Abstract
To evaluate potential enhancement of Zika virus (ZIKV) infection among patients with prior dengue virus (DENV) infection, we compared loads of viral RNA among patients infected with ZIKV (n = 1070), DENV-2 (n = 312), or DENV-3 (n = 260). Compared to patients without prior DENV infection, patients with prior DENV infection had significantly higher mean loads of viral RNA if infected with DENV-2 (10.6 vs 11.6 log10 GCE/mL, respectively; t test, P < .0001) or DENV-3 (10.3 vs 10.9 log10 GCE/mL; P < .0001), but not ZIKV (4.7 vs 4.7 log10 GCE/mL; P = .959). These findings provide evidence against in vivo enhancement of ZIKV by anti-DENV antibodies.
Keywords: antibody-dependent enhancement, Puerto Rico, viral load, Zika virus
Prior dengue virus infection is associated with higher viral load in patients with dengue, but not Zika virus, disease.
INTRODUCTION
Following the emergence of dengue hemorrhagic fever (DHF) in Southeast Asia in the 1950s, antibody-dependent enhancement (ADE) was proposed as a pathophysiologic mechanism of increased disease severity [1]. Subsequent analyses demonstrated that ADE occurs when an antibody generated during an initial (primary) infection with 1 of the 4 dengue viruses (DENV-1–4) binds to virus during a subsequent (secondary) infection with a heterologous DENV; however, the antibody weakly neutralizes or fails to neutralize the infecting virus. Bound antibody then facilitates entry of DENV into Fc receptor-expressing immune cells (eg, macrophages, dendritic cells), resulting in more efficient virus uptake and replication and ultimately higher viral loads in serum. Studies suggest that high viral loads are associated with an imbalanced immune response and that these factors together may result in increased vascular leakage and, consequently, DHF [1]. In line with this explanation, the magnitude of viremia during DENV infection has been associated with increased disease severity, and secondary infection has been associated with increased risk of developing DHF; however, magnitude of risk differs by a variety of factors, including time between and sequence of DENV infections, anti-DENV IgG antibody titer, and the infecting DENV [1].
Following the emergence of Zika virus (ZIKV) from the Pacific islands to the Americas and concurrent detection of unexpected clinical syndromes [2], various in vitro and animal studies reported evidence of ADE of ZIKV mediated by anti-DENV antibodies [3]. Findings include increased levels of ZIKV replication in vitro and production of pro-inflammatory cytokines and an increase in ZIKV pathogenesis in immunodeficient mice [3]. Conversely, ZIKV pathogenesis in nonhuman primates was unaffected by pre-existing anti-DENV antibodies, and it instead resulted in modulation of innate and cellular immune responses to ZIKV infection, blunted duration of viremia, decreased liver injury, and increased activity of cytotoxic T cells [4]. Therefore, data from humans is needed to evaluate potential ADE of ZIKV mediated by anti-DENV antibodies in vivo [5].
The objective of this analysis was to seek evidence for 1 indicator of ADE—viral RNA levels—by assessing if prior DENV infection was associated with increased load of viral RNA among patients symptomatically infected with DENV-2, DENV-3, or ZIKV.
METHODS
Serum specimens were collected from febrile patients presenting for care with acute febrile illness (AFI) identified by the following: (1) the Enhanced Dengue Surveillance System (EDSS), which operated in the emergency department of a secondary care hospital from 2005–2006 [6]; or (2) the Sentinel Enhanced Dengue Surveillance System (SEDSS), which has operated in the emergency department of 1 tertiary care hospital and 1 out-patient clinic since 2012 [7]. Both EDSS and SEDSS were approved by the Institutional Review Board at the Centers for Disease Control and Prevention. Written informed consent or assent was obtained from all study participants.
