Abstract
Despite being a yellow pigment visible to the human eye, coelimycin (CPK) remained to be an undiscovered secondary metabolite for over 50 years of Streptomyces research. Although the function of this polyketide is still unclear, we now know that its “cryptic” nature is attributed to a very complex and precise mechanism of cpk gene cluster regulation in the model actinomycete S. coelicolor A3(2). It responds to the stringent culture density and timing of the transition phase by the quorum-sensing butanolide system and to the specific nutrient availability/uptake signals mediated by the global (pleiotropic) regulators; many of which are two-component signal transduction systems. The final effectors of this regulation cascade are predicted to be two cluster-situated Streptomyces antibiotic regulatory proteins (SARPs) putatively activating the expression of type I polyketide synthase (PKS I) genes. After its synthesis, unstable, colorless antibiotic coelimycin A reacts with specific compounds in the medium losing its antibacterial properties and giving rise to yellow coelimycins P1 and P2. Here we review the current knowledge on coelimycin synthesis regulation in Streptomyces coelicolor A3(2). We focus on the regulatory feedback loop which interconnects the butanolide system with other cpk cluster-situated regulators. We also present the effects exerted on cpk genes expression by the global, pleiotropic regulators, and the regulatory connections between cpk and other biosynthetic gene clusters.
Keywords: Actinomycetes, Coelimycin, Streptomyces coelicolor, Secondary metabolism, Antibiotics, Type I polyketide synthase
Introduction
Streptomyces are Gram-positive, filamentous bacteria that are potent producers of secondary metabolites—specialized compounds with adaptive functions (Traxler and Kolter 2015)—many of which have antibiotic, immunosuppressant, antitumor, and other biological activities (Hopwood 2007). In the past two decades, the availability of complete genome sequences led to the development of over 20 biosynthetic gene cluster detection tools (www.secondarymetabolites.org/mining/) and revealed that the model organism Streptomyces coelicolor A3(2) could synthesize more than 20 secondary metabolites, many of them being still unidentified products of so-called cryptic or silent biosynthetic gene clusters (BGCs) (Bentley et al. 2002; Blin et al. 2017). Typical BGCs contain regulatory, tailoring, precursor supply, and transport genes organized around the main synthase subunit genes. In case of modular polyketide synthases and non-ribosomal peptide synthetases, they usually span over several tens of kilobases (Medema et al. 2015).
Among wide repertoire of S. coelicolor A3(2) chromosomally encoded bioactive molecules, there are 4 antimicrobial compounds: coelimycin A (CPK A, precursor of yellow coelimycins P1 and P2), calcium-dependent antibiotic (CDA), red-pigmented undecylprodigiosin (RED), and blue-colored actinorhodin (ACT) (Liu et al. 2013). Their production is induced by environmental, physiological, or nutrient limitation signals (Van Der Heul et al. 2018) coupled with vegetative mycelium autolysis and subsequent salvage of its constituents in order to form aerial mycelium that allows sporulation (Bibb 2005). Each biosynthetic gene cluster encodes its own pathway-specific Streptomyces antibiotic regulatory proteins (SARPs): CpkO (formerly KasO) and CpkN (cpk cluster), CdaR (cda cluster), RedZ and RedD (red cluster), and ActII-orf4 (act cluster) (Liu et al. 2013). Initially, regulatory roles of SARP cluster-situated regulators (CSRs) were believed not to extend beyond the borders of their respective metabolite biosynthetic gene clusters but this paradigm was shifted by mutational/overexpression studies suggesting that they may also control other BGCs indirectly by modulating global regulators such as AfsR2/AfsS (Huang et al. 2005). Nevertheless, it was found that cellular levels of actII-orf4 and redD transcripts correlate with the production levels of respective secondary metabolites (Takano et al. 1992; Gramajo et al. 2014). Global (pleiotropic) regulators act on numerous, often distant genes in the chromosome and orchestrate multiple pathways to proceed with major cellular events such as morphogenesis, development, and antibiotic production. For many years, they have been believed to exert their functions on biosynthetic genes via cluster-situated regulators (McKenzie and Nodwell 2007) but later findings have demonstrated their ability to bind to promoters of biosynthetic genes (Ryding et al. 2002) or even within the coding sequences, implying their direct role in the regulation of secondary metabolism. In view of these findings, the definitions of “pathway-specific” and “pleiotropic” regulators as well as “higher-level” and “lower-level” may need revision. Until today, products of more than 50 genes were identified to directly or indirectly affect secondary metabolite production in S. coelicolor A3(2), most of them acting on multiple biosynthetic pathways (Van Wezel and McDowall 2011; Van Der Heul et al. 2018).
Biosynthetic gene coding for coelimycin type I polyketide synthase (PKS I) was first identified in 1997 by DNA probe hybridization to acyltransferase domain specific for malonyl-CoA (Kuczek et al. 1997). S. coelicolor A3(2) genome sequence publication in 2002 allowed to annotate cpk cluster (Pawlik et al. 2007). It wasn’t until 2010 when its products were detected as a yellow pigment excreted to the medium (yCPK) (Gottelt et al. 2010; Pawlik et al. 2010) later identified as coelimycins P1 and P2 (Gomez-Escribano et al. 2012), and mycelium-associated, colorless, antimicrobial compound (abCPK) (Gottelt et al. 2010) deduced to be coelimycin A (Challis 2014). As a matter of fact, biosynthesis of the yellow pigment was first reported in 1978 by Rudd in his PhD dissertation. In his work, Rudd successfully mapped the genetic locus responsible for the synthesis of the compound (Rudd 1978). CPK production is dependent on the medium composition and the density of the inoculum (Gottelt et al. 2010; Pawlik et al. 2010) which can be attributed to both the transcription regulation and participation of some molecules from the medium in the final biosynthetic steps. Expression of cpk genes at the very early transition phase of culture growth is an early event of the metabolic switch from primary to secondary metabolism (Nieselt et al. 2010).
Up to now, “big data” from numerous transcriptomic, proteomic, and chromatin immunoprecipitation (ChIP) experiments have provided fragmentary and scattered information about cpk cluster regulation in the overwhelming picture of S. coelicolor A3(2) secondary metabolism regulatory networks. The aim of this work is to extract, summarize, and comment on this information from the perspective of coelimycin synthesis regulation.
Regulation within the cpk cluster
Originally the cpk cluster annotation was limited to SCO6269–SCO6288 genes (Pawlik et al. 2007). Currently, the neighboring genes SCO6265–SCO6268 coding the butanolide system proteins are also included in the cluster. The 58-kb coelimycin biosynthesis cpk cluster contains 24 genes functionally belonging to 5 groups: core biosynthetic (cpkA, cpkB, cpkC, scoT), precursor supply (cpkPα, cpkPβ, accA1, cpkK), post-polyketide tailoring (scF, cpkD, cpkE, cpkG, cpkH, cpkI), export (cpkF), and regulatory (scbR, scbA, scbB, orfB, cpkO, scbR2, cpkN) genes (Pawlik et al. 2007; Gomez-Escribano et al. 2012). Two genes (cpkJ, cpkL) have not been assigned a function. Eight transcriptional units (cpkPβ/cpkPα/accA1, scF, cpkA/cpkB/cpkC, cpkD/cpkE/cpkF/cpkG/cpkO/cpkH, cpkI, cpkJ/cpkK/cpkL, scbR2, scoT/cpkN) (Chen et al. 2016) and ten promoter regions binding different transcription factors have been identified (pscbR, pscbA, porfB, paccA1/pscF, pcpkA/pcpkD, pcpkO, pcpkI/pcpkJ, pscbR2/pscoT, pcpkN) (Takano et al. 2001; Takano et al. 2005; Gottelt et al. 2010; Li et al. 2015) but more promoters are predicted to be found in the intergenic regions of cpk cluster. Transcription start sites corresponding to transcripts of scbR, scbA, cpkA, cpkC, cpkD, cpkI, cpkO, cpkH, and scbR2 have been determined (Takano et al. 2001; Romero et al. 2014; Jeong et al. 2016).
