Low-cost, high-throughput nucleic acid sequencing ushered the field of microbial ecology into a new era in which the microbial composition of nearly every conceivable environment on the planet is under examination. However, static “screenshots” derived from sequence-only approaches belie the underlying complexity of the microbe-microbe and microbe-host interactions occurring within these systems.
KEYWORDS: interactions, microbial communities, microbiome, model systems
ABSTRACT
Low-cost, high-throughput nucleic acid sequencing ushered the field of microbial ecology into a new era in which the microbial composition of nearly every conceivable environment on the planet is under examination. However, static “screenshots” derived from sequence-only approaches belie the underlying complexity of the microbe-microbe and microbe-host interactions occurring within these systems. Reductionist experimental models are essential to identify the microbes involved in interactions and to characterize the molecular mechanisms that manifest as complex host and environmental phenomena. Herein, we focus on three models (Bacillus-Streptomyces, Aliivibrio fischeri-Hawaiian bobtail squid, and gnotobiotic mice) at various levels of taxonomic complexity and experimental control used to gain molecular insight into microbe-mediated interactions. We argue that when studying microbial communities, it is crucial to consider the scope of questions that experimental systems are suited to address, especially for researchers beginning new projects. Therefore, we highlight practical applications, limitations, and tradeoffs inherent to each model.
PERSPECTIVE
Microbiomes shape the fundamental biology of environments and can have substantial impacts on macroscopic ecosystems. Within their hosts, microbiomes alter metabolism, behavior, and disease. Experimental insight into the molecular mechanisms underlying microbiome interactions remains elusive. High complexity, variable plasticity, and low manipulability of natural systems remain barriers to recapitulating microbiomes in the laboratory.
Distilling the extreme complexity of biology into discrete, functional units remains a difficult challenge. As early as 1662, René Descartes posited that biology could be explained as collectives of self-operating machinery termed “automata” (1). We have dissected the molecular nature of these “machines” into their constituent parts. For example, forward genetic screens, reverse genetics, and complementation aim to connect genomic loci with organism-level effects and are invaluable in understanding how genes function and phenotypes manifest. As we increasingly appreciate how microbes influence ecology and host fitness, models are essential to limit complexity and maximize experimental control, such that we can begin to understand how interactions within microbial communities influence biology (2). From a microbial perspective, understanding the influences of fitness can resolve common and distinct features of microbial interactions in different systems. While microbial fitness is often conceived of as a static property, the dynamics of microbial interactions are shaped by environmental and temporal plasticity and competition. Thus, the phenotypes that shape microbial fitness are the sum of many variables, including but not limited to the presence and regulation of genes, the interspecies interactions of a microbial community, and chemical gradients (3). Further, emergent properties of microbial communities can confound the simplest studies. For example, different combinations of relatively simple ≤5 member communities in Drosophila can mediate changes in host life span and fecundity, with some members influencing these traits only in the presence of certain other community members (4). Considering this complexity, model systems integrating reductionist experimental frameworks are necessary to link the underlying interaction networks of microbiomes to host biology.
For early career researchers and researchers embarking on new projects, it is important to understand the kinds of questions that certain models address well and where reduction can maximize experimental control with minimal loss of biological relevance. Herein, we describe three model systems with different levels of manipulability and complexity which have been used to uncover molecular mechanisms of interactions. First, we discuss Bacillus-Streptomyces pairwise interactions to highlight the high experimental control and manipulability of this system used to uncover molecular mechanisms of microbial competition. We then discuss the Aliivibrio-squid system, which is uniquely suited for studies of microbial colonization. Finally, we discuss gnotobiotic mice as a model system that can be used to investigate mammalian gut interactions. We highlight where each of these models excels (Fig. 1) and describe limitations within each system to underscore the importance of selecting an appropriate model to address the scientific question at hand.
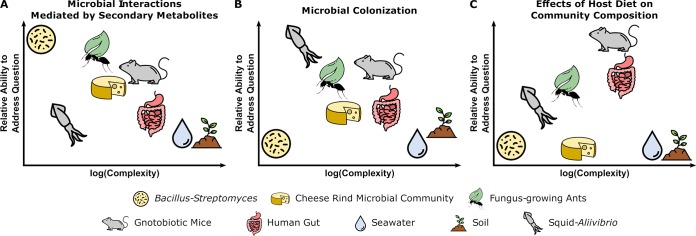
FIG 1.
