Summary
Dietary restriction is known to extend the lifespan and reduce fat stores in most species tested to date, but the molecular mechanisms linking these events remain unclear. Here, we found that bacterial deprivation of Caenorhabditis elegans leads to lifespan extension with concomitant mobilization of fat stores. We find that LIPL-5 expression is induced by starvation and that the LIPL-5 lipase is present in coelomocyte cells and regulates fat catabolism and longevity during the bacterial deprivation response. Either LIPL-5 or coelomocyte deficiency prevents the rapid mobilization of intestinal triacylglycerol and enhanced lifespan extension in response to bacterial deprivation, whereas the combination of both defects has no additional or synergistic effect. Thus, the capacity to mobilize fat via LIPL-5 is directly linked to an animal’s capacity to withstand long-term nutrient deprivation. Our data establish a role for LIPL-5 and coelomocytes in regulating fat consumption and lifespan extension upon DR.
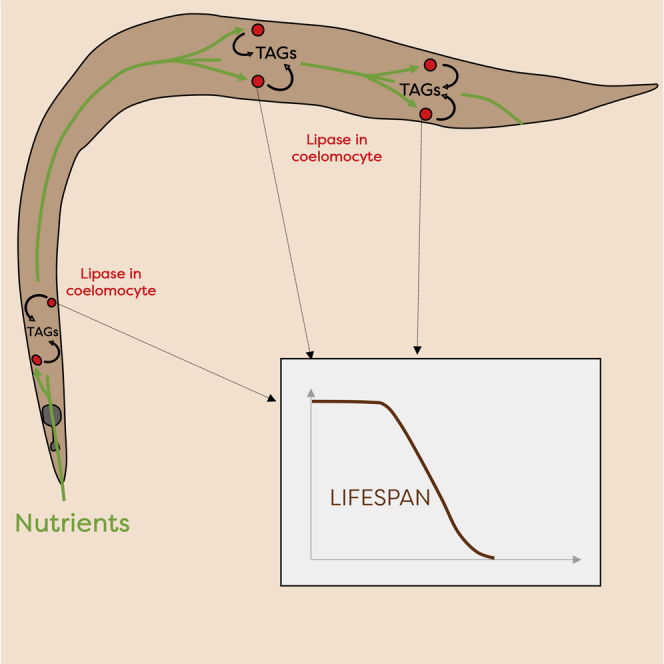
Graphical Abstract
Highlights
-
•
The lipase LIPL-5 is required for triacylglyceride catabolism under starvation
-
•
LIPL-5 limits lifespan extension under starvation
-
•
LIPL-5 predominantly localizes in coelomocytes
-
•
LIPL-5 controls catabolism and lifespan in coelomocytes
Buis et al. demonstrate that, under conditions of food scarcity, mobilization of triacylglycerides in the gut and lifespan extension of C. elegans are regulated by a lipase called LIPL-5 detected exclusively in coelomocytes. The work shows that LIPL-5 links an animal’s capacity to withstand nutrient deprivation to fat mobilization.
Introduction
A large body of evidence supports links among dietary restriction (DR), fat metabolism, and longevity in numerous species (Masoro et al., 1982, Hansen et al., 2013). DR, defined as diminished food intake without malnutrition, not only decreases fat stores but also modestly (20%–40%) but significantly extends the lifespan of a range of species (Masoro et al., 1982). The capacity to mobilize fat has been shown to play an important role in the ability of mice to withstand food restriction. Various aspects of fat metabolism have been linked lifespan in various species (Bustos and Partridge, 2017, Gonzalez-Covarrubias, 2013, Schroeder and Brunet, 2015), and in humans, although associations between body mass index and morbidity have been established (Afzal et al., 2016), the nature of the molecular link between fat content and longevity remains largely unknown. Altogether, these data suggest that the way fat is mobilized or used may affect the lifespan of wild-type animals subjected to DR.
Here, we investigated the relationship between DR-induced lifespan extension and fat catabolism in Caenorhabditis elegans. To date, lipases, that degrade triacylglycerol were shown to affect longevity, but there is no clear evidence for a role for lipases in DR-mediated longevity. For example, in the nematode C. elegans, adipose triglyceride lipase-1 (ATGL-1) activity affects lipid storage during nutrient deprivation but does not affect lifespan (Narbonne and Roy, 2009, Noble et al., 2013, Zhang et al., 2010). In contrast, the lipoprotein lipase LIPL-4 is required for lifespan extension of C. elegans through ablation of the germline, but modulation of its activity does not affect fat storage or metabolism (O’Rourke et al., 2013). Similarly, fatty acid desaturation plays a key role in promoting lifespan extension of C. elegans via effects on autophagy (Goudeau et al., 2011, Lapierre et al., 2011). Although LIPL-1 and LIPL-3 are induced by starvation and their overexpression increases lifespan under normal nutritional conditions, their effect on lifespan occurs independently of DR (O’Rourke and Ruvkun, 2013).
Here, we have explored this question in detail by analyzing fat catabolism in C. elegans subjected to DR via bacterial deprivation. We find that expression of the lipase LIPL-5/LIPF is induced by starvation, and animals carrying a mutation in the gene encoding for the lipase LIPL-5/LIPF show decreased triacylglycerol (TAG) consumption and exhibit enhanced lifespan compared with wild-type animals in response to bacterial deprivation. LIPL-5 is expressed in and/or taken up by coelomocytes, which are known as scavenger cells in the body cavity, and we show that functional coelomocytes contribute to both the enhanced TAG metabolism and the extended lifespan induced by bacterial deprivation. Our data are consistent with the idea that coelomocytes fulfill integrative functions (Fares and Greenwald, 2001) and that LIPL-5 is a link for nutrition, fat catabolism, and lifespan that acts in these cells.