The core polyketide chain of coelimycin is assembled by the main subunits of the modular polyketide synthase—CpkA, CpkB, and CpkC (Gomez-Escribano et al. 2012). During synthesis, type II thioesterase ScoT maintains PKS activity by removal of non-reactive acyl residues blocking the “assembly line” and was shown to be mandatory for coelimycin synthesis (Kotowska et al. 2014). It was proposed that the intermediate is released from PKS as a hydroxyaldehyde and subsequently transformed by post-polyketide tailoring enzymes and presumably transported outside of the bacterial cell by CpkF membrane efflux protein where it undergoes epoxidation to coelimycin A. Its weak antibiotic activity can be attributed to two reactive epoxide rings. Spontaneous reactions of the epoxides with N-acetylcysteine or glutamate present in the medium lead to formation of yellow-pigmented coelimycins P1 or P2, respectively (Gottelt et al. 2010; Gomez-Escribano et al. 2012). It is likely, that other coelimycins, also colorless, are formed as a result of the reaction of the epoxide rings with other substrates.
The functions of the regulatory proteins encoded within cpk cluster are as follows. ScbA is a γ-butyrolactone (GBL) synthase (Hsiao et al. 2007) accompanied by ScbB which also participates in GBL synthesis (Sidda et al. 2016). ScbR is a TetR-like GBL receptor, its homolog ScbR2 is a pseudo-GBL receptor not affected by GBLs, but shown to bind RED, ACT, and other antibiotics (Xu et al. 2010a; Wang et al. 2014). OrfB is a homolog of histidine protein kinases (Takano et al. 2005) but its target for phosphorylation has not been found. CpkO and CpkN are two SARP proteins. Their binding sites are not known. CpkO is an activator necessary for cpk gene expression as shown by transcriptomic and qRT-PCR analysis of ΔcpkO mutant strain (Gottelt et al. 2010; Takano et al. 2005). Since SARPs generally activate their target genes, CpkN is also likely an activator.
If the conditions of growth of wild-type S. coelicolor A3(2) are suitable for CPK production, the yellow pigment is observed about 24 h earlier than the typical time when other colored metabolites appear (Gottelt et al. 2010; Pawlik et al. 2010). In a high-resolution time-series transcriptomic study of a fermenter-grown culture in minimal medium with glucose and glutamate, all the cpk cluster genes showed a strong transient expression peak around 22–24 h and many of them remained at constantly elevated expression levels afterwards (Nieselt et al. 2010). Transcription peaks of regulatory genes scbR, scbA, scbB, and cpkO (22 h) preceded those of other cpk cluster genes. Next was orfB (peak at 23 h time point) followed by the rest of the cpk cluster genes including regulators scbR2 and cpkN (24 h). Transcription of scoT needed to maintain the enzymatic activity of the modular PKS subunits started to increase 1 h later than that of core biosynthetic genes. Interestingly, this sharp peak of cpk cluster regulatory gene transcription preceded the moment traditionally seen as the onset of secondary metabolism correlated with phosphate depletion and upregulation of the pho regulon (35 h in the same study) followed by the expression of red (38 h) and act (43 h) gene clusters (Nieselt et al. 2010).
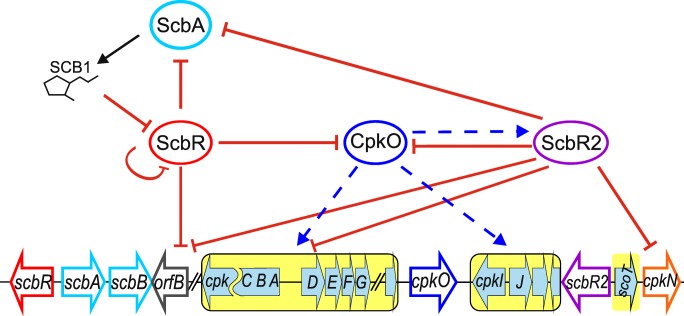
The rapid increase and sharp decline of the transcription of cpk genes are governed by the butanolide system (Fig. 1). During the exponential-growth phase, the synthesis of γ-butyrolactone SCB1 (by ScbA protein) and its receptor protein ScbR is on the basal level. At this time point, ScbR exists mainly as a DNA-protein complex—it binds promoter regions of scbR, scbA (Takano et al. 2001), cpkO (Takano et al. 2005), and orfB (Li et al. 2015) genes, acting as a transcription inhibitor. The SCB1 level rises proportionally to the number of dividing bacterial cells. After its concentration reaches the threshold, SCB1 binding to ScbR results in its dissociation from DNA and subsequent derepression of its own, scbA and that of cpkO gene expression (Takano et al. 2001; Takano et al. 2005). The elevated level of CpkO directly or indirectly activates the transcription of cpk genes, including that of scbR2 encoding pseudo-GBL receptor. Inhibitory interaction of ScbR2 with scbA promoter consequently blocks SCB1 biosynthesis. Since ScbR2 is known to bind to cpkO, cpkN, and orfB promoters and to inhibit their transcription (Gottelt et al. 2010; Li et al. 2015), it has been proposed that it may serve as a switch turning off coelimycin synthesis.
Fig. 1.
Coelimycin biosynthetic gene cluster regulation by the cluster-situated regulators. CPK biosynthetic genes are marked with a yellow background. The lines ending with arrows indicate transcription activation, with the exception of an arrow indicating γ-butyrolactone SCB1 production. The lines ending with bars indicate repression of transcription or inhibition of ScbR by SCB1. The solid lines indicate promoter binding while dashed lines imply an indirect or unknown regulatory mechanism. See the text for further information and references
A number of deletion mutants of the butanolide system genes were analyzed, leading to the following observations: (i) in ΔscbA mutant transcription of scbR, scbR2, cpkO, and scbA is diminished (D’Alia et al. 2011) and addition of exogenous SCB1 to ΔscbA mutant restores transcription of both scbR and scbR2 but not scbA (Takano et al. 2005), (ii) in ΔcpkO mutant transcription of scbR2 is diminished (Gottelt et al. 2010), (iii) ∆scbR2 mutant is upregulated in scbR, scbA, (Wang et al. 2011; Li et al. 2015) and cpkO transcription (Gottelt et al. 2010), (iv) in ΔscbR mutant expression of scbR2 (Gottelt et al. 2010) and cpkO (Takano et al. 2005) is constitutive, transcription of scbR is upregulated and transcription of scbA abolished (Takano et al. 2001; Li et al. 2015).
Inactivation of either scbA or scbR was shown to abolish SCB1 production (Takano et al. 2001). Addition of exogenous SCB1 to ΔscbA mutant-restored transcription of scbR, but not that of scbA which led to a proposal of these proteins forming a complex required to activate scbA transcription (Takano et al. 2005; Mehra et al. 2008). In an excellent review paper, it was speculated that ScbR, although being an autorepressor and the repressor of cpkO, may act as an activator upon binding to scbA promoter and that the concentration of exogenous SCB1 is high enough to saturate all ScbR molecules thus preventing activation of scbA transcription (Van Wezel and McDowall 2011). Our explanation for the mandatory (“activatory”) role of ScbR in scbA transcription follows (see Fig. 1).