Tradeoffs between experimental questions and complexity of microbiome systems. Each microbiome system is suited to address different types of questions based on the culturability of microbes, genetic tractability of microbes and host (where relevant), ability to maintain system in laboratory setting, and ability to make host/environment germfree. Three different systems are shown in this figure as examples. (A) Pairwise interactions between B. subtilis and Streptomyces spp. are well-suited for characterizing the functions of secondary metabolites in microbial interactions. (B) The symbiosis between bobtail squid and A. fischeri is fundamental to understanding host and microbial factors that influence colonization. (C) The use of gnotobiotic mice is crucial for making links between host diet and the effects on specific microbial taxa in a community (see the text for specific details). Specific original image credit from the Noun Project (https://thenounproject.com/): Fertile Soil by Ben Davis; Droplet by Focus; Mouse by Iconic; Cheese Wheel by Anniken & Andreas; Bacteria by Arthur Shlain; Squid by Artem Kovyazin; ant by Yugudesign; leaf by Saeful Muslim; all used and modified under the Creative Commons License, Attribution 3.0.
UNCOVERING MOLECULAR MECHANISMS OF INTERACTIONS USING BACILLUS AND STREPTOMYCES
Among the simplest model systems for exploring microbial interactions are pairwise interactions between culturable bacteria. Importantly, these systems intrinsically offer high experimental control to study the molecular underpinnings of interactions that occur between and within microbial communities. As an example, coculture of the soil bacteria Bacillus subtilis and Streptomyces spp. demonstrates the power to dissect the molecular mechanisms of competition. Both B. subtilis and Streptomyces species are amenable to genetic manipulation, produce antibiotics and other secondary metabolites, and undergo multicellular development (e.g., biofilm formation, motility, and sporulation) on agar plates, providing macroscopic visualization of interactions. Together, the ability to perform mutagenesis screens, generate targeted gene deletions and complements, extract secondary metabolites in isolation, and easily adjust medium and plating configurations to uncover new macroscopic phenotypes all contribute to this system’s high level of experimental manipulability.
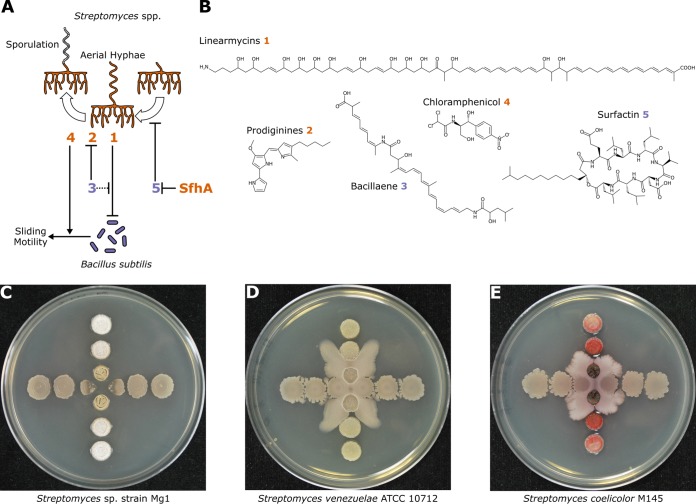
Pairwise interactions between Bacillus and Streptomyces demonstrate that secondary metabolites have multiple roles mediating competition (Fig. 2). For instance, B. subtilis produces the lipopeptide surfactin, which triggers its own biofilm formation and multicellular motility (5–7). In contrast, surfactin interferes with the aerial development and sporulation of many Streptomyces spp. (8). However, Streptomyces sp. strain Mg1 produces a secreted hydrolase that detoxifies surfactin and allows this bacterium to sporulate when cultured with B. subtilis (9). Similarly, B. subtilis produces bacillaene that interferes with prodigiosin pigment production in Streptomyces coelicolor and Streptomyces lividans (10, 11) and protects B. subtilis from lysis by linearmycins produced by strain Mg1 (12–14) (Fig. 2C). In addition to bacillaene, B. subtilis may protect itself from linearmycin-induced lysis by activating a linearmycin-induced, coupled signaling system and exporter that are necessary and sufficient for linearmycin resistance (12, 15). Finally, as an additional means to escape competition, subinhibitory concentrations of chloramphenicol and several other ribosome-targeting antibiotics induce directional sliding motility in B. subtilis away from Streptomyces (16) (Fig. 2D).
