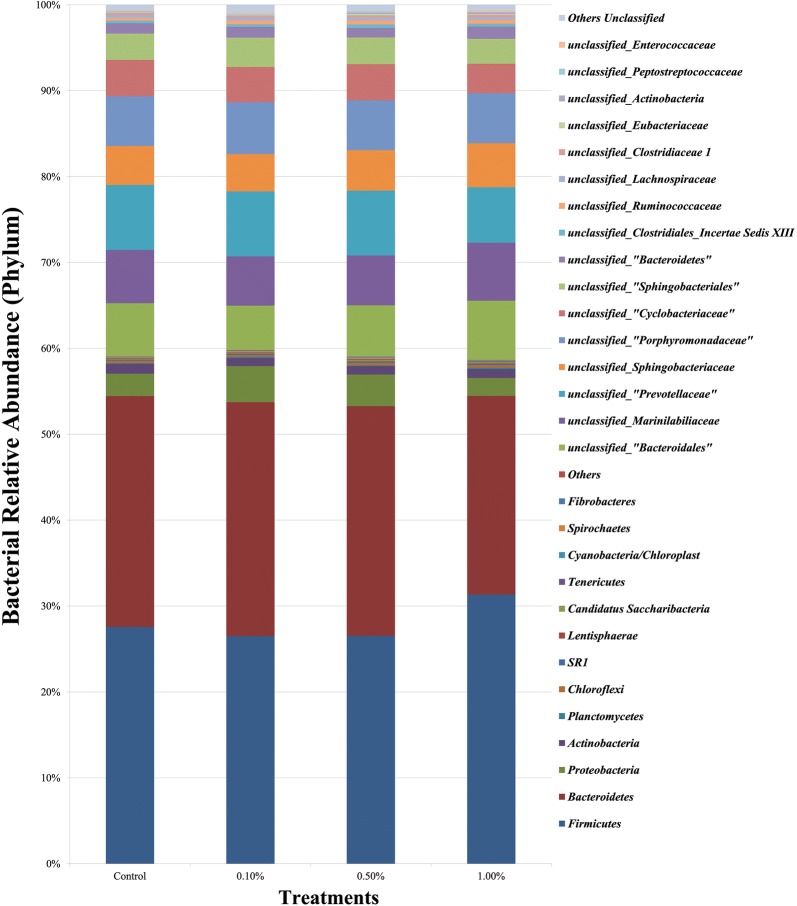
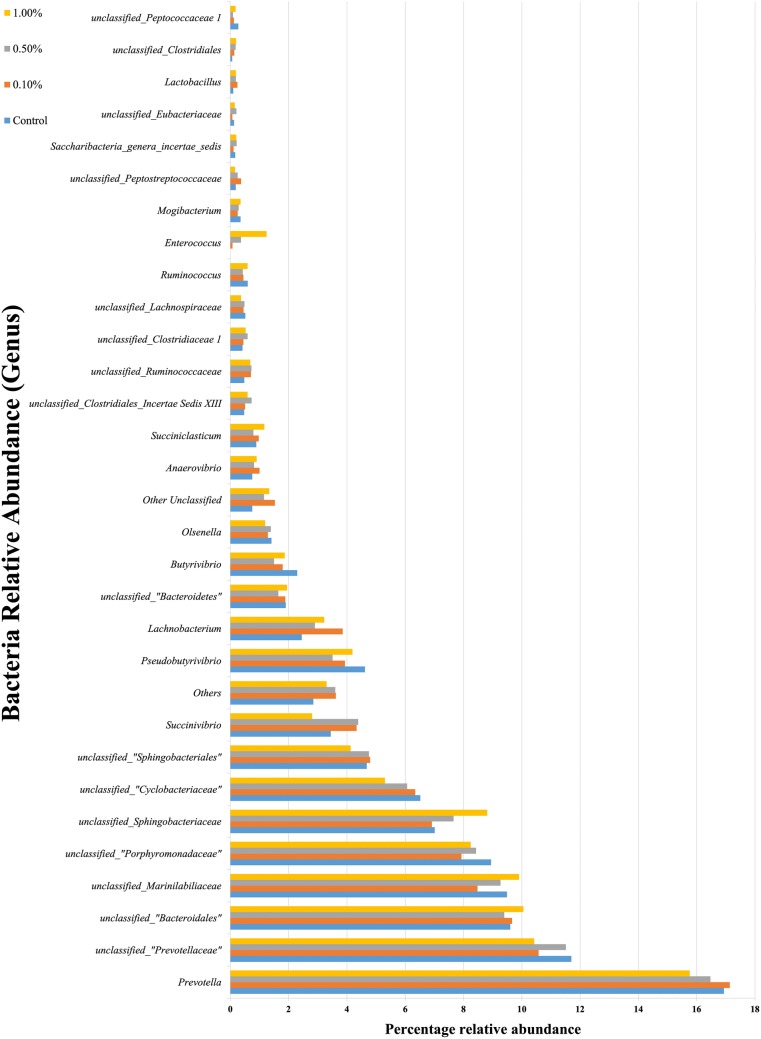
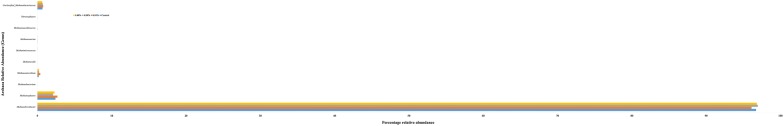
Bioinformatics analysis revealed that the bacterial sequences were predominantly affiliated with two phyla, Bacteroidetes and Firmicutes (Fig. 1), while the archaeal sequences were predominantly affiliated with phylum Euryarchaeota. Notably, a very low abundance of Thaumarchaeota was observed only in the group supplemented with 0.1% E. faecium SROD. At the genus level, Prevotella (15–17%) and Methanobrevibacter (96%) dominated the bacterial and archaeal communities’ composition of the in vitro rumen fermenta, respectively (Figs. 2 and 3). The relative abundance of Anaerovibrio, Enterococcus, Lachnobacterium, unclassified Clostridiales Incertae Sedis XII, unclassified Clostridiaceae 1, unclassified Clostridiales, and unclassified Ruminococcaceae also increased with supplementation of E. faecium SROD. Notably, the Enterococcus relative abundance increased with increasing inclusion rate of E. faecium SROD from 0% for the control, to 0.075%, 0.366%, and 1.240%, respectively, in each treatment group (Fig. 2). In addition, supplementation of 0.1% E. faecium SROD increased the relative abundance of Methanomicrobium to the greatest extent (0.386%), followed by 0.5% E. faecium SROD (0.211%); similar relative abundances were detected with 1.0% E. faecium SROD and the control of 0.184% and 0.175%, respectively.
Fig. 1.
Bacterial phylum-level compositions of the control and Enterococcus faecium SROD-supplemented rumen fermenta. The data portray phylum-level 16S rRNA pyrotagged gene sequences. Sequences were classified using the RDP naive Bayesian rRNA Classifier with an 80% confidence threshold
Fig. 2.