In scbR deletion mutant, there is no ScbR necessary for cpkO repression. Upregulated CpkO may raise the concentration of ScbR2 protein which is a repressor of scbA (Wang et al. 2011). The same mechanism may account for upregulation of scbR/scbR2 and failure to restore scbA transcription in scbA null mutant by exogenous SCB1 (Takano et al. 2005). Concentration of exogenous SCB1 is probably high enough to complex whole cellular ScbR bound to scbR and cpkO promoters (thus creating conditions similar to those in ∆scbR mutant) resulting in derepression of both genes. This leads to upregulation of scbR2 transcript level by CpkO and repression of scbA by ScbR2. The reason for abolished transcription of scbA in ΔscbA mutant may be simply repression of its promoter by ScbR devoid of SCB1.
Not assigning ScbR an activatory role for scbA transcription but rather attributing its reduction to the upregulated ScbR2 level as an indirect consequence of scbR deletion/inactivation allows to provide a mechanism accounting for seemingly contradictory observations (Fig. 1). It would be intriguing to unravel the exact pathway (direct or non-direct) of CpkO activatory impact on scbR2 transcription.
Influence of the cpk cluster-situated regulators on other metabolic pathways
The butanolide ScbA/ScbR/ScbR2 system was thought to primarily target coelimycin biosynthetic gene cluster in S. coelicolor A3(2) but the turning point was publication of 16 and 58 genome-scale, confirmed binding sites of ScbR and ScbR2, respectively, along with their respective null mutants’ transcriptomic data. It revealed that 30.1% and 42.3% of all genes are at least 20% differentially expressed in ∆scbR and ∆scbR2, respectively, in comparison with the parent strain M145 (Li et al. 2015). Taken together, these data shifted ScbR and ScbR2 impact range from pathway-specific to pleiotropic regulators. It also provided experimental proof for the key role of ScbR2 in molecular cross-regulation of secondary metabolite synthesis.
Mutants ∆scbA and ∆cpkO, which both lack ScbR2 synthesis, showed precocious actII-orf4 and redD transcription but only the former strain was accordingly advanced in production of ACT and RED (Takano et al. 2001; Gottelt et al. 2010; D’Alia et al. 2011). No obvious ACT and RED synthesis phenotype was observed in the ∆cpkO strain (Takano et al. 2005). This discrepancy can be attributed to different levels of ScbA protein in both mutants. The lack of ScbR2 may be the cause of precocious scbA transcription in ∆cpkO (Takano et al. 2005) but it cannot affect the complete lack of ScbA protein in the mutant ∆scbA for the absence of the gene. Deletion of scbA was indeed associated with earlier expression of the genes encoding primary metabolism enzymes that ensure antibiotic precursor supply (D’Alia et al. 2011) required for ACT and RED synthesis.
CPK overproducing ∆scbR2 strain was dramatically reduced in ACT, RED, and CDA synthesis. This phenotypic effect was reflected in transcript levels of respective activators (increased cpkO, decreased actII-orf4, redZ, cdaR), all of which were shown to be direct targets for binding by ScbR2 (Li et al. 2015). As discussed earlier, ScbR2 is a repressor of cpkO gene; hence, the effect of scbR2 deletion on transcription of ACT, RED, and CDA activators is surprising, suggesting a potential activatory role of ScbR2 or another regulatory mechanism. Such a mechanism would downregulate the transcription of actII-orf4, redZ, and cdaR perhaps in response to ∆scbR2-uninhibited coelimycin production or highly elevated levels of scbA transcription in the mutant (Wang et al. 2011). ScbR2 DNA-binding activity was found to be relieved upon binding of RED and ACT further underscoring its antibiotic cross-regulatory role (Xu et al. 2010a). As for now, CPK binding to ScbR2 wasn’t shown in S. coelicolor A3(2) but studies conducted in S. lividans TK24 suggest the existence of such interaction (Sun et al. 2017).
An important target repressed by both ScbR and ScbR2 is accA2 gene (Li et al. 2015). AccA2 is an essential subunit α of acyl-CoA carboxylases, enzymatic complexes providing carboxylated precursors for fatty acid and polyketide biosynthesis. Its close homolog, AccA1 encoded within the cpk cluster, was shown by in vitro reconstitution experiment to cooperate with the same acyl-CoA carboxylase β subunits as AccA2 (Rodríguez and Gramajo 1999; Rodríguez et al. 2001). CpkK is another putative β subunit of acyl-CoA carboxylases, homologous to an essential AccB protein (Rodríguez et al. 2001). It is possible that CpkK may form complexes with both AccA1 and AccA2.
ScbR and ScbR2 proteins were also shown to interact forming a heterodimer able to bind a novel target, the promoter of SCO5158, a gene involved in metal transport. The heterodimer was also shown to coexist with ScbR and ScbR2 homodimers which is how these proteins most commonly bind DNA (Li et al. 2017).
Effects of pleiotropic regulators on coelimycin synthesis
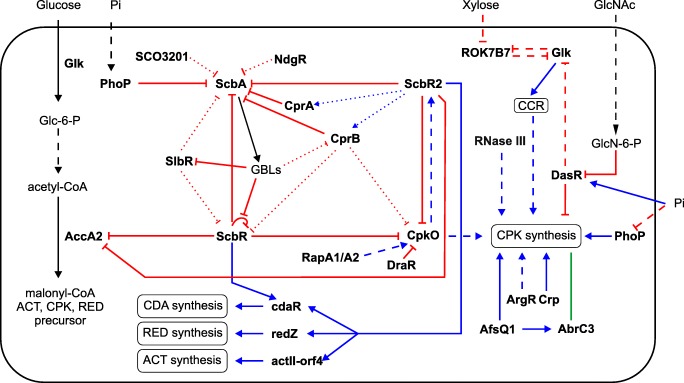
The network of interactions between the cpk gene cluster, butanolide system, and other regulatory proteins of S. coelicolor A3(2) is outlined in Fig. 2.
Fig. 2.
The regulatory pathways interconnected with cpk cluster regulation. The diagram links nutrient signals with pleiotropic and pathway-specific regulators affecting coelimycin synthesis and the feedback effect of cpk cluster genes. The blue lines ending with an arrow indicate activation. The red lines ending with a bar indicate repression. The green line with no ending indicates evidence of DNA binding, but the effect on transcription is unknown. The solid lines indicate the direct effect (DNA or ligand binding), dashed lines indicate indirect effect, and dotted lines indicate proposed but direct interactions. The thin black lines indicate transport or transformation of chemical compounds. Only the effects of butanolide system proteins ScbR and ScbR2 on other biosynthetic gene clusters were included in the diagram as they most accurately represent the impact of cpk cluster transcription on the production of other antibiotics. CCR–carbon catabolite repression. See the text for further information and references
Two-component systems
Two-component systems (TCSs) are key signal transduction mechanisms in bacteria, acting as sensors of environmental condition changes and response modulators of transcription. These functions are accomplished through their membrane-bound histidine kinases (HKs) and response regulators (RRs), respectively (Rodríguez et al. 2013). S. coelicolor A3(2) genome encodes an impressive number of 67 typical TCSs, of which only a few have been shown to regulate secondary metabolite synthesis.