FIG 2.
Secondary metabolites mediate interactions between B. subtilis and Streptomyces spp. (A) Summary schematic of interactions between B. subtilis and Streptomyces spp. The secondary metabolites produced by B. subtilis and Streptomyces spp. are represented by the purple and orange numbers, respectively, and the chemical structures are shown in panel B. SfhA refers to surfactin hydrolase produced by Streptomyces sp. strain Mg1 that specifically hydrolyzes the ester linkage in surfactin (compound 5). (C to E) Streptomyces spp. (vertical) and B. subtilis (horizontal) spotted in a perpendicular pattern on agar plates. (C) B. subtilis colonies proximal to Streptomyces sp. strain Mg1 colonies are lysed by linearmycins (compound 1). (Republished from Frontiers in Microbiology [3].) (D) Subinhibitory concentrations of chloramphenicol (compound 4) produced by Streptomyces venezuelae induce sliding motility of proximal B. subtilis colonies. (E) Production of the red pigment prodiginine (compound 2) is strongly induced in Streptomyces coelicolor colonies proximal to sliding B. subtilis colonies, which do not produce bacillaene (compound 3). (Images in panels D and E courtesy of Yongjin Liu and Paul Straight, reproduced with permission.)
We highlight the above as examples of multifaceted interactions that can occur between one pair of microbes. Further, even by simply substituting one member of the pair, new interaction dynamics may emerge. For instance, recent work on interactions between Streptomyces venezuelae and Saccharomyces cerevisiae uncovered a new type of “exploration” motility in S. venezuelae induced by the production of volatile trimethylamine (17). However, it is important to consider the artificial abstraction when microbes are transplanted into the laboratory. Compared to microbes in their natural environments, microbes in growth medium encounter atypical nutrients at inordinate concentrations and grow at unnaturally high cell densities. Consequently, microbes may produce extracellular products (e.g., antibiotics) at concentrations that elicit nonphysiological/hormetic responses in interacting partners (18, 19). Furthermore, the evolutionary implications from pairwise interactions are often unknown or unclear. Nevertheless, microbial coculture allows us to infer mechanisms that are impossible to uncover from sequencing studies alone. Therefore, to gain similar mechanistic insight into interactions occurring in communities, model systems where microbes can be isolated in pure culture and investigated in simplified pairwise interactions are invaluable.
COLONIZATION OF THE LIGHT ORGAN BY ALIIVIBRIO FISCHERI TO INVESTIGATE HOST-MICROBE INTERACTIONS
The bacterium Aliivibrio fischeri (formerly Vibrio fischeri) specifically establishes a symbiosis within the light organ of newly hatched Hawaiian bobtail squid (Euprymna scolopes). This symbiosis has proven an excellent system to investigate colonization dynamics and specificity: though the ocean harbors an incredibly complex microbial community (>106 bacterial cells/ml), the relatively rare A. fischeri (<1 in 5,000 cells) specifically colonizes the light organ (20).
Specialized cilia and mucus recruit A. fischeri during early squid development. Bacteria within the mucus are chemotactically attracted toward pores and swim into light organ crypts (21). During the earliest stages of colonization, A. fischeri expresses a suite of genes under the “symbiotic colonization-sensor” RscS regulator (22, 23), which promotes polysaccharide production and biofilm formation (24–26) essential for colonization. The bacterially produced, diaminopimelic acid (DAP) type peptidoglycan tracheal cytotoxin (TCT) and lipid A cause apoptosis of ciliated cells (20). The squid subsequently detoxifies TCT (27) and lipid A (28), followed by hemocyte infiltration and tissue regeneration to form the mature light organ (20). Further, squid nitric oxide (NO) signaling (29, 30) and detoxification (31) are tuned in response to colonization, modulating A. fischeri populations and excluding competitors from the light organ (20). When RscS is introduced into A. fischeri MJ11, a fish symbiont that naturally lacks RscS, the bacteria gain the ability to colonize E. scolopes (23), despite more than 400 unique genes in the laboratory squid strain ES114 compared to MJ11. Aside from biofilm formation and RscS-controlled responses, bacterial motility (20), type VI secretion systems (32), bacterial stress responses (33), other A. fischeri regulatory cascades (34), and host genetic factors (35) play key roles in colonization success.