Results
Bacterial Deprivation Promotes Fat Consumption and Extends Lifespan
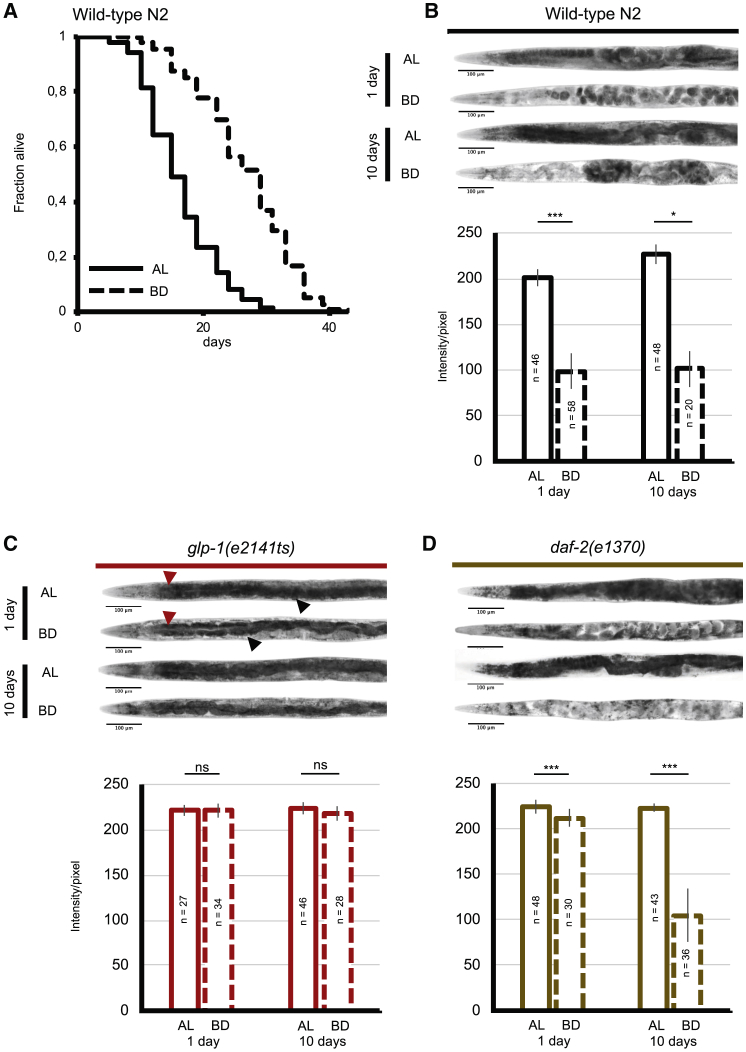
To explore the relationship between fat metabolism and lifespan extension, we measured whole-body TAG fat content and lifespan in animals fed ad libitum or subjected to DR for 24 h and until 10 days (Figures 1A and 1B). Although DR can be achieved through various feeding and genetic protocols in C. elegans (Greer and Brunet, 2009), we elected to use bacterial deprivation because it is the severest method of DR (Kaeberlein et al., 2006, Sutphin and Kaeberlein, 2008). To test this, we measured TAG content of wild-type animals using oil red O (ORO) staining, Sudan black staining, and thin-layer chromatography (Figures 1B, S1, and S2). We found that animals harbored larger TAG stores than they did under ad libitum feeding conditions using ORO staining and thin-layer chromatography (Figures 1B and S1). Using Sudan black staining, we also found wild-type animals lost intestinal TAG, but this difference was at the limit of statistical significance. It reached statistical difference using one statistical test (two-tailed Student’s t test), but not another test (two-tailed Mann-Whitney U test) (Figure S1). Altogether, our results are in line with previous observations (Lapierre et al., 2013, O’Rourke et al., 2009). We detected a significant and rapid reduction in fat stores (within 24 h) when animals were subjected to bacterial deprivation (Figure 1B). Long-lived animals such as the germline-less glp-1(e2141ts) mutants or the daf-2(e1370) mutants, which have defective insulin-insulin growth factor 1 (IGF-1)-like receptor signaling, have larger TAG stores than wild-type animals when fed ad libitum, and they consume them at a slower pace when subjected to DR (Figures 1C and 1D). Altogether, these data suggest that the consumption of fat stores is associated with lifespan and may therefore alter the effect of DR on longevity. We therefore searched for enzymes capable of affecting fat stores during bacterial deprivation.
Figure 1.
Caenorhabditis elegans Loses Fat Stores and Its Lifespan Is Extended upon Bacterial Deprivation
Lifespan analyses of wild-type (N2) C. elegans (n > 100 worms) fed ad libitum (AL) or subjected to bacterial deprivation. p < 0.0001. Mantel-Cox log-rank test. Representative of at least 3 replicate experiments (see Table S2 for number of replicates). Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) for the indicated C. elegans strains fed AL or subjected to bacterial deprivation (BD) for 1 or 10 days for wild-type animals (A), glp-1(e2141ts) mutant animals (B), and daf-2(e1370) (D). (C) Red arrows show intestinal fat, while black arrows show hypodermis with or without fat. Mean ± SD of n = at least 20 worms. ns, not significant; ∗∗p < 0.01 and ∗∗∗p < 0.001 by two-tailed Mann-Whitney U test. Representative of 3 biological replicates. See also Figures S1–S3.
LIPL-5 Mediates Triacylglyceride Catabolism in Adult C. elegans
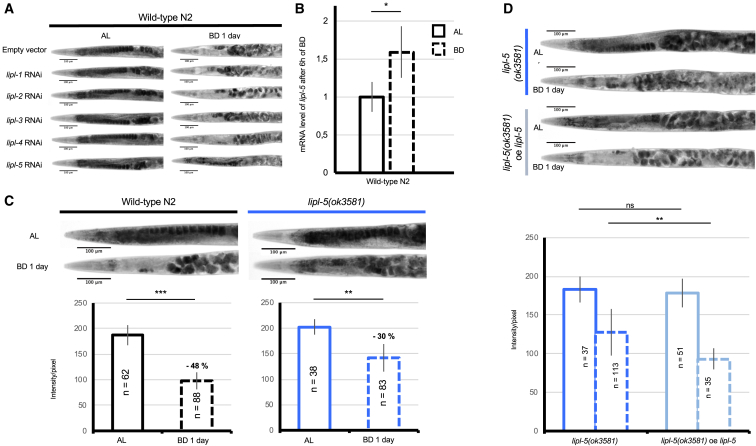
TAG is catabolized to free fatty acids and glycerol by the action of lipases. Five lipase genes (lipl-1–lipl-5) are known to be rapidly and transiently induced by DR in C. elegans (O’Rourke and Ruvkun, 2013), which is consistent with the observation that wild-type animals consume most of their TAG stores within 24 h of bacterial deprivation (Figure 1B) (McKay et al., 2003). To test the involvement of these enzymes in bacterial deprivation-induced fat catabolism in fertile adults, we inactivated each gene using RNAi. We found that RNAi efficiently suppressed all genes except lipl-4 (Figure S3A). The relative inefficiency of lipl-4 RNAi may be because of the particularly strong induction of this enzyme by bacterial deprivation (data not shown). However, because lipl-4(tm4417) mutants contain normal TAG levels under ad libitum and DR conditions (O’Rourke et al., 2009), we considered LIPL-4 unlikely to be relevant to our investigation. Only lipl-5 inactivation by RNAi in fertile wild-type C. elegans reduced TAG catabolism (Figures 2A and S3B). LIPL-5 is highly homologous to isoform 1 of the human gastric triacylglycerol lipase LIPF. Thus, we confirmed that lipl-5 is induced by nutrient shortage (Figure 2B) and its inhibition prevents TAG catabolism upon bacterial deprivation (Figure 2A). The induction of lipl-5 we measured was robust but modest and transient; we detected significant induction after 6 h (Figure 2B), but this induction failed to reach statistical significance after 12 h (data not shown). These results confirmed data previously published by the Ruvkun lab (O’Rourke et al., 2009). To confirm the results obtained with lipl-5 RNAi data, we examined lipl-5(ok3581) mutants, which carry a 300 bp deletion in the lipl-5 gene. These animals exhibited an inability to catabolize fat similar to that of wild-type animals subjected to lipl-5 RNAi (Figure 2C). We also generated an independent lipl-5(bab-8) deletion strain by CRISPR/Cas9-mediated editing (MCP12), which showed a similarly reduced capacity to catabolize fat upon bacterial deprivation (Figure S4). Finally, overexpression of lipl-5 under the control of the endogenous lipl-5 promoter was sufficient to increase fat catabolism in lipl-5(ok3581) mutants upon bacterial deprivation (Figure 2C). These data establish a central role for LIPL-5 in bacterial deprivation-induced fat catabolism in C. elegans.
Figure 2.
LIPL-5/LIPF Mediates Fat Catabolism Induced by Bacterial Deprivation
(A) Oil red O (ORO) staining of fat stores in wild-type C. elegans subjected to lipl-1, lipl-2, lipl-3, lipl-4, or lipl-5 RNAi. Animals were fed ad libitum (AL) or subjected to bacterial deprivation (BD) for 1 day.
(B) qRT-PCR analysis of lipl-5 mRNA in wild-type C. elegans subjected to BD for 6 h. Representative of 3 biological replicates. Mean ± SD of n > 20 worms. ∗p < 0.05 by Student’s t test.
(C) Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) for wild-type C. elegans and lipl-5(ok3581) mutants fed AL or subjected to BD for 1 day. Mean ± SD of n > 20 worms. ∗∗p < 0.01 and ∗∗∗p < 0.001 by two-tailed Mann-Whitney U test. Representative of 6 biological replicates.