AfsQ1/Q2
AfsQ1/Q2 two-component system is involved in the regulation of carbon, phosphate, and nitrogen metabolism along with antibiotic synthesis. AfsQ1 was shown to activate synthesis of ACT, RED, and CDA by directly binding to the promoters of actII-orf4, redZ (but not redD), and cdaR (Wang et al. 2013). Deletion of afsQ1/Q2 revealed a dramatically decreased expression of cpk cluster and abolished CPK synthesis. AfsQ1 binds to the cpkA promoter and not to any other promoter in the cluster, including that of cpkO or cpkN. Interestingly, when AfsQ1 DNA-binding motif was mutated, CPK synthesis was abolished as a result of the decrease in cpkA/B/C transcript; however, expression of other cpk genes was enhanced. Authors concluded that AfsQ1 binding to the cpkA promoter may help recruit RNA polymerase and activate cpkA/B/C transcription. They also suggested that another regulatory cascade mechanism is involved in regulation of cpk cluster by AfsQ1/Q2 (Chen et al. 2016).
DraR/K
DraR/K TCS regulates physiological and morphological differentiation in S. coelicolor A3(2) along with secondary metabolism in a medium-dependent manner (Yu et al. 2012; Yu et al. 2014). DraR represses coelimycin and undecylprodigiosin and activates actinorhodin production. Its regulatory role in CPK and ACT biosynthesis was shown to be mediated by binding to cpkO and actII-orf4 promoters, respectively. On the contrary, DraR regulates RED synthesis independently of redD or redZ promoter binding. A possible cooperation of DraR with AfsQ1 was proposed in the regulation of actII-orf4, adding another level of complexity to secondary metabolism coordination in S. coelicolor A3(2) (Yu et al. 2012). Transcriptomic studies revealed a general drastic upregulation of cpk cluster genes in ∆draR/K mutant over 60 h time. The effect was most pronounced in 42 h time point and involved regulatory genes cpkO and scbR2. Butanolide system genes scbR, scbA, and scbB transcripts were upregulated in 36 h and 42 h time points but showed downregulation after 60 h in the mutant when compared with the parent strain. The decrease in scbA and scbR transcript abundance in 60 h time point might be a consequence of scbR2 transcription, which peaks after 42 h and decreases until 60 h time point (Yu et al. 2014). Concluding from the available data, DraR/K two-component system is a cpk cluster repressor.
PhoR/P
PhoR/P two-component system senses phosphate starvation and suppresses central metabolism and development and secondary metabolism until enough phosphate is salvaged for morphological differentiation. From ChIP on chip and transcriptomic studies, PhoP was shown to bind hundreds of genomic regions and affect transcription of corresponding genes. The majority of target genes are repressed rather than activated by PhoP. Downregulation of all detected cpk cluster transcripts in ∆phoP mutant clearly suggested that PhoP is a cpk cluster activator. Interestingly, cpk cluster was found to be the most PhoP-enriched region of S. coelicolor A3(2) genome due to intensive binding of the regulator within coding sequences of cpkB (2 sites) and cpkC (1 site) but not in promoter regions. PhoP was also shown to bind to scbA promoter and the observed enhancement of scbA transcription in ∆phoP mutant led the authors to the conclusion that it is scbA repressor (Allenby et al. 2012). In another study, scbR transcript and protein were more abundant in ∆phoP than in the wild-type strain (Thomas et al. 2012). To our knowledge, decreased transcription of cpk genes and at the same time upregulation of scbA and scbR transcripts in ∆phoP mutant could be a consequence of drastically downregulated scbR2 transcription that was indeed observed (Allenby et al. 2012).
RapA1/A2
Deletion of rapA1/A2 did not cause any change in growth or morphology of S. coelicolor A3(2); however, it reduced ACT production. Further proteomic studies revealed decreased abundance of CpkI protein in the ∆rapA1/A2 mutant. Reverse-transcriptase PCR showed downregulation in both pathway-specific regulatory genes actII-orf4 and cpkO transcripts providing a reason for reduced ACT production and indicating downregulation of coelimycin synthesis in ∆rapA1/A2 mutant, although the product of cpk cluster was not characterized at that time (Lu et al. 2007).
AbrC1/C2/C3
AbrC1/C2/C3 is an atypical TCS comprised of two histidine kinases AbrC1/C2 and a response regulator AbrC3. The system is highly conserved among all sequenced Streptomyces species. AbrC3 is an activator of morphological development along with ACT, RED, and CDA production (Yepes et al. 2011). It was shown to bind to actII-orf4 but not to redD or redZ promoters. AbrC3 intragenic binding to cpkC was detected, although no significant changes in the expression of cpk genes was demonstrated (Rico et al. 2014). Interestingly, AbrC3 is positively regulated by AfsQ1 through the direct binding in the promoter region (Wang et al. 2013). Further studies using different growth conditions may be necessary to induce the regulatory effect of AbrC1/C2/C3 on coelimycin synthesis.
TetR family and γ-butyrolactone binding proteins
TetR family regulators are widely distributed among bacteria. They are transcription repressors with a helix-turn-helix DNA-binding motif that dissociate from their target sequences and derepress them upon ligand binding. The ligand molecules are often quorum-sensing signaling compounds and end-products or intermediates of antibiotic biosynthetic pathways. S. coelicolor A3(2) genome encodes at least 150 TetR-like regulators (Ramos et al. 2005); two of which, ScbR and ScbR2 were described earlier in this work.
SCO3201
S. coelicolor A3(2) TetR family SCO3201 protein was identified in an attempt to find a major repressor of ACT synthesis in S. lividans that accounts for its very weak actinorhodin production capacity. SCO3201 gene disruption in S. coelicolor A3(2) did not produce any distinct phenotype. However, it was demonstrated to inhibit ACT, RED, and CDA synthesis along with morphological differentiation and sporulation upon protein overexpression. SCO3201 is a negative regulator and was shown to bind to the promoter region of scbA but not that of scbR or cpkO. SCO3201 overexpression led to downregulation of scbR and cpkO transcription in exponential but not in the stationary growth phase while the transcription of scbA was upregulated in the stationary growth phase. Authors suggested that SCO3201 may lose its repressory activity upon binding an unknown ligand in the stationary phase (Xu et al. 2010b). We hypothesize that SCO3201 binding to scbA promoter is repressive (Fig. 2) and leads to decreased SCB1 production in the exponential phase and in turn—decreased scbR, cpkO, and scbR2 transcripts. After dissociation of SCO3201 from scbA promoter in the stationary phase, low abundance of ScbR2 production is insufficient to fully repress scbA. In order to gain a better view of the regulation mechanism, it would be interesting to measure the scbR2 expression pattern in the same conditions. Functionally, SCO3201 might mimic ScbR; thus, it would be intriguing to assess its GBL binding activity.
CprA/CprB
CprA and CprB share 90.7% amino acid sequence identity and they are close homologs of ScbR. CprA was shown to be the activator of sporulation and ACT and RED production, while CprB was demonstrated to inhibit sporulation and ACT synthesis but had no effect on RED production (Onaka et al. 1998). Interestingly, CprA and CprB were found to repress scbA promoter and, consequently, GBL production (Li et al. 2015). Binding of CprB to the promoters of scbR and cpkO was also demonstrated. In the same studies, addition of the extract from S. coelicolor A3(2) liquid culture but not the exogenous SCB2 γ-butyrolactone caused dissociation of CprB from an artificially synthesized consensus sequence (Bhukya et al. 2014). Interestingly, the promoter of cprA was bound by ScbR and ScbR2 while the promoter region of cprB was bound by ScbR2, suggesting the existence of a feedback loop (Li et al. 2015).