An implicit and unique strength of the squid-A. fischeri light organ system is its simplicity, as one-host, one-microbe studies are experimentally manageable and yield ecologically relevant insights into the molecular mechanisms of this symbiosis. Historically, the majority of mechanistic research describing both host and microbe in the squid-Aliivibrio symbiosis has focused on a single strain, A. fischeri ES114. As such, assessing the extent to which the molecular insights of ES114 colonization apply to other A. fischeri strains remains an ongoing effort in this system. Notably, multiple strains of ecologically and phylogenetically distinct A. fischeri have been experimentally evolved within the squid host, selecting for alleles of the regulator binK that coordinate symbiosis traits and enhance colonization and growth within the light organ (36). Thus, to better understand how specificity relates to the diversity of both A. fischeri and E. scolopes that exists in nature, future studies are needed to address the impact of strain- and population-level diversity on colonization success and host-microbe fidelity. Nevertheless, the many molecular interactions between one host species and one bacterial strain in this system, even when restricting focus to interactions surrounding colonization, make it a promising research area. Furthermore, whether the specialized physical, chemical, and genetic interactions between squid and A. fischeri during colonization have broader implications across different microbes and hosts is unknown. However, a newly emerging system involves the squid nidamental gland, which is situated next to the light organ and harbors a more complex community that consists of Roseobacter, Flavobacteriales, Rhizobiales, and Verrucomicrobia (37). We envision that comparison between these two adjacent organs within the same animal that recruit a different set of microbial symbionts from the same seawater environment will provide further insight into how host selection affects microbiome composition and function.
LEVELS OF COMPLEXITY IN GERMFREE MICE
In humans, the gut microbiota is a complex community containing hundreds of species that impact a variety of health outcomes (38, 39). The microbiota is critical for normal development, as germfree animals possess immune, digestive, and behavioral differences compared to conventional counterparts (40). Germfree animals offer a platform for characterizing interactions with the host and defined communities of microbes (together known as gnotobiotics), ranging from monoassociations to complex communities. Arguably, monocolonized and germfree animals represent vast oversimplification. Defined synthetic communities simplify complex microbiotas while maintaining diversity, and the use of genome-sequenced strains facilitates multi-omics studies (41, 42). Further, using a simplified core microbiota with a genetically tractable strain of interest offers a compromise between creating a well-controlled experiment and not relying on monoassociation studies. For example, to determine the role of the microbial conversion of choline to trimethylamine, mice were colonized with a simplified, six-member gut microbiota containing a single member that could metabolize choline or a mutant of the same strain that was unable to use choline. This approach demonstrates that choline-metabolizing bacteria compete with their hosts for choline and can exacerbate diet-induced metabolic disease in hosts and alter DNA methylation patterns in the brains of offspring (43). Notably, the choline utilization pathway is not taxonomically conserved, and it would be impossible to infer this phenotype from sequencing the 16S rRNA gene from gut communities (44).
To study entire communities, germfree mice can be colonized with complex communities, often from fecal samples. Donor communities can demonstrate a proof of principle of microbiota-mediated effects on a particular phenotype, such as linking the microbiota to obesity (45, 46). However, with increasing community complexity, more reproducibility issues arise. For instance, though donor communities reduce the artificial nature of gnotobiotics, rare strains may be stochastically lost in the transplanted community. When human fecal microbiota are transplanted into germfree mice, 10 to 30% of operational taxonomic units fail to colonize the mouse (47). Strains present at 0.15% of the community can impact phenotypes like choline conversion to trimethylamine (44). Alternatively, using donor microbiota derived from the same species as the germfree animal can be more appropriate for certain ecological questions and better retain members (48). Reproducibility is also an issue for studying some emergent phenotypes of complex communities, as maintenance of certain members may depend on diet or even water pH (49), and social, coprophagous animals like mice may necessitate cages as biological units of replication, rather than individuals (50, 51). Though reproducibility issues also arise in simplified communities, troubleshooting whether small changes in abiotic or biotic factors influence phenotypes is more challenging in complex communities and could be limiting in a mouse system with a relatively slow generation time and ethical constraints on animal usage.