Bacterial genus-level compositions of the control and Enterococcus faecium SROD-supplemented rumen fermenta. The data portray genus-level 16S rRNA pyrotagged gene sequences. Sequences were classified using the RDP naive Bayesian rRNA Classifier with an 80% confidence threshold. The minor group in the panel is composed of genera with a percentage of reads < 0.4% of the total reads in all samples
Fig. 3.
Archaeal genus-level compositions of the control and Enterococcus faecium SROD-supplemented rumen fermenta. The data portray genus-level 16S rRNA pyrotagged gene sequences. Sequences were classified using the RDP naive Bayesian rRNA Classifier with an 80% confidence threshold. The minor group in the panel is composed of genera showing a percentage of reads < 0.4% of the total reads in all samples
Discussion
Effects of E. faecium SROD supplementation on rumen fermentation in vitro
The ruminal microbiome plays an important role not only in animal health and productivity but also in food and environmental safety. Enhancing the rumen microflora through probiotic supplementation stimulates fermentation. With prolonged culturing, our newly isolated probiotic strain E. faecium SROD displays full activity after 12 h of incubation (Kim et al. 2016); hence, in the present study, we collected and analyzed the fermentation parameters after 12 h of culture. The total gas production level is an indication of the fermentation rate. Since supplementation of 0.1% E. faecium SROD increased the gas production compared to the control, this probiotic appears to affect the fermentation rate of the rumen in vitro. This result was supported by Shi et al. (2017) claims that fermentation by inoculating E. faecium is an effective approach in improving the quality of corn-soybean meal mixed feed. The lower tendency of the pH with supplementation of E. faecium SROD could be related to the production of organic acids by the bacterium. Indeed, a previous study on E. faecium demonstrated that it increased the levels of organic acids such as acetate, propionate, and succinate (Ribeiro et al. 2009).