SlbR
SlbR does not belong to any characterized regulator family but is capable of binding γ-butyrolactone SCB1. It was shown to bind to the promoter of scbR and scbA, and the DNA binding was relieved in the presence of SCB1, but no profile of butanolide system and cpk cluster gene transcription was provided. Pattern of expression of slbR gene resembles that of scbR but contrary to ScbR; SlbR binds adpA promoter and affects morphology (Yang et al. 2012). It represses ACT and RED synthesis along with spore formation in rich media. Its properties make it more similar to ArpA, ScbR homolog in S. griseus. Further studies are anticipated to reveal the pleiotropic effects of SlbR on secondary metabolism and differentiation.
ROK-signature proteins
Glk and Rok7B7 belong to the ROK (regulators, ORFs, and kinases) family of proteins that contains transcriptional factors and kinases associated mainly with primary metabolism. However, both Glk and Rok7B7 were shown to affect not only carbon catabolite repression (CCR) but also growth and secondary metabolism (Świątek et al. 2013; Romero-Rodríguez et al. 2016).
Glk
Glucose kinase (Glk, GlkA) plays a major role in carbon catabolite repression (CCR), a mechanism ensuring sequential utilization of carbon sources, from the most to the least preferred, when a mixture of them is available in the cell environment (Titgemeyer and Brückner 2002). Glk has two distinct activities: (i) enzymatic (phosphorylation of glucose to glucose-6-phosphate which can enter central carbon catabolism pathways) and (ii) regulatory (possible interactions with transcription factors) (Gubbens et al. 2012). In order to distinguish between those 2 effects of S. coelicolor A3(2) Glk, glkA null mutant complemented with heterologous Glk from Zymomonas mobilis, which complements only enzymatic activity, was constructed (ScoZm strain) (Romero-Rodríguez et al. 2016). Transcriptomic profiles of S. coelicolor A3(2) parent strain on media with both glucose and agar (repressive conditions) or agar as the sole carbon source (non-repressive conditions) were compared. Moreover, transcriptomic profiles of ScoZm and the parent strain grown in repressive conditions were analyzed. The putative kinase orfB and cpkN transcripts were downregulated, and that of scbA was upregulated in ScoZm when compared with the M145 wild-type strain. These effects were not observed in M145 grown in repressive relative to non-repressive conditions, suggesting the dependence of these genes on the regulatory activity of GlkA. On the other hand, the presence of glucose upregulated transcription of genes scF–cpkG (SCO6272–SCO6279) and cpkI–cpkL (SCO6282–SCO6285) in the wild-type strain. Interestingly, the effect of glucose on cpk cluster expression depends on other constituents of the medium—in minimal medium NMMP (Romero-Rodríguez et al. 2016)—addition of glucose enhanced CPK synthesis, while on rich medium 79, it was prevented (Pawlik et al. 2010). It was also reported that different additional carbon sources in the NMMP medium—mannitol or fructose—can elicit contrary effects on cpk cluster protein levels upon glucose addition—activation or repression, respectively (Gubbens et al. 2012).
Rok7B7
Rok7B7 is a repressor of the xylose operon and an activator of the sugar phosphotransferase system (PTS) genes, involved among others in N-acetylglucosamine (GlcNAc) internalization. The protein was shown to be a direct repressor of RED production and an ACT synthesis activator (Park et al. 2009; Świątek et al. 2013). The only trace information on Rok7B7 regulatory effect on coelimycin synthesis is found in proteomic studies, which demonstrated that CpkH protein is much less abundant in ∆rok7B7 mutant than in the parent strain (Świątek et al. 2013).
Other regulators
DasR
DasR is a global regulator linking nutrient stress to antibiotic synthesis. High concentration of GlcNAc in minimal growth medium, resembling its accumulation after autolytic vegetative mycelium degradation in nature, triggers the onset of secondary metabolism. DasR also connects carbon, nitrogen, and phosphate metabolism. The protein dissociates from its targets upon binding of glucosamine-6-phosphate, an intermediate of GlcNAc metabolism (Rigali et al. 2006), while at the same time, high phosphate concentrations enhance its DNA-binding activity (Tenconi et al. 2015). DasR controls RED and ACT biosynthesis through the respective pathway-specific regulators. Indeed, DasR was shown to bind to the promoters of redD, redZ, and actII-orf4 and repress them (Rigali et al. 2008; Świątek-Połatyńska et al. 2015). ∆dasR mutant showed more abundant scbA and cpkO transcripts which resulted in enhanced transcription of the cpk cluster, clearly showing that DasR is coelimycin synthesis repressor. The regulator was demonstrated to bind to intragenic regions of three biosynthetic genes cpkA/B/C but no direct interaction with cpk or butanolide system gene promoters was observed implying a more complex mechanism of cpk cluster regulation (Świątek-Połatyńska et al. 2015).
ArgR
In bacteria, ArgR is a transcriptional repressor of arginine and pyrimidine biosynthetic genes that uses l-arginine as co-repressor. However, it was also demonstrated to activate ACT and RED production (Pérez-Redondo et al. 2012). Interestingly, ∆argR mutant exhibited the formation of spore-like chains when grown in liquid culture (Botas et al. 2018). ArgR was found to be an activator of butanolide system and cpk gene cluster. Together with post-PKS and precursor supply, regulatory genes scbA, scbB, scbR, scbR2, cpkO, and cpkN were all downregulated in ∆argR mutant (Pérez-Redondo et al. 2012; Botas et al. 2018). ∆argR mutant cpk cluster genes followed a profile of downregulation and delayed expression in the stationary phase, probably as a result of additional control mechanisms to those of ArgR (Botas et al. 2018).
Crp
Crp (cyclic AMP receptor protein) is a transcription regulator controlling colony development, precursor synthesis/flux, and production of at least 8 secondary metabolites in S. coelicolor A3(2) (Derouaux et al. 2005). Unlike in Gram-negative bacteria, Crp does not play a role in CCR in Streptomyces (Romero-Rodríguez et al. 2016). The regulator was demonstrated to activate ACT and CDA synthesis. ∆crp mutant was hypersporulating and was noticeably delayed in germination, growth, and RED synthesis although ultimately produced the same amount of antibiotic as the parent strain (Derouaux et al. 2005; Gao et al. 2012). Indeed, Crp was shown to directly bind in or upstream of the coding regions of actII-orf4, redZ, and cdaR along with cpkA and some precursor supply and post-PKS cpk genes. The transcription of accA1, scF, and cpkA was upregulated upon Crp overexpression, suggesting its activatory role in coelimycin production in S. coelicolor A3(2) (Gao et al. 2012).
NdgR
NdgR (regulator for nitrogen source-dependent growth and antibiotic production) is a direct transcriptional activator of leucine and methionine biosynthesis (Kim et al. 2012); a glycerol utilization operon repressor (Lee et al. 2017); and a regulator of growth, morphological differentiation, and nitrogen source-dependent ACT production (Yang et al. 2009). It targets scbR/A promoter region. In proteomic studies CpkE and CpkJ were found to be more abundant in ∆ndgR mutant than in the parent strain (Yang et al. 2009); thus, we speculate that NdgR is cpk cluster inhibitor.
GntR
GntR (SCO1678) is a FadR subfamily gluconate-binding repressor of the gluconate operon SCO1679–SCO1682. ACT synthesis was shown to be decreased in gluconate-supplemented in comparison with the glucose-supplemented SMM medium in S. coelicolor A3(2). What is more, in ∆gntR mutant, gluconate induced the production of coelimycin, suggesting a connection between gluconate metabolism and the production of this compound. The authors speculated the existence of an unknown protein acting as a switch from ACT and RED synthesis to minor or cryptic secondary metabolite production in the absence of alternative carbon sources and GntR protein (Tsypik et al. 2017).