Overall, gnotobiotic animals provide an approach to interrogate the role of complex microbiota in emergent phenotypes of interest by reducing the complexity to controllable independent variables (e.g., a single bacterial strain or product). Experimenting with multiple levels of community complexity applies to germfree hosts beyond mice (e.g., Arabidopsis, Danio, and Drosophila), but specific mechanisms may differ. For example, facultatively anaerobic pathogens exploiting inflammation-associated oxidation in the typically anaerobic mouse gut would not be readily apparent in aerobic Drosophila guts (52, 53). Further, although mice are often sought as medically relevant models, the ease of producing large numbers of gnotobiotic animals and availability of tools in other models, such as imaging in translucent zebrafish, can reveal alternative mechanisms for microbial proteins mediating mutualism that may have remained obscure in a mouse model (54). Ultimately, shared insights from different models support broad ecological principles of microbiome interactions.
CONCLUSION
By leveraging the unique features of experimental microbiome systems, important and outstanding questions can be addressed (Fig. 1). Chief among these questions is understanding how interactions between microbes and hosts influence behavior and health and how communities respond to perturbations, such as invasion or abiotic stresses. Although it is well understood that microbiomes influence the health of hosts and macroscopic ecosystems, the specific molecular mechanisms remain elusive. For instance, what interactions differentiate “healthy” and “dysbiotic” microbial communities are often unresolved. Further, communities can exhibit emergent phenotypes that are not seen when members are grown in isolation, such as catabolism of recalcitrant materials (55, 56), biofilm formation (57), or antibiotic production (58–60).
As microbiome research continues, new frameworks for characterizing the interactions that occur within microbial communities will emerge from novel systems spanning the spectra of complexity and tractability and developments enabling established systems to address new questions. As examples, two particular systems that we are especially interested in are the cheese rind microbial community and the gardens of fungus-growing ants. The cheese rind microbial community is an emerging system particularly suitable for characterization of multipartite interactions and simulating ecological phenomena through control of abiotic factors (61–64), yet the unclear evolutionary relationships between members may limit its applicability to coevolved, natural systems. In contrast, because the microbial symbionts of fungus-growing ants provide a coevolutionary framework from which to investigate microbial population dynamics (65, 66), nutrient flow (67), host-pathogen interactions (68–70), and defensive symbiosis (71), further characterizations of these microbiomes may provide broader implications for other natural systems (59, 60).
In conclusion, delineating community states that contribute to emergent properties and complex interactions will require experimental models, and the ideal balance between a model’s complexity, ease of manipulation, and overall biological relevance will depend upon the scientific questions posed.
ACKNOWLEDGMENTS
We apologize to those whose work we have not discussed due to the space constraints.
We thank Mark Mandel for critical appraisal of the manuscript.
This project was supported through National Institutes of Health (NIH) U19 Al109673 and NIH U19 TW009872. Additional support was provided to M.G.C. by NIH T32 GM008505, to R.M.S. by an NLM training grant to the Computation and Informatics in Biology and Medicine Training Program (NLM 5T15LM007359), and to J.R.B. by NIH T32 AI55397.