(D) Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) for lipl-5(ok3581) either expressing an empty vector or expressing lipl-5 (expression drive by its endogenous promoter) fed AL or subjected to BD for 1 day. Solid bars, BD; dashed bars, AL. Mean ± SD of n = at least 20 worms. ns, not significant; ∗∗p < 0.01 by two-tailed Mann-Whitney U test. Representative of 3 biological replicates.
See also Figures S4 and S5.
LIPL-5, Predominantly Localized in Coelomocytes, Limits Bacterial Deprivation-Induced Longevity
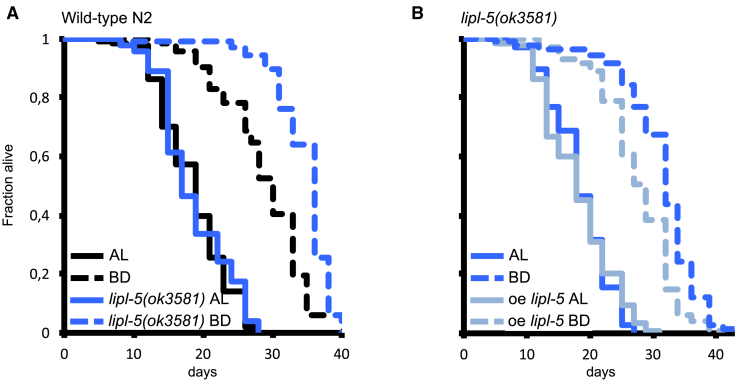
Because the lipl-5(ok3581) mutants fail to catabolize fat in response to bacterial deprivation, we examined the influence of lipl-5 deletion and restoration on lifespan extension in response to bacterial deprivation. The lipl-5(ok3581) mutants exposed to bacterial deprivation experienced a lifespan extension nearly twice that of wild-type animals (Figure 3A; Table S2), and conversely, re-expression of lipl-5 in the mutants reversed the bacterial deprivation-extended lifespan to the duration seen with wild-type animals (Figure 3B; Table S2). Altogether, these experiments establish that LIPL-5 regulates both fat catabolism and DR-induced lifespan extension in C. elegans.
Figure 3.
LIPL-5/LIPF Limits Lifespan Extension in Response to Bacterial Deprivation
Lifespan analyses of C. elegans fed ad libitum (AL) or subjected to bacterial deprivation (BD).
(A) BD extended lifespan by 56% for wild-type animals (p < 0.0001) and 86% for lipl-5(ok3581) mutant animals (p < 0.0001). The mean lifespan of wild-type animals was similar to that of lipl-5(ok3581) mutant animals in similar conditions (p = 0.515). In BD, the lifespan of wild-type animals was 16% lower than that of lipl-5(ok3581) mutant animals (p < 0.001).
(B) BD extended lifespan by 72% for lipl-5(ok3581) mutant animals (p < 0.0001) and 50% for lipl-5(ok3581) mutant animals overexpressing lipl-5 behind its own promoter (p < 0.0001). The mean lifespan of lipl-5(ok3581) mutant animals was similar to that of lipl-5(ok3581) mutant animals overexpressing lipl-5 behind its own promoter in AL (p = 0.651). In BD, the lifespan of lipl-5(ok3581) mutant animals was 12% higher than that of lipl-5(ok3581) mutant animals overexpressing lipl-5 behind its own promoter (p < 0.001). Mean ± SD of n = at least 100 worms. Mantel-Cox log-rank test (see Table S2 for number of replicates).
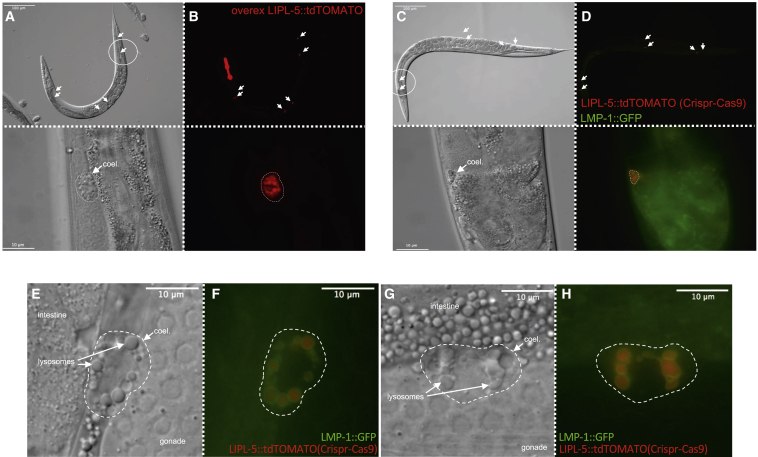
We next investigated the cellular localization of LIPL-5 in C. elegans. To this end, we expressed a fusion protein consisting of LIPL-5 and the fluorescent dye tdTOMATO, which emits light at high wavelengths, allowing ready visualization of LIPL-5 against the relatively high green autofluorescent background exhibited by C. elegans. Animals were injected with plasmids containing extrachromosomal arrays of LIPL-5::tdTOMATO under the control of the endogenous promoter, which typically leads to significant overexpression of the protein. LIPL-5::tdTOMATO fluorescence was only detected in the six coelomocytes of C. elegans (Figure 4A), which are specialized endocytotic cells of mesodermal origin located in the pseudocoelom (Fares and Greenwald, 2001, Zhang et al., 2001). Coelomocytes are thought to be involved in detoxification and have been proposed to be proto-hepatocytes (Fares and Greenwald, 2001). To ensure that this unexpected finding was not caused by artifactual uptake of supra-physiological levels of LIPL-5::tdTOMATO, we generated additional mutants using CRISPR/Cas9 editing to place a single copy of LIPL-5::tdTOMATO at the endogenous locus. Because some lipases are known to be more potent at acidic pH and are usually lysosomal, we also introduced a lysosome-associated membrane protein 1 (LMP-1)::GFP marker to test for LIPL-5 co-localization in this organelle. We found that LIPL-5 was localized specifically in the lysosomes of coelomocytes, even in the strain expressing physiological levels of the reporter protein (Figures 4B and 4C). To determine whether LIPL-5 is expressed by coelomocytes or produced elsewhere and then endocytosed, we used the intestinal-specific promoter ges-1 to enforce LIPL-5 expression in the gut, where lipases are generally abundant. LIPL-5::tdTOMATO was also found predominantly in coelomocytes in these animals (Figure S5A), indicating that LIPL-5 can be transported to or endocytosed by coelomocytes. Gut-specific overexpression of LIPL-5 was able to increase fat catabolism and reduce the enhanced lifespan response to bacterial deprivation exhibited by lipl-5(ok3581) mutants, suggesting that coelomocyte-localized LIPL-5 plays an important role in modulating these phenotypes (Figures S5B and S5C).
Figure 4.
LIPL-5/LIPF Is Localized to Coelomocyte Lysosomes
(A and B) (A) Differential interference contrast images and (B) fluorescence images of adult wild-type C. elegans expressing tdTOMATO-tagged LIPL-5 driven by the endogenous promoter. Upper and lower panels show the same animal at different magnifications (scale bars: 100 μm and 10 μm, respectively). The LIPL-5::tdTOMATO signal is confined to coelomocytes (white arrows and dashed circles).
(C and D) (C) Images are as described for (A) and (B) except that (D) animals co-expressed LIPL-5::tdTOMATO and GFP-tagged LMP-1 (lysosome-associated membrane protein) to visualize lysosomes. White arrows and dashed circle indicate coelomocytes.
(E–H) (E and G) Differential interference contrast and (F and H) fluorescence images similar to (A) and (B), respectively, except at a higher magnification to show coelomocytes. Each experiment was replicated at least 3 times (biological replicates).