RNase III
RNase III is a double-stranded RNA-specific endoribonuclease involved in rRNA processing, sense/antisense RNA degradation, and other gene expression regulation mechanisms (Blomberg et al. 1990; Babitzke et al. 1993; Drider and Condon 2004). S. coelicolor A3(2) rnc deletion mutant was unable to produce ACT, RED, and CDA in a set of different solid media and SMM liquid medium, optimized for antibiotic production (Sello and Buttner 2008). RNA-seq analysis of S. coelicolor A3(2) rnc mutant transcriptome revealed strong downregulation of cpkD, cpkE, cpkF, cpkG, cpkO, cpkI, cpkJ, and cpkK along with cdaR and other cda gene transcription in comparison with the wild-type strain. In the same study, RNase III was demonstrated to immunoprecipitate with mRNA of cpkE and cpkI transcripts suggesting RNase III can regulate these gene transcripts’ levels through mRNA binding without cleavage mechanism (Gatewood et al. 2012). A scenario has been proposed in which RNase III could bind and/or cleave the double-stranded mRNA-cutoRNA (convergent untranslated overlapping RNA—a species of antisense RNAs) pair that resulted from the overlap of 3′ untranslated regions from convergently oriented genes. The implication of this could be altered stability of one or both transcripts; however, the overall functions of RNase III and cutoRNAs are not clear yet (Moody et al. 2013). Beside one cutoRNA localized in SCO6268, other antisense RNAs have been identified in cpk cluster genes (scbR, scbA, accA1, cpkC, cpkB, cpkE, cpkH) along with potential small non-coding RNAs localized in the cpk intergenic regions (scr6287–6286, scr6280) (Moody et al. 2013; Romero et al. 2014), suggesting an additional regulatory mechanism to that of cluster-situated and global regulators.
Coelimycin biosynthetic gene cluster as part of the global regulatory network
The physiological role of coelimycin in S. coelicolor A3(2) is not known. It is unlikely to be a “chemical weapon” against other microorganisms, as the weak antibiotic compound coelimycin A is unstable. We speculate that due to the presence of two reactive epoxide rings, coelimycin A may act as a detector of certain compounds in the environment and take part in an unknown signaling pathway. Whatever its biological function, it apparently requires stringent control and is released only transiently at the beginning of the metabolic switch from exponential growth to antibiotic production.
In view of the available data, we describe a consistent mechanism of coelimycin biosynthetic gene cluster regulation by the butanolide system proteins and CPK biosynthesis pathway-specific activator CpkO (Fig. 1). Binding sites of both SARPs from this cluster, CpkO and CpkN, have not been found so far. Finding differences between their direct targets might clarify why transcription of some cpk genes remains elevated after the sharp activation peak (Nieselt et al. 2010). Nevertheless it is clear that CpkO activates the synthesis of ScbR2 protein which in turn acts as a switch turning off the expression of cpk gene cluster. This connection (activation of scbR2 by CpkO) allows to explain some previously unclear experimental results. Moreover, ScbR2 appears to be a key player in cross-regulation of other biosynthetic clusters. As opposed to being the transcription repressor of cpk cluster, it seems to activate ACT, RED, and CDA clusters (Li et al. 2015).
The coupling of cpk and butanolide system genes in the cluster and the existence of CpkO-ScbR2-ScbA feedback loop interconnecting cpk and butanolide system may indicate their co-evolution (Medema et al. 2014) and their co-influence on other biosynthetic gene clusters. The outcome of the interplay within the cpk cluster is especially important when we take into account that coelimycin production is an early secondary metabolism event in the life cycle of the cell and therefore, it is involved in shaping the rest of it. Indeed, both ScbR and ScbR2 were shown to participate in multiple regulatory pathways (Li et al. 2015). The impact of cpk cluster is also reinforced by the existence of only one GBL synthase (ScbA/ScbB), producing at least 8 different GBLs (Sidda et al. 2016), and several GBL-receptor homolog genes encoded in S. coelicolor A3(2) genome (Nishida et al. 2007). SlbR, a protein non-resembling typical GBL receptors, has also been demonstrated to bind GBL, further expanding the range of controlled pathways.
We made an attempt to bring together a diagram (Fig. 2) of pleiotropic regulatory systems that influence cpk cluster expression including the links to other antibiotic BGCs, but the different cross-regulatory connections were difficult to integrate. Some regulators are well characterized and their description is firmly based on different experimental techniques. Most of available information comes from transcriptomic and proteomic studies as well as genome-scale detection of binding sites. Differences in experimental conditions, data acquisition, and data analysis between omics-based studies make it difficult to compare their results and hinder the creation of a unified regulatory-network model.
Concerted action of global regulators is responsible for the ability of streptomycetes to utilize diverse and often fluctuating food sources. Nutrient limitation leads to vegetative mycelium autolysis and metabolic switch towards secondary metabolite production and morphological differentiation. Through the action of DasR, S. coelicolor A3(2) may sense the availability of GlcNAc (a preferred carbon and nitrogen source) and hold or induce secondary metabolism according to other signals indicating if the environment is rich in nutrients or not (Rigali et al. 2008). DasR, as well as other regulators involved in the intertwining nutrient-sensory networks, such as Glk, PhoR/P, AfsQ1/Q2, Crp (Urem et al. 2016) and many others, influences transcription of coelimycin biosynthetic gene cluster showing a connection between the nutrient sensing pathways and the butanolide system, although their exact interactions remain to be established. Data presented in this work suggest that pleiotropic regulators control cpk cluster on multiple levels by binding to promoters and affecting transcription of (i) the butanolide system genes, (ii) the cpk pathway-specific activator cpkO and (iii) the core biosynthetic genes. The reported observations of binding of global regulators within the coding sequences of core biosynthetic genes and emerging evidence of non-coding RNAs’ involvement in the control of secondary metabolism set new challenges in deciphering the multi-level regulation of secondary metabolite synthesis in Streptomyces coelicolor A3(2).