REFERENCES
- 1.Descartes R. 1662. De homine figuris et latinatate donatus a Florentio Schuyl. P. Leffen & F. Moyardum, Leyden, The Netherlands. [Google Scholar]
- 2.Tecon R, Mitri S, Ciccarese D, Or D, van der Meer JR, Johnson DR. 2019. Bridging the holistic-reductionist divide in microbial ecology mSystems 4:e00265-18. doi: 10.1128/mSystems.00265-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stubbendieck RM, Vargas-Bautista C, Straight PD. 2016. Bacterial communities: interactions to scale. Front Microbiol 7:1234. doi: 10.3389/fmicb.2016.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, Ludington WB. 2018. Microbiome interactions shape host fitness. Proc Natl Acad Sci U S A 115:E11951–E11960. doi: 10.1073/pnas.1809349115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. [DOI] [PubMed] [Google Scholar]
- 6.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gestel J, Vlamakis H, Kolter R. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straight PD, Willey JM, Kolter R. 2006. Interactions between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J Bacteriol 188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoefler BC, Gorzelnik KV, Yang JY, Hendricks N, Dorrestein PC, Straight PD. 2012. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc Natl Acad Sci U S A 109:13082–13087. doi: 10.1073/pnas.1205586109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. 2007. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci U S A 104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vargas-Bautista C, Rahlwes K, Straight P. 2014. Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J Bacteriol 196:717–728. doi: 10.1128/JB.01022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbendieck RM, Straight PD. 2015. Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genet 11:e1005722. doi: 10.1371/journal.pgen.1005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stubbendieck RM, Brock DJ, Pellois J-P, Gill JJ, Straight PD. 2018. Linearmycins are lytic membrane-targeting antibiotics. J Antibiot 71:372. doi: 10.1038/s41429-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 14.Hoefler BC, Stubbendieck RM, Josyula NK, Moisan SM, Schulze EM, Straight PD. 2017. A link between linearmycin biosynthesis and extracellular vesicle genesis connects specialized metabolism and bacterial membrane physiology. Cell Chem Biol 24:1238–1249.e7. doi: 10.1016/j.chembiol.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Stubbendieck RM, Straight PD. 2017. Linearmycins activate a two-component signaling system involved in bacterial competition and biofilm morphology. J Bacteriol 199:e00186-17. doi: 10.1128/JB.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Kyle S, Straight PD. 2018. Antibiotic stimulation of a Bacillus subtilis migratory response. mSphere 3:e00586-17. doi: 10.1128/mSphere.00586-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA. 2017. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 6:1–21. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies J. 2006. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol 33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 19.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Mandel MJ, Dunn AK. 2016. Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Front Microbiol 7:1982. doi: 10.3389/fmicb.2016.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 23.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh P, Brooks JF, Ray VA, Mandel MJ, Visick KL. 2015. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol 81:5223–5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visick KL, Skoufos LM. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. 2010. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol 12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rader BA, Kremer N, Apicella MA, Goldman WE, McFall-Ngai MJ. 2012. Modulation of symbiont lipid A signaling by host alkaline. Mar Biol 3:e00093-12. doi: 10.1128/mBio.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. 2010. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A 107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Ruby EG. 2011. The roles of NO in microbial symbioses. Cell Microbiol 13:518–526. doi: 10.1111/j.1462-5822.2011.01576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker NP, Le Brun NE, Dixon R, Hutchings MI. 2010. There’s NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol 18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN. 2018. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A 115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-Vibrio model. PLoS Biol 12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabrina Pankey M, Foxall RL, Ster IM, Perry LA, Schuster BM, Donner RA, Coyle M, Cooper VS, Whistler CA. 2017. Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria. Elife 6:e24414. doi: 10.7554/eLife.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins AJ, Fullmer MS, Gogarten JP, Nyholm SV. 2015. Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front Microbiol 6:123. doi: 10.3389/fmicb.2015.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, et al. . 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 40.Wostmann BS. 1996. Germfree and gnotobiotic animal models: background and applications, 1st ed CRC Press, Boca Raton, FL. [Google Scholar]