Coelomocytes Play a Central Role in Regulating Fat Metabolism and Lifespan in C. elegans
To test the importance of coelomocytes in the fat catabolism and longevity phenotypes induced by bacterial deprivation, we employed the coelomocyte-deficient strain NP717, which expresses a variant of the diphtheria toxin A fragment (E148D) under the control of the coelomocyte-specific unc-122 promoter (Fares and Greenwald, 2001). This strain also expresses GFP fused to a signal sequence from SEL-1, which is expressed in the body wall under the myo-3 promoter. In normal animals, ssGFP is secreted into the pseudocoelom and is endocytosed by and accumulates in coelomocytes. In coelomocyte-deficient animals, however, ssGFP accumulates in the pseudocoelom, giving rise to the distinctive coelomocyte uptake (cup) phenotype (Figure S6).
As previously described (Park et al., 2015), the NP717 strain had a shorter lifespan than wild-type animals under ad libitum feeding conditions (Figure 5A; Table S2); however, in line with our hypothesis that coelomocytes may play an important role in bacterial deprivation-induced fat and longevity phenotypes, NP717 animals showed diminished fat catabolism and enhanced longevity compared with wild-type animals when subjected to bacterial deprivation, similar to the phenotype of lipl-5 mutants (Figure 5; Table S2). These results are in apparent contradiction with findings that coelomocyte-deficient animals have a normal lifespan when fed ad libitum and are unaffected by various DR protocols (Park and Park, 2017). To our knowledge, the only difference between the experimental protocols was our use of Escherichia coli strain ht115 for ad libitum feeding, compared with OP50 used by Park and Park (2017); thus, the reason for this discrepancy remains unclear. However, our results with the NP717 strain and lipl-5(ok3581) mutants, together with the LIPL-5 localization study, are consistent with a crucial role for coelomocyte-localized LIPL-5 as a mediator of fat catabolism and lifespan extension induced by bacterial deprivation.
Figure 5.
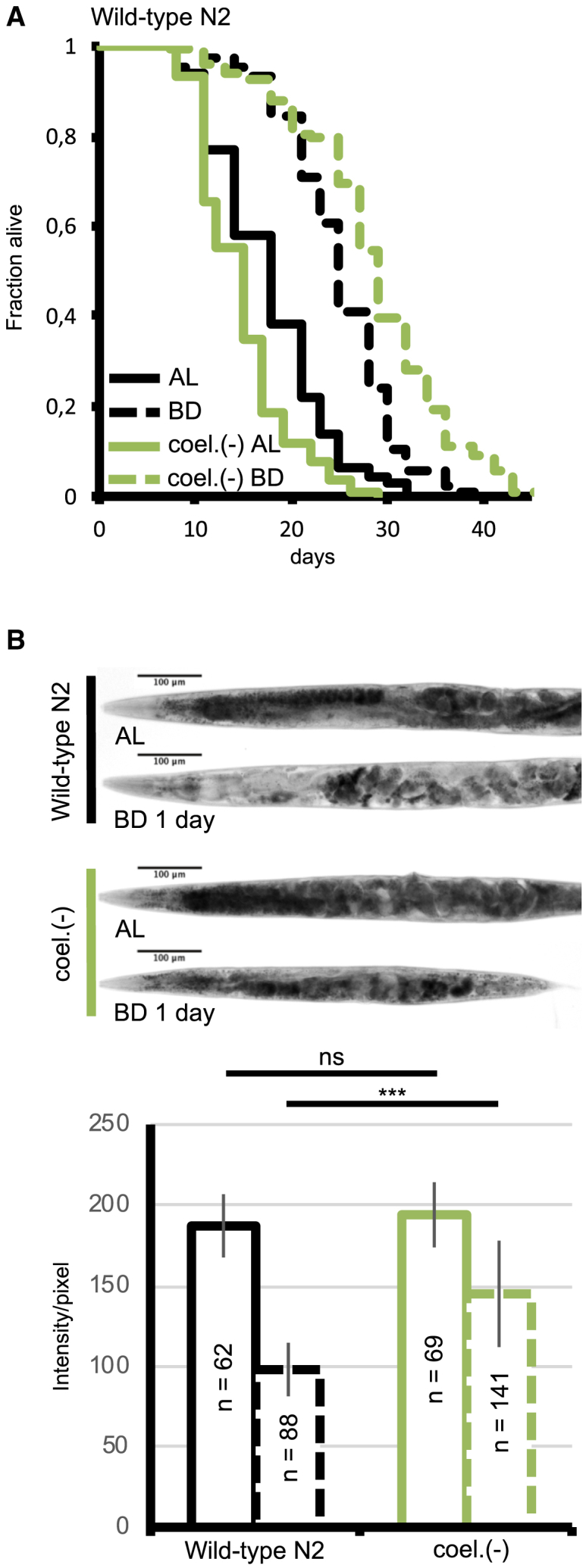
Coelomocyte-Deficient C. elegans Exhibit Decreased Fat Catabolism and Enhanced Lifespan Extension in Response to Bacterial Deprivation
(A) Lifespan analyses of C. elegans fed ad libitum (AL) or subjected to bacterial deprivation (BD). BD extended lifespan by 89% for coelomocyte-deprived animals (p < 0.0001) and by 42% for wild-type animals (p < 0.0001). In AL, the lifespan of coelomocyte-deprived animals was 16% shorter than that of wild-type animals (p < 0.001). In BD, the lifespan of coelomocyte-deprived animals was 12% longer than that of wild-type animals (p < 0.001). Mantel-Cox log-rank test. Representative of 6 replicate experiments (see Table S2 for number of replicates).
(B) Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) of wild-type C. elegans and coelomocyte-deficient animals (coel.(−)) fed AL or subjected to BD for 1 day. Mean ± SD of n = at least 30 worms. ns, not significant; ∗∗∗p < 0.001 by two-tailed Mann-Whitney U test. Representative of 6 biological replicates.
See also Figure S6.