Acknowledgements
This work was supported by the Hirszfeld Institute of Immunology and Experimental Therapy, PAS and Wroclaw Centre of Biotechnology programme “The Leading National Research Centre (KNOW) for years 2014-2018”.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allenby NEE, Laing E, Bucca G, Kierzek AM, Smith CP. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res. 2012;40:9543–9556. doi: 10.1093/nar/gks766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Granger L, Olszewski J, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Thomson NR, James KD, Harris DE, Quail MA, Harper D, Bateman A, Brown S, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Larke L, Murphy L, Oliver K, Rabbinowitsch E, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Woodward J, Barrell BG, Parkhill J. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bhukya H, Bhujbalrao R, Bitra A, Anand R. Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2) Nucleic Acids Res. 2014;42:10122–10133. doi: 10.1093/nar/gku587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, De Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. AntiSMASH 4.0 - improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P, Wagner EG, Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9(7):2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botas A, Pérez-Redondo R, Rodríguez-García A, Álvarez-álvarez R, Yagüe P, Manteca A, Liras P. ArgR of Streptomyces coelicolor is a pleiotropic transcriptional regulator: effect on the transcriptome, antibiotic production, and differentiation in liquid cultures. Front Microbiol. 2018;9:1–18. doi: 10.3389/fmicb.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J Ind Microbiol Biotechnol. 2014;41:219–232. doi: 10.1007/s10295-013-1383-2. [DOI] [PubMed] [Google Scholar]
- Chen S, Zheng G, Zhu H, He H, Chen L, Zhang W, Jiang W, Lu Y. Roles of two-component system AfsQ1/Q2 in regulating biosynthesis of the yellow-pigmented coelimycin P2 in Streptomyces coelicolor. FEMS Microbiol Lett. 2016;363:1–8. doi: 10.1093/femsle/fnw160. [DOI] [PubMed] [Google Scholar]
- D’Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2) Microb Biotechnol. 2011;4:239–251. doi: 10.1111/j.1751-7915.2010.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouaux A, Gerkens P, Noens EEE, Mazzucchelli G, Koerten HK, Titgemeyer F, De Pauw E, Leprince P, Van Wezel GP, Galleni M, Piette A, Vion S, De Pauw E, van Wezel GP, Rigali S. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J Proteome Res. 2005;4:1699–1708. doi: 10.1021/pr050155b. [DOI] [PubMed] [Google Scholar]
- Drider D, Condon C. The continuing story of endoribonuclease III. J Mol Microbiol Biotechnol. 2004;8:195–200. doi: 10.1159/000086700. [DOI] [PubMed] [Google Scholar]
- Gao C, Hindra MD, Yin C, Elliot MA. Crp is a global regulator of antibiotic production in Streptomyces. MBio. 2012;3:1–12. doi: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood ML, Bralley P, Weil MR, Jones GH. RNA-Seq and RNA immunoprecipitation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J Bacteriol. 2012;194:2228–2237. doi: 10.1128/JB.06541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Song L, Fox DJ, Yeo V, Bibb MJ, Challis GL. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem Sci. 2012;3:2716–2720. [Google Scholar]
- Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2) Microbiology. 2010;156:2343–2353. doi: 10.1099/mic.0.038281-0. [DOI] [PubMed] [Google Scholar]
- Gramajo HC, Takano E, Bibb MJ. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Solid Fuel Chem. 2014;48:272–274. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- Gubbens J, Janus M, Florea BI, Overkleeft HS, Van Wezel GP. Identification of glucose kinase-dependent and -independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol Microbiol. 2012;86:1490–1507. doi: 10.1111/mmi.12072. [DOI] [PubMed] [Google Scholar]
- Hopwood DA. Streptomyces in nature and medicine: the antibiotic makers. New York: Oxford University Press; 2007. [Google Scholar]
- Hsiao NH, Söding J, Linke D, Lange C, Hertweck C, Wohlleben W, Takano E. ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize γ-butyrolactones. Microbiology. 2007;153:1394–1404. doi: 10.1099/mic.0.2006/004432-0. [DOI] [PubMed] [Google Scholar]
- Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Kim JN, Kim MW, Bucca G, Cho S, Yoon YJ, Kim BG, Roe JH, Kim SC, Smith CP, Cho BK. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat Commun. 2016;7:1–11. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lee BR, Kim JN, Kim BG. NdgR, a common transcriptional activator for methionine and leucine biosynthesis in Streptomyces coelicolor. J Bacteriol. 2012;194:6837–6846. doi: 10.1128/JB.00695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotowska M, Ciekot J, Pawlik K. Type II thioesterase ScoT is required for coelimycin production by the modular polyketide synthase Cpk of Streptomyces coelicolor A3(2) Acta Biochim Pol. 2014;61:141–147. [PubMed] [Google Scholar]
- Kuczek K, Pawlik K, Kotowska M, Mordarski M. Streptomyces coelicolor DNA homologous with acyltransferase domains of type I polyketide synthase gene complex. FEMS Microbiol Lett. 1997;157:195–200. doi: 10.1111/j.1574-6968.1997.tb12773.x. [DOI] [PubMed] [Google Scholar]
- Lee BR, Bhatia SK, Song H-S, Yang YH. The role of NdgR in glycerol metabolism in Streptomyces coelicolor. Bioprocess Biosyst Eng. 2017;40:7717–7724. doi: 10.1007/s00449-017-1813-z. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Li S, Ji J, Wang W, Yang K (2015) ScbR-and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci Rep 5. 10.1038/srep14831 [DOI] [PMC free article] [PubMed]
- Li X, Wang J, Shi M, Wang W, Corre C, Yang K. Evidence for the formation of ScbR/ScbR2 heterodimers and identification of one of the regulatory targets in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2017;101:5333–5340. doi: 10.1007/s00253-017-8275-8. [DOI] [PubMed] [Google Scholar]
- Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang W, Shu D, Zhang W, Chen L, Qin Z, Yang S, Jiang W. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2007;77:625–635. doi: 10.1007/s00253-007-1184-5. [DOI] [PubMed] [Google Scholar]
- McKenzie NL, Nodwell JR. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol. 2007;189:5284–5292. doi: 10.1128/JB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol. 2014;10:e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, De Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gómez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, Van Der Donk WA, Dorrestein PC, Doyle S, Driessen AJM, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Höfte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Méndez C, Metsä-Ketelä M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Müller R, Neilan BA, Nett M, Nielsen J, O’Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Süssmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, Van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glöckner FO. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra S, Charaniya S, Takano E, Hu WS. A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS One. 2008;3:e2724. doi: 10.1371/journal.pone.0002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MJ, Young RA, Jones SE, Elliot MA. Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics. 2013;14(1):558. doi: 10.1186/1471-2164-14-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen ØM, Sletta H, Alam MT, Merlo ME, Moore J, Omara WAM, Morrissey ER, Juarez-Hermosillo MA, Rodríguez-García A, Nentwich M, Thomas L, Iqbal M, Legaie R, Gaze WH, Challis GL, Jansen RC, Dijkhuizen L, Rand DA, Wild DL, Bonin M, Reuther J, Wohlleben W, Smith MCM, Burroughs NJ, Martín JF, Hodgson DA, Takano E, Breitling R, Ellingsen TE, Wellington EMH. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics. 2010;11:1–9. doi: 10.1186/1471-2164-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Ohnishi Y, Beppu T, Horinouchi S. Evolution of γ-butyrolactone synthases and receptors in Streptomyces. Environ Microbiol. 2007;9:1986–1994. doi: 10.1111/j.1462-2920.2007.01314.x. [DOI] [PubMed] [Google Scholar]
- Onaka H, Nakagawa T, Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol. 1998;28:743–753. doi: 10.1046/j.1365-2958.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2) J Ind Microbiol Biotechnol. 2009;36:1073–1083. doi: 10.1007/s10295-009-0591-2. [DOI] [PubMed] [Google Scholar]
- Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E. A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2) Arch Microbiol. 2007;187:87–99. doi: 10.1007/s00203-006-0176-7. [DOI] [PubMed] [Google Scholar]