- 41.Bratburd JR, Keller C, Vivas E, Gemperline E, Li L, Rey FE, Currie CR. 2018. Gut microbial and metabolic responses to Salmonella enterica serovar Typhimurium and Candida albicans. mBio 9:e02032-18. doi: 10.1128/mBio.02032-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romano KA, Martinez-del Campo A, Kasahara K, Chittim CL, Vivas EI, Amador-Noguez D, Balskus EP, Rey FE. 2017. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22:279–290.e7. doi: 10.1016/j.chom.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. 2015. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6:e02481-14. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 46.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arrieta M-C, Walter J, Finlay BB. 2016. Human microbiota-associated mice: a model with challenges. Cell Host Microbe 19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 49.Barnett JA, Gibson DL. 2019. H2Oh No! The importance of reporting your water source in your in vivo microbiome studies. Gut Microbes 10:261–269. doi: 10.1080/19490976.2018.1539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hugenholtz F, de Vos WM. 2018. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazic SE, Clarke-Williams CJ, Munafò MR. 2018. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol 16:e2005282. doi: 10.1371/journal.pbio.2005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun 75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ. 2016. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rolig AS, Sweeney EG, Kaye LE, DeSantis MD, Perkins A, Banse AV, Hamilton MK, Guillemin K. 2018. A bacterial immunomodulatory protein with lipocalin-like domains facilitates host–bacteria mutualism in larval zebrafish. Elife 7:e37172. doi: 10.7554/eLife.37172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewin GR, Johnson AL, Soto RDM, Perry K, Book AJ, Horn HA, Pinto-Tomás AA, Currie CR. 2016. Cellulose-enriched microbial communities from leaf-cutter ant (Atta colombica) refuse dumps vary in taxonomic composition and degradation ability. PLoS One 11:e0151840. doi: 10.1371/journal.pone.0151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlos C, Fan H, Currie CR. 2018. Substrate shift reveals roles for members of bacterial consortia in degradation of plant cell wall polymers. Front Microbiol 9:364. doi: 10.3389/fmicb.2018.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lozano GL, Bravo JI, Garavito Diago MF, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, Handelsman J. 2019. Introducing THOR, a model microbiome for genetic dissection of community behavior. mBio 10:e02846-18. doi: 10.1128/mBio.02846-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, Braun DR, Nelson J, Simpkins SW, McDonald BR, Myers CL, Piotrowski JS, Thompson CJ, Currie CR, Li L, Rajski SR, Bugni TS. 2017. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem Biol 12:3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevrette MG, Currie CR. 2019. Emerging evolutionary paradigms in antibiotic discovery. J Ind Microbiol Biotechnol 46:257–271. doi: 10.1007/s10295-018-2085-6. [DOI] [PubMed] [Google Scholar]
- 60.Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE, Barns KJ, Book AJ, Cagnazzo J, Carlos C, Flanigan W, Grubbs KJ, Horn HA, Hoffmann FM, Klassen JL, Knack JJ, Lewin GR, McDonald BR, Muller L, Melo WGP, Pinto-Tomás AA, Schmitz A, Wendt-Pienkowski E, Wildman S, Zhao M, Zhang F, Bugni TS, Andes DR, Pupo MT, Currie CR. 2019. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun 10:516. doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfe BE, Button JE, Santarelli M, Dutton RJ. 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monnet C, Landaud S, Bonnarme P, Swennen D. 2015. Growth and adaptation of microorganisms on the cheese surface. FEMS Microbiol Lett 362:1–9. doi: 10.1093/femsle/fnu025. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Kastman EK, Guasto JS, Wolfe BE. 2018. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat Commun 9:336. doi: 10.1038/s41467-017-02522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kastman EK, Kamelamela N, Norville JW, Cosetta CM, Dutton RJ, Wolfe BE. 2016. Biotic interactions shape the ecological distributions of Staphylococcus species. mBio 7:e01157-16. doi: 10.1128/mBio.01157-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McDonald BR, Chevrette MG, Klassen JL, Horn HA, Caldera EJ, Wendt-Pienkowski E, Cafaro MJ, Ruzzini AC, Arnam EV, Weinstock GM, Gerardo NM, Poulsen M, Suen G, Clardy J, Currie CR. 2019. Biogeography and microscale diversity shape the biosynthetic potential of fungus-growing ant-associated Pseudonocardia. bioRxiv doi: 10.1101/545640. [DOI]
- 66.Caldera EJ, Currie CR. 2012. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: impacts of coevolution and multipartite symbiosis. Am Nat 180:604–617. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steffan SA, Chikaraishi Y, Currie CR, Horn H, Gaines-Day HR, Pauli JN, Zalapa JE, Ohkouchi N. 2015. Microbes are trophic analogs of animals. Proc Natl Acad Sci U S A 112:15119–15124. doi: 10.1073/pnas.1508782112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Currie CR, Mueller UG, Malloch D. 1999. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci U S A 96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birnbaum SSL, Gerardo NM. 2016. Patterns of specificity of the pathogen Escovopsis across the fungus-growing ant symbiosis. Am Nat 188:52–65. doi: 10.1086/686911. [DOI] [PubMed] [Google Scholar]
- 70.Gerardo NM, Mueller UG, Currie CR. 2006. Complex host-pathogen coevolution in the Apterostigma fungus-growing ant-microbe symbiosis. BMC Evol Biol 6:88. doi: 10.1186/1471-2148-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704. doi: 10.1038/19519. (Erratum, 423:461, 2003.) doi: 10.1038/19519. [DOI] [Google Scholar]