The lack of an effect on the ammonia–N concentration with supplementation of E. faecium SROD at all inclusion rates indicates that the probiotic does not influence the ruminal N-metabolism level (Pang et al. 2014). However, the methane concentration was significantly reduced with the lowest inclusion rate of 0.1% E. faecium SROD, which comparable to 0.5% E. faecium SROD inclusion rate. Moreover, we found that the group with the highest inclusion rate of 1.0% E. faecium SROD had a comparable methane concentration with that of the control, which is in opposition to our previous result of Kim et al. (2016) wherein inoculation of 1.0% E. faecium SROD significantly reduced the methane concentration. The difference in these results might be due to the different substrate used, which was maize silage in the previous study but was feed concentrate and rice straw at a 60:40 ratio in the present study. E. faecium SROD is a fumarate reductase-producing bacteria (Kim et al. 2016), which competes with methanogens in utilizing H2 in the rumen. H2 serves as electron donor for reduction of fumarate to succinate. Hence, lower methane concentration was observed in 0.1% E. faecium SROD inclusion.
Supplementation of E. faecium SROD increased the concentration of VFAs, and increasing inclusion rates of E. faecium SROD increased the acetate concentration. E. faecium SROD converts fumarate to succinate and thus, increase in available succinate for propionate production. Also, fumarate can be converted into propionate and acetate via different pathways (Demeyer and Henderickz 1967). Acetate is a precursor of milk constituents (Kleiber et al. 1952). Nocek and Kautz (2006) demonstrated an increase in milk yield by 2.3 L per cow per day following dietary supplementation with 5 × 109 CFU of E. faecium and 2 × 109 yeast (Saccharomyces cerevisiae) cells per cow per day. Moreover, the increased acetate concentration observed in the E. faecium SROD-supplemented treatment groups compared to the control was comparable that observed in our previous study on E. faecium SROD (Kim et al. 2016). However, a higher acetate concentration was observed in the present study. These results indicate that supplementation of E. faecium SROD as a probiotic can improve the milk fat and milk yield in dairy animals.
Higher concentrations of propionate, butyrate, and total VFAs concentrations were observed in the E. faecium SROD-supplemented groups than the control, which is line with the results reported by Kim et al. (2016) and Pang et al. (2014). It has been reported by Ribeiro et al. that E. faecium increased the levels of organic acids such as acetate, propionate, and succinate that made E. faecium as dietary supplement for domestic animals worldwide. Oetzel et al. (2007) reported that E. faecium plus S. cerevisiae increased milk fat percentages when used as direct fed microbe (DFM) for first lactation cows and increased milk protein percentages for second and greater lactation cows. Moreover, E. faecium with yeast as DFM increased dry matter intake, milk yield, and milk protein content during the postpartum period (Nocek et al. 2003).
Volatile fatty acids are important contributors to the overall performance of the animal because they improve growth, production, and health simultaneously. With these increase in VFAs means also increase in energy available for the animal, which explains the improvement of breast and legs yield, as well as the water holding capacity of meat but low abdominal fat deposition in dietary supplementation of E. faecium (Zampiga et al. 2018). The significant increase in butyrate concentration (Table 3) during fermentation is well known for many regulatory and immunological functions in cattle. Also, decreased in acetate to propionate ratio in this study indicates increased in the positive energy balance. Apás et al. (2010) reported that inclusion of a probiotic containing a mixture of E. faecium DDE 39, Lactobacillus reuteri DDL 19, L. alimentarius DDL 48, and Bifidobacterium bifidum DDBA resulted in improvement in average body weight by 9% when fed to goats for 8 weeks, commencing at 75 days of age, and increased body weight gain and improved feed use efficiency were observed with supplementation of E. faecium, L. acidophilus, L. plantarum, L. salivarius, L. casei/paracasei, or Bifidobacterium spp. to young calves compared with control groups (Frizzo et al. 2011).