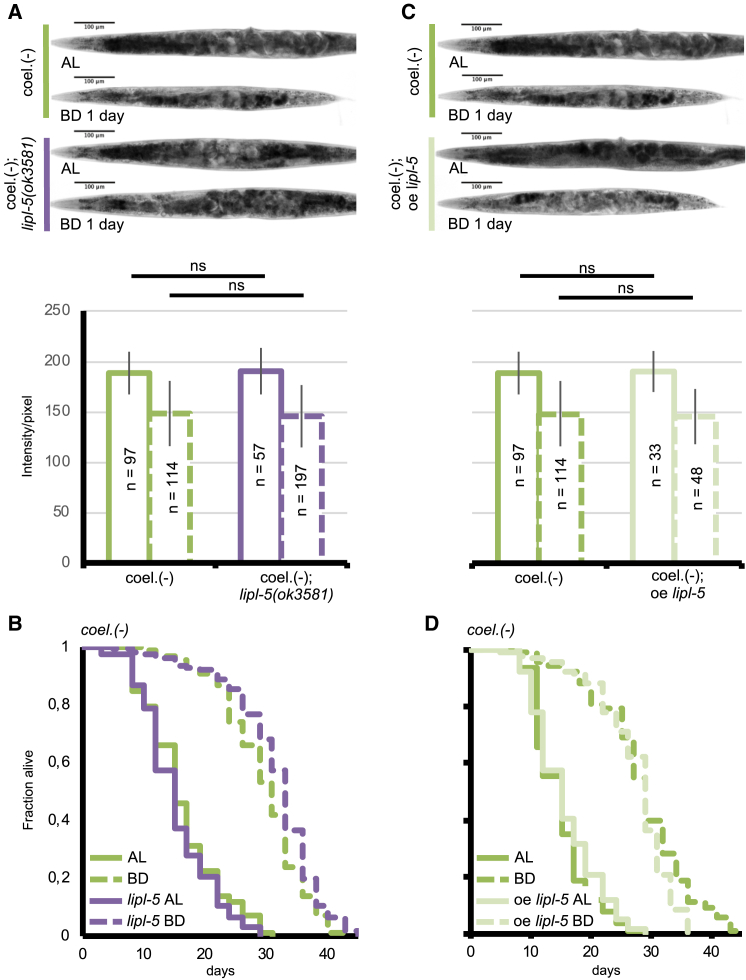
To test this directly, we deleted or overexpressed lipl-5 in the coelomocyte-deficient NP717 strain. We found that concomitant genetic deletion of lipl-5 had no additive effect on the enhanced lifespan and reduced fat catabolism phenotypes exhibited by coelomocyte-deficient animals subjected to bacterial deprivation (Figures 6A and 6B; Table S2), suggesting that lipl-5 and coelomocytes act via a similar and/or overlapping mechanism in mediating these phenotypes. Similarly, overexpression of lipl-5 in coelomocyte-deficient animals (driven by the endogenous lipl-5 promoter) failed to influence fat catabolism or lifespan extension in response to starvation (Figures 6C and 6D; Table S2). These data suggest that these functions of LIPL-5 are mediated predominantly in coelomocytes. To test this directly, we restored lipl-5 expression only in the coelomocytes of lipl-5(ok3581) animals by using the coelomocyte-specific unc-122 promoter. This was sufficient to reduce lifespan extension upon bacterial deprivation (Figure 7; Table S2). The ability to catabolize fat was also restored, although to a lesser extent. Intestinal TAG was lost more intensely upon bacterial deprivation, but this difference reached a statistical difference using one statistical test (two-tailed Student’s t test), but not the other (two-tailed Mann-Whitney U test) (Figure 7A). These data suggest that the functions of coelomocytes in regulating fat catabolism and lifespan in response to bacterial deprivation are mostly mediated by LIPL-5. Our lifespan data do not exclude the involvement of other factors. Collectively, the results presented here uncover a role for coelomocytes in mediating both fat catabolism and lifespan phenotypes of C. elegans subjected to DR. The lipase LIPL-5 mediates both phenotypes, with the coelomocyte-localized enzyme playing a predominant role.
Figure 6.
LIPL-5/LIPF Functions in Coelomocytes
(A and C) Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) for coelomocyte-deficient single mutants (coel.(−)) (represented twice; A and C) and coel.(−);lipl-5(ok3581) double mutants (A) or coel.(−) mutants overexpressing lipl-5 (C). Animals were fed ad libitum (AL) or subjected to bacterial deprivation (BD) for 1 day. Mean ± SD of n = 30. ns, not significant; ∗p < 0.05 by two-tailed Mann-Whitney U test. Representative of 3 biological replicates.
(B and D) Lifespan analyses of C. elegans fed AL or subjected to BD. (B) BD extended lifespan by 89% for coelomocyte-deprived animals (p < 0.0001) and by 106% for coelomocyte-deficient mutants for lipl-5(ok3581) (p < 0.0001). These two strains have similar lifespan in AL (p = 0.185), but coelomocyte-deprived mutants for lipl-5(ok3581) had lifespan increased by 12% in BD (p < 0.05) when compared with that of coelomocyte-deficient animals with a wild-type copy of lipl-5. (D) BD extended lifespan by 82% for coelomocyte-deficient animals (p < 0.0001) and by 93% for coelomocyte-deficient animals overexpressing lipl-5 driven by its own promoter (p < 0.0001). These two strains have similar lifespan in AL (p = 0.234) and in BD (p = 0.309). Mantel-Cox log-rank test (see Table S2 for number of replicates).
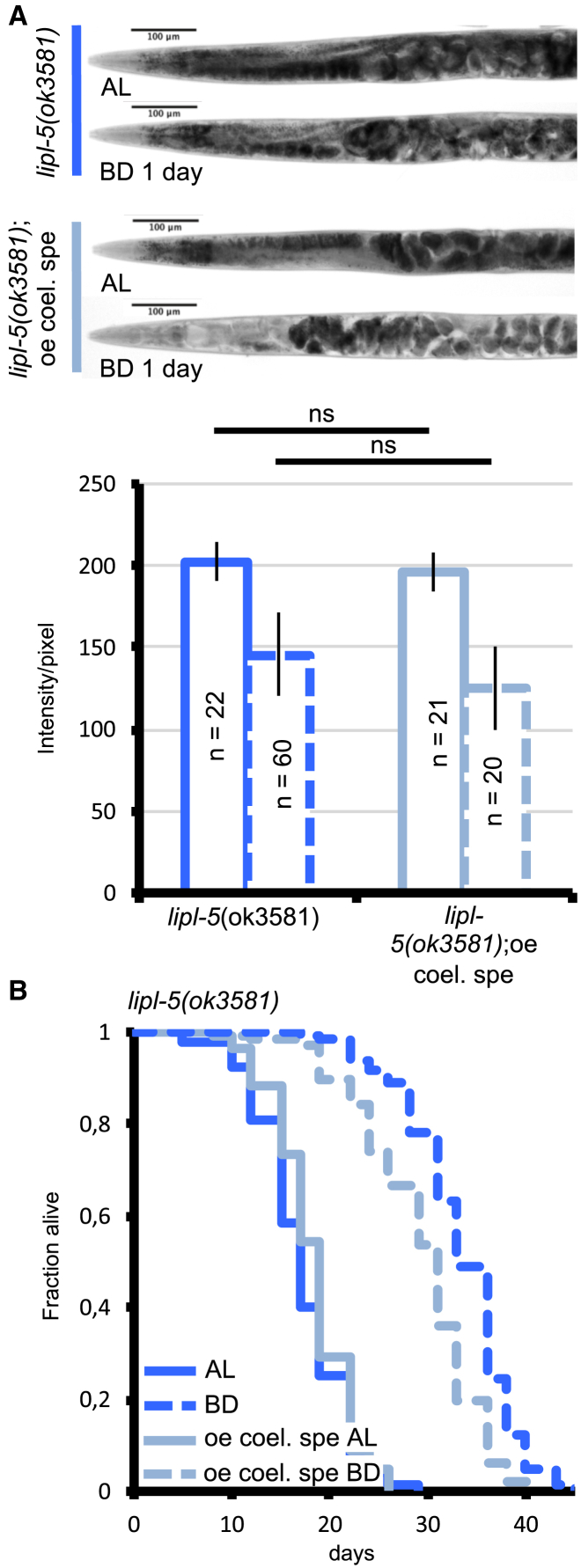
Figure 7.
Coelomocyte-Localized LIPL-5/LIPF Rescues Bacterial Deprivation-Induced Fat Catabolism and Lifespan Extension in lipl-5(ok3581) Mutants
(A) Light micrographs of representative oil red O (ORO)-stained whole animals (upper panels) and densitometric quantification of staining of the two first intestinal cells after background removal (lower panels) for lipl-5(ok3581) mutants with or without coelomocyte-specific (cc1-driven) overexpression of lipl-5. Animals were fed ad libitum (AL) or subjected to bacterial deprivation (BD) for 1 day. Mean ± SD of n = at least 20 worms. ns, not significant; two-tailed Mann-Whitney U test. Representative of 3 biological replicates.
(B) Lifespan analyses of C. elegans fed AL or subjected to BD. BD extended lifespan by 83% for lipl-5(ok3581) mutants (p < 0.0001) and by 70% for lipl-5(ok3581) mutants overexpressing lipl-5 specifically in coelomocytes (p < 0.0001). These two strains exhibit similar lifespan in AL (p = 0.143), but in BD, lipl-5(ok3581) mutant animals had a lifespan 12% longer when compared with lipl-5(ok3581) mutants overexpressing lipl-5 specifically in coelomocytes (p < 0.001). Mantel-Cox log-rank test (see Table S2 for number of replicates).