Specimens included in this analysis had been collected within 5 days of illness onset and tested positive by reverse transcription-polymerase chain reaction (RT-PCR) for infection with DENV-2 or DENV-3 during 2005–2006 or ZIKV during 2016. Load of viral RNA was quantified using a standard curve method based on RT-PCR assays to detect DENV or ZIKV nucleic acid [8, 9]. Briefly, the same primer set used to detect DENV or ZIKV was used to generate virus-specific target amplicons, which were cloned individually into a TOPO TA vector system (Invitrogen, Carlsbad, CA). Insert-containing plasmids were expanded, purified, and used as templates to generate linear RNA transcripts by in vitro transcription (Lucigen, Middleton, WI). RNA transcripts were purified and the remaining DNA template was digested with Turbo DNaseI (Invitrogen, Carlsbad, CA). Quality control and quantification of the linear RNA transcripts were performed by spectrophotometry and real time RT-PCR with and without reverse transcriptase to account for the presence of template DNA in the RNA solution. Serial dilutions with known concentrations of RNA transcript were performed for each target virus and tested with the corresponding RT-PCR assay. Load of viral RNA, expressed as genome copy equivalents per milliliter of specimen (GCE/mL), were determined by linear regression calculated from CT values of each dilution and the corresponding RNA mass (converted to genome copies) detected per PCR reaction; this was further converted to copies per milliliter considering the RNA extraction procedure and the volume of specimen tested in each step of the process.
Status of primary versus secondary flavivirus infection (ie, serostatus) was defined by detection of anti-DENV immunoglobulin (Ig)G antibody by enzyme-linked immunosorbent assay (ELISA) [10]. Secondary infection was defined by detection of anti-DENV IgG antibody, whereas primary infection was defined by lack of detection of anti-DENV IgG antibody. DHF and severe dengue were defined according to the World Health Organization’s 1997 and 2009 guidelines, respectively [11, 12].
We used SAS version 9.3 (Cary, North Carolina) to compare the distribution of participant demographics, DPO, and serostatus between patients infected with ZIKV, DENV-2, and DENV-3, conducting t and χ 2 tests as appropriate. For each virus, the mean log10 GCE/mL for patients with primary or secondary infection was computed with unadjusted comparisons made using t tests. To facilitate adjusted comparisons between virus and serostatus, a multivariable linear regression model of log10 GCE/mL was fit with predictors for days post-illness onset (DPO; treated as categorical), virus, serostatus, and their 2- and 3-way product interaction terms. Using a backward selection approach, terms with a P value < .05 were retained in the model. From this model, heterogeneity in differences in mean log10 GCE/mL by serostatus for each virus were estimated controlling for DPO.
RESULTS
We identified patients infected with ZIKV (n = 1070), DENV-2 (n = 312), and DENV-3 (n = 260) (Table 1). Patients with ZIKV infection were older and more frequently female than those infected with DENV-2 or DENV-3. The distribution of DPO of specimen collection was similar for patients infected with ZIKV, DENV-2, and DENV-3; however, no specimens collected on DPO 0 were available for patients infected with DENV-2 or DENV-3. The majority (>80%) of patients infected with DENV-2 or ZIKV were experiencing secondary flavivirus infection, whereas those infected with DENV-3 were more evenly distributed between primary and secondary infection.
Table 1.
Characteristics of Patients Infected With ZIKV (n = 1070), DENV-2 (n = 312), or DENV-3 (n = 260), Puerto Rico, 2005–2006 and 2016
| ZIKV n = 1070 | DENV-2 n = 312 | DENV-3 n = 260 | |
|---|---|---|---|
| Participant characteristics | |||
| Age in years, median (range)a | 28 (0.08–88) | 25 (0.5–81) | 24 (0–84) |
| Female sex, n (%)b | 623 (58.2) | 127 (40.8) | 129 (49.6) |
| Pregnant, n (%) | 49 (4.6) | 5 (1.6) | 5 (1.9) |
| Hospitalized, n (%) | 18 (1.7) | 158 (50.6) | 114 (43.8) |
| Day post-illness onset of specimen collection, n (%) | |||
| 0 | 89 (8.3) | 0c | 0c |
| 1 | 332 (31.0) | 60 (19.2) | 55 (21.2) |
| 2 | 294 (27.4) | 63 (20.2) | 77 (30.0) |
| 3 | 245 (22.9) | 84 (26.8) | 62 (23.9) |
| 4 | 110 (10.3) | 66 (21.0) | 39 (15.1) |
| 5 | 0c | 39 (12.4) | 27 (10.4) |
| Serostatus | |||
| Primary, n (%) | 143 (13.4) | 39 (12.4) | 116 (44.8) |
| Secondary, n (%) | 927 (86.6) | 274 (87.8) | 144 (55.4) |
Abrreviations: DENV, dengue virus; ZIKV, Zika virus.