Effects of E. faecium SROD supplementation on rumen in vitro microbial abundance and community composition
Microorganisms inhabiting in the rumen contribute directly or indirectly to dietary organic matter degradation (Wang et al. 2017). F. succinogenes and R. flavefaciens are two of the major cellulolytic bacterial species found in the rumen, and F. succinogenes was reported to be present in greater quantities than R. flavefaciens (Koike and Kobayashi 2001), which was confirmed in the present study. Moreover, the lower quantities of methanogens observed with supplementation of E. faecium SROD, along with the increase of cellulolytic bacteria, F. succinogenes and R. flavefaciens, and general fungi quantities with supplementation of 0.1% E. faecium SROD support that the reduction in methane concentration was directly due to the activity of E. faecium SROD in reducing the abundance of methanogenic bacteria. Lower levels of supplementation of E. faecium SROD enhanced the cellulolytic bacteria F. succinogenes and R. flavefaciens, and the general fungi, which could explain the significant decrease in methane concentration. This decrease in methane production is similar in Chaucheyras-Durand et al. (2010) study when F. succinogenes was inoculated in lambs. F. succinogenes is a non-H2-producing species (Chaucheyras-Durand et al. 2010), which is a substrate for methane production and hence, lower methane concentration was observed in this study. However, increased supplementation of E. faecium SROD (1%) had comparable methane concentration to control but it tended decrease. This might be due to increase in population of R. flavefaciens, which might increase available H2 and electrons for methanogenesis.
Analysis of the rumen microbiome is of great importance for understanding the microbial ecosystem at large, which could be accomplished through determination of the microbial communities and their symbiosis. To best correlate and describe the results of in vitro rumen fermentation parameters with microbial abundance and community composition at greater resolution, we utilized a new and high-throughput molecular technique. Further adoption of high-throughput techniques can lay the foundation for new advancements in ruminant production by gaining a deeper-level microbial understanding of proven nutritional strategies (McCann et al. 2014). We conducted a sample-based rarefaction test to assess whether the samples and sequences provided efficient OTU coverage. The OTU is an operational definition of a species or a group of species that is often used when only DNA sequencing data are available. The alpha rarefaction curve constructed in this study became flattered to the right of the axis, which indicates that an efficient and reasonable number of reads had been used in the analysis; thus, additional sequencing was not necessary.
Through calculation of the number of OTUs and the measure of species richness estimators, we estimated the diversity within samples. The Chao index estimates the richness of the diversity, while the Shannon–Weaver index takes into account the number and evenness of species present. On the other hand, the Simpson index depicts probability of the that two randomly selected individuals in the habitat will belong to the same species. In this study, the pyrosequencing data demonstrated comparable bacterial or archaeal communities, in terms of diversity, richness, number, and evenness of species, among treatments. However, higher communities, diversity, richness, number and evenness of species were observed in bacteria than in archaea.
Bacteroidetes and Firmicutes were present at the highest relative abundance at the phylum level for all groups, which is consistent with the findings of Jami et al. (2013) and Wang et al. (2016). Naas et al. (2014) reported that Bacteroidetes specialize in lignocellulose degradation and are associated with butyrate production; however, Firmicutes represent the major butyrate-producing group of microbes (Naas et al. 2014). The dominance of Euryarchaeota in this study is in line with the results of Wang et al. (2016). Thaumarchaeota, which was observed only under supplementation with 0.1% E. faecium SROD, represents a group of chemolithoautotrophic ammonia-oxidizers (Spang et al. 2010) and is likely a dominant producer of the critical vitamin B12 (Doxey et al. 2015). Supplementation of E. faecium SROD also enhanced the growth of Anaerovibrio, Enterococcus, Lachnobacterium, unclassified Clostridiales Incertae Sedis XII, unclassified Clostridiaceae 1, unclassified Clostridiales, and unclassified Ruminococcaceae by increasing their relative abundances. The increased relative abundance of Enterococcus with increasing inclusion rates of E. faecium SROD indicates that E. faecium SROD grew well anaerobically and in symbiosis with other microbes.