- Pawlik K, Kotowska M, Kolesiński P. Streptomyces coelicolor A3(2) produces a new yellow pigment associated with the polyketide synthase Cpk. J Mol Microbiol Biotechnol. 2010;19:147–151. doi: 10.1159/000321501. [DOI] [PubMed] [Google Scholar]
- Pérez-Redondo R, Rodríguez-García A, Botas A, Santamarta I, Martín JF, Liras P. ArgR of Streptomyces coelicolor is a versatile regulator. PLoS One. 2012;7:e32697. doi: 10.1371/journal.pone.0032697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martı M, Molina-henares AJ, Tera W, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico S, Santamaría RI, Yepes A, Rodríguez H, Laing E, Bucca G, Smith CP, Díaz M. Deciphering the regulon of Streptomyces coelicolor AbrC3, a positive response regulator of antibiotic production. Appl Environ Microbiol. 2014;80:2417–2428. doi: 10.1128/AEM.03378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S, Nothaft H, Noens EEE, Schlicht M, Colson S, Müller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, Van Wezel GP. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. 2006;61:1237–1251. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez E, Gramajo H. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- Rodríguez E, Banchio C, Diacovich L, Bibb MJ, Gramajo H. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor A3(2) Appl Environ Microbiol. 2001;67:4166–4176. doi: 10.1128/AEM.67.9.4166-4176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez H, Rico S, Díaz M, Santamaría RI. Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb Cell Factories. 2013;12:1–10. doi: 10.1186/1475-2859-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DA, Hasan AH, fei LY, Kime L, Ruiz-Larrabeiti O, Urem M, Bucca G, Mamanova L, Laing EE, van Wezel GP, Smith CP, Kaberdin VR, Mcdowall KJ. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol Microbiol. 2014;94:963–987. doi: 10.1111/mmi.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Rodríguez A, Rocha D, Ruiz-Villafan B, Tierrafría V, Rodríguez-Sanoja R, Segura-González D, Sánchez S. Transcriptomic analysis of a classical model of carbon catabolite regulation in Streptomyces coelicolor. BMC Microbiol. 2016;16:1–16. doi: 10.1186/s12866-016-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd BAM (1978) Genetics of pigmented secondary metabolites in Streptomyces coelicolor. PhD thesis. University of East Anglia, Norwich
- Ryding J, Todd B, Anderson WCC. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J Bacteriol. 2002;184:794–805. doi: 10.1128/JB.184.3.794-805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sello JK, Buttner MJ. The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J Bacteriol. 2008;190:4079–4083. doi: 10.1128/JB.01889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidda JD, Poon V, Song L, Wang W, Yang K, Corre C. Overproduction and identification of butyrolactones SCB1-8 in the antibiotic production superhost: Streptomyces M1152. Org Biomol Chem. 2016;14:6390–6393. doi: 10.1039/c6ob00840b. [DOI] [PubMed] [Google Scholar]
- Sun YQ, Busche T, Rückert C, Paulus C, Rebets Y, Novakova R, Kalinowski J, Luzhetskyy A, Kormanec J, Sekurova ON, Zotchev SB. Development of a biosensor concept to detect the production of cluster-specific secondary metabolites. ACS Synth Biol. 2017;6:1026–1033. doi: 10.1021/acssynbio.6b00353. [DOI] [PubMed] [Google Scholar]
- Świątek MA, Gubbens J, Bucca G, Song E, Yang YH, Laing E, Kim BG, Smith CP, Van Wezel GP. The ROK family regulator Rok7B7 pleiotropically affects xylose utilization, carbon catabolite repression, and antibiotic production in Streptomyces coelicolor. J Bacteriol. 2013;195:1236–1248. doi: 10.1128/JB.02191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Świątek-Połatyńska MA, Bucca G, Laing E, Gubbens J, Titgemeyer F, Smith CP, Rigali S, Van Wezel GP. Genome-wide analysis of in vivo binding of the master regulator DasR in Streptomyces coelicolor identifies novel non-canonical targets. PLoS One. 2015;10:1–24. doi: 10.1371/journal.pone.0122479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E, Gramajo HC, Strauch E, Andres N, White J, Bibb MJ. Transcriptional regulation of the redD transcriptional activator gene accounts for growth phase dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2) Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- Takano E, Kinoshita H, Mersinias V, Bucca G, Hotchkiss G, Nihira T, Smith CP, Bibb M, Wohlleben W, Chater K. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol. 2005;56:465–479. doi: 10.1111/j.1365-2958.2005.04543.x. [DOI] [PubMed] [Google Scholar]
- Tenconi E, Urem M, Świątek-Połatyńska MA, Titgemeyer F, Muller YA, Van Wezel GP, Rigali S. Multiple allosteric effectors control the affinity of DasR for its target sites. Biochem Biophys Res Commun. 2015;464:324–329. doi: 10.1016/j.bbrc.2015.06.152. [DOI] [PubMed] [Google Scholar]
- Thomas L, Hodgson DA, Wentzel A, Nieselt K, Ellingsen TE, Moore J, Morrissey ER, Legaie R, Wohlleben W, Rodríguez-García A, Martín JF, Burroughs NJ, Wellington EMH, Smith MCM. Metabolic switches and adaptations deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol Cell Proteomics. 2012;11:M111.013797. doi: 10.1074/mcp.M111.013797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Brückner R. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32:956–970. doi: 10.1039/c5np00013k. [DOI] [PubMed] [Google Scholar]
- Tsypik O, Makitrynskyy R, Bera A, Song L, Wohlleben W, Fedorenko V, Ostash B. Role of GntR family regulatory gene SCO1678 in gluconate metabolism in Streptomyces coelicolor M145. Biomed Res Int. 2017;2017(9529501):1–9. doi: 10.1155/2017/9529501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urem M, Świątek-Połatyńska MA, Rigali S, van Wezel GP. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol Microbiol. 2016;102:183–195. doi: 10.1111/mmi.13464. [DOI] [PubMed] [Google Scholar]
- Van Der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, Van Wezel GP. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep. 2018;35:575–604. doi: 10.1039/c8np00012c. [DOI] [PubMed] [Google Scholar]
- Van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011;28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Wang L, Zhang G, Fan K, Tan H, Yang K. A novel role of ‘pseudo’γ-butyrolactone receptors in controlling γ-butyrolactone biosynthesis in Streptomyces. Mol Microbiol. 2011;82:236–250. doi: 10.1111/j.1365-2958.2011.07811.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Mast Y, Wang J, Zhang W, Zhao G, Wohlleben W, Lu Y, Jiang W. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol. 2013;87:30–48. doi: 10.1111/mmi.12080. [DOI] [PubMed] [Google Scholar]
- Wang W, Ji J, Li X, Wang J, Li S, Pan G, Fan K, Yang K. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci. 2014;111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Wang J, Wang L, Tian X, Yang H, Fan K, Yang K, Tan H. “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem. 2010;285:27440–27448. doi: 10.1074/jbc.M110.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Seghezzi N, Esnault C, Virolle MJ. Repression of antibiotic production and sporulation in Streptomyces coelicolor by overexpression of a TetR family transcriptional regulator. Appl Environ Microbiol. 2010;76:7741–7753. doi: 10.1128/AEM.00819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Song E, Kim EJ, Lee K, Kim WS, Park SS, Hahn JS, Kim BG. NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2009;82:501–511. doi: 10.1007/s00253-008-1802-x. [DOI] [PubMed] [Google Scholar]
- Yang YH, Song E, Kim JN, Lee BR, Kim EJ, Park SH, Kim WS, Park HY, Jeon JM, Rajesh T, Kim YG, Kim BG. Characterization of a new ScbR-like γ-butyrolactone binding regulator (SlbR) in Streptomyces coelicolor. Appl Microbiol Biotechnol. 2012;96:113–121. doi: 10.1007/s00253-011-3803-4. [DOI] [PubMed] [Google Scholar]
- Yepes A, Rico S, Rodríguez-García A, Santamaría RI, Díaz M. Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PLoS One. 2011;6:e19980. doi: 10.1371/journal.pone.0019980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W (2012) Differential regulation of antibiotic biosynthesis by DraR-K , a novel two-component system in Streptomyces coelicolor. 85:535–556. 10.1111/j.1365-2958.2012.08126.x [DOI] [PubMed]
- Yu Z, Zhu H, Zheng G (2014) A genome-wide transcriptomic analysis reveals diverse roles of the two-component system DraR-K in the physiological and morphological differentiation of Streptomyces coelicolor. 10.1007/s00253-014-6102-z [DOI] [PubMed]