aAge was unavailable for 6 and 3 patients infected with DENV-2 and DENV-3, respectively.
bSex was unavailable for 1 patient infected with DENV-2.
cExcluded from analysis.
Patients with ZIKV infection were hospitalized significantly less frequently than those infected with DENV-2 or DENV-3 (1.7% vs 50.6% and 43.8%, respectively; P < .0001) (Table 1). Patients with secondary infection with DENV-2 were hospitalized more frequently than those with primary infection (53.3% vs 31.6%, respectively; P = .01). Frequency of hospitalization among patients infected with DENV-3 did not differ by status of primary versus secondary infection (39.6% vs 49.1%, respectively; P = .13). Only 1 patient met the case definition for DHF and had secondary infection with DENV-2. Among 16 patients who met the case definition for severe dengue, 5 had secondary and 0 had primary infection with DENV-2, and 7 had secondary and 4 had primary infection with DENV-3; these differences were not statistically significant.
Frequency of hospitalization was not different among patients with secondary versus primary infection with ZIKV (2.0% vs 1.4%, respectively; P = .62). Arthralgia was reported significantly more frequently among patients with secondary as compared to primary infection with ZIKV (82.3% vs 52.4%, respectively; P < .01); frequency of reported fever, rash, conjunctivitis, or all 4 symptoms was not significantly different by serostatus.
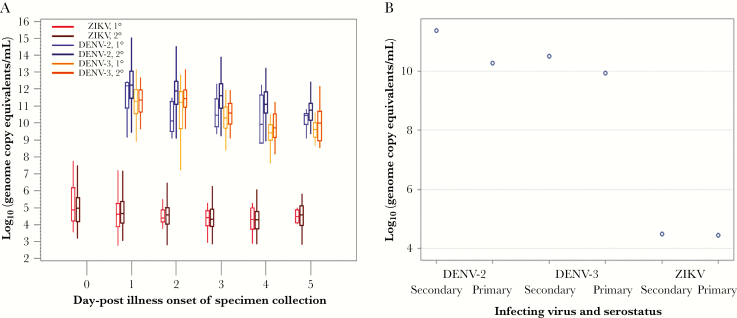
Load of viral RNA for each virus stratified by serostatus and DPO is shown in Figure 1A. Overall, load of viral RNA was significantly higher among patients with secondary compared to primary infection with DENV-2 (mean = 11.6 vs 10.6 log10 GCE/mL respectively [difference: +1.0]; P < .0001) or DENV-3 (mean = 10.9 vs 10.3 GCE/mL [difference: +0.6]; P < .0001), but not ZIKV (mean = 4.7 vs 4.7 log10 GCE/mL [difference: +0.0]; P = .96).
Figure 1.
Load of Viral RNA in Patients Infected With ZIKV (n = 1070), DENV-2 (n = 312), or DENV-3 (n = 260), Puerto Rico, 2005–2006 and 2016
A, Viral load by serostatus and day post-illness onset of specimen collection. Upper and lower limits of plots indicate the maximum and minimum values, top and bottom limits of the boxes indicate the third and first quartiles, and the horizontal bar inside the box indicates the median. B, Median viral load by serostatus. DENV indicates dengue virus; ZIKV, Zika virus.