The inclusion of 0.1% E. faecium SROD increased the concentrations of propionate and total VFAs but decreased the methane concentration during in vitro rumen fermentation. Also, a significant increase in butyrate concentration indicates regulatory and immunological functions in cattle. These findings were validated by the determination of the quantities of specific microbes related to the production of these components. In particular, the quantities of general fungi, F. succinogenes, and R. flavefaciens increased with the inclusion of 0.1% E. faecium SROD, while lower quantities of methanogens were observed in the treatment groups compared to the control. Using a pyrosequencing technique, we further demonstrated that supplementation of E. faecium SROD enhances the growth of Anaerovibrio, Enterococcus, Lachnobacterium, unclassified Clostridiales Incertae Sedis XII, unclassified Clostridiaceae 1, unclassified Clostridiales, and unclassified Ruminococcaceae by increasing their relative abundances.
Overall, these results demonstrate that E. faecium SROD is a potentially valuable feed additive for ruminal methane mitigation and to enhance the productivity of the ruminant. This will further help to reduce the use of harmful chemicals and antibiotics. To further evaluate the potential of E. faecium SROD, in vivo trial will be done to determine the growth performance, efficiency, and their effect on rumen microbiome and population of the animals as well as the methane production using Greenfeed technology.
Acknowledgements
Not applicable.
Abbreviations
- NH3–N
ammonia–nitrogen
- CFU
colony-forming units
- GC
gas chromatography
- OUT
operational taxonomic units
- qRT-PCR
quantitative real-time polymerase chain reaction
- SEM
standard error of the mean
- VFAs
total volatile fatty acids
- Con
Control
- T1
0.1% E. faecium
- T2
0.5% E. faecium
- T3
1.0% E. faecium
Authors’ contributions
LLM, SSL and SHK conceived and designed the study. LLM and AAB conducted research work and acquisition of data. LLM, AAB, and SHK analyzed and/or interpreted of data. LLM drafted the manuscript. ZY, KKC, SBK, and KL reviewed and revised of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry, (Project No. 317016-03-2-WT011), Republic of Korea.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
The experimental design and procedures in this study were reviewed and approved by the Animal Care and Use Committee of the Department of Animal Science and Technology, Sunchon National University Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Apás AL, Dupraz J, Ross R, González SN, Arena ME. Probiotic administration effect on fecal mutagenicity and microflora in the goat’s gut. J Biosci Bioeng. 2010;110:537–540. doi: 10.1016/j.jbiosc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Arena MP, Capozzi V, Russo P, Drider D, Spano G, Fiocco D. Immunobiosis and probiosis: antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl Microbiol Biotechnol. 2018;102:9949–9958. doi: 10.1007/s00253-018-9403-9. [DOI] [PubMed] [Google Scholar]
- Asanuma N, Iwamoto M, Hino T. Effect of the addition of fumarate on methane production by ruminal microorganisms in vitro. J Dairy Sci. 1999;82:780–787. doi: 10.3168/jds.S0022-0302(99)75296-3. [DOI] [PubMed] [Google Scholar]
- Chaney L, Marbach P. Modified reagents of urea and for determination ammonia. Clin Chem. 1962;8:130–132. doi: 10.1021/AC60252A045. [DOI] [PubMed] [Google Scholar]
- Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. doi: 10.2307/2531532. [DOI] [PubMed] [Google Scholar]
- Chaucheyras-Durand F, Masséglia S, Fonty G, Forano E. Influence of the composition of the cellulolytic flora on the development of hydrogenotrophic microorganisms, hydrogen utilization, and methane production in the rumens of gnotobiotically reared lambs. Appl Environ Microbiol. 2010;76:7931–7937. doi: 10.1128/AEM.01784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Kim KY, Lee J-H, Choi Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 2010;10:101. doi: 10.1186/1471-2180-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeyer D, Henderickz H. Competitive inhibition of in vitro methane production by mixed rumen bacteria. Arch Int Physiol Biochim. 1967;75:157. [PubMed] [Google Scholar]
- Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol. 2007;62:313–322. doi: 10.1111/j.1574-6941.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9:461–471. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzo LS, Zbrun MV, Soto LP, Signorini ML. Effects of probiotics on growth performance in young calves: a meta-analysis of randomized controlled trials. Anim Feed Sci Technol. 2011;169:147–156. doi: 10.1016/j.anifeedsci.2011.06.009. [DOI] [Google Scholar]
- Fuller R. A review-probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. doi: 10.1111/j.1365-2672.1989.tb05105.x. [DOI] [PubMed] [Google Scholar]
- Han SK, Kim SH, Shin HS. UASB treatment of wastewater with VFA and alcohol generated during hydrogen fermentation of food waste. Process Biochem. 2005 doi: 10.1016/j.procbio.2005.01.005. [DOI] [Google Scholar]
- Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013;7:1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Mamuad LL, Kim D-W, Kim S-K, Lee S-S. Fumarate reductase-producing enterococci reduce methane production in rumen fermentation in vitro. J Microbiol Biotechnol. 2016;26:558–566. doi: 10.4014/jmb.1512.12008. [DOI] [PubMed] [Google Scholar]
- Kleiber M, Smith AH, Black AL, Brown MA, Tolbert BM. Acetate as a precursor of milk constituents in the intact dairy cow. J Biol Chem. 1952;197:371–379. [PubMed] [Google Scholar]
- Koike S, Kobayashi Y. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol Lett. 2001;204:361–366. doi: 10.1016/S0378-1097(01)00428-1. [DOI] [PubMed] [Google Scholar]
- Lauková A, Mareková M, Javorský P. Detection and antimicrobial spectrum of a bacteriocin-like substance produced by Enterococcus faecium CCM4231. Lett Appl Microbiol. 1993;16:257–260. doi: 10.1111/j.1472-765X.1993.tb01413.x. [DOI] [Google Scholar]
- Lee HJ, Jung JY, Oh YK, Lee SS, Madsen EL, Jeon CO. Comparative survey of rumen microbial communities and metabolites across one caprine and three bovine groups, using bar-coded pyrosequencing and 1H nuclear magnetic resonance spectroscopy. Appl Environ Microbiol. 2012;78:5983–5993. doi: 10.1128/AEM.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Gu Q, Wang Y, Yu Y, Yang L, Chen JV. Novel vitamin B12-producing Enterococcus spp. and preliminary in vitro evaluation of probiotic potentials. Appl Microbiol Biotechnol. 2017;101:6155–6164. doi: 10.1007/s00253-017-8373-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu T, Zhao M, Feng F, Luo W, Zhong H. In vitro and in vivo investigations of probiotic properties of lactic acid bacteria isolated from Chinese traditional sourdough. Appl Microbiol Biotechnol. 2019;103:1893–1903. doi: 10.1007/s00253-018-9554-8. [DOI] [PubMed] [Google Scholar]
- Mamuad L, Kim SH, Jeong CD, Choi YJ, Jeon CO, Lee S-S. Effect of fumarate reducing bacteria on in vitro rumen fermentation, methane mitigation and microbial diversity. J Microbiol. 2014;52:120–128. doi: 10.1007/s12275-014-3518-1. [DOI] [PubMed] [Google Scholar]
- Mansour NM, Elkhatib WF, Aboshanab KM, Bahr MMA. Inhibition of Clostridium difficile in mice using a mixture of potential probiotic strains Enterococcus faecalis NM815, E. faecalis NM915, and E. faecium NM1015: novel candidates to control C. difficile infection (CDI) Probiotics Antimicrob Proteins. 2018;10:511–522. doi: 10.1007/s12602-017-9285-7. [DOI] [PubMed] [Google Scholar]
- McCann JC, Wickersham TA, Loor JJ. High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinform Biol Insights. 2014;8:109–125. doi: 10.4137/BBI.S15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas AE, Mackenzie AK, Mravec J, Schückel J, Willats WGT, Eijsink VGH, Pope PB. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio. 2014 doi: 10.1128/mbio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocek JE, Kautz WP. Direct-fed microbial supplementation on ruminal digestion, health, and performance of pre- and postpartum dairy cattle. J Dairy Sci. 2006;89:260–266. doi: 10.3168/jds.S0022-0302(06)72090-2. [DOI] [PubMed] [Google Scholar]
- Nocek JE, Kautz WP, Leedle JAZ, Block E. Direct-fed microbial supplementation on the performance of dairy cattle during the transition period. J Dairy Sci. 2003;86:331–335. doi: 10.3168/jds.S0022-0302(03)73610-8. [DOI] [PubMed] [Google Scholar]