Discussion
DR is a well-established longevity paradigm in numerous species, including humans (Redman et al., 2018). In this work, we tested the role of fat catabolism in regulating lifespan extension upon DR. We found that DR-induced lifespan extension in wild-type C. elegans is accompanied by a marked induction of LIPL-5 and a concomitant increase in catabolism of TAG stores. Animals subjected to lipl-5 RNAi and lipl-5(ok3581) mutants showed diminished fat catabolism and extended longevity compared with wild-type animals upon bacterial deprivation. Thus, LIPL-5 may represent a nexus linking nutrient deprivation, fat catabolism, and longevity.
We found that LIPL-5 was mainly detectable in coelomocytes. Although this result was unexpected, we are confident that it was not an artifact induced by endocytosis of supra-physiological protein levels, because coelomocyte-specific staining was also observed when the endogenous lipl-5 gene was replaced by a single copy of tagged LIPL-5 and expressed under the control of its own promoter. Whether LIPL-5 is produced within coelomocytes or is endocytosed from the pseudocoelom remains unclear. However, our data show that LIPL-5 can affect fat catabolism and lifespan when its expression is restricted to coelomocytes. These data therefore suggest that, independent of where lipl-5 is expressed, its action mainly takes place in coelomocytes. We cannot firmly exclude that LIPL-5 may function marginally in other tissues as well. Animals lacking coelomocytes behaved similarly to lipl-5 mutants under bacterial deprivation conditions, and concomitant loss of both coelomocytes and lipl-5 did not have an additive effect on either fat catabolism or lifespan extension. However, coelomocyte-specific restoration of lipl-5 expression in lipl-5(ok3581) mutants was sufficient to restore fat catabolism and longevity, although not quite fully. This result could imply a requirement for higher levels of LIPL-5 or expression in tissues other than coelomocytes to restore the wild-type longevity phenotype.
In summary, we show here that coelomocytes can integrate nutrient-derived signals to influence both fat metabolism and lifespan in C. elegans. Our data thus provide another example of inter-tissue communication and support the notion that coelomocytes integrate nutritional signals (Fares and Greenwald, 2001). How coelomocytes communicate with the functions of other tissues, such as the intestine, remains unknown, and identifying new factors involved in this communication is an exciting prospect for future studies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli HT115 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Escherichia coli RNAi lipl-1 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Escherichia coli RNAi lipl-2 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Escherichia coli RNAi lipl-3 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Escherichia coli RNAi lipl-4 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Escherichia coli RNAi lipl-5 | Ahringer library Nature 408, 325–330 (2000) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRI Reagent ® | Sigma-Aldrich | T9424-100ML |
| 2X MRWB (2% formaldehyde [methanol free], 160 mM KCl; 40 mM NaCl, 4 mM EGTA, 1 mM spermidine, 0.4 mM spermine, 30 mM PIPES, pH 7.4, and 0.2% β-mercaptoethanol) | O’Rourke et al., 2009 | N/A |
| Critical Commercial Assays | ||
| Gibson Assembly Kit | NEB | E5510S |
| RNeasy Mini Kits | QIAGEN | 74104 |
| iScript cDNA Synthesis kit | Bio-Rad | 1708890 |
| Experimental Models: Organisms/Strains | ||
| N2 Bristol | CGC | N2 |
| glp-1(e2141ts)III | Gift from Kenyon Lab | CF1903 |
| daf-2(e1370)III | CGC | CB1370 |
| lipl-5(ok3581)V | CGC | RB2573 |
| lynEx03 = [(pAB02(lipl-5p::lipl-5::td-tomato) and co-injection marker myo-2p::mCherry] | UMS3421 | MCP39 |
| lipl-5(ok3581)V;lynEx03 | Our lab | HGA2912 |
| unc-119(ed3)III;pwIs50 | CGC | RT258 |
| pwIs50 = [lmp-1::GFP + Cbr-unc-119(+)] | ||
| lipl-5::td-tomato | UMS3421 | MCP34 |
| lipl-5::td-tomato;pwIs50 | Our lab | HGA2914 |
| coel.(-) | Gift from Fares Lab | NP717 |
| coel.(-);lipl-5(ok3581)V | Our lab | HGA2915 |
| coel.(-);lynEx03 | Our lab | HGA2916 |
| lipl-5(ok3581)V;lynEx04 | Our lab | HGA2917 |
| lynEx04 = [(pAB02(pcc1::lipl-5::td-tomato) and co-injection marker myo-2p::mCherry] | ||
| lipl-5(bab-8)V | UMS3421 | MCP12 |
| lipl-5(ok3581)V;lynEx02 | Our lab |
HGA8021 |
| lynEx02 = [(pAB01(ges-1p::lipl-5::gfp) and co-injection marker myo-2p::mCherry] | ||
| Oligonucleotides | ||
| See Table S3 for oligonucleotide information | N/A | |
| Recombinant DNA | ||
| pHD43 plasmid | kindly provided by Hanna Fares, University of Arizona Cancer Center | N/A |
| pPD95.75-Pges-1 plasmid | kindly provided by Hideki Inoue, Pittsburgh University | N/A |
| pCFJ90: myo-2p::mCherry | Addgene | http://www.addgene.org/19327/ |
| Software and Algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/ | |
| two-tailed Mann-Whitney U test | This paper | https://www.socscistatistics.com/tests/mannwhitney/default2.aspx |
| two-tailed Student’s t test | This paper | https://www.xlstat.com/fr/ |
| Mantel–Cox log-rank | This paper | https://www.xlstat.com/fr/ |
| WinCATS software | This paper | https://www.camag.com/en/tlc_hptlc/supportservices/software/wincats_releases.cfm?command=archive |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hugo Aguilaniu (haguilaniu@gmail.com).
Experimental Model and Subject Details
Worm Strains
Nematodes were grown and maintained under standard conditions. N2 Bristol was used as the wild-type strain. All other strains and their origins are listed in Table S1.
Strain Construction
Double mutant strains were generated using standard genetic procedures. lipl-5 (ok3581) mutants were backcrossed five times, and the presence of the lipl-5(ok3581) mutation was confirmed by PCR using allele-specific primers. We confirmed that the NP717 strain lacked coelomocytes by testing for the coelomocyte uptake (cup) phenotype where GFP is taken up within the pseudoceolom. To generate transgenic animals, pAB01(ges-1p::lipl-5::gfp) was injected at 50 ng/μL, pAB02(lipl-5p::lipl-5::tdTOMATO) and pAB03 (pcc1::lipl-5::tdTOMATO) were injected at 10 ng/μL, as previously described (ref?), and the myo-2p::mCherry (pCFJ90) marker was co-injected at 2.5 ng/μL. These transgenes were named lynEx02, lynEx03, and lynEx04, respectively. The myo-2p::mCherry marker and empty vectors used here were confirmed to have no effect on lifespan (data not shown). CRISPR insertions for lipl-5(bab-8) and lipl-5::tdTOMATO were generated by the UMS3421. The lipl-5(bab-8) mutant was created by inserting an ED10-Hygro-HisClo cassette into the lipl-5 locus and then removing it to leave a 20 bp scar in place of the lipl-5 gene. The lipl-5::tdTOMATO strain was generated by insertion of a tdTOMATO-tagged version of LIPL-5 instead of the cassette.