The multivariable model of log10 GCE/mL retained terms for DPO, virus, serostatus, and the interactions between virus and DPO (P < .0001), suggesting virus-specific differences in load of viral RNA over time. We observed interaction between virus and serostatus, indicating virus-specific differences in effect of serostatus on load of viral RNA. These 2 factors together contributed nearly all of the observed variation in load of viral RNA (r2 = 0.92). Although there was an overall significant interaction between virus and serostatus (Figure 1B), adjusted differences in load of viral RNA by serostatus were significant for DENV-2 (adjusted difference = +1.1; 95% CI, +0.7–1.5; log10 GCE/mL higher for secondary vs primary infection) and DENV-3 (+0.6; 95% CI, +0.3–0.8), but not ZIKV (+0.0; 95% CI, -0.2–+0.3). Similarly, the effect of serostatus on load of viral RNA was significantly different for DENV-2 and DENV-3 combined compared to ZIKV (P < .0001).
DISCUSSION
Utilization of 2 facility-based surveillance systems to identify patients with dengue or ZIKV disease enabled evaluation of load of viral RNA in humans as a proxy for ADE of ZIKV and DENV. After quantitation of load of viral RNA among patients experiencing primary or secondary infection with ZIKV, DENV-2, or DENV-3, we did not observe evidence to support in vivo enhancement of ZIKV mediated by anti-DENV antibodies; however, increased load of viral RNA was observed to enhance DENV-2 and DENV-3. Similarly, risk of hospitalization was higher among patients experiencing secondary infection with DENV-2, but not DENV-3 or ZIKV.
Evaluation of in vivo enhancement of ZIKV mediated by anti-DENV antibodies is challenging for several reasons. First, the large majority of pediatric and adult patients with ZIKV disease do not require in-patient care, among whom there is no apparent clinical phenotype by which to measure enhancement of acute ZIKV disease. Therefore, load of viral RNA or quantitation of cytokine levels may be the most appropriate in vivo markers of potential ADE. One study from Colombia utilized these measures to evaluate ADE among 45 patients infected with ZIKV and 20 infected with DENV-2 [13]. Although no association was observed between serostatus and load of viral RNA in patients infected with either ZIKV or DENV-2, interpretation of the findings was limited by a small sample size and lack of stratification of specimens by DPO, which is relevant as viremia decreases over time. Similar findings were observed in a case-control study of pregnant women with ZIKV infection in Brazil [14].
The magnitude of enhancement of DENV can differ according to the infecting DENV, as observed here, as well as the sequence of and time between DENV infections [1]. Consequently, we cannot rule out that enhancement of ZIKV may occur among other populations with a different history of prior infection with DENV. Moreover, even if anti-DENV antibodies do not increase Zika viral loads to cause severe disease, prior DENV infection may still be associated with the virulence or pathogenesis, or both, of ZIKV. For example, although there is no evidence to date to associate maternal serostatus with congenital Zika syndrome [15], initial findings suggest that having neutralizing antibodies against DENV-2 is associated with developing Guillain-Barré syndrome after ZIKV infection [16]. Therefore, further investigation of possible enhancement of ZIKV infection mediated by anti-DENV antibodies is merited, as findings are relevant to understanding the pathophysiology of severe complications of ZIKV infection as well as evaluation of ZIKV and DENV vaccines.
Though strengthened by evaluation of a robust number of patients infected with ZIKV, DENV-2, or DENV-3, this investigation still was subjected to limitations. First, different molecular diagnostic tests were used to screen patients with AFI for infection with DENV or ZIKV; however, because the performance of both tests is similar [8, 9], it is unlikely that individuals with low loads of viral RNA were missed. Second, although dengue patients experiencing secondary infection with DENV-2 were more often hospitalized than those with primary infection, hospitalization is an imperfect measure of disease severity due to variable clinical practices. Because of the small number of dengue patients that met the case definitions for DHF or severe dengue, we were unable to reliably assess the proportion of patients with severe disease by primary versus secondary infection with DENV-2 or -3. Last, in classifying patients with secondary flavivirus infection, we did not quantify anti-DENV IgG titers or identify the specific DENV(s) with which patients had been previously infected. Consequently, we were unable to determine if a specific range of anti-DENV IgG titer or sequence of past DENV infection may have been associated with differential increases in load of viral RNA.