- Oetzel GR, Emery KM, Kautz WP, Nocek JE. Direct-fed microbial supplementation and health and performance of pre- and postpartum dairy cattle: a field trial. J Dairy Sci. 2007;90:2058–2068. doi: 10.3168/jds.2006-484. [DOI] [PubMed] [Google Scholar]
- Ørskov ER, McDonald I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J Agric Sci. 1979;92:499. doi: 10.1017/S0021859600063048. [DOI] [Google Scholar]
- Pang DG, Yang HJ, Cao BB, Wu TT, Wang JQ. The beneficial effect of Enterococcus faecium on the in vitro ruminal fermentation rate and extent of three typical total mixed rations in northern China. Livest Sci. 2014;167:154–160. doi: 10.1016/j.livsci.2014.06.008. [DOI] [Google Scholar]
- Ribeiro M, Pereira J, Ac Queiroz, Bettero V, Mantovani H, Cj Silva. Influence of intraruminal infusion of propionic acid and forage to concentrate levels on intake, digestibility and rumen characteristics in young bulls. R Bras Zootec. 2009;38:948–955. doi: 10.1590/S1516-35982009000500023. [DOI] [Google Scholar]
- Roesch L, Fulthorpe R, Riva A, Casella G, Hadwin A, Kent A, Daroub S, Camargo F, Farmerie W, Triplett E. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi EA, Vendramini RC, Carlos IZ, De Oliveira MG, De Valdez GF. Efeito de um novo produto fermentado de soja sobre os lípides séricos de homens adultos normocolesterolêmicos. Arch Latinoam Nutr. 2003;53:47–51. [PubMed] [Google Scholar]
- Sarantinopoulos P, Andrighetto C, Georgalaki MD, Rea MC, Lombardi A, Cogan TM, Kalantzopoulos G, Tsakalidou E. Biochemical properties of enterococci relevant to their technological performance. Int Dairy J. 2001;11:621–647. doi: 10.1016/S0958-6946(01)00087-5. [DOI] [Google Scholar]
- SAS Institute . Base SAS 9.3 procedures guide: statistical procedures. Cary: SAS Institute; 2012. [Google Scholar]
- Shannon CE, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1964. [Google Scholar]
- Shi C, Zhang Y, Lu Z, Wang Y. Solid-state fermentation of corn-soybean meal mixed feed with Bacillus subtilis and Enterococcus faecium for degrading antinutritional factors and enhancing nutritional value. J Anim Sci Biotechnol. 2017 doi: 10.1186/s40104-017-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen KB, Teske A. Stratified communities of active archaea in deep marine subsurface sediments. Appl Environ Microbiol. 2006;72:4596–4603. doi: 10.1128/AEM.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Tabaru H, Kadota E, Yamada H, Sasaki N, Takeuchi A. Determination of volatile fatty acids and lactic acid in bovine plasma and ruminal fluid by high performance liquid chromatography. Nihon Juigaku Zasshi. 1988;50:1124–1126. doi: 10.1292/jvms1939.50.1124. [DOI] [PubMed] [Google Scholar]
- Uyeno Y, Shigemori S, Shimosato T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 2015;30:126–132. doi: 10.1264/jsme2.ME14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu Q, Kong F, Yang Y, Wu D, Mishra S, Li Y. Exploring the goat rumen microbiome from seven days to two years. PLoS ONE. 2016 doi: 10.1371/journal.pone.0154354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Giller K, Kreuzer M, Ulbrich SE, Braun U, Schwarm A. Contribution of ruminal fungi, archaea, protozoa, and bacteria to the methane suppression caused by oilseed supplemented diets. Front Microbiol. 2017;8:1864. doi: 10.3389/fmicb.2017.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shi C, Zhang Y, Song D, Lu Z, Wang Y. Microbiota in fermented feed and swine gut. Appl Microbiol Biotechnol. 2018;102:2941–2948. doi: 10.1007/s00253-018-8829-4. [DOI] [PubMed] [Google Scholar]
- Yang E, Fan L, Jiang Y, Doucette C, Fillmore S. Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express. 2012;2:48. doi: 10.1186/2191-0855-2-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Fan L, Yan J, Jiang Y, Doucette C, Fillmore S, Walker B. Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Express. 2018;8:10. doi: 10.1186/s13568-018-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampiga M, Flees J, Meluzzi A, Dridi S, Sirri F. Application of omics technologies for a deeper insight into quali-quantitative production traits in broiler chickens: a review. J Anim Sci Biotechnol. 2018;9:61. doi: 10.1186/s40104-018-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.