Method Details
Plasmid Construction
The plasmid pAB01(ges-1p::lipl-5::gfp), encoding GFP-tagged LIPL-5 driven by the intestine-specific ges-1 promoter, was constructed by amplifying the lipl-5 open reading frame (ORF) from genomic DNA (from the start codon to the end of lipl-5 gene before the stop codon) using SalI and KpnI primers (SalI/lipl-5 F: 5′-CTGGTCGACATGTGGC GGTTTGCCGTTTTTC-3′; lipl-5/KpnI R: 5′-GTGGTACCTTTCCCAAATAATCGTCAG TGCACAGC-3′). The fragment was inserted into the worm expression vector pPD95.75-Pges-1 (kindly provided by Hideki Inoue, Pittsburgh University) upstream and in-frame with the GFP sequence and downstream and in-frame with the ges-1 promoter. Essential parts of the final plasmid pAB01 were sequenced to verify that the lipl-5 sequence was in-frame with the ges-1 promoter and GFP sequences.
The plasmid pAB02 (lipl-5p::lipl-5::tdTOMATO), encoding tdTOMATO-tagged LIPL-5 driven by the endogenous promoter, was constructed by amplifying a fragment starting 1.5 kb upstream of and extending through the lipl-5 ORF, excluding the last stop codon, from genomic DNA using a Gibson Assembly Kit (primers: lipl-5p/lipl-5 gibson F: 5′-CAC AAC GAT GGA TAC GCT AAC AAC TTG GAA ATG AAA TAA GCT TCT AAA ACA ATT ATT TTC AGA CGA TAC TTG GTC-3′; lipl-5p/lipl-5 gibson R: 5′-GAT GAC CTC CTC GCC CTT GCT CAC CAT TAC CGG TAC CGA TTT TCC CAA ATA ATC GTC AGT GC-3′). The fragment was inserted into the worm expression vector sur-5::td-tomato (kindly provided by Dr. Ehud Cohen, Hebrew University) upstream and in-frame with the td-tomato sequence. Essential parts of the final plasmid pAB02 were sequenced to confirm that the lipl-5 promoter and gene sequences were in-frame with td-tomato.
The plasmid pAB03 (pcc1::lipl-5::tdTOMATO), encoding tdTOMATO-tagged LIPL-5 driven by the coelomocyte-specific promoter pcc-1, was constructed by amplifying a 180 bp sequence upstream of unc-122 from the pHD43 plasmid (kindly provided by Hanna Fares, University of Arizona Cancer Center), which was previously described as the coelomocyte promoter (Fares and Greenwald, 2001), and the lipl-5 gene, excluding the last stop codon, from genomic DNA. The two fragments were inserted into the pAB02 plasmid to replace the lipl-5 promoter and gene, upstream and in-frame with the td-tomato sequence, using a Gibson Assembly Kit (primers PCR1 F1 (Plasmid+Pcc1): 5′-CGC TAA CAA CTT GGA AAT GAA ATA AGC TTG TTG ACA CGC AGT TTC CCT-3′; PCR1 R1F2 (Pcc1+lipl-5): 5′-AAC GGC AAA CCG CCA CAT ATT GTG AGC CCA ATG AAG TAA AAT TTC-3′; PCR2 R1F2 (Pcc1+lipl-5): 5′-GAA ATT TTA CTT CAT TGG GCT CAC AAT ATG TGG CGG TTT GCC GTT-3′; PCR2 R2 (lipl-5+Plasmid): 5′-CCC TTG CTC ACC ATT ACC GGT ACC GAT TTT CCC AAA TAA TCG TCA GTG CAC-3′). Essential parts of the final plasmid pAB03 were sequenced to confirm that the coelomocyte promoter sequence was in-frame with the lipl-5 gene and td-tomato.
Lifespan Assays
Lifespan assays were conducted according to standard protocols (Goudeau et al., 2011) at 20°C, starting from day 1 of adulthood. Our assay was performed at 20°C. Lifespan assays were conducted on plates supplemented with 15 μM 5-fluorouracil to prevent progeny from hatching. For BD experiments, plates contained 50 μM kanamycin, which was confirmed to not affect lifespan (unpublished data), and worms were washed 3 times in M9 buffer prior to experiments. Contaminated worms, or those crawling off the plate, exploding, or bagging were censored at the time of their loss.
RT-qPCR
Primer sequences are provided in the STAR Methods. In brief, synchronized L1 larvae were grown at 25°C on ht115 E. coli until they reached day 1 of adulthood. For RNAi experiments, worms were grown on ht115 E. coli bacteria carrying empty vector or expressing dsRNA against the gene of interest. The BD experiments, worms were transferred to bacteria-free plates and suppression of target gene expression was verified at the end of the experiment. Animals were collected, washed, and frozen at −80°C for at least 24 h before RNA was extracted using TRI Reagent ® (MRC Molecular Research Center) and RNeasy Mini Kits (QIAGEN). The concentration and purity of RNA samples were determined using a DropSense (Trinean) spectrophotometer. cDNA was synthesized from aliquots of 1 μg of RNA using an iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed using the StepOnePlus System (Applied Biosystems) according to the manufacturer’s recommended protocol. Relative mRNA levels were normalized to the housekeeping genes pmp-3, act-1, and cdc-42. Primers are listed in the Supplemental Information.
Primers were designed with Primer 3. PMP-3 F: 5′-GTT CCC GTG TTC ATC ACT CAT-3′ and R: 5′-ACA CCG TCC AGA AGC TGT AGA-3′; CDC-42 F: 5′-CTG CTG GAC AGG AAG ATT ACG-3′ and R: 5′-CTC GGA CAT TCT CGA ATG AAG-3′; ACT-1 F: 5′-GCT GGA CGT GAT CTT ACT GAT TAC C-3′ and ACT-1 R: 5′-GTA GCA GAG CTT CTC CTT GAT GTC-3′; LILP-1 F: 5′-CGG TTT GCG CTG GAC TTA-3′ and R: 5′-GAA CAC GAG TTG CGT TAA-3′; LIPL-2 F: 5′-TGG ATG CAG ATG GTT CGC AA-3′ and R: 5′-GCC CTT GAT GGC TCC GAA AT-3′; LIPL-3 F: 5′-GCT CGT GAT TCT TGC GGT TC-3′ and R: 5′-CGG ATA ACC CCA TCG CTC AA-3′; LIPL-4 F: 5′-GAA ACG TTG TTC GCG CAG TT-3′ and R: 5′-AAC TTG GCT GGC TGC ATT TG-3′; LIPL-5 F: 5′-ACT GGG GAA CCA AGA CGA AC-3′ and R: 5′-TAT CAG CCA ACC AAT CGG CA-3′.
Oil Red O Staining of Neutral Lipids
Approximately 200 worms per sample were collected in phosphate-buffered saline (PBS) without MgCl2 and resuspended in a buffer composed of 120 μL of PBS 1X and 120 μL of 2X MRWB (2% formaldehyde [methanol free], 160 mM KCl; 40 mM NaCl, 4 mM EGTA, 1 mM spermidine, 0.4 mM spermine, 30 mM PIPES, pH 7.4, and 0.2% β-mercaptoethanol). Samples were shaken for 1 h at 20°C, washed twice with PBS, incubated in 1 mL 60% isopropanol for 15 min at 20°C, and transferred to 1mL 60% Oil Red O solution overnight. After two washes with PBS, worms were resuspended in 200 μL PBS containing 0.001% Triton X-100, mounted between a slide and coverslip, and observed on a Nikon Eclipse 80i microscope with a 10 × objective. Images were captured with a monochrome digital camera (DS-Qi1Mc). Densitometric quantification of staining (expressed as intensity/pixel) was performed with ImageJ software on the two first intestinal cells after background removal. All experiments analyzed at least 20 worms per condition per point.
Coelomocyte Imaging
Worms were grown and maintained under standard conditions. Images of at least 25 worms were captured at 100 × , 400 × , and 1000 × magnification using 800 ms, 400 ms, and 800 ms exposure times, respectively. At least 4 of the 6 coelomocytes present in each worm were imaged. Worms were imaged on a Nikon Eclipse 80i microscope equipped with a monochrome digital camera (DS-Qi1Mc), and artificial color was added with ImageJ.