In summary, our findings provide evidence against in vivo enhancement of ZIKV infection mediated by anti-DENV antibodies. Additional studies in other jurisdictions in populations with different histories of DENV infection prior to ZIKV infection should be conducted to further these findings.
Acknowledgments
We thank Laura Adams for assistance with data queries and management. We also thank the patients that enrolled in the Enhanced Dengue Surveillance System and Sentinel Enhanced Dengue Surveillance System for their participation in these studies, as well as the study and clinical staff at Saint Luke’s Episcopal Hospital in Ponce and Guayama that made the studies possible.
Disclaimer. The views expressed in this article are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention or the US Public Health Service.
Financial support. This study was supported by the US Centers for Disease Control and Prevention.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Wilder-Smith A, Ooi EE, Horstick O, Wills B. Dengue. Lancet 2019; 393:350–63. [DOI] [PubMed] [Google Scholar]
- 2. Pierson TC, Diamond MS. The emergence of Zika virus and its new clinical syndromes. Nature 2018; 560:573–81. [DOI] [PubMed] [Google Scholar]
- 3. Sariol CA, Nogueira ML, Vasilakis N. A tale of two viruses: does heterologous flavivirus immunity enhance Zika disease? Trends Microbiol 2018; 26:186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pantoja P, Pérez-Guzmán EX, Rodríguez IV, et al. . Zika virus pathogenesis in rhesus macaques is unaffected by pre-existing immunity to dengue virus. Nat Commun 2017; 8:15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halstead SB. Biologic evidence required for Zika disease enhancement by dengue antibodies. Emerg Infect Dis 2017; 23:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramos MM, Arguello DF, Luxemburger C, et al. . Epidemiological and clinical observations on patients with dengue in Puerto Rico: results from the first year of enhanced surveillance--June 2005-May 2006. Am J Trop Med Hyg 2008; 79:123–7. [PubMed] [Google Scholar]
- 7. Tomashek KM, Lorenzi OD, Andujar-Perez DA, et al. . Clinical and epidemiologic characteristics of dengue and other etiologic agents among patients with acute febrile illness, Puerto Rico, 2012–2015. PLOS Negl Trop Dis 2017; 11:e0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chien LJ, Liao TL, Shu PY, et al. . Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J Clin Microbiol 2006; 44:1295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santiago GA, Vázquez J, Courtney S, et al. . Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat Commun 2018; 9:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson AJ, Martin DA, Karabatsos N, Roehrig JT. Detection of anti-arboviral immunoglobulin G by using a monoclonal antibody-based capture enzyme-linked immunosorbent assay. J Clin Microbiol 2000; 38:1827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Health Organization. Dengue Haemorrhagic Fever: Diagnosis, Treatment, and Control. Geneva, Switzerland: World Health Organization, 1997. [Google Scholar]
- 12. World Health Organization. Dengue Guidelines for Diagnosis, Treatment, Prevention, and Control. Geneva, Switzerland: World Health Organization, 2009. [PubMed] [Google Scholar]
- 13. Terzian ACB, Schanoski AS, Mota MTO, et al. . Viral load and cytokine response profile does not support antibody-dependent enhancement in dengue-primed Zika virus-infected patients. Clin Infect Dis 2017; 65:1260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pedroso C, Fischer C, Feldmann M, et al. . Cross-protection of dengue virus infection against congenital Zika syndrome, Northeastern Brazil. Emerg Infect Dis 2019; 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halai UA, Nielsen-Saines K, Moreira ML, et al. . Maternal Zika virus disease severity, virus load, prior dengue antibodies, and their relationship to birth outcomes. Clin Infect Dis 2017; 65:877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lynch RM, Mantus G, Encinales L, et al. . Augmented Zika and dengue neutralizing antibodies are associated with Guillain-Barre syndrome. J Infect Dis 2019; 219:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]