Lipid Analysis by Thin Layer Chromatography
Each sample consisted of a worm extract equivalent to 25μg soluble protein adjusted to 2 mL with distilled water. Cholesterol (2 μg) was added to each sample as an internal standard. Samples were mixed with 100 μL acetic acid, 2.3 mL methanol, and 2.3 mL chloroform and incubated for 5 min. The upper phase was discarded, and the organic phase was washed twice with 1 mL water/methanol/chloroform (2/2.5/2.5 v/v), dried under nitrogen, dissolved in 50 μL chloroform/methanol (1/1), and analyzed. A 10 μL aliquot of each sample was spotted onto 20 × 10 cm HPTLC Silicagel 60 plates (Merck, Germany) alongside six dilutions of a standard solution (50 ng/μL squalene, cholesterol palmitate, triolein, 1,3-diolein, 1,2-diolein, cholesterol, 25-hydroxy cholesterol, monoolein, and 1,2-dipalmitoyl-phosphocholine), and subjected to a multistep development procedure: (i) chloroform/ diethyl ether/ethyl acetate/methanol/acetic acid (76/6/20/2:1) to 4.5 cm from the lower edge of the plate, (ii) n-hexane/diethyl ether/acetic acid (80/20/1) to 6 cm, and (iii) n-hexane/acetic acid (100/1) to 80 cm. Plates were dried for 5 min after each development step. Bands were detected by immersion in a charring solution and drying at 250°C for 1 min and quantified densitometrically using a CAMAG TLC Scanner 4 and WinCATS software in absorption mode at 560 nm using a tungsten lamp. Samples were quantified by comparison with the standard curve spotted on each plate. Each analysis was performed in triplicate.
Sudan Black B Staining
Sudan Black B staining of lipids was performed on animals fed ad libitum (AL) or starved (BD) for 48 h. Worms were grown at 25°C from the L1 to the L4 stage and then shifted at 20°C. About 400 worms per sample were washed 4 times in M9 buffer and once with PBS + 20% Tween. The supernatant was removed, and 200 μL of PBS was added to the BD samples and 200 μL PBS plus 20 μL of 0.5 mg/mL 4′,6-diamidino-2-phenylindole (DAPI) was added to the AL samples. Fresh 1% paraformaldehyde in PBS was added to the worm pellet and incubated for 30 min at room temperature with stirring. Worms were then subjected to three cycles of freeze-thawing in liquid nitrogen and a 37°C water bath, incubated for 10 min on ice, and washed with M9. AL and BD worms were pooled, washed sequentially with 25%, 50%, and 70% ethanol, and then incubated overnight in Sudan Black B solution (50% saturated solution; 15 mg/mL in 70% ethanol). Animals were washed again in 70%, 50%, and 25% ethanol, resuspended in glycerol, and mounted animals on slides. Worms were imaged and photographed using a Nikon Eclipse 80i (Zeiss) microscope equipped with a monochrome digital camera (DS-Qi1Mc).
Quantification and Statistical Analysis
Statistical Analysis
Results are expressed as the mean ± standard deviation (SD) of triplicates, and all experiments were performed at least three times. For ORO experiments, value of n can be found in figure. Group differences were analyzed using a two-tailed Student’s t test (parametric) or a two-tailed Mann-Whitney U test (non-parametric). Tests used are indicated in the figure legend. For lifespan assays, Excel XLStat was used to calculate the mean and maximal lifespans and differences were analyzed using the Mantel–Cox log-rank test. Value of n (number of worms) can be found in the Table S2. A p value of < 0.05 was considered statistically significant.
Acknowledgments
We thank all members of the Aguilaniu lab for fruitful discussions; the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440); Marie Delattre, Johnny Fares, and Cynthia Kenyon for providing strains and reagents; and Anne O’Rourke for helpful comments on the manuscript. We also thank the PLATIM (Plateau Technique Imagerie/Microscopie IFR 128). This work was supported by an European Research Council grant to H.A. (H2020/2014-2019-N°647003).
Author Contributions
A.B., J.G., and S.B. performed the experiments; L.M. helped with the staining and lifespan experiments; N.L. and H.G. performed the thin-layer chromatography (TLC) experiments; and H.A. conceived the study and wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: July 23, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.06.064.
Supplemental Information
References
- Afzal S., Tybjærg-Hansen A., Jensen G.B., Nordestgaard B.G. Change in Body Mass Index Associated With Lowest Mortality in Denmark, 1976–2013. JAMA. 2016;315:1989–1996. doi: 10.1001/jama.2016.4666. [DOI] [PubMed] [Google Scholar]
- Bustos V., Partridge L. Good Ol’ Fat: Links between Lipid Signaling and Longevity. Trends Biochem. Sci. 2017;42:812–823. doi: 10.1016/j.tibs.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Covarrubias V. Lipidomics in longevity and healthy aging. Biogerontology. 2013;14:663–672. doi: 10.1007/s10522-013-9450-7. [DOI] [PubMed] [Google Scholar]
- Goudeau J., Bellemin S., Toselli-Mollereau E., Shamalnasab M., Chen Y., Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L., Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Flatt T., Aguilaniu H. Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 2013;17:10–19. doi: 10.1016/j.cmet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T.L., Smith E.D., Tsuchiya M., Welton K.L., Thomas J.H., Fields S., Kennedy B.K., Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Lapierre L.R., Gelino S., Meléndez A., Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L.R., Silvestrini M.J., Nuñez L., Ames K., Wong S., Le T.T., Hansen M., Meléndez A. Autophagy genes are required for normal lipid levels in C. elegans. Autophagy. 2013;9:278–286. doi: 10.4161/auto.22930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro E.J., Yu B.P., Bertrand H.A. Action of food restriction in delaying the aging process. Proc. Natl. Acad. Sci. USA. 1982;79:4239–4241. doi: 10.1073/pnas.79.13.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R.M., McKay J.P., Avery L., Graff J.M. C. elegans: a model for exploring the genetics of fat storage. Dev. Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- Noble T., Stieglitz J., Srinivasan S. An integrated serotonin and octopamine neuronal circuit directs the release of an endocrine signal to control C. elegans body fat. Cell Metab. 2013;18:672–684. doi: 10.1016/j.cmet.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E.J., Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E.J., Soukas A.A., Carr C.E., Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E.J., Kuballa P., Xavier R., Ruvkun G. ω-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27:429–440. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-K., Park S.-K. Coelomocytes are required for lifespan extension via different methods of dietary restriction in C. elegans. Toxicol. Environ. Health Sci. 2017;9:59–63. [Google Scholar]
- Park J.-K., Hwang J.-K., Song K.-H., Park S.-K. Role of Coelomocytes in Stress Response and Fertility in Caenorhabditis elegans. J. Life Sci. 2015;25:263–268. [Google Scholar]
- Redman L.M., Smith S.R., Burton J.H., Martin C.K., Il’yasova D., Ravussin E. Metabolic Slowing and Reduced Oxidative Damage with Sustained Caloric Restriction Support the Rate of Living and Oxidative Damage Theories of Aging. Cell Metab. 2018;27:805–815.e4. doi: 10.1016/j.cmet.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder E.A., Brunet A. Lipid Profiles and Signals for Long Life. Trends Endocrinol. Metab. 2015;26:589–592. doi: 10.1016/j.tem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin G.L., Kaeberlein M. Dietary restriction by bacterial deprivation increases life span in wild-derived nematodes. Exp. Gerontol. 2008;43:130–135. doi: 10.1016/j.exger.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Grant B., Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.O., Box A.C., Xu N., Le Men J., Yu J., Guo F., Trimble R., Mak H.Y. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010;107:4640–4